Simple Summary
Developing cultivars with resistance genes (R genes) is an effective strategy to support high yield and quality in Brassica crops. The availability of clone R gene and genomic sequences in Brassica species and Arabidopsis thaliana provide the opportunity to compare genomic regions and survey R genes across genomic databases. In this paper, we aim to identify genes related to cloned genes through sequence identity, providing a repertoire of species-wide related R genes in Brassica crops. The comprehensive list of candidate R genes can be used as a reference for functional analysis.
Abstract
Various diseases severely affect Brassica crops, leading to significant global yield losses and a reduction in crop quality. In this study, we used the complete protein sequences of 49 cloned resistance genes (R genes) that confer resistance to fungal and bacterial diseases known to impact species in the Brassicaceae family. Homology searches were carried out across Brassica napus, B. rapa, B. oleracea, B. nigra, B. juncea, B. carinata and Arabidopsis thaliana genomes. In total, 660 cloned disease R gene homologs (CDRHs) were identified across the seven species, including 431 resistance gene analogs (RGAs) (248 nucleotide binding site-leucine rich repeats (NLRs), 150 receptor-like protein kinases (RLKs) and 33 receptor-like proteins (RLPs)) and 229 non-RGAs. Based on the position and distribution of specific homologs in each of the species, we observed a total of 87 CDRH clusters composed of 36 NLR, 16 RLK and 3 RLP homogeneous clusters and 32 heterogeneous clusters. The CDRHs detected consistently across the seven species are candidates that can be investigated for broad-spectrum resistance, potentially providing resistance to multiple pathogens. The R genes identified in this study provide a novel resource for the future functional analysis and gene cloning of Brassicaceae R genes towards crop improvement.
1. Introduction
The Brassicaceae family consists of 44 tribes, 372 genera and 4060 species [1,2]. Among these, there are two prominent genera: Arabidopsis, which contains the model organism A. thaliana, and Brassica, which contains species such as B. napus, B. oleracea, B. nigra, B. rapa, B. carinata, and B. juncea, which are cultivated as a source of vegetables, condiments and oil [3,4,5,6].
The Brassicaceae ancestral genome has undergone three rounds of whole genome duplication/triplication, leading to the evolution of Arabidopsis and Brassica species [7,8,9,10]. A lineage separation occurred between the 2 genera~29.50 million years ago (MYA) [11], followed by the divergence of the Brassica A, B, and C sub-genomes. The diploid B. nigra (BB, 2n = 16) diverged from B. rapa (AA, 2n = 20) and B. oleracea (CC, 2n = 18)~6.2 to 7.9 MYA [9,12] and the divergence between the A and C sub-genomes occurred 4.6 MYA later [13]. Interspecific hybridisation, followed by polyploidisation, in Brassica species resulted in 3 allotetraploids; B. juncea (AABB, 2n = 4x = 36), which evolved~0.039–0.055 MYA [14], B. carinata (BBCC, 2n = 4x = 34) which evolved~0.047 MYA [11], and B. napus (AACC, 2n = 4x = 38), being the most recent species to evolve some~7500 years ago [15]. The gene content of these allopolyploids represents a history of gene loss and duplication due to polyploidisation and whole genome duplication [16,17].
The large demand and intensified cultivation of Brassica crops have made them vulnerable to abiotic and biotic stresses, particularly to diseases. While the most common control methods for managing pathogens are specific cultural practises and chemical application, the deployment of disease-resistant crops is more environmentally friendly and cost-effective. Brassica crops have two types of disease resistance: qualitative and quantitative. While quantitative relies on several minor genes with partial resistance expressed at the later crop stages, qualitative resistance is governed by major genes or resistance genes (R genes), largely expressed in the early crop stages through to maturity. Both resistance types are useful, however, qualitative is widely utilised in Brassica cultivar development because its effect is easily manifested and can be easily identified at the cotyledon stage. For instance, a set of differential blackleg isolates containing avirulence (Avr) genes is used to screen R genes in B. napus lines via the assessment of a hypersensitive response (HR) observed in the cotyledons [18,19].
Resistance gene analogs (RGAs) play an important role in host resistance [20] and are generally categorized into three main classes, nucleotide-binding site -leucine rice repeats (NLRs), receptor-like protein kinases (RLKs), and receptor-like proteins (RLPs). The NLR family, which is the most common class of RGAs, carries cytoplasmic receptors for recognising specific pathogens and are involved in effector-triggered plant immunity (ETI) [21,22,23,24]. RLKs and RLPs are involved in pattern-triggered immunity (PTI), which relies on pattern recognition receptors (PRRs) to elicit the first line of defence by recognising pathogen elicitors [25,26].
Examining gene homology among plant species is important to obtain the possible functions of a gene. Several studies have exploited gene homology for crop improvement. For instance, the homolog of an A. thaliana R gene, At_NDR1, was cloned and functionally characterised in Coffea arabica, conferring R-gene-mediated resistance to coffee leaf rust caused by Hemileia vastatrix [27]. Further, homologs of the Triticum aestivum Mla gene, TmMla in Hordeum vulgare, Sr33 in Secale cereale, and Sr50 in Aegilops tauschii were introgressed in T. aestivum, providing disease resistance [28,29,30].
Here, we used the sequences of 49 cloned R genes with a confirmed function against fungal and bacterial diseases to identify cloned disease resistance gene homologs (CDRHs) across six Brassica crops and A. thaliana. The evolutionary events including the loss, retention, and diversification of RGA domains in the CDRHs were also investigated. The outcome of this study could facilitate the identification and cloning of functional RGAs and their application in Brassica breeding programs towards disease resistance improvement.
2. Materials and Methods
2.1. Collection of Gene and Genomic Data
A comprehensive search was conducted to identify cloned R genes that provide qualitative resistance to fungal and bacterial diseases in all six Brassica species and Arabidopsis. A total of 49 cloned R genes were identified and included in this study (42 in Arabidopsis and 7 in the Brassica species) based on the following 3 criteria: (1) has a known gene-for-gene interaction with a corresponding pathogen Avr gene or (2) confers resistance in the form of a HR, indicating that it is involved in a gene-for-gene interaction or (3) acts as a helper or accessory gene necessary for the gene-for-gene interaction (Table 1). The complete protein (amino acid, aa) sequence of each gene was extracted from the UniProtKb (https://www.uniprot.org/uniprot/, accessed on 10 October 2020) [31] or NCBI (https://www.ncbi.nlm.nih.gov/, accessed on 10 October 2020) website (Table 1). The genome used for each of the seven species is listed in Table 2.

Table 1.
The 49 cloned R genes from Arabidopsis thaliana (At), Brassica juncea (Bju), Brassica napus (Bna) and Brassica rapa (Bra) used for homology searches.

Table 2.
Seven Brassicaceae species with their corresponding genome version and size used in this study.
2.2. Homolog Identification and Classification
To perform the homology search, the protein sequence of each of the cloned genes was aligned across the seven genomes using translated Basic Local Alignment Search Tool (tBLASTn) using CoGeBlast [93]. Following the criteria used by previous studies identifying homologous genes in plants, tBLASTn hits with an E value range outside E0 to E-45 [94,95,96] or which did not have >70% similarity [96,97,98,99] were removed from further analyses. Since the smallest reference gene used in the study, At_Rpw8.1, has 148 aa [75], any tBLASTn hits with <148 aa, were also removed from further analyses.
The list of predicted RGAs derived from the RGAugury pipeline [100] in A. thaliana, B. rapa, B. nigra, B. oleracea, B. juncea and B. napus were extracted from a previous study [101] and used to classify homologs. The RGAugury pipeline was also used to predict B. carinata RGAs in this study. The total number of CDRHs and their RGA classification including Nucleotide-binding site (N), Coiled-coil (CC)-NBS-Leucine rich repeats (LRR) (CNL), CC-NBS (CN), NBS-LRR (NL), Toll/Interleukin-1 receptor (TIR)-NBS-LRR (TNL), TIR-NBS (TN), TIR with unknown domains (TX), NLR with other domains (Other-NLR), Receptor-like kinase protein (RLK) with LRR (LRR-RLK), RLK with Lysin motif (LsyM) (LysM-RLK), RLK with other receptor (Other-RLK), Receptor-like protein (RLP) with LRR (LRR-RLP) and RLP with LysM (LysM-RLP) were identified for each species. We further classified the RGAs according to whether they had the same predicted domain to their homologous counterpart, or whether it was different.
Homolog types, such as paralog (homologous genes within the same species) or ortholog (homologous genes in different species), were also determined for each of the 49 cloned R genes. Paralogs were further classified as tandem, when a paralog exists within 5 Mb of the cloned R gene, or segmented, when a paralog is >5 Mb away from the cloned gene or the paralog is located on another chromosome [102]. Lastly, genes that were homologous to two or more cloned RGAs were also identified.
2.3. Gene Cluster Analysis
Two types of gene clusters were identified in this study. The first was a homogenous RGA cluster which is defined as a cluster with at least 2 or more (but no more than 8) RGAs of the same class, either NLR, RLK or RLP, located within a 200 kb region on the same chromosome [103,104]. The second was a heterogeneous cluster which refers to clusters containing different classes of RGAs or containing both an RGA and a Non-RGA (for example, a homolog that has not been identified using the RGAugury pipeline).
3. Results
3.1. Distribution of CDRHs
We used the sequences of the 49 cloned R genes to obtain CDRHs across the 7 species (Table 2). A total of 660 CDRHs, including 248 NLRs, 150 RLKs, 33 RLPs and 229 Non-RGAs (genes without RGA-related domains) were identified (Figure 1, Tables S1 and S2). B. juncea contained the highest number of CDRHs with 136, followed by B. carinata with 119, B. napus with 101, B. rapa with 80, B. oleracea and B. nigra with 78, and A. thaliana with 68 (Figure 1). The total CDRHs identified in Brassica polyploids was 356, with an average of 119, while the total CDRHs identified in Brassica diploids was 236, with an average of 79 (Figure 1). On the other hand, A. thaliana contained less CDRHs, 68, in comparison to Brassica species with an average of 99 CDRHs (Figure 1).
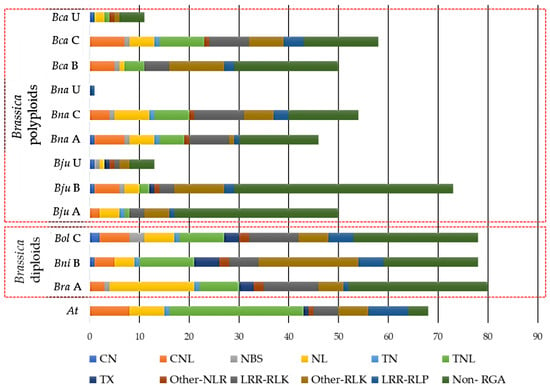
Figure 1.
Distribution of cloned disease resistance gene homologs (CDRHs) with their resistance gene analogs (RGA) classes/subclasses (CN = coiled-coil (CC)-nucleotide-binding site (NBS), CNL = CC-NBS-leucine rice repeats (LRR), NL = NBS-LRR, TN = Toll/Interleukin-1 receptor (TIR)-LRR, TNL = Toll/Interleukin-1 receptor (TIR)-NBS-LRR, TX = Toll/Interleukin-1 receptor (TIR) with other domains, Other-NLR= NBS-LRR with other domains, LRR-RLK= LRR-receptor-like kinase proteins (RLK), Other-RLK = RLK with other domains, LRR-RLP = LRR-receptor-like proteins) and Non-RGA (CDRHs identified without RGA domain based on the RGAugury pipeline) in Brassica rapa (Bra), B. nigra (Bni), B. oleracea (Bol), B. juncea (Bju), B. napus (Bna), B. carinata (Bca), and Arabidopsis thaliana (At). A, B, C, and U (unplaced) refer to the genome/sub-genome of Brassica species.
The individual sub-genomes of each of the polyploid species contained fewer homologs and RGAs of the cloned genes than their respective A, B and C genome Brassica progenitors. B. napus and B. juncea had 46 CDRHs (30 RGAs, 16 non-RGAs) and 50 CDRHs (17 RGAs, 33 non-RGAs), respectively, while their A sub-genome had 80 CDRHs (52 RGAs, 28 non-RGAs). B. juncea and B. carinata had 73 CDRHs (29 RGAs, 44 non-RGAs) and 50 CDRHs (29 RGAs, 21 non-RGAs), respectively, while their B sub-genome progenitor had 78 CDRHs (59 RGAs, 19 non-RGAs) (Table S2). B. carinata and B. napus had 58 CDRHs (43 RGAs, 15 non-RGAs) and 54 CDRHs (40 RGAs, 14 non-RGAs), respectively, while their C sub-genome progenitor had 78 CDRHs (53 RGAs, 25 non-RGAs) (Table S2).
The total number of CDRHs for each disease was counted in this study. For the bacterial disease, the cloned R genes against Pseudomonas syringae, the causal agent of black leaf spot (BLS) disease, (At_ADR1, At_BAK1, At_FLS2, At_NDR1, At_NRG1a, At_NRG1b, At_PBS1, At_RIN4, At_RLP30, At_RLP32, At_RPM1, At_RPS2, At_RPS4, At_RPS5, At_RRS1, and At_SOBIR1) had the highest number of CDRHs; 239 (153 RGAs, 86 non-RGAs) (Table 3 and Table 4, Table S1). B. napus had the highest number of RGAs related to BLS resistance, 33, followed by 31 and 25 RGAs in B. carinata and B. juncea, respectively. RGAs related to BLS resistance were most observed in the C genome/sub-genome with 49 RGAs, followed by 43 and 42 RGAs in A and B genome/sub-genome, respectively. For the clone R gene At_RLP1 against Xanthomonas spp., the causal agent of black rot (BR), 6 RGAs were only obtained in Brassica C genome (Table S1).

Table 3.
Cloned genes in Brassica crops and Arabidopsis thaliana and their corresponding paralogs (cloned disease R gene homolog type).

Table 4.
Cloned genes in Brassica crops and Arabidopsis thaliana and their corresponding orthologs (cloned disease R gene homolog type).
For the fungal disease, the cloned R genes against Leptosphaeria maculans, the causal agent of blackleg (BL) disease, (Bna_MAPk, Bna_LepR3/Rlm2, Bna_Rlm9/4/7, At_RLM1a, At_RLM1b, and At_RLM3), had a total of 165 CDRHs (86 RGAs, 79 non-RGAs) (Table 3 and Table 4, Table S1). Of these, 20 RGAs were found in B. nigra, followed by 16 RGAs in B. carinata (8 in B sub-genome, 7 in C sub-genome, and 1 unplaced) and 15 RGAs in B. juncea (4 in A sub-genome, 9 in B sub-genome, and 2 unplaced). On the other hand, the cloned R genes against Albugo candida, the causal agent of white rust (WR) disease, (Bju_WRR1, At_RAC1, At_WRR4a, At_WRR4b, At_WRR8, At_WRR9 and At_WRR12), 123 CDRHs (102 RGAs, 21 non-RGAs) were identified (Table 3 and Table 4, Table S1). Of these RGAs, 22 were observed in B. napus (13 in the A sub-genome and 8 in the C sub-genome), followed by 19 RGAs in B. rapa (Table S1). A total of 109 CDRHs (95 RGAs, 14 non-RGAs) were identified to be related with cloned R genes against fungal pathogen Hyaloperonospora arabidopsidis, the causal agent of downy mildew (DM) disease, (At_ADR1, NRG1a, NRG1b, At_RLP42, At_RPP1, At_RPP2a, At_RPP2B, At_RPP4, At_RPP5, At_RPP7, At_RPP8, At_RPP13, and At_RPP39) (Table 3 and Table 4, Table S1). Of these, the 27 RGAs obtained in A. thaliana was the highest number of RGAs observed compared to the RGAs in other studied species (Table S1). On the other hand, the cloned R genes against Plasmodiophora brassicae (also a fungus), the causal agent of clubroot (CR) disease, (Bra_Crr1a and cRa/cRb) had a total of 48 CDRHs (38 RGAs) (Table 3 and Table 4, Table S1). B. napus and B. oleracea had the highest counts with 9 RGAs (4 in the A sub-genome and 5 in the C sub-genome) and 8 RGAs, respectively. While B. nigra had 7 RGAs, B. rapa had 6 RGAs (Table S1).
We recorded a total of 75 CDRHs, including 60 RGAs, for the cloned R genes (At_BAK1, At_RLP23, At_RLP30, and At_SOBIR1) against fungal pathogen Sclerotinia sclerotiorum, the causal agent of Sclerotinia stem rot (SSR) disease (Table 3 and Table 4, Table S1). The 22 and 18 RGAs in the C and A genome/sub-genomes of the Brassica species, respectively, were higher compared to that of other genome/sub-genomes and A. thaliana (Table S1). For the cloned R genes against Fusarium oxysporum (also a fungus), the causal agent of Fusarium wilt (FW) disease (Bol_FocBo1, At_RFO1, At_RFO2, and At_RFO3), 50 CDRHs (34 RGAs, 16 non-RGAs) were obtained (Table 3 and Table 4, Table S1). B. carinata with 9 RGAs (4 in the B sub-genome and 5 in the C sub-genome) had the highest number, while B. juncea with 2 RGAs (1 in each B sub-genome and unplaced contigs) had the lowest RGA count across the studied species (Table S1).
The cloned R genes against fungal pathogens Erysiphe cichoracearum the causal agent of powdery mildew (PW) (At_RPW8.1, At_RPW8.2 and At_ADR1) and Botrytis cinerea the causal agent of grey mould (GM) (At_RLP42 and At_RLM3) were observed as having 9 CDRHs (all RGAs) and 5 CDRHs (3 RGAs, 2 non-RGAs), respectively (Table 4 and Table S1). Only the Brassica B and C genomes had RGAs with 3 and 2 genes, respectively for PW resistance (Table S1), while A. thaliana contained all 3 RGAs for GM resistance (Table S1).
3.2. Identification of CDRH Types
This study identified 68 CDRHs that are homologous to more than 1 of the cloned R genes (Table S1). Of these, 12 RGAs were previously identified and functionally characterised disease resistance genes such as At_NRG1a, At_NRG1b, At_RAC1, At_WRR4b, At_WRR9, At_RLM1a, At_RPP4, At_RPP5, At_RPP2a, At_WRR8, Bra_Crr1a, and Bra_cRa/cRb (Table S1). For instance, At_WRR4b, a WR R gene, and At_RLM1b, a BL R gene, were homologous to each other in this study. This was also the case with At_RPP2a, a DM R gene, and Bol_FocBo1, a FW R gene.
For the paralogous CDRHs, a total of 62 paralogs, including 43 tandem (69%) and 18 segmented (31%), were observed to the cloned R genes in A. thaliana (At_ADR1, At_BAK1, At_FLS2, At_NDR1, NRG1a, NRG1b, At_PBS1, At_RAC1, At_RIN4, At_RFO1, At_RFO2, At_RFO3, At_RLM1a, At_RLM1b, At_RLM3, At_RPM1, At_RPP1, At_RPP2a, At_RPP2b, At_RPP4, At_RPP5, At_RPP8, At_RPP13, At_RPP39, At_RPS2, At_RPS4, At_RPS5, At_Rpw8.1, At_Rpw8.2, At_RRS1, At_SOBIR1, At_WRR4a, At_WRR4b, At_WRR8, At_WRR9, and At_WRR12) (Table 3). On the other hand, 23 paralogs, including 19 segmented (83%) and 4 tandem (17%), were observed to the cloned R genes in Brassica species (Bju_WRR1, Bna_MAPk, Bra_Crr1a, Bra_cRa/cRb, Bol_FocBo1, Bna_LepR3/Rlm2, and Bna_Rlm9/4/7) (Table 3).
In terms of RGA domain retention and losses, this study found that 431 and 229 out of 660 CDRHs have retained (as RGA) and lost (as Non-RGA) resistance domains and motifs from the original gene, respectively (Table 3 and Table 4). In some cases, the RGA class of CDRHs tend to be different from their corresponding cloned gene because the RGA domain has been contracted/truncated. For example, At_RPP8 encoding a CNL, had 4 NL and 1 CN CDRHs, while At_WRR8 encoding a TNL, had 1 TN, 1 NL, and 1 NBS CDRHs (Table 3 and Table 4). At_RPP8 and its CDRHs had a common NBS domain, while At_WRR8 and its CDRHs had a common domain of either TIR, NBS or LRR (Table 3 and Table 4). On the other hand, 2 CDRHs did not have a common RGA domain with their homologous cloned R gene. These included Bra_cRa/cRb (TNL) and Bju_WRR1 (CNL), which both had at least one Other-RLK CDRH (Table 3 and Table 4).
3.3. Identifying Clusters of CDRHs across Brassica Crops and Arabidopsis thaliana
The position of CDRHs across chromosomes of each studied species creates an opportunity to determine whether these genes form clusters. This study identified a total of 87 RGA clusters across the seven species, with the highest number of clusters observed in A. thaliana, 21, followed by 14, 13, 12, 11, 9, and 7 clusters in B. carinata, B. napus, B. nigra, B. juncea, B. rapa, and B. oleracea, respectively (Figure 2). B. carinata, B. napus and B. nigra had the highest total number of CDRH RGAs with 78, 71, and 59, respectively, while B. rapa and B. oleracea had the lowest total numbers of CDRH RGAs with 52 and 53, respectively (Table 3).
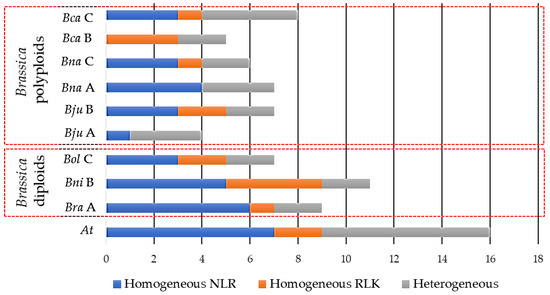
Figure 2.
Distribution of gene clusters of cloned disease resistance homologs in Brassica rapa (Bra), B. nigra (Bni), B. oleracea (Bol), B. juncea (Bju), B. napus (Bna), B. carinata (Bca), and Arabidopsis thaliana (At). NLR is nucleotide-binding site (NBS)- leucine rice repeats (LRR) while an RLK is receptor-like kinase proteins. A, B, and C refer to the genome/sub-genome of Brassica species.
Across the studied species, 55 homogeneous RGA clusters, including 36 NLR, 16 RLK and 3 RLP homogeneous clusters, were identified (Figure 2). Within the homogeneous cluster, the cloned R genes At_BAK1, Bra_cRa/cRb, At_FLS2, Bna_LepR3/Rlm2, At_NDR1, At_NRG1a, At_NRG1b, At_RFO1, At_RFO2, At_RLM1a, At_RLM1b, At_RPP1, At_RPP2a, At_RPP2b, At_RPP4, At_RPP5, At_RPP39, At_RPS4, At_RRS1, At_WRR4a, At_WRR4b, At_WRR8 and At_WRR9 were observed to form a cluster either to their corresponding tandem paralog/s or to other functionally characterised R gene/s (Table S1). On the other hand, 32 heterogeneous RGA clusters were obtained (Figure 2). Aside from having cluster members with different RGA domains, a different heterogenous cluster with RGA and non-RGA members was observed (Table S1). However, the non-RGA may have a resistance-related function or partial RGA domain structure (missing some of the key domains). For example, in this study, a non-RGA AT3G20590 which is a non-specific disease resistance protein-like gene [105] clustered with the At_NDR1 gene. Furthermore, a non-RGA, B08g104510.1, which is a mitogen-activated protein kinase [91] clustered with NLRs Bo8g104700.1, Bo8g104710.1 and Bo8g104730.1 (Table S1). Finally, the non-RGA BnaA02g24430D, which has an LRR domain [15], clustered with BnaA02g24440D, a LRR-RLP.
4. Discussion
RGAs are the most important genes that need to be discovered and cloned for the improvement of Brassica crop disease resistance. The availability of Brassica genomic resources, along with the model species A. thaliana, and the aid of computational and bioinformatic tools have led to their widespread identification. Across A. thaliana and Brassica species, an approach utilising homology can reveal associations between functionally characterised R genes and RGAs, and how each species’ genetic repertoire differs (for example, RGA and non-RGA content).
The larger number of total CDRHs in Brassica polyploids over Brassica diploids is likely due to polyploidisation [11,14,15]. It has previously been shown that the total number of genes, RGAs and glucosinolate-related genes in Brassica polyploids were higher than in the Brassica diploid/progenitor species [11,101]. The number of DNA transposable elements, a major factor in plant genome expansion [106], was also found to be higher in polyploid B. napus compared to the diploids B. rapa and B. oleracea [107], which could likely be the case for B. carinata and B. juncea when compared to their corresponding diploid progenitors. On the other hand, the fewer counts of CDRHs in Arabidopsis than Brassica species could probably be due to whole genome triplication events which did not happen in ancestral Arabidopsis while it occurred in ancestral Brassica [7,108]. As a result, it is expected that the increased genome size of Brassicas also increased their gene number compared to A. thaliana [11]. Furthermore, Brassica crops undergone long history of extensive breeding to improve disease resistance which may have led to an increase in their RGA content [109].
The fewer RGAs in the individual sub-genomes of the Brassica polyploids compared to their diploid genome progenitor that we found in this study is consistent with the other Brassica RGA studies [11,96,109,110,111,112]. Duplicated disease R genes or RGAs are favourably lost in the sub-genomes of polyploid Brassicas after a duplication event compared to their diploid genome progenitors [109,110,113,114]. This event was also observed in other species such as legumes [115], maize [116,117], and wheat [118]. In B. napus, the loss of RGAs is thought to be a result of homoeologous exchange between the A and C sub-genomes [107,119].
The total number of CDRHs, particularly the RGAs, per disease was also determined. Limited genetic resistance towards BLS, PW, and GM disease has been identified in Brassica species, making the RGAs obtained here a valuable starting point for future studies to explore potential BLS, PW, and GM R genes. For WR, it has been reported that B. rapa and the A sub-genome of B. napus are a good source of resistance [120,121,122]. The majority of markers associated with WR resistance that have been utilised for resistance exploration were also derived from the A-genome [80,123,124,125,126]. For BL, previous investigations showed that B-genome Brassica species have high levels of phenotypic resistance to BL compared to Brassicas containing the A and C genomes, and A. thaliana [127,128,129]. However, the association of phenotypic BL resistance to the identified RGAs in this study is yet to be confirmed. Another, QTL against FW and BR have previously been identified in Brassica C genome/sub-genome [89,130,131,132,133,134].
Among Brassicaceae species, R genes conferring resistance to DM have only been cloned in A. thaliana [63,64,65,66,69,70,71,79], while other R genes and quantitative trait loci (QTL) identified in B. rapa and B. oleracea are yet to be functionally confirmed [135,136,137,138,139]. For CR, recent studies have also shown QTL to be associated with resistance in the A and C genome of Brassica species [140,141,142,143,144]. B. nigra have also been noted with high phenotypic resistance to CR pathotypes in Canada [145], however, the RGAs obtained here need to be functionally verified with phenotypic CR resistance. Lastly, for SSR, Brassica A and C genomes have both been reported to harbour QTL linked to possible resistance against the disease [146,147,148,149].
Our results showed that there are a considerable number of CDRHs throughout Brassica crops and A. thaliana. CDRHs, especially those with resistance domains (RGAs), play important roles in disease resistance responses, and their subsequent application in breeding programs will help to improve disease resistance. However, RGAs are not the only genes that may confer disease resistance in Brassica crops and this is particularly true for diseases whose resistance response is quantitatively controlled, such as SSR [147]. Therefore, the non-RGAs identified in this study may be useful, but still need further analyses and confirmation.
A CDRH can be homologous to more than one cloned R gene because some of the genes may share the same resistance domains. Considerable number of collinear genes were obtained between Arabidopsis and Brassica species as they originated from one ancestral species [11,150]. It is also possible that the homology to one more gene could imply multiple resistance function. The At_RPP8 gene, causing resistance to DM disease, was later found to contain two alleles; HRT and RCY1, which confer resistance to turnip crinkle virus and yellow strain cucumber mosaic virus, respectively [151,152,153]. The At_RRS1 gene, initially associated with the avirulence gene popP2, which triggers resistance against Ralstonia solanacearum [154,155], was later found to also mediate a resistant response against P. syringae and Colletotrichum higginsianum [77]. However, this assumption needs thorough investigation and multiple functional characterisation to be confirmed.
The large number of tandem duplicates or paralogs in A. thaliana over Brassica species is consistent with findings in previous studies [13,156,157]. Tandem duplication may have occurred more frequently in A. thaliana because its ancestors did not undergo whole genome triplication, hence there was no extensive genome fractionation [108]. Conversely, the large number of segmented paralogs in Brassica species over A. thaliana could also be due to genome fractionation and block reshuffling which separated the homologous RGAs during the process of these evolutionary events [108,158]. Segmented paralogs act as gene-buffers in forms of structural variation such as copy number variation (CNV), which has been found abundantly in B. napus and B. oleracea, and is associated with SSR, CR and BL resistance [109,112].
Homology analysis is useful in elucidating gene gains and losses, and verifying retained resistance domains or function of genes [159]. From an ancestral gene, homologs could undergo neofunctionalisation, subfunctionalisation or duplication-degeneration-complementation (DDC), non-functionalisation or pseudogenisation, escape from adaptive conflict (EAC) and other routes involving gene dosage and redundancy [160,161,162,163,164,165,166]. In plants, RGAs are prone to rapid gene expansion during evolutionary events, as well as gene loss and contraction, as they respond to environmental stress such as disease pressure [167,168]. Nevertheless, truncated RGAs such as NL and TN have been cloned and functionally characterised with disease resistance in A. thaliana [53,61,62]. While a NBS gene has been reported as a signalling component in disease resistance [20], genes with TX domains were able to interact with different R and Avr genes to elicit disease resistance in A. thaliana [169,170], and CC domain has been reported as a candidate for the blackleg R genes Rlm1, LepR2, and LepR4 in B. oleracea [171,172,173].
In gene clustering, the greater number of clusters in A. thaliana could possibly be due to its smaller genome size, compared to the Brassica species (Table 2), where the position of the genes or RGAs tend to be closer to each other. However, between Brassica species, the presence of RGAs could be a factor to gene clustering as it was observed in this study that the higher the total number of CDRHs with an RGA domain in each species, the higher the likelihood that these specific RGAs were part of a gene clusters.
Earlier studies have suggested homogeneous gene cluster may have evolved via tandem duplication [174,175]. The existence of tandem paralogs is yet to be functionally confirmed, however, their co-existence with cloned genes in a cluster suggests a “balancing” model in which genetic variation in disease resistance is maintained despite the presence of selection pressure [176]. In previous A. thaliana studies, a NLR Suppressor of Non-expressor of Pathogenesis-Related Genes 1-1, CONSTITUTIVE 1 (SNC1) requires its co-clustered NLR SIDEKICK SNC1 1 (SIKIC1), SIKIC2 and SIKIC3 to mediate defence signalling [177], while the NLRs Chilling Sensitive 1 (CHS1) or TN2 pairs to Suppressors of CHS (CHS1 or CHS2) gene 3 (SOC3) to monitor the homeostasis of Senescence-Associated E3 Ubiquitin Ligase 1 (SAUL1) [178], which is a positive regulator of PTI in plants [179]. Thus, clustering of these RGAs may be maintaining variation in disease resistance but when selection pressure occurs, RGAs could act as either accessory or helper or sensor needed in disease resistance [180,181].
The “birth and death” model could also be the fate of the RGAs in a gene cluster. The “birth and death” model indicates that when a RGA function is overcome by a pathogen, the duplication process facilitates DNA sequence exchanges of homologous genes via cross-over, leading to sequence mispairing, loss of the original sequence, converting the gene, and eventually generating a novel RGA with possible altered pathogen specificities [182]. The emergence of cloned genes At_RPP13 in A. thaliana, Pm3 in wheat, L in flax and elF4E in capsicum was said to follow the “birth and death” model [183,184,185,186]. The same mechanism of “birth and death” has likely occurred within a blackleg resistance gene Bna_Rlm9/4/7 which contains three alleles Rlm9, Rlm4 and Rlm7 on chromosome A07 of B. napus [85,187] because their corresponding avirulence genes have been found to have an epistatic interaction, indicating an evolutionary arms race between the host and pathogen [188,189,190].
For the heterogeneous clusters, this clustering occurs because of random ectopic recombination, chromosomal translocation, gene transposition and co-localisation of the genes [191,192,193]. However, the genes with different domains in a cluster is yet to be functionally confirmed. The same can be said for homogenous clusters, where there is a high chance that the distribution and position of CDRHs is not random; however, this assumption requires further research for confirmation.
5. Conclusions
The identification of RGAs throughout the genome and underlying QTL is one of the breakthroughs that has accelerated disease resistance improvement in crops. While the process of identifying and functionally testing R genes has shortened, QTLs can have numerous candidates which results in time consuming validation. Hence, the use of cloned R gene sequences to search for RGA homologs can provide a basis for narrowing down candidates for functional characterisation.
The findings in this study can also be useful in studying the evolution and mechanisms of resistance in these genes, which can later help to guide appropriate crop methodologies to develop disease resistant and resilient Brassica cultivars. Additionally, gene-specific markers from these specific RGAs can be used as diagnostic markers in determining Brassica lines with disease resistance and possibly explore new QTL not only in Brassica species but in other members of the Brassicaceae family.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/biology11060821/s1, Table S1: Results from the BLAST analysis (Comparative Genomics website) along with the identification of resistance gene analogs (RGAs) using RGAugury pipeline in Brassicaceae species and the underlying types of homolog; Table S2: List of resistance gene analogs (RGAs) in Brassica carinata zd-1 v1.0 identified by RGAugury pipeline.
Author Contributions
A.Y.C. and J.B. conceptualized the paper; A.Y.C. wrote the original draft along with formal analyses; T.X.N., S.T. and W.J.W.T. helped improve the paper by suggesting additional ideas and by thorough revision/editing; P.E.B. analysed the Brassica carinata genes using the RGAugury pipeline; D.E. and J.B. supervised, reviewed, and suggested revisions to the paper. All authors have read and agreed to the published version of the manuscript.
Funding
This study is funded by the Australian Research Council projects (DP200100762, and DP210100296) and Grains Research and Development Corporation UWA1905-006RTX.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data used in this research is publicly available. The gene sequences can be found at https://www.uniprot.org/uniprot/ (accessed on 10 October 2020) and https://www.ncbi.nlm.nih.gov/ (accessed on 10 October 2020) while the genomic databases can be found at https://www.arabidopsis.org/ (accessed on 27 December 2020), http://brassicadb.org/ (accessed on 27 December 2020), http://bigd.big.ac.cn/gwh (accessed 27 December 2020), and http://brassicadb.bio2db.com/ (accessed on 10 April 2021). The data (results) presented in this research are available in the Supplementary Materials.
Acknowledgments
All authors acknowledge the University of Western Australia Research Training Program economic support during A.Y.C. and W.J.W.T. respective doctoral studies. A.Y.C. and W.J.W.T. would also like to acknowledge the support of the Philippine Rice Research Institute (by granting study-leave) and Grains Research and Development Corporation, respectively.
Conflicts of Interest
The authors declare no known competing financial interests or personal relationships that could have influenced the work reported here.
References
- Tamokou, J.D.D.; Mbaveng, A.T.; Kuete, V. Chapter 8—Antimicrobial activities of african medicinal spices and vegetables. In Medicinal Spices and Vegetables from Africa; Kuete, V., Ed.; Academic Press: Cambridge, MA, USA, 2017; pp. 207–237. [Google Scholar] [CrossRef]
- Warwick, S.I.; Mummenhoff, F.; Sauder, C.A.; Koch, M.A.; Al-Shehbaz, I.A. Closing the gaps: Phylogenetic relationships in the Brassicaceae based on DNA sequence data of nuclear ribosomal ITS region. Plant Syst. Evol. 2010, 285, 209–232. [Google Scholar] [CrossRef]
- Fahey, J.W. Brassicas. In Encyclopedia of Food Sciences and Nutrition, 2nd ed.; Caballero, B., Ed.; Academic Press: Oxford, UK, 2003; pp. 606–615. [Google Scholar]
- Gupta, S.K. Biology and Breeding of Crucifers; CRC Press: Boca Raton, FL, USA, 2016. [Google Scholar]
- Sun, R. Economic/Academic Importance of Brassica rapa. In The Brassica rapa Genome; Springer: Berlin/Heidelberg, Germany, 2015; pp. 1–15. [Google Scholar]
- Kumar, A.; Banga, S.S.; Meena, P.D.; Kumar, P.R. Brassica Oilseeds: Breeding and Management; CABI: Wallingford, Oxfordshire, UK, 2015. [Google Scholar]
- Beilstein, M.A.; Nagalingum, N.S.; Clements, M.D.; Manchester, S.R.; Mathews, S. Dated molecular phylogenies indicate a Miocene origin for Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2010, 107, 18724–18728. [Google Scholar] [CrossRef] [PubMed]
- Bowers, J.; Chapman, B.; Rong, J.; Paterson, A.H. Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 2003, 422, 433–438. [Google Scholar] [CrossRef]
- Lysak, M.A.; Koch, M.A.; Pecinka, A.; Schubert, I. Chromosome triplication found across the tribe Brassiceae. Genome Res. 2005, 15, 516–525. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.-W.; Lai, K.-N.; Tai, P.-Y.; Li, W.-H. Rates of Nucleotide Substitution in Angiosperm Mitochondrial DNA Sequences and Dates of Divergence between Brassica and Other Angiosperm Lineages. J. Mol. Evol. 1999, 48, 597–604. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Wei, Y.; Xiao, D.; Gong, K.; Sun, P.; Ren, Y.; Yuan, J.; Wu, T.; Yang, Q.; Li, X.; et al. Brassica carinata genome characterization clarifies U’s triangle model of evolution and polyploidy in Brassica. Plant Physiol. 2021, 186, 388–406. [Google Scholar] [CrossRef]
- Navabi, Z.-K.; Huebert, T.; Sharpe, A.G.; O’Neill, C.M.; Bancroft, I.; Parkin, I.A. Conserved microstructure of the Brassica B Genome of Brassica nigra in relation to homologous regions of Arabidopsis thaliana, B. rapa and B. oleracea. BMC Genom. 2013, 14, 250. [Google Scholar] [CrossRef]
- Liu, S.; Liu, Y.; Yang, X.; Tong, C.; Edwards, D.; Parkin, I.A.P.; Zhao, M.; Ma, J.; Yu, J.; Huang, S.; et al. The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nat. Commun. 2014, 5, 3930. [Google Scholar] [CrossRef]
- Yang, J.; Liu, D.; Wang, X.; Ji, C.; Cheng, F.; Liu, B.; Hu, Z.; Chen, S.; Pental, D.; Ju, Y.; et al. The genome sequence of allopolyploid Brassica juncea and analysis of differential homoeolog gene expression influencing selection. Nat. Genet. 2016, 48, 1225–1232. [Google Scholar] [CrossRef]
- Chalhoub, B.; Denoeud, F.; Liu, S.; Parkin, I.A.P.; Tang, H.; Wang, X.; Chiquet, J.; Belcram, H.; Tong, C.; Samans, B.; et al. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 2014, 345, 950–953. [Google Scholar] [CrossRef]
- Yang, T.-J.; Kim, J.S.; Kwon, S.-J.; Lim, K.-B.; Choi, B.-S.; Kim, J.-A.; Jin, M.; Park, J.Y.; Lim, M.-H.; Kim, H.-I.; et al. Sequence-Level Analysis of the Diploidization Process in the Triplicated FLOWERING LOCUS C Region of Brassica rapa. Plant Cell 2006, 18, 1339–1347. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Tehrim, S.; Wang, L.; Dossa, K.; Zhang, X.; Ke, T.; Liao, B. Evolutionary history and functional divergence of the cytochrome P450 gene superfamily between Arabidopsis thaliana and Brassica species uncover effects of whole genome and tandem duplications. BMC Genom. 2017, 18, 733. [Google Scholar] [CrossRef] [PubMed]
- Balesdent, M.H.; Barbetti, M.J.; Li, H.; Sivasithamparam, K.; Gout, L.; Rouxel, T. Analysis of Leptosphaeria maculans Race Structure in a Worldwide Collection of Isolates. Phytopathology 2005, 95, 1061. [Google Scholar] [CrossRef]
- Marcroft, S.J.; Elliott, V.L.; Cozijnsen, A.J.; Salisbury, P.A.; Howlett, B.J.; Van de Wouw, A.P. Identifying resistance genes to Leptosphaeria maculans in Australian Brassica napus cultivars based on reactions to isolates with known avirulence genotypes. Crop Pasture Sci. 2012, 63, 338–350. [Google Scholar] [CrossRef]
- Sekhwal, M.K.; Li, P.; Lam, I.; Wang, X.; Cloutier, S.; You, F.M. Disease Resistance Gene Analogs (RGAs) in Plants. Int. J. Mol. Sci. 2015, 16, 19248–19290. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.D.G.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef]
- Le Roux, C.; Huet, G.; Jauneau, A.; Camborde, L.; Trémousaygue, D.; Kraut, A.; Zhou, B.; Levaillant, M.; Adachi, H.; Yoshioka, H.; et al. A Receptor Pair with an Integrated Decoy Converts Pathogen Disabling of Transcription Factors to Immunity. Cell 2015, 161, 1074–1088. [Google Scholar] [CrossRef]
- Ravensdale, M.; Bernoux, M.; Ve, T.; Kobe, B.; Thrall, P.H.; Ellis, J.G.; Dodds, P.N. Intramolecular Interaction Influences Binding of the Flax L5 and L6 Resistance Proteins to their AvrL567 Ligands. PLOS Pathog. 2012, 8, e1003004. [Google Scholar] [CrossRef]
- Whitham, S.; Dinesh-Kumar, S.; Choi, D.; Hehl, R.; Corr, C.; Baker, B. The product of the tobacco mosaic virus resistance gene N: Similarity to toll and the interleukin-1 receptor. Cell 1994, 78, 1101–1115. [Google Scholar] [CrossRef]
- Afzal, A.J.; Wood, A.J.; Lightfoot, D.A. Plant receptor-like serine threonine kinases: Roles in signaling and plant defense. Mol. Plant Microbe Interact. 2008, 21, 507–517. [Google Scholar] [CrossRef]
- Chisholm, S.T.; Coaker, G.; Day, B.; Staskawicz, B.J. Host-Microbe Interactions: Shaping the Evolution of the Plant Immune Response. Cell 2006, 124, 803–814. [Google Scholar] [CrossRef] [PubMed]
- Cacas, J.-L.; Petitot, A.-S.; Bernier, L.; Estevan, J.; Conejero, G.; Mongrand, S.; Fernandez, D. Identification and characterization of the Non-race specific Disease Resistance 1 (NDR1) orthologous protein in coffee. BMC Plant Biol. 2011, 11, 144. [Google Scholar] [CrossRef] [PubMed]
- Mago, R.; Zhang, P.; Vautrin, S.; Šimková, H.; Bansal, U.; Luo, M.-C.; Rouse, M.; Karaoglu, H.; Periyannan, S.; Kolmer, J.; et al. The wheat Sr50 gene reveals rich diversity at a cereal disease resistance locus. Nat. Plants 2015, 1, 15186. [Google Scholar] [CrossRef] [PubMed]
- Periyannan, S.; Moore, J.; Ayliffe, M.; Bansal, U.; Wang, X.; Huang, L.; Deal, K.; Luo, M.; Kong, X.; Bariana, H.; et al. The Gene Sr33, an Ortholog of Barley Mla Genes, Encodes Resistance to Wheat Stem Rust Race Ug99. Science 2013, 341, 786–788. [Google Scholar] [CrossRef]
- Jordan, T.; Seeholzer, S.; Schwizer, S.; Töller, A.; Somssich, I.E.; Keller, B. The wheat Mla homologue TmMla1 exhibits an evolutionarily conserved function against powdery mildew in both wheat and barley. Plant J. 2011, 65, 610–621. [Google Scholar] [CrossRef]
- The UniProt Consortium. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef]
- Grant, J.J.; Chini, A.; Basu, D.; Loake, G.J. Targeted Activation Tagging of the Arabidopsis NBS-LRR gene, ADR1, Conveys Resistance to Virulent Pathogens. Mol. Plant-Microbe Interact. 2003, 16, 669–680. [Google Scholar] [CrossRef]
- Castel, B.; Ngou, P.-M.; Cevik, V.; Redkar, A.; Kim, D.-S.; Yang, Y.; Ding, P.; Jones, J.D.G. Diverse NLR immune receptors activate defence via the RPW8-NLR NRG 1. New Phytol. 2019, 222, 966–980. [Google Scholar] [CrossRef]
- Saile, S.C.; Jacob, P.; Castel, B.; Jubic, L.M.; Salas-Gonzáles, I.; Bäcker, M.; Jones, J.D.G.; Dangl, J.L.; El Kasmi, F. Two unequally redundant “helper” immune receptor families mediate Arabidopsis thaliana intracellular “sensor” immune receptor functions. PLOS Biol. 2020, 18, e3000783. [Google Scholar] [CrossRef]
- Gao, M.; Wang, X.; Wang, D.; Xu, F.; Ding, X.; Zhang, Z.; Bi, D.; Cheng, Y.T.; Chen, S.; Li, X.; et al. Regulation of Cell Death and Innate Immunity by Two Receptor-like Kinases in Arabidopsis. Cell Host Microbe 2009, 6, 34–44. [Google Scholar] [CrossRef]
- Li, J.; Wen, J.; Lease, K.A.; Doke, J.T.; Tax, F.; Walker, J.C. BAK1, an Arabidopsis LRR Receptor-like Protein Kinase, Interacts with BRI1 and Modulates Brassinosteroid Signaling. Cell 2002, 110, 213–222. [Google Scholar] [CrossRef]
- Kim, M.H.; Kim, Y.; Kim, J.W.; Lee, H.-S.; Lee, W.S.; Kim, S.-K.; Wang, Z.-Y.; Kim, S.-H. Identification of Arabidopsis BAK1-Associating Receptor-Like Kinase 1 (BARK1) and Characterization of its Gene Expression and Brassinosteroid-Regulated Root Phenotypes. Plant Cell Physiol. 2013, 54, 1620–1634. [Google Scholar] [CrossRef] [PubMed]
- Albert, I.; Böhm, H.; Albert, M.; Feiler, C.E.; Imkampe, J.; Wallmeroth, N.; Brancato, C.; Raaymakers, T.M.; Oome, S.; Zhang, H.; et al. An RLP23–SOBIR1–BAK1 complex mediates NLP-triggered immunity. Nat. Plants 2015, 1, 15140. [Google Scholar] [CrossRef] [PubMed]
- Tabata, S.; Kaneko, T.; Nakamura, Y.; Kotani, H.; Kato, T.; Asamizu, E.; Miyajima, N.; Sasamoto, S.; Kimura, T.; Hosouchi, T.; et al. Sequence and analysis of chromosome 5 of the plant Arabidopsis thaliana. Nature 2000, 408, 823–826. [Google Scholar] [CrossRef]
- Gómez-Gómez, L.; Boller, T. FLS2: An LRR Receptor–like Kinase Involved in the Perception of the Bacterial Elicitor Flagellin in Arabidopsis. Mol. Cell 2000, 5, 1003–1011. [Google Scholar] [CrossRef]
- Century, K.S.; Shapiro, A.D.; Repetti, P.P.; Dahlbeck, D.; Holub, E.; Staskawicz, B.J. NDR1, a Pathogen-Induced Component Required for Arabidopsis Disease Resistance. Science 1997, 278, 1963–1965. [Google Scholar] [CrossRef]
- Swiderski, M.R.; Innes, R.W. The Arabidopsis PBS1 resistance gene encodes a member of a novel protein kinase subfamily. Plant J. 2001, 26, 101–112. [Google Scholar] [CrossRef]
- Borhan, M.H.; Holub, E.B.; Beynon, J.L.; Rozwadowski, K.; Rimmer, S.R. The Arabidopsis TIR-NB-LRR Gene RAC1 Confers Resistance to Albugo candida (White Rust) and Is Dependent on EDS1 but not PAD4. Mol. Plant-Microbe Interact. 2004, 17, 711–719. [Google Scholar] [CrossRef]
- Axtell, M.J.; Staskawicz, B.J. Initiation of RPS2-Specified Disease Resistance in Arabidopsis Is Coupled to the AvrRpt2-Directed Elimination of RIN4. Cell 2003, 112, 369–377. [Google Scholar] [CrossRef]
- Day, B.; Dahlbeck, D.; Staskawicz, B.J. NDR1 Interaction with RIN4 Mediates the Differential Activation of Multiple Disease Resistance Pathways in Arabidopsis. Plant Cell 2006, 18, 2782–2791. [Google Scholar] [CrossRef]
- Liu, J.; Elmore, J.M.; Lin, Z.-J.D.; Coaker, G. A Receptor-like Cytoplasmic Kinase Phosphorylates the Host Target RIN4, Leading to the Activation of a Plant Innate Immune Receptor. Cell Host Microbe 2011, 9, 137–146. [Google Scholar] [CrossRef] [PubMed]
- Mackey, D.; Belkhadir, Y.; Alonso, J.; Ecker, J.; Dangl, J.L. Arabidopsis RIN4 Is a Target of the Type III Virulence Effector AvrRpt2 and Modulates RPS2-Mediated Resistance. Cell 2003, 112, 379–389. [Google Scholar] [CrossRef]
- Mackey, D.; Holt, B.F.; Wiig, A.; Dangl, J.L. RIN4 Interacts with Pseudomonas syringae Type III Effector Molecules and Is Required for RPM1-Mediated Resistance in Arabidopsis. Cell 2002, 108, 743–754. [Google Scholar] [CrossRef]
- Diener, A.C.; Ausubel, F.M. Resistance to Fusarium Oxysporum 1, a Dominant Arabidopsis Disease-Resistance Gene, Is Not Race Specific. Genetics 2005, 171, 305–321. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Diener, A.C. Arabidopsis thaliana resistance to Fusarium oxysporum 2 Implicates Tyrosine-Sulfated Peptide Signaling in Susceptibility and Resistance to Root Infection. PLoS Genet. 2013, 9, e1003525. [Google Scholar] [CrossRef] [PubMed]
- Cole, S.J.; Diener, A.C. Diversity in receptor-like kinase genes is a major determinant of quantitative resistance to Fusarium oxysporum f. sp. matthioli. New Phytol. 2013, 200, 172–184. [Google Scholar] [CrossRef] [PubMed]
- Staal, J.; Kaliff, M.; Bohman, S.; Dixelius, C. Transgressive segregation reveals two Arabidopsis TIR-NB-LRR resistance genes effective against Leptosphaeria maculans, causal agent of blackleg disease. Plant J. 2006, 46, 218–230. [Google Scholar] [CrossRef] [PubMed]
- Staal, J.; Kaliff, M.; Dewaele, E.; Persson, M.; Dixelius, C. RLM3, a TIR domain encoding gene involved in broad-range immunity of Arabidopsis to necrotrophic fungal pathogens. Plant J. 2008, 55, 188–200. [Google Scholar] [CrossRef]
- Jehle, A.K.; Fürst, U.; Lipschis, M.; Albert, M.; Felix, G. Perception of the novel MAMP eMax from different Xanthomonas species requires the Arabidopsis receptor-like protein ReMAX and the receptor kinase SOBIR. Plant Signal. Behav. 2013, 8, e27408. [Google Scholar] [CrossRef]
- Jehle, A.K.; Lipschis, M.; Albert, M.; Fallahzadeh-Mamaghani, V.; Fürst, U.; Mueller, K.; Felix, G. The Receptor-Like Protein ReMAX of Arabidopsis Detects the Microbe-Associated Molecular Pattern eMax from Xanthomonas. Plant Cell 2013, 25, 2330–2340. [Google Scholar] [CrossRef]
- Albert, I.; Zhang, L.; Bemm, H.; Nürnberger, T. Structure-Function Analysis of Immune Receptor AtRLP23 with Its Ligand nlp20 and Coreceptors AtSOBIR1 and AtBAK1. Mol. Plant-Microbe Interact. 2019, 32, 1038–1046. [Google Scholar] [CrossRef]
- Wang, G.; Ellendorff, U.; Kemp, B.; Mansfield, J.W.; Forsyth, A.; Mitchell, K.; Bastas, K.; Liu, C.-M.; Woods-Tör, A.; Zipfel, C.; et al. A Genome-Wide Functional Investigation into the Roles of Receptor-Like Proteins in Arabidopsis. Plant Physiol. 2008, 147, 503–517. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Fraiture, M.; Kolb, D.; Löffelhardt, B.; Desaki, Y.; Boutrot, F.F.G.; Tor, M.; Zipfel, C.; Gust, A.A.; Brunner, F. Arabidopsis RECEPTOR-LIKE PROTEIN30 and Receptor-Like Kinase SUPPRESSOR OF BIR1-1/EVERSHED Mediate Innate Immunity to Necrotrophic Fungi. Plant Cell 2013, 25, 4227–4241. [Google Scholar] [CrossRef] [PubMed]
- Fan, L.; Fröhlich, K.; Melzer, E.; Pruitt, R.N.; Albert, I.; Zhang, L.; Joe, A.; Hua, C.; Song, Y.; Albert, M.; et al. Genotyping-by-sequencing-based identification of Arabidopsis pattern recognition receptor RLP32 recognizing proteobacterial translation initiation factor IF1. BioRxiv 2021. [Google Scholar] [CrossRef]
- Zhang, L.; Kars, I.; Essenstam, B.; Liebrand, T.W.H.; Wagemakers, L.; Elberse, J.; Tagkalaki, P.; Tjoitang, D.; Van den Ackerveken, G.; Van Kan, J.A.L. Fungal Endopolygalacturonases Are Recognized as Microbe-Associated Molecular Patterns by the Arabidopsis Receptor-Like Protein responsiveness to botrytis polygalacturonases1. Plant Physiol. 2014, 164, 352–364. [Google Scholar] [CrossRef] [PubMed]
- Grant, M.R.; Godiard, L.; Straube, E.; Ashfield, T.; Lewald, J.; Sattler, A.; Innes, R.W.; Dangl, J.L. Structure of the Arabidopsis RPM1 Gene Enabling Dual Specificity Disease Resistance. Science 1995, 269, 843–846. [Google Scholar] [CrossRef]
- Tornero, P.; Chao, R.A.; Luthin, W.N.; Goff, S.A.; Dangl, J.L. Large-Scale Structure –Function Analysis of the Arabidopsis RPM1 Disease Resistance Protein. Plant Cell 2002, 14, 435–450. [Google Scholar] [CrossRef]
- Botella, M.A.; Parker, J.E.; Frost, L.N.; Bittner-Eddy, P.D.; Beynon, J.L.; Daniels, M.J.; Holub, E.B.; Jones, J.D.G. Three Genes of the Arabidopsis RPP1 Complex Resistance Locus Recognize Distinct Peronospora parasitica Avirulence Determinants. Plant Cell 1998, 10, 1847–1860. [Google Scholar] [CrossRef]
- Sinapidou, E.; Williams, K.; Nott, L.; Bahkt, S.; Tör, M.; Crute, I.; Bittner-Eddy, P.; Beynon, J. Two TIR:NB:LRR genes are required to specify resistance to Peronospora parasitica isolate Cala2 in Arabidopsis. Plant J. 2004, 38, 898–909. [Google Scholar] [CrossRef]
- Van Der Biezen, E.A.; Freddie, C.T.; Kahn, K.; Parker, J.E.; Jones, J. Arabidopsis RPP4 is a member of the RPP5 multigene family of TIR-NB-LRR genes and confers downy mildew resistance through multiple signalling components. Plant J. 2002, 29, 439–451. [Google Scholar] [CrossRef]
- Parker, J.E.; Coleman, M.J.; Szabò, V.; Frost, L.N.; Schmidt, R.; Van Der Biezen, E.A.; Moores, T.; Dean, C.; Daniels, M.J.; Jones, J. The Arabidopsis downy mildew resistance gene RPP5 shares similarity to the toll and interleukin-1 receptors with N and L6. Plant Cell 1997, 9, 879–894. [Google Scholar] [CrossRef] [PubMed]
- Barragan, A.C.; Wu, R.; Kim, S.-T.; Xi, W.; Habring, A.; Hagmann, J.; Van De Weyer, A.-L.; Zaidem, M.; Ho, W.W.H.; Wang, G.; et al. RPW8/HR repeats control NLR activation in Arabidopsis thaliana. PLoS Genet. 2019, 15, e1008313. [Google Scholar] [CrossRef] [PubMed]
- Tsuchiya, T.; Eulgem, T. An alternative polyadenylation mechanism coopted to the Arabidopsis RPP7 gene through intronic retrotransposon domestication. Proc. Natl. Acad. Sci. USA 2013, 110, E3535–E3543. [Google Scholar] [CrossRef] [PubMed]
- McDowell, J.M.; Dhandaydham, M.; Long, T.A.; Aarts, M.G.M.; Goff, S.; Holub, E.B.; Dangl, J.L. Intragenic Recombination and Diversifying Selection Contribute to the Evolution of Downy Mildew Resistance at the RPP8 Locus of Arabidopsis. Plant Cell 1998, 10, 1861–1874. [Google Scholar] [CrossRef]
- Bittner-Eddy, P.D.; Crute, I.R.; Holub, E.B.; Beynon, J.L. RPP13 is a simple locus in Arabidopsis thaliana for alleles that specify downy mildew resistance to different avirulence determinants in Peronospora parasitica. Plant J. 2000, 21, 177–188. [Google Scholar] [CrossRef]
- Goritschnig, S.; Krasileva, K.; Dahlbeck, U.; Staskawicz, B.J. Computational Prediction and Molecular Characterization of an Oomycete Effector and the Cognate Arabidopsis Resistance Gene. PLoS Genet. 2012, 8, e1002502. [Google Scholar] [CrossRef]
- Bent, A.F.; Kunkel, B.N.; Dahlbeck, D.; Brown, K.L.; Schmidt, R.; Giraudat, J.; Leung, J.; Staskawicz, B.J. RPS2 of Arabidopsis thaliana: A Leucine-Rich Repeat Class of Plant Disease Resistance Genes. Science 1994, 265, 1856–1860. [Google Scholar] [CrossRef]
- Gassmann, W.; Hinsch, M.E.; Staskawicz, B.J. The Arabidopsis RPS4 bacterial-resistance gene is a member of the TIR-NBS-LRR family of disease-resistance genes. Plant J. 1999, 20, 265–277. [Google Scholar] [CrossRef]
- Deslandes, L.; Olivier, J.; Theulières, F.; Hirsch, J.; Feng, D.X.; Bittner-Eddy, P.; Beynon, J.; Marco, Y. Resistance to Ralstonia solanacearum in Arabidopsis thaliana is conferred by the recessive RRS1-R gene, a member of a novel family of resistance genes. Proc. Natl. Acad. Sci. USA 2002, 99, 2404–2409. [Google Scholar] [CrossRef]
- Xiao, S.; Ellwood, S.; Calis, O.; Patrick, E.; Li, T.; Coleman, M.; Turner, J.G. Broad-Spectrum Mildew Resistance in Arabidopsis thaliana Mediated by RPW8. Science 2001, 291, 118–120. [Google Scholar] [CrossRef]
- Warren, R.F.; Henk, A.; Mowery, P.; Holub, E.; Innes, R.W. A Mutation within the Leucine-Rich Repeat Domain of the Arabidopsis Disease Resistance Gene RPS5 Partially Suppresses Multiple Bacterial and Downy Mildew Resistance Genes. Plant Cell 1998, 10, 1439–1452. [Google Scholar] [CrossRef]
- Sarris, P.F.; Duxbury, Z.; Huh, S.U.; Ma, Y.; Segonzac, C.; Sklenar, J.; Derbyshire, P.; Cevik, V.; Rallapalli, G.; Saucet, S.B.; et al. A Plant Immune Receptor Detects Pathogen Effectors that Target WRKY Transcription Factors. Cell 2015, 161, 1089–1100. [Google Scholar] [CrossRef]
- Borhan, M.H.; Gunn, N.; Cooper, A.; Gulden, S.; Tör, M.; Rimmer, S.R.; Holub, E.B. WRR4 Encodes a TIR-NB-LRR Protein That Confers Broad-Spectrum White Rust Resistance in Arabidopsis thaliana to Four Physiological Races of Albugo candida. Mol. Plant-Microbe Interact. 2008, 21, 757–768. [Google Scholar] [CrossRef] [PubMed]
- Cevik, V.; Boutrot, F.; Apel, W.; Robert-Seilaniantz, A.; Furzer, O.J.; Redkar, A.; Castel, B.; Kover, P.X.; Prince, D.C.; Holub, E.B.; et al. Transgressive segregation reveals mechanisms of Arabidopsis immunity to Brassica-infecting races of white rust ( Albugo candida). Proc. Natl. Acad. Sci. USA 2019, 116, 2767–2773. [Google Scholar] [CrossRef] [PubMed]
- Arora, H.; Padmaja, K.L.; Paritosh, K.; Mukhi, N.; Tewari, A.K.; Mukhopadhyay, A.; Gupta, V.; Pradhan, A.K.; Pental, D. BjuWRR1, a CC-NB-LRR gene identified in Brassica juncea, confers resistance to white rust caused by Albugo candida. Theor. Appl. Genet. 2019, 132, 2223–2236. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Djavaheri, M.; Wang, H.; Larkan, N.J.; Haddadi, P.; Beynon, E.; Gropp, G.; Borhan, M.H. Leptosphaeria maculans Effector Protein AvrLm1 Modulates Plant Immunity by Enhancing MAP Kinase 9 Phosphorylation. iScience 2018, 3, 177–191. [Google Scholar] [CrossRef]
- Larkan, N.J.; Lydiate, D.J.; Parkin, I.A.P.; Nelson, M.N.; Epp, D.J.; Cowling, W.A.; Rimmer, S.R.; Borhan, M.H. The B rassica napus blackleg resistance gene LepR3 encodes a receptor-like protein triggered by the L eptosphaeria maculans effector AVRLM 1. New Phytol. 2013, 197, 595–605. [Google Scholar] [CrossRef]
- Larkan, N.J.; Ma, L.; Borhan, M.H. The Brassica napus receptor-like protein RLM2 is encoded by a second allele of the LepR3/Rlm2 blackleg resistance locus. Plant Biotechnol. J. 2015, 13, 983–992. [Google Scholar] [CrossRef]
- Larkan, N.J.; Ma, L.; Haddadi, P.; Buchwaldt, M.; Parkin, I.A.P.; Djavaheri, M.; Borhan, M.H. The Brassica napus wall-associated kinase-like (WAKL) gene Rlm9 provides race-specific blackleg resistance. Plant J. 2020, 104, 892–900. [Google Scholar] [CrossRef]
- Haddadi, P.; Larkan, N.J.; Van de Wouw, A.; Zhang, Y.; Neik, T.X.; Beynon, E.; Bayer, P.; Edwards, D.; Batley, J.; Borhan, M.H. Brassica napus genes Rlm4 and Rlm7, conferring resistance to Leptosphaeria maculans, are alleles of the Rlm9 wall-associated kinase-like resistance locus. bioRxiv 2021. [Google Scholar] [CrossRef]
- Hatakeyama, K.; Niwa, T.; Kato, T.; Ohara, T.; Kakizaki, T.; Matsumoto, S. The tandem repeated organization of NB-LRR genes in the clubroot-resistant CRb locus in Brassica rapa L. Mol. Genet. Genom. 2017, 292, 397–405. [Google Scholar] [CrossRef] [PubMed]
- Ueno, H.; Matsumoto, E.; Aruga, D.; Kitagawa, S.; Matsumura, H.; Hayashida, N. Molecular characterization of the CRa gene conferring clubroot resistance in Brassica rapa. Plant Mol. Biol. 2012, 80, 621–629. [Google Scholar] [CrossRef] [PubMed]
- Hatakeyama, K.; Suwabe, K.; Tomita, R.N.; Kato, T.; Nunome, T.; Fukuoka, H.; Matsumoto, S. Identification and Characterization of Crr1a, a Gene for Resistance to Clubroot Disease (Plasmodiophora brassicae Woronin) in Brassica rapa L. PLoS ONE 2013, 8, e54745. [Google Scholar] [CrossRef]
- Shimizu, M.; Pu, Z.-J.; Kawanabe, T.; Kitashiba, H.; Matsumoto, S.; Ebe, Y.; Sano, M.; Funaki, T.; Fukai, E.; Fujimoto, R.; et al. Map-based cloning of a candidate gene conferring Fusarium yellows resistance in Brassica oleracea. Theor. Appl. Genet. 2015, 128, 119–130. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.Y.; Krishnakumar, V.; Chan, A.P.; Thibaud-Nissen, F.; Schobel, S.; Town, C.D. Araport11: A complete reannotation of the Arabidopsis thaliana reference genome. Plant J. 2017, 89, 789–804. [Google Scholar] [CrossRef] [PubMed]
- Ap Parkin, I.; Koh, C.; Tang, H.; Robinson, S.J.; Kagale, S.; Clarke, W.E.; Town, C.D.; Nixon, J.; Krishnakumar, V.; Bidwell, S.L.; et al. Transcriptome and methylome profiling reveals relics of genome dominance in the mesopolyploid Brassica oleracea. Genome Biol. 2014, 15, R77. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Cai, X.; Wu, J.; Liu, M.; Grob, S.; Cheng, F.; Liang, J.; Cai, C.; Liu, Z.; Liu, B.; et al. Improved Brassica rapa reference genome by single-molecule sequencing and chromosome conformation capture technologies. Hortic. Res. 2018, 5, 50. [Google Scholar] [CrossRef]
- Lyons, E.; Pedersen, B.; Kane, J.; Alam, M.; Ming, R.; Tang, H.; Wang, X.; Bowers, J.; Paterson, A.; Lisch, D.; et al. Finding and Comparing Syntenic Regions among Arabidopsis and the Outgroups Papaya, Poplar, and Grape: CoGe with Rosids. Plant Physiol. 2008, 148, 1772. [Google Scholar] [CrossRef]
- Rameneni, J.J.; Lee, Y.; Dhandapani, V.; Yu, X.; Su, R.C.; Oh, M.H.; Yong, P.L. Genomic and Post-Translational Modification Analysis of Leucine-Rich-Repeat Receptor-Like Kinases in Brassica rapa. PLoS ONE 2015, 10, e0142255. [Google Scholar] [CrossRef]
- Wei, Z.; Wang, J.; Yang, S.; Song, Y. Identification and expression analysis of the LRR-RLK gene family in tomato (Solanum lycopersicum) Heinz 1706. Genome 2015, 58, 121–134. [Google Scholar] [CrossRef]
- Yang, H.; Bayer, P.E.; Tirnaz, S.; Edwards, D.; Batley, J. Genome-Wide Identification and Evolution of Receptor-Like Kinases (RLKs) and Receptor like Proteins (RLPs) in Brassica juncea. Biology 2021, 10, 17. [Google Scholar] [CrossRef]
- Singh, S.; Chand, S.; Singh, N.K.; Sharma, T.R. Genome-Wide Distribution, Organisation and Functional Characterization of Disease Resistance and Defence Response Genes across Rice Species. PLoS ONE 2015, 10, e0125964. [Google Scholar] [CrossRef]
- Alamery, S.; Tirnaz, S.; Bayer, P.; Tollenaere, R.; Chaloub, B.; Edwards, D.; Batley, J. Genome-wide identification and comparative analysis of NBS-LRR resistance genes in Brassica napus. Crop Pasture Sci. 2017, 69, 79–93. [Google Scholar] [CrossRef]
- Jiang, M.; Dong, X.; Lang, H.; Pang, W.; Zhan, Z.; Li, X.; Piao, Z. Mining of Brassica-Specific Genes (BSGs) and Their Induction in Different Developmental Stages and under Plasmodiophora brassicae Stress in Brassica rapa. Int. J. Mol. Sci. 2018, 19, 2064. [Google Scholar] [CrossRef]
- Li, P.; Quan, X.; Jia, G.; Xiao, J.; Cloutier, S.; You, F.M. RGAugury: A pipeline for genome-wide prediction of resistance gene analogs (RGAs) in plants. BMC Genom. 2016, 17, 852. [Google Scholar] [CrossRef]
- Tirnaz, S.; Bayer, P.; Inturrisi, F.; Neik, T.; Yang, H.; Dolatabadian, A.; Zhang, F.; Severn-Ellis, A.; Patel, D.; Pradhan, A.; et al. Resistance gene analogs in the Brassicaceae: Identification, characterization, distribution and evolution. Plant Physiol. 2020, 184, 909–922. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, G.; Garg, V.; Kudapa, H.; Doddamani, D.; Pazhamala, L.T.; Khan, A.W.; Thudi, M.; Lee, S.-H.; Varshney, R.K. Genome-wide dissection of AP2/ERF and HSP90 gene families in five legumes and expression profiles in chickpea and pigeonpea. Plant Biotechnol. J. 2016, 14, 1563–1577. [Google Scholar] [CrossRef] [PubMed]
- Holub, E.B. The arms race is ancient history in Arabidopsis, the wildflower. Nat. Rev. Genet. 2001, 2, 516–527. [Google Scholar] [CrossRef]
- Jupe, F.; Pritchard, L.; Etherington, G.J.; MacKenzie, K.; Cock, P.J.; Wright, F.; Sharma, S.K.; Bolser, D.; Bryan, G.J.; Jones, J.D.; et al. Identification and localisation of the NB-LRR gene family within the potato genome. BMC Genom. 2012, 13, 75. [Google Scholar] [CrossRef]
- Zheng, M.S.; Takahashi, H.; Miyazaki, A.; Hamamoto, H.; Yamaguchi, I.; Kusano, T.; Shah, J. Up-regulation of Arabidopsis thaliana NHL10 in the hypersensitive response to Cucumber mosaic virus infection and in senescing leaves is controlled by signalling pathways that differ in salicylate involvement. Planta 2004, 218, 740–750. [Google Scholar] [CrossRef]
- Fedoroff, N. Transposons and genome evolution in plants. Proc. Natl. Acad. Sci. USA 2000, 97, 7002. [Google Scholar] [CrossRef]
- Bayer, P.E.; Scheben, A.; Golicz, A.A.; Yuan, Y.; Faure, S.; Lee, H.; Chawla, H.S.; Anderson, R.; Bancroft, I.; Raman, H.; et al. Modelling of gene loss propensity in the pangenomes of three Brassica species suggests different mechanisms between polyploids and diploids. Plant Biotechnol. J. 2021, 19, 2488–2500. [Google Scholar] [CrossRef] [PubMed]
- Cheng, F.; Wu, J.; Wang, X. Genome triplication drove the diversification of Brassica plants. Hortic. Res. 2014, 1, 14024. [Google Scholar] [CrossRef] [PubMed]
- Dolatabadian, A.; Bayer, P.E.; Tirnaz, S.; Hurgobin, B.; Edwards, D.; Batley, J. Characterization of disease resistance genes in the Brassica napus pangenome reveals significant structural variation. Plant Biotechnol. J. 2019, 18, 969–982. [Google Scholar] [CrossRef] [PubMed]
- Inturrisi, F.; Bayer, P.; Yang, H.; Tirnaz, S.; Edwards, D.; Batley, J. Genome-wide identification and comparative analysis of resistance genes in Brassica juncea. Mol. Breed. 2020, 40, 78. [Google Scholar] [CrossRef]
- Zhang, Y.; Xia, R.; Kuang, H.; Meyers, B.C. The Diversification of Plant NBS-LRR Defense Genes Directs the Evolution of MicroRNAs That Target Them. Mol. Biol. Evol. 2016, 33, 2692–2705. [Google Scholar] [CrossRef] [PubMed]
- Bayer, P.E.; Golicz, A.A.; Tirnaz, S.; Chan, C.K.; Edwards, D.; Batley, J. Variation in abundance of predicted resistance genes in the Brassica oleracea pangenome. Plant Biotechnol. J. 2019, 17, 789–800. [Google Scholar] [CrossRef]
- Wu, P.; Shao, Z.-Q.; Wu, X.-Z.; Wang, Q.; Wang, B.; Chen, J.-Q.; Hang, Y.-Y.; Xue, J.-Y. Loss/retention and evolution of NBS-encoding genes upon whole genome triplication of Brassica rapa. Gene 2014, 540, 54–61. [Google Scholar] [CrossRef]
- Yu, J.; Tehrim, S.; Zhang, F.; Tong, C.; Huang, J.; Cheng, X.; Dong, C.; Zhou, Y.; Qin, R.; Hua, W.; et al. Genome-wide comparative analysis of NBS-encoding genes between Brassica species and Arabidopsis thaliana. BMC Genom. 2014, 15, 3. [Google Scholar] [CrossRef]
- Zheng, F.; Wu, H.; Zhang, R.; Li, S.; He, W.; Wong, F.-L.; Li, G.; Zhao, S.; Lam, H.-M. Molecular phylogeny and dynamic evolution of disease resistance genes in the legume family. BMC Genom. 2016, 17, 402. [Google Scholar] [CrossRef]
- Schnable, J.C.; Springer, N.M.; Freeling, M. Differentiation of the maize subgenomes by genome dominance and both ancient and ongoing gene loss. Proc. Natl. Acad. Sci. USA 2011, 108, 4069. [Google Scholar] [CrossRef] [PubMed]
- Woodhouse, M.R.; Schnable, J.C.; Pedersen, B.S.; Lyons, E.; Lisch, D.; Subramaniam, S.; Freeling, M. Following Tetraploidy in Maize, a Short Deletion Mechanism Removed Genes Preferentially from One of the Two Homeologs. PLoS Biol. 2010, 8, e1000409. [Google Scholar] [CrossRef] [PubMed]
- Gu, L.; Si, W.; Zhao, L.; Yang, S.; Zhang, X. Dynamic evolution of NBS–LRR genes in bread wheat and its progenitors. Mol. Genet. Genom. 2014, 290, 727–738. [Google Scholar] [CrossRef] [PubMed]
- Hurgobin, B.; Golicz, A.A.; Bayer, P.E.; Chan, C.-K.K.; Tirnaz, S.; Dolatabadian, A.; Schiessl, S.V.; Samans, B.; Montenegro, J.D.; Parkin, I.A.P.; et al. Homoeologous exchange is a major cause of gene presence/absence variation in the amphidiploid Brassica napus. Plant Biotechnol. J. 2018, 16, 1265–1274. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, M. Mapping of a Locus Controlling Resistance to Albugo candida in Brassica napus Using Molecular Markers. Phytopathology 1995, 85, 218–220. [Google Scholar] [CrossRef]
- Kole, C.; Teutonico, R.; Mengistu, A.; Williams, P.; Osborn, T. Molecular mapping of a locus controlling resistance to Albugo candida in Brassica rapa. Phytopathology 1996, 86, 367–369. [Google Scholar] [CrossRef]
- Kole, C.; Williams, P.; Rimmer, S.; Osborn, T. Linkage mapping of genes controlling resistance to white rust (Albugo candida) in Brassica rapa (syn. campestris) and comparative mapping to Brassica napus and Arabidopsis thaliana. Genome 2002, 45, 22–27. [Google Scholar] [CrossRef]
- Bhayana, L.; Paritosh, K.; Arora, H.; Yadava, S.K.; Singh, P.; Nandan, D.; Mukhopadhyay, A.; Gupta, V.; Pradhan, A.K.; Pental, D. A Mapped Locus on LG A6 of Brassica juncea Line Tumida Conferring Resistance to White Rust Contains a CNL Type R Gene. Front. Plant Sci. 2020, 10, 1690. [Google Scholar] [CrossRef]
- Panjabi-Massand, P.; Yadava, S.K.; Sharma, P.; Kaur, A.; Kumar, A.; Arumugam, N.; Sodhi, Y.S.; Mukhopadhyay, A.; Gupta, V.; Pradhan, A.K.; et al. Molecular mapping reveals two independent loci conferring resistance to Albugo candida in the east European germplasm of oilseed mustard Brassica juncea. Theor. Appl. Genet. 2010, 121, 137–145. [Google Scholar] [CrossRef]
- Singh, B.K.; Nandan, D.; Ambawat, S.; Ram, B.; Kumar, A.; Singh, T.; Meena, H.S.; Kumar, V.; Singh, V.V.; Rai, P.K.; et al. Validation of molecular markers for marker-assisted pyramiding of white rust resistance loci in Indian Mustard (Brassica juncea L.). Can. J. Plant Sci. 2015, 95, 939–945. [Google Scholar] [CrossRef]
- Somers, D.; Rakow, G.; Rimmer, S. Brassica napus DNA markers linked to white rust resistance in Brassica juncea. Theor. Appl. Genet. 2002, 104, 1121–1124. [Google Scholar] [CrossRef] [PubMed]
- Fredua-Agyeman, R.; Coriton, O.; Huteau, V.; Parkin, I.A.P.; Chèvre, A.-M.; Rahman, H. Molecular cytogenetic identification of B genome chromosomes linked to blackleg disease resistance in Brassica napus × B. carinata interspecific hybrids. Theor. Appl. Genet. 2014, 127, 1305–1318. [Google Scholar] [CrossRef] [PubMed]
- Roy, N.N. Interspecific transfer of Brassica juncea-type high blackleg resistance to Brassica napus. Euphytica 1984, 33, 295–303. [Google Scholar] [CrossRef]
- Saal, B.; Struss, D. RGA- and RAPD-derived SCAR markers for a Brassica B-genome introgression conferring resistance to blackleg in oilseed rape. Theor. Appl. Genet. 2005, 111, 281–290. [Google Scholar] [CrossRef] [PubMed]
- Lv, H.; Fang, Z.; Yang, L.; Zhang, Y.; Wang, Q.; Liu, Y.; Zhuang, M.; Yang, Y.; Xie, B.; Liu, B.; et al. Mapping and analysis of a novel candidate Fusarium wilt resistance gene FOC1 in Brassica oleracea. BMC Genom. 2014, 15, 1094. [Google Scholar] [CrossRef] [PubMed]
- Pu, Z.; Ino, Y.; Kimura, Y.; Tago, A.; Shimizu, M.; Natsume, S.; Sano, Y.; Fujimoto, R.; Kaneko, K.; Shea, D.J.; et al. Changes in the Proteome of Xylem Sap in Brassica oleracea in Response to Fusarium oxysporum Stress. Front. Plant Sci. 2016, 7, 31. [Google Scholar] [CrossRef][Green Version]
- Saha, P.; Kalia, P.; Sonah, H.; Sharma, T.R. Molecular mapping of black rot resistance locus X ca1bo on chromosome 3 in I ndian cauliflower (Brassica oleracea var. botrytis L.). Plant Breed. 2014, 133, 268–274. [Google Scholar] [CrossRef]
- Sharma, B.B.; Kalia, P.; Singh, D.; Sharma, T.R. Introgression of Black Rot Resistance from Brassica carinata to Cauliflower (Brassica oleracea botrytis Group) through Embryo Rescue. Front. Plant Sci. 2017, 8, 1255. [Google Scholar] [CrossRef]
- Sharma, B.B.; Kalia, P.; Yadava, D.K.; Singh, D.; Sharma, T.R. Genetics and Molecular Mapping of Black Rot Resistance Locus Xca1bc on Chromosome B-7 in Ethiopian Mustard (Brassica carinata A. Braun). PLoS ONE 2016, 11, e0152290. [Google Scholar] [CrossRef]
- Farinhó, M.; Coelho, P.; Carlier, J.; Svetleva, D.; Monteiro, A.; Leitão, J.M. Mapping of a locus for adult plant resistance to downy mildew in broccoli (Brassica oleracea convar. italica). Theor. Appl. Genet. 2004, 109, 1392–1398. [Google Scholar] [CrossRef]
- Li, J.; Ding, Q.; Wang, F.; Li, H.; Zhang, Y.; Liu, L.; Jiao, Z.; Gao, J. Genome-wide gene expression profiles in response to downy mildew in Chinese cabbage (Brassica rapa L. ssp. pekinensis). Eur. J. Plant Pathol. 2018, 151, 861–873. [Google Scholar] [CrossRef]
- Singh, S.; Sharma, S.R.; Kalia, P.; Deshmukh, R.; Kumar, V.; Sharma, P. Molecular mapping of the downy mildew resistance gene Ppa3 in cauliflower (Brassica oleracea var. botrytis L.). J. Hortic. Sci. Biotechnol. 2012, 87, 137–143. [Google Scholar] [CrossRef]
- Yu, S.; Zhang, F.; Yu, R.; Zou, Y.; Qi, J.; Zhao, X.; Yu, Y.; Zhang, D.; Li, L. Genetic mapping and localization of a major QTL for seedling resistance to downy mildew in Chinese cabbage (Brassica rapa ssp. pekinensis). Mol. Breed. 2009, 23, 573–590. [Google Scholar] [CrossRef]
- Zhang, B.; Li, P.; Su, T.; Li, P.; Xin, X.; Wang, W.; Zhao, X.; Yu, Y.; Zhang, D.; Yu, S.; et al. BrRLP48, Encoding a Receptor-Like Protein, Involved in Downy Mildew Resistance in Brassica rapa. Front. Plant Sci. 2018, 9, 1708. [Google Scholar] [CrossRef]
- Ce, F.; Mei, J.; He, H.; Zhao, Y.; Hu, W.; Yu, F.; Li, Q.; Ren, X.; Si, J.; Song, H.; et al. Identification of Candidate Genes for Clubroot-Resistance in Brassica oleracea Using Quantitative Trait Loci-Sequencing. Front. Plant Sci. 2021, 12, 2569. [Google Scholar] [CrossRef]
- Dakouri, A.; Zhang, X.; Peng, G.; Falk, K.C.; Gossen, B.; Strelkov, S.E.; Yu, F. Analysis of genome-wide variants through bulked segregant RNA sequencing reveals a major gene for resistance to Plasmodiophora brassicae in Brassica oleracea. Sci. Rep. 2018, 8, 17657. [Google Scholar] [CrossRef]
- Farid, M.; Yang, R.-C.; Kebede, B.; Rahman, H. Evaluation of Brassica oleracea accessions for resistance to Plasmodiophora brassicae and identification of genomic regions associated with resistance. Genome 2019, 63, 91–101. [Google Scholar] [CrossRef]
- Mehraj, H.; Akter, A.; Miyaji, N.; Miyazaki, J.; Shea, D.J.; Fujimoto, R.; Doullah, A.-U. Genetics of Clubroot and Fusarium Wilt Disease Resistance in Brassica Vegetables: The Application of Marker Assisted Breeding for Disease Resistance. Plants 2020, 9, 726. [Google Scholar] [CrossRef]
- Lv, H.; Fang, Z.; Yang, L.; Zhang, Y.; Wang, Y. An update on the arsenal: Mining resistance genes for disease management of Brassica crops in the genomic era. Hortic. Res. 2020, 7, 34. [Google Scholar] [CrossRef]
- Hasan, M.J.; Strelkov, S.E.; Howard, R.J.; Rahman, H. Screening of Brassica germplasm for resistance to Plasmodiophora brassicae pathotypes prevalent in Canada for broadening diversity in clubroot resistance. Can. J. Plant Sci. 2012, 92, 501–515. [Google Scholar] [CrossRef]
- Qasim, M.U.; Zhao, Q.; Shahid, M.; Samad, R.A.; Ahmar, S.; Wu, J.; Fan, C.; Zhou, Y. Identification of QTLs Containing Resistance Genes for Sclerotinia Stem Rot in Brassica napus Using Comparative Transcriptomic Studies. Front. Plant Sci. 2020, 11, 776. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.; Jian, H.; Lu, K.; Filardo, F.; Yin, N.; Liu, L.; Qu, C.; Li, W.; Du, H.; Li, J. Genome-wide association analysis and differential expression analysis of resistance to Sclerotinia stem rot in Brassica napus. Plant Biotechnol. J. 2016, 14, 1368–1380. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Cai, G.; Tu, J.; Li, L.; Liu, S.; Luo, X.; Zhou, L.; Fan, C.; Zhou, Y. Identification of QTLs for Resistance to Sclerotinia Stem Rot and BnaC.IGMT5.a as a Candidate Gene of the Major Resistant QTL SRC6 in Brassica napus. PLoS ONE 2013, 8, e67740. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Zhao, Q.; Liu, S.; Shahid, M.; Lan, L.; Cai, G.; Zhang, C.; Fan, C.; Wang, Y.; Zhou, Y. Genome-wide Association Study Identifies New Loci for Resistance to Sclerotinia Stem Rot in Brassica napus. Front. Plant Sci. 2016, 7, 1418. [Google Scholar] [CrossRef]
- Murat, F.; Louis, A.; Maumus, F.; Armero, A.; Cooke, R.; Quesneville, H.; Crollius, H.R.; Salse, J. Understanding Brassicaceae evolution through ancestral genome reconstruction. Genome Biol. 2015, 16, 262. [Google Scholar] [CrossRef]
- Cooley, M.B.; Pathirana, S.; Wu, H.-J.; Kachroo, P.; Klessig, D.F. Members of the Arabidopsis HRT/RPP8 Family of Resistance Genes Confer Resistance to Both Viral and Oomycete Pathogens. Plant Cell 2000, 12, 663–676. [Google Scholar] [CrossRef]
- Takahashi, H.; Miller, J.; Nozaki, Y.; Takeda, M.; Shah, J.; Hase, S.; Ikegami, M.; Ehara, Y.; Dinesh-Kumar, S.P. RCY1, an Arabidopsis thaliana RPP8/HRT family resistance gene, conferring resistance to cucumber mosaic virus requires salicylic acid, ethylene and a novel signal transduction mechanism. Plant J. 2002, 32, 655–667. [Google Scholar] [CrossRef]
- Kachroo, P.; Yoshioka, K.; Shah, J.; Dooner, H.K.; Klessig, D.F. Resistance to Turnip Crinkle Virus in Arabidopsis Is Regulated by Two Host Genes and Is Salicylic Acid Dependent but NPR1, Ethylene, and Jasmonate Independent. Plant Cell 2000, 12, 677–690. [Google Scholar] [CrossRef]
- Warren, R.F.; Merritt, P.M.; Holub, E.; Innes, R.W. Identification of Three Putative Signal Transduction Genes Involved in R Gene-Specified Disease Resistance in Arabidopsis. Genetics 1999, 152, 401–412. [Google Scholar] [CrossRef]
- Tasset, C.; Bernoux, M.; Jauneau, A.; Pouzet, C.; Brière, C.; Kieffer-Jacquinod, S.; Rivas, S.; Marco, Y.; Deslandes, L. Autoacetylation of the Ralstonia solanacearum Effector PopP2 Targets a Lysine Residue Essential for RRS1-R-Mediated Immunity in Arabidopsis. PLoS Pathog. 2010, 6, e1001202. [Google Scholar] [CrossRef]
- Qiao, X.; Li, Q.; Yin, H.; Qi, K.; Li, L.; Wang, R.; Zhang, S.; Paterson, A.H. Gene duplication and evolution in recurring polyploidization–diploidization cycles in plants. Genome Biol. 2019, 20, 38. [Google Scholar] [CrossRef] [PubMed]
- Rizzon, C.; Ponger, L.; Gaut, B.S. Striking Similarities in the Genomic Distribution of Tandemly Arrayed Genes in Arabidopsis and Rice. PLoS Comput. Biol. 2006, 2, e115. [Google Scholar] [CrossRef] [PubMed]
- Tong, C.; Gill, R.A.; Xiang, Y.; Ma, L.; Cheng, X.; Huang, J.; Liu, S. Fractionization of polyploid duplicated genes: Gene loss, expression divergence, and epigenetic regulation in Brassica napus. In The Brassica napus Genome; Springer: Cham, Switzerland, 2018; pp. 149–158. [Google Scholar] [CrossRef]
- Glover, N.; Dessimoz, C.; Ebersberger, I.; Forslund, S.K.; Gabaldón, T.; Huerta-Cepas, J.; Martin, M.-J.; Muffato, M.; Patricio, M.; Pereira, C.; et al. Advances and Applications in the Quest for Orthologs. Mol. Biol. Evol. 2019, 36, 2157–2164. [Google Scholar] [CrossRef] [PubMed]
- Panchy, N.; Lehti-Shiu, M.; Shiu, S.-H. Evolution of Gene Duplication in Plants. Plant Physiol. 2016, 171, 2294–2316. [Google Scholar] [CrossRef] [PubMed]
- Baker, C.R.; Hanson-Smith, V.; Johnson, A.D. Following Gene Duplication, Paralog Interference Constrains Transcriptional Circuit Evolution. Science 2013, 342, 104–108. [Google Scholar] [CrossRef] [PubMed]
- Das, M.; Haberer, G.; Panda, A.; Das Laha, S.; Ghosh, T.C.; Schäffner, A.R. Expression Pattern Similarities Support the Prediction of Orthologs Retaining Common Functions after Gene Duplication Events. Plant Physiol. 2016, 171, 2343. [Google Scholar] [CrossRef]
- Marais, D.L.D.; Rausher, M.D. Escape from adaptive conflict after duplication in an anthocyanin pathway gene. Nature 2008, 454, 762–765. [Google Scholar] [CrossRef]
- Force, A.; Lynch, M.; Pickett, F.B.; Amores, A.; Yan, Y.-L.; Postlethwait, J. Preservation of Duplicate Genes by Complementary, Degenerative Mutations. Genetics 1999, 151, 1531–1545. [Google Scholar] [CrossRef]
- Freeling, M.; Scanlon, M.J.; Fowler, J.E. Fractionation and subfunctionalization following genome duplications: Mechanisms that drive gene content and their consequences. Curr. Opin. Genet. Dev. 2015, 35, 110–118. [Google Scholar] [CrossRef]
- Ohno, S. Evolution by Gene Duplication, 1st ed.; Springer: Berlin/Heidelberg, Germany, 1970; p. 160. [Google Scholar] [CrossRef]
- Faulkner, C.; Robatzek, S. Plants and pathogens: Putting infection strategies and defence mechanisms on the map. Curr. Opin. Plant Biol. 2012, 15, 699–707. [Google Scholar] [CrossRef]
- Nepal, M.P.; Benson, B.V. CNL Disease Resistance Genes in Soybean and Their Evolutionary Divergence. Evol. Bioinform. 2015, 11, 49–63. [Google Scholar] [CrossRef] [PubMed]
- Kato, H.; Saito, T.; Ito, H.; Komeda, Y.; Kato, A. Overexpression of the TIR-X gene results in a dwarf phenotype and activation of defense-related gene expression in Arabidopsis thaliana. J. Plant Physiol. 2014, 171, 382–388. [Google Scholar] [CrossRef] [PubMed]
- Nandety, R.S.; Caplan, J.L.; Cavanaugh, K.; Perroud, B.; Wroblewski, T.; Michelmore, R.W.; Meyers, B.C. The Role of TIR-NBS and TIR-X Proteins in Plant Basal Defense Responses. Plant Physiol. 2013, 162, 1459–1472. [Google Scholar] [CrossRef]
- Ferdous, M.J.; Hossain, M.R.; Park, J.-I.; Robin, A.H.K.; Jesse, D.M.I.; Jung, H.-J.; Kim, H.-T.; Nou, I.-S. Inheritance Pattern and Molecular Markers for Resistance to Blackleg Disease in Cabbage. Plants 2019, 8, 583. [Google Scholar] [CrossRef] [PubMed]
- Ferdous, M.J.; Hossain, M.R.; Park, J.-I.; Robin, A.H.K.; Natarajan, S.; Jesse, D.M.I.; Jung, H.-J.; Kim, H.-T.; Nou, I.-S. In-silico identification and differential expressions of LepR4-syntenic disease resistance related domain containing genes against blackleg causal fungus Leptosphaeria maculans in Brassica oleracea. Gene Rep. 2020, 19, 100598. [Google Scholar] [CrossRef]
- Hossain, M.R.; Ferdous, M.J.; Park, J.-I.; Robin, A.H.K.; Natarajan, S.; Jung, H.-J.; Kim, H.-T.; Nou, I.-S. In-silico identification and differential expression of putative disease resistance-related genes within the collinear region of Brassica napus blackleg resistance locus LepR2’ in Brassica oleracea. Hortic. Environ. Biotechnol. 2020, 61, 879–890. [Google Scholar] [CrossRef]
- Leister, D. Tandem and segmental gene duplication and recombination in the evolution of plant disease resistance genes. Trends Genet. 2004, 20, 116–122. [Google Scholar] [CrossRef]
- Michelmore, R.W.; Meyers, B.C. Clusters of Resistance Genes in Plants Evolve by Divergent Selection and a Birth-and-Death Process. Genome Res. 1998, 8, 1113–1130. [Google Scholar] [CrossRef]
- Stahl, E.A.; Dwyer, G.; Mauricio, R.; Kreitman, M.; Bergelson, J. Dynamics of disease resistance polymorphism at the Rpm1 locus of Arabidopsis. Nature 1999, 400, 667–671. [Google Scholar] [CrossRef]
- Dong, O.X.; Ao, K.; Xu, F.; Johnson, K.C.M.; Wu, Y.; Li, L.; Xia, S.; Liu, Y.; Huang, Y.; Rodriguez, E.; et al. Individual components of paired typical NLR immune receptors are regulated by distinct E3 ligases. Nat. Plants 2018, 4, 699–710. [Google Scholar] [CrossRef]
- Liang, W.; Wersch, S.; Tong, M.; Li, X. TIR-NB-LRR immune receptor SOC 3 pairs with truncated TIR-NB protein CHS 1 or TN 2 to monitor the homeostasis of E3 ligase SAUL 1. New Phytol. 2019, 221, 2054–2066. [Google Scholar] [CrossRef] [PubMed]
- Tong, M.; Kotur, T.; Liang, W.; Vogelmann, K.; Kleine, T.; Leister, D.; Brieske, C.; Yang, S.; Lüdke, D.; Wiermer, M.; et al. E3 ligase SAUL1 serves as a positive regulator of PAMP-triggered immunity and its homeostasis is monitored by immune receptor SOC3. New Phytol. 2017, 215, 1516–1532. [Google Scholar] [CrossRef] [PubMed]
- Van Wersch, S.; Li, X. Stronger When Together: Clustering of Plant NLR Disease resistance Genes. Trends Plant Sci. 2019, 24, 688–699. [Google Scholar] [CrossRef]
- De Araújo, A.C.; Fonseca, F.C.D.A.; Cotta, M.G.; Alves, G.S.C.; Miller, R.N.G. Plant NLR receptor proteins and their potential in the development of durable genetic resistance to biotic stresses. Biotechnol. Res. Innov. 2019, 3, 80–94. [Google Scholar] [CrossRef]
- Friedman, A.R.; Baker, B.J. The evolution of resistance genes in multi-protein plant resistance systems. Curr. Opin. Genet. Dev. 2007, 17, 493–499. [Google Scholar] [CrossRef] [PubMed]
- Charron, C.; Nicolaï, M.; Gallois, J.-L.; Robaglia, C.; Moury, B.; Palloix, A.; Caranta, C. Natural variation and functional analyses provide evidence for co-evolution between plant eIF4E and potyviral VPg. Plant J. 2008, 54, 56–68. [Google Scholar] [CrossRef] [PubMed]
- Dodds, P.N.; Lawrence, G.J.; Catanzariti, A.-M.; Teh, T.; Wang, C.-I.A.; Ayliffe, M.A.; Kobe, B.; Ellis, J.G. Direct protein interaction underlies gene-for-gene specificity and coevolution of the flax resistance genes and flax rust avirulence genes. Proc. Natl. Acad. Sci. USA 2006, 103, 8888–8893. [Google Scholar] [CrossRef]
- Rose, L.E.; Bittner-Eddy, P.D.; Langley, C.H.; Holub, E.B.; Michelmore, R.W.; Beynon, J.L. The Maintenance of Extreme Amino Acid Diversity at the Disease Resistance Gene, RPP13, in Arabidopsis thaliana. Genetics 2004, 166, 1517–1527. [Google Scholar] [CrossRef]
- Yahiaoui, N.; Brunner, S.; Keller, B. Rapid generation of new powdery mildew resistance genes after wheat domestication. Plant J. 2006, 47, 85–98. [Google Scholar] [CrossRef]
- Delourme, R.; Pilet-Nayel, M.-L.; Archipiano, M.; Horvais, R.; Tanguy, X.; Rouxel, T.; Brun, H.; Renard, M.; Balesdent, M.H. A Cluster of Major Specific Resistance Genes to Leptosphaeria maculans in Brassica napus. Phytopathology 2004, 94, 578–583. [Google Scholar] [CrossRef]
- Ghanbarnia, K.; Ma, L.; Larkan, N.J.; Haddadi, P.; Fernando, W.G.D.; Borhan, M.H. Leptosphaeria maculans AvrLm9: A new player in the game of hide and seek with AvrLm4-7. Mol. Plant Pathol. 2018, 19, 1754–1764. [Google Scholar] [CrossRef] [PubMed]
- Plissonneau, C.; Daverdin, G.; Ollivier, B.; Blaise, F.; Degrave, A.; Fudal, I.; Rouxel, T.; Balesdent, M. A game of hide and seek between avirulence genes AvrLm4-7 and AvrLm3 in Leptosphaeria maculans. New Phytol. 2016, 209, 1613–1624. [Google Scholar] [CrossRef] [PubMed]
- Lazar, N.; Mesarich, C.H.; Petit-Houdenot, Y.; Talbi, N.; De la Sierra-Gallay, I.L.; Zélie, E.; Blondeau, K.; Gracy, J.; Ollivier, B.; Blaise, F.; et al. A new family of structurally conserved fungal effectors displays epistatic interactions with plant resistance proteins. bioRxiv 2021. [Google Scholar] [CrossRef]
- Meyers, B.C.; Kozik, A.; Griego, A.; Kuang, H.; Michelmore, R.W. Genome-Wide Analysis of NBS-LRR–Encoding Genes in Arabidopsis. Plant Cell 2003, 15, 809–834. [Google Scholar] [CrossRef]
- Richly, E.; Kurth, J.; Leister, D. Mode of Amplification and Reorganization of Resistance Genes During Recent Arabidopsis thaliana Evolution. Mol. Biol. Evol. 2002, 19, 76–84. [Google Scholar] [CrossRef]
- Perazzolli, M.; Malacarne, G.; Baldo, A.; Righetti, L.; Bailey, A.; Fontana, P.; Velasco, R.; Malnoy, M. Characterization of Resistance Gene Analogues (RGAs) in Apple (Malus × domestica Borkh.) and Their Evolutionary History of the Rosaceae Family. PLoS ONE 2014, 9, e83844. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).