Arbuscular Mycorrhizal Symbiosis Leads to Differential Regulation of Genes and miRNAs Associated with the Cell Wall in Tomato Leaves
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Growth and Tissue Collection
2.2. Sclerotinia Sclerotiorum Inoculum
2.3. Infection Assays of Tomato Leaves with the Foliar Pathogen S. sclerotiorum
2.4. RNA Extraction
2.5. cDNA Synthesis and Quantitative RT-PCR (qPCR)
2.6. Determination of Mycorrhiza Colonization
2.7. Cellulose Quantification
3. Results
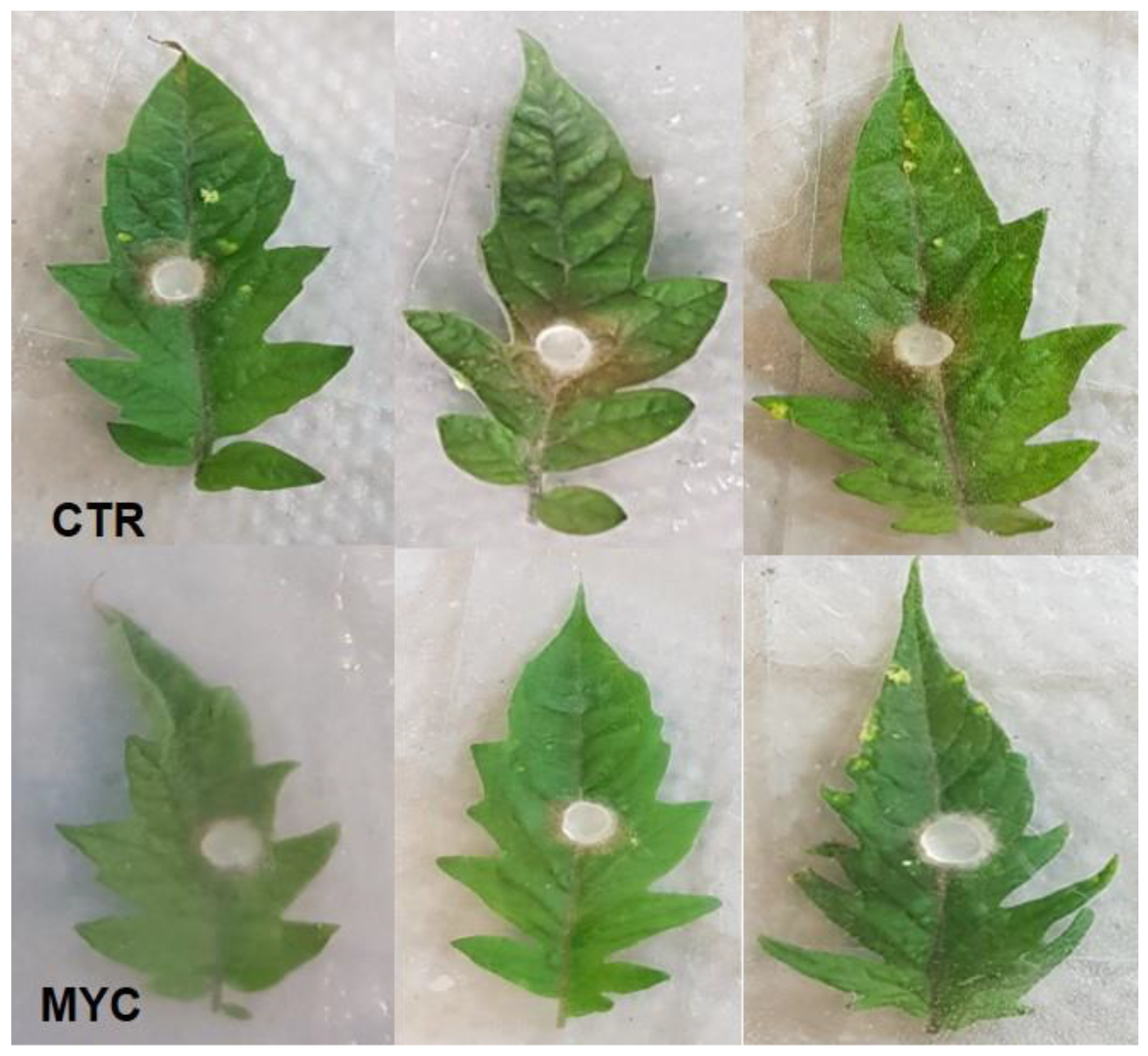
3.1. Leaves of AM Tomato Plants Are Less Susceptible to the Foliar Pathogen S. sclerotiorum
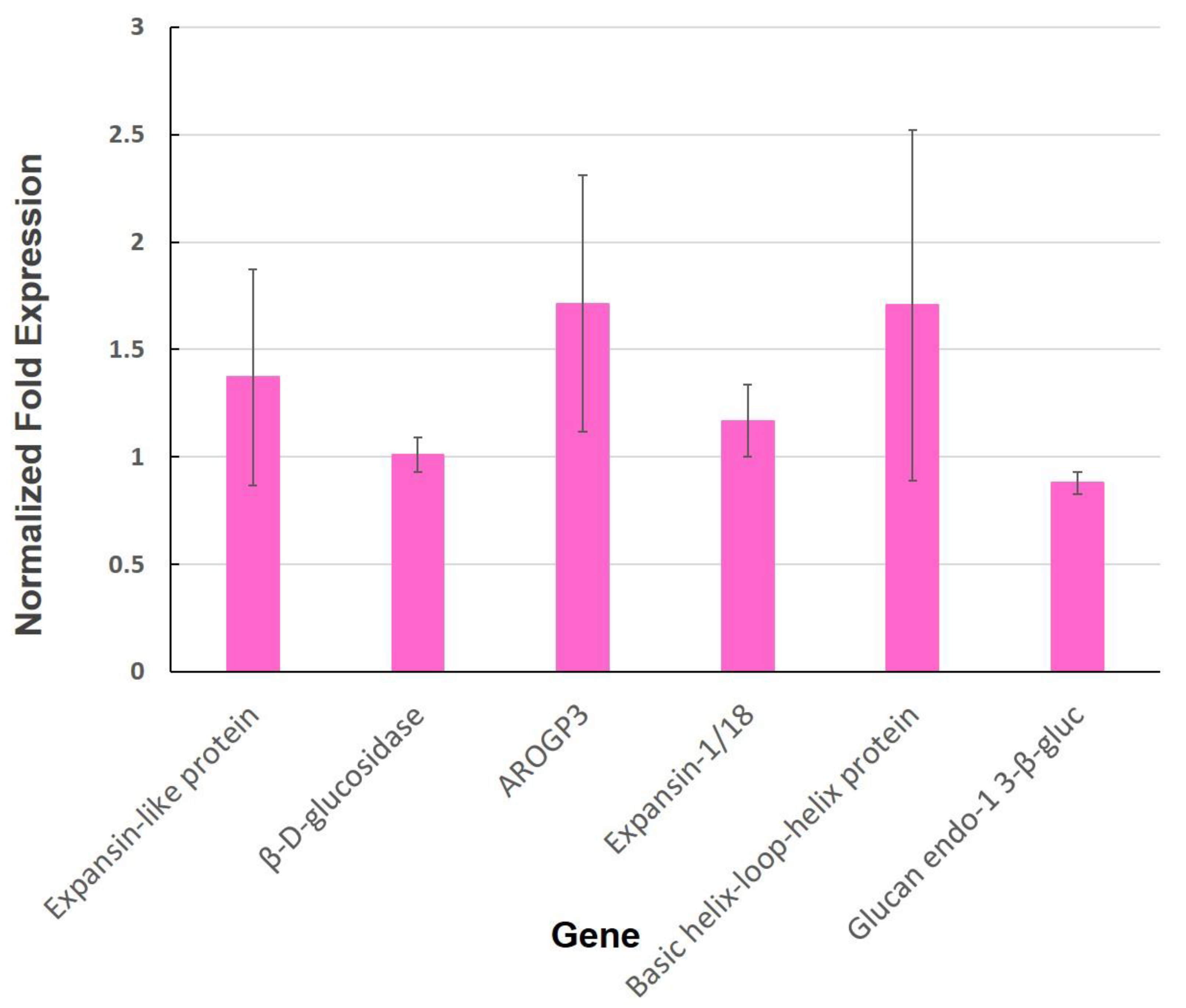
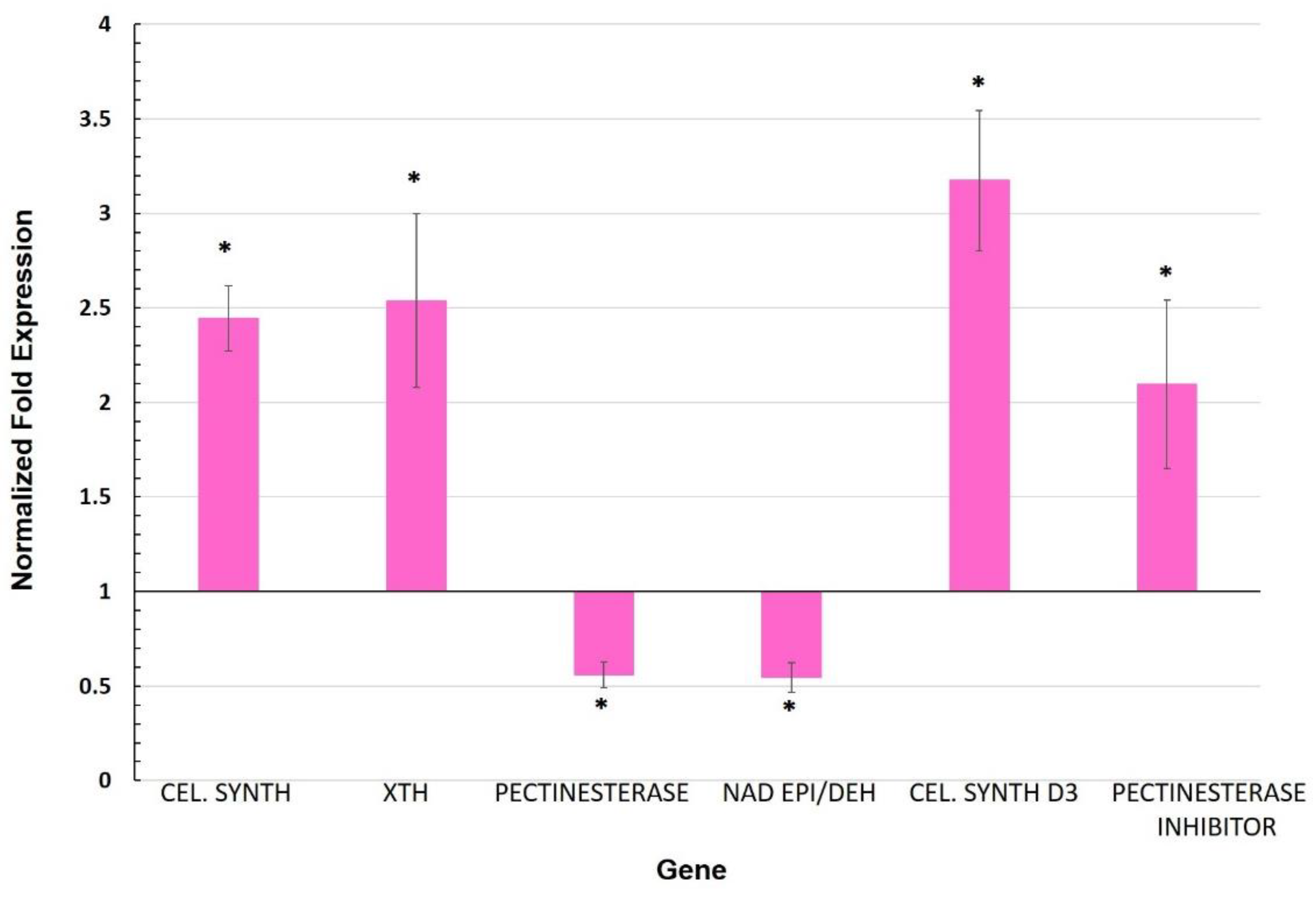
3.2. Expression of Genes Involved in the Cell Wall
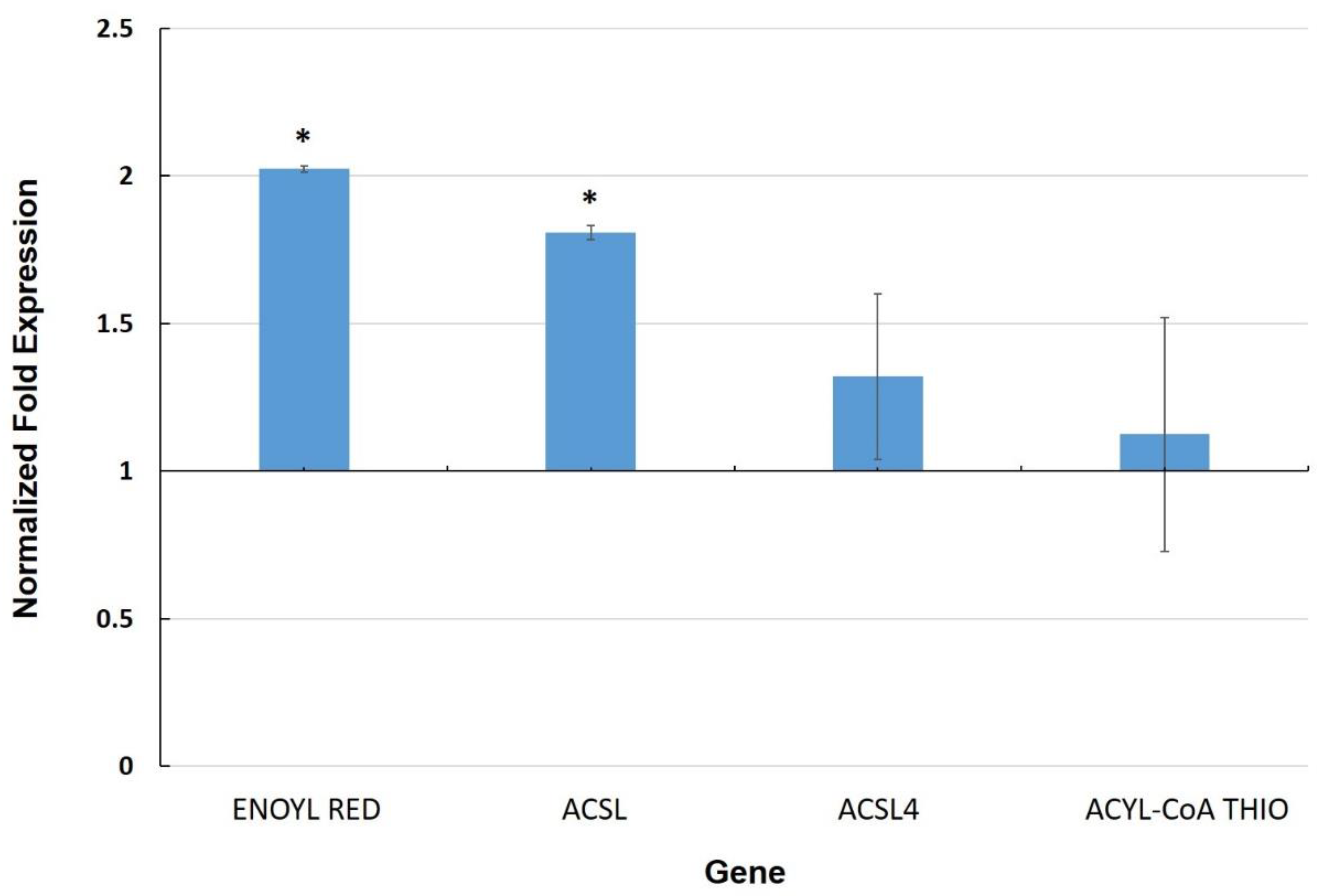
3.3. Expression of Genes Involved in the Cuticle
3.4. Cellulose Content in Leaves of Mycorrhizal Tomato Plants
3.5. Expression Analysis of Selected miRNAs and Their Target Genes in Leaves of Mycorrhiza-Colonized Tomato
4. Discussion
4.1. Mycorrhiza Colonization Primes Tomato Plants for a Better Defense against a Foliar Pathogen
4.2. Cell Wall and Cuticle Genes in Mycorrhiza Defense Priming
4.3. miRNAs as Potential Players in Mycorrhiza Defense Priming
5. Conclusions
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Gene | Accession ID | Fwd | Rv |
|---|---|---|---|
| Cellulose synthase | Solyc07g051820 | TGGGGTCAAAAGTTAGGCTGG | CGGGTCGGGTAAACAGTAGG |
| Expansin-like protein | Solyc08g077330 | AACTCTCAAACATGCCCGGA | TCCACAACTCCCAGTTTCTGT |
| β-D-glucosidase | Solyc11g071640 | TCGGTAGTCAGGAGCATAGAGA | GAGCAGTGGGTGACTAGGTG |
| AROGP3 | Solyc05g005540 | CTTCCTTCCTGCATCTTACTTCT | CATCTTTAGCCACAACAACATCC |
| Expansin-1/18 | Solyc06g076220 | CCCTGGTGTTTTTACTGCCG | GCTCCACCCATAGTACCAGA |
| Xyloglucan endotransglucosydase/hydrolase | Solyc09g008320 | TGGGGTCCTAATCACCAGAGT | CGACTTGAATCCACTGCCTGA |
| Pectinesterase | Solyc06g009190 | CAAGGCGGGAACGTATTTCG | CATTTCCCACCACAGCAACG |
| NAD-dependent epimerase | Solyc12g010540 | CGGTGTTTCGCTTCAACGAG | TGTGGAGGAGAGATACCGGG |
| Basic helix-loop-helix | Solyc09g083360 | AACTAAGAGTGGAGCAGCAGA | TCAATGGGCACGAAGGTTCC |
| Cellulose synthase-like protein D3 | Solyc03g097050 | TGCTGGAATAATACAGGTGATGT | AACAAGCATGGGAAGACGGA |
| Glucan endo-1,3-β glucosidase | Solyc07g047710 | AAATTTCAGTGAATGGGCAGC | TTGCTACAATCTGCACCCCC |
| Pectinesterase inhibitor | Solyc07g042390 | TCGAGCAGGTAAAGCGTCTG | TCCTCCATCGAGTCACCCAT |
| Enoyl reductase | Solyc10g078740 | AGTGACACAAAAGTCCTGGCA | CTTTTGCTGCACGACTTCCC |
| Long-chain-fatty-acid-CoA ligase | Solyc03g025720 | AGCATTTGGGGCTCCTGTTT | CGACCCGGGAATATGAGGTC |
| Long-chain-fatty-acid-CoA ligase 4 | Solyc12g009040 | TGGGAGAGTTACGGGCAAAC | GCGGAAGCATCCCTGAAATG |
| Acyl-CoA thioesterase | Solyc01g094550 | AGGGAGAGCTCGTAGCGT | ACTGCTAATGAACCTGTAGTGAC |
Appendix B
| miRNA | Primer |
|---|---|
| sly-miR156d-3p | GCTCACTGCTCTATCTGTCACC |
| sly-miR164a-3p | CATGTGCCTGTTTTCCCCATC |
| sly-miR164a-5p | TGGAGAAGCAGGGCACGTGCA |
| sly-miR171e-5p | AGATATTGATGCGGTTCAATC |
| sly-miR397-5p | ATTGAGTGCAGCGTTGATGA |
| sly-miR398a | TATGTTCTCAGGTCGCCCCTG |
Appendix C
| Gene | Accession ID | Fwd | Rv |
|---|---|---|---|
| Calmodulin-binding protein | Solyc09g082560.2.1 | GCGATCCAATTCACTGCTGC | TCAGGGCTTTTCTTGCCAAAT |
| Cyclin B1 | Solyc01g009040.2.1 | GGCATCAGACAATCTTGCACC | AACTCCACAAGCAGCCTTGC |
| UDP-glucuronic acid decarboxylase 4 | Solyc01g066710.2.1 | CAGTGCTTCTGTGTCCGTTG | GGCACCTTTCCACCTGCATTA |
| UDP-glucuronic acid decarboxylase 5 | Solyc11g066150.1.1 | GTGACAGAGCCCTTGTTGGT | GCAGAATCCTTGCTCCGACA |
| NAC domain containing protein | Solyc07g066330.2.1 | GTGGAATCCAAATTACCACCAGG | CAACACATGCCACTTCAGGA |
| Glucan endo-1,3 β-glucosidase B | Solyc01g059980.2.1 | CAACATTCACATAACAGAGGCTCA | ATGTGATGGCAAGTTGTTCCC |
| Tubulin β-1 chain | Solyc03g025730.2.1 | CGGAACTTATCGACTCGGTTATG | CCTGAAAACCTTGTAAGCAATCA |
| Laccase | Solyc07g049460.2.1 | CCCTTGCTCCGTTAATCAAACA | TCCGTGACGTAGGGATCAGT |
Appendix D. SlPT End-Point PCR

Appendix E. Mycorrhiza-Induced Resistance at 27 h
Appendix F. Cell Wall-Related Genes
References
- Parniske, M. Arbuscular mycorrhiza: The mother of plant root endosymbioses. Nat. Rev. Microbiol. 2008, 6, 763–775. [Google Scholar] [CrossRef] [PubMed]
- Bonfante, P. At the interface between mycorrhizal fungi and plants: The structural organization of cell wall, plasma membrane and cytoskeleton. In The Mycota; Hock, B., Ed.; Springer: Berlin, Germany, 2001; pp. 45–61. [Google Scholar]
- Aroca, R.; Vernieri, P.; Ruiz-Lozano, J.M. Mycorrhizal and non-mycorrhizal Lactuca sativa plants exhibit contrasting responses to exogenous ABA during drought stress and recovery. J. Exp. Bot. 2008, 59, 2029–2041. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Evelin, H.; Kapoor, R.; Giri, B. Arbuscular mycorrhizal fungi in alleviation of salt stress: A review. Ann. Bot. 2009, 104, 1263–1280. [Google Scholar] [CrossRef] [PubMed]
- Alguacil, M.M.; Torrecillas, E.; Caravaca, F.; Fernández, D.A.; Azcón, R.; Roldán, A. The application of an organic amendment modifies the arbuscular mycorrhizal fungal communities colonizing native seedlings grown in a heavy-metal-polluted soil. Soil Biol. Biochem. 2011, 43, 1498–1508. [Google Scholar] [CrossRef]
- Salam, E.A.; Alatar, A.; El-Sheikh, M.A. Inoculation with arbuscular mycorrhizal fungi alleviates harmful effects of drought stress on damask rose. Saudi J. Biol. Sci. 2017, 25, 1772–1780. [Google Scholar] [CrossRef]
- Oyewole, B.O.; Olawuyi, O.J.; Odebode, A.C.; Abiala, M.A. Influence of Arbuscular mycorrhiza fungi (AMF) on drought tolerance and charcoal rot disease of cowpea. Biotechnol. Rep. 2017, 14, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Whipps, J.M. Prospects and limitations for mycorrhizas in biocontrol of root pathogens. Can. J. Bot. 2004, 82, 1198–1227. [Google Scholar] [CrossRef]
- Harrier, L.A.; Watson, C.A. The potential role of arbuscular mycorrhizal (AM) fungi in the bioprotection of plants against soil-borne pathogens in organic and/or other sustainable farming systems. Pest. Manag. Sci. 2004, 60, 149–157. [Google Scholar] [CrossRef]
- Liu, J.; Maldonado-Mendoza, I.E.; Lopez-Meyer, M.; Cheung, F.; Town, C.D.; Harrison, M.J. Arbuscular mycorrhizal symbiosis is accompanied by local and systemic alterations in gene expression and an increase in disease resistance in the shoots. Plant J. 2007, 50, 529–544. [Google Scholar] [CrossRef]
- Pozo, M.J.; Jung, S.C.; López-Ráez, J.A.; Azcón-Aguilar, C. Impact of arbuscular mycorrhizal symbiosis on plant response to biotic stress: The role of plant defense mechanisms. In Arbuscular Mycorrhizas: Physiology and Function; Springer: Dordrecht, The Netherlands, 2010; pp. 193–207. [Google Scholar]
- Cameron, D.D.; Neal, A.L.; van Wees, S.C.M.; Tonemail, J. Mycorrhiza-induced resistance: More than the sum of its parts? Trends Plant Sci. 2013, 18, 539–545. [Google Scholar] [CrossRef]
- Hildebrandt, U.; Regvar, M.; Bothe, H. Arbuscular mycorrhiza and heavy metal tolerance. Phytochemistry 2007, 68, 139–146. [Google Scholar] [CrossRef]
- Mora-Romero, G.A.; Cervantes-Gámez, R.G.; Galindo-Flores, H.; González-Ortíz, M.A.; Félix-Gastélum, R.; Maldonado-Mendoza, I.E.; Salinas-Pérez, R.; León-Félix, J.; Martínez-Valenzuela, M.C.; López-Meyer, M. Mycorrhiza-induced protection against pathogens is both genotype-specific and graft-transmissible. Symbiosis 2015, 66, 55–64. [Google Scholar] [CrossRef]
- Cervantes-Gámez, R.G.; Bueno-Ibarra, M.A.; Cruz-Mendívil, A.; Calderón-Vázquez, C.L.; Ramírez-Douriet, C.M.; Maldonado-Mendoza, I.E.; López-Meyer, M. Arbuscular mycorrhizal symbiosis-induced expression changes in Solanum lycopersicum leaves revealed by RNA-seq analysis. Plant Mol. Biol. Rep. 2016, 34, 89–102. [Google Scholar] [CrossRef]
- Pozo, M.J.; Verhage, A.; García-Andrade, J.; García, J.M.; Azcón-Aguilar, C. Priming plant defence against pathogens by arbuscular mycorrhizal fungi. In Mycorrhizas-Functional Processes and Ecological Impact; Springer: Berlin/Heidelberg, Germany, 2008; pp. 123–135. [Google Scholar]
- Pozo, M.J.; Azcón-Aguilar, C. Unraveling mycorrhiza-induced resistance. Curr. Opin. Plant Biol. 2007, 10, 393–398. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Medina, A.; Flors, V.; Heil, M.; Mauch-Mani, B.; Pieterse, C.; Pozo, M.J.; Ton, J.; van Dam, N.M.; Conrath, U. Recognizing plant defense priming. Trends Plant Sci. 2016, 21, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Hao, Z.; Fayolle, L.; van Tuinen, D.; Chatagnier, O.; Li, X.; Gianinazzi, S.; Gianinazzi-Pearson, V. Local and systemic mycorrhiza-induced protection against the ectoparasitic nematode Xiphinema index involves priming of defence gene responses in grapevine. J. Exp. Bot. 2012, 63, 3657–3672. [Google Scholar] [CrossRef]
- Jung, S.C.; Martinez-Medina, A.; Lopez-Raez, J.A.; Pozo, M.J. Mycorrhiza-induced resistance and priming of plant defenses. J. Chem. Ecol. 2012, 38, 651–664. [Google Scholar] [CrossRef]
- Hohmann, P.; Messmer, M.M. Breeding for mycorrhizal symbiosis: Focus on disease resistance. Euphytica 2017, 213, 113. [Google Scholar] [CrossRef]
- Cordier, C.; Pozo, M.J.; Barea, J.M.; Gianinazzi, S.; Gianinazzi-Pearson, V. Cell defense responses associated with localized and systemic resistance to Phytophthora parasitica induced in tomato by an arbuscular mycorrhizal fungus. Mol. Plant Microbe Interact. 1998, 11, 1017–1028. [Google Scholar] [CrossRef]
- Marro, N.; Caccia, M.; Doucet, M.E.; Cabello, M.; Becerra, A.; Lax, P. Mycorrhizas reduce tomato root penetration by false root-knot nematode Nacobbus aberrans. Appl. Soil Ecol. 2018, 124, 262–265. [Google Scholar] [CrossRef]
- Pozo, M.J.; Cordier, C.; Dumas-Gaudot, E.; Gianinazzi, S.; Barea, J.M.; Azcón-Aguilar, C. Localized versus systemic effect of arbuscular mycorrhizal fungi on defence responses to infection in tomato plants. J. Exp. Bot. 2002, 53, 525–534. [Google Scholar] [CrossRef] [PubMed]
- Fritz, M.; Jakobsen, I.; Lyngkjaer, M.F.; Thordal-Christensen, H.; Pons-Kühnemann, J. Arbuscular mycorrhiza reduces susceptibility of tomato to Alternaria solani. Mycorrhiza 2006, 16, 413–419. [Google Scholar] [CrossRef] [PubMed]
- Sanmartín, N.; Pastor, V.; Pastor-Fernández, J.; Flors, V.; Pozo, M.J.; Sánchez-Bel, P. Role and mechanisms of callose priming in mycorrhiza-induced resistance. J. Exp. Bot. 2020, 71, 2769–2781. [Google Scholar] [CrossRef] [PubMed]
- Campos-Soriano, L.; García-Martínez, J.; Segundo, B.S. The arbuscular mycorrhizal symbiosis promotes the systemic induction of regulatory defence-related genes in rice leaves and confers resistance to pathogen infection. Mol. Plant Pathol. 2012, 13, 579–592. [Google Scholar] [CrossRef] [PubMed]
- Goddard, M.L.; Belval, L.; Martin, I.R.; Roth, L.; Laloue, H.; Deglène-Benbrahim, L.; Valat, L.; Bertsch, C.; Chong, J. Arbuscular Mycorrhizal Symbiosis Triggers Major Changes in Primary Metabolism Together with Modification of Defense Responses and Signaling in Both Roots and Leaves of Vitis vinifera. Front. Plant Sci. 2021, 12, 721614. [Google Scholar] [CrossRef]
- Voinnet, O. Origin, biogenesis, and activity of plant microRNAs. Cell 2009, 136, 669–687. [Google Scholar] [CrossRef] [PubMed]
- Vidigal, J.A.; Ventura, A. The biological functions of miRNAs: Lessons from in vivo studies. Trends Cell Biol. 2015, 25, 137–147. [Google Scholar] [CrossRef]
- Song, X.; Li, Y.; Cao, X.; Qi, Y. MicroRNAs and their regulatory roles in plant-environment interactions. Annu. Rev. Plant Biol. 2019, 70, 489–525. [Google Scholar] [CrossRef]
- Sunkar, R.; Kapoor, A.; Zhu, J.K. Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in is mediated by downregulation of miR398 and important for oxidative stress tolerance. Plant Cell 2006, 18, 2051–2065. [Google Scholar] [CrossRef]
- Navarro, L.; Dunoyer, P.; Jay, F.; Arnold, B.; Dharmasiri, N.; Estelle, M.; Voinnet, O.; Jones, J.D. A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 2006, 312, 436–439. [Google Scholar] [CrossRef]
- Lu, Y.D.; Gan, Q.H.; Chi, X.Y.; Qin, S. Roles of microRNA in plant defense and virus offense interaction. Plant Cell Rep. 2008, 27, 1571–1579. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.G.; Shan, J.X.; Shi, M.; Gao, J.P.; Lin, H.X. The miR156-SPL9-DFR pathway coordinates the relationship between development and abiotic stress tolerance in plants. Plant J. 2014, 80, 1108–1117. [Google Scholar] [CrossRef] [PubMed]
- Naya, L.; Paul, S.; Valdés-López, O.; Mendoza-Soto, A.B.; Nova-Franco, B.; Sosa-Valencia, G.; Reyes, J.L.; Hernández, G. Regulation of copper homeostasis and biotic interactions by microRNA 398b in common bean. PLoS ONE 2014, 9, e84416. [Google Scholar] [CrossRef] [PubMed]
- Mendoza-Soto, A.B.; Naya, L.; Leija, A.; Hernández, G. Responses of symbiotic nitrogen-fixing common bean to aluminum toxicity and delineation of nodule responsive microRNAs. Front. Plant Sci. 2015, 6, 587. [Google Scholar] [CrossRef]
- Xie, M.; Zhang, S.; Yu, B. microRNA biogenesis, degradation and activity in plants. Cell Mol. Life Sci. 2015, 72, 87–99. [Google Scholar] [CrossRef]
- Kouhi, F.; Sorkheh, K.; Ercisli, S. MicroRNA expression patterns unveil differential expression of conserved miRNAs and target genes against abiotic stress in safflower. PLoS ONE 2020, 15, e0228850. [Google Scholar] [CrossRef]
- Axtell, M.J.; Westholm, J.O.; Lai, E.C. Vive la différence: Biogenesis and evolution of microRNAs in plants and animals. Genome Biol. 2011, 12, 221. [Google Scholar] [CrossRef]
- Ziv, C.; Zhao, Z.; Gao, Y.G.; Xia, Y. Multifunctional Roles of Plant Cuticle During Plant-Pathogen Interactions. Front. Plant Sci. 2018, 9, 1088. [Google Scholar] [CrossRef]
- Ho-Plágaro, T.; Huertas, R.; Tamayo-Navarrete, M.I.; Ocampo, J.A.; García-Garrido, J.M. An improved method for Agrobacterium rhizogenes-mediated transformation of tomato suitable for the study of arbuscular mycorrhizal symbiosis. Plant Methods 2018, 14, 34. [Google Scholar] [CrossRef]
- Updegraff, D.M. Semimicro determination of cellulose inbiological materials. Anal. Biochem. 1969, 32, 420–424. [Google Scholar] [CrossRef]
- Engelsdorf, T.; Will, C.; Hofmann, J.; Schmitt, C.; Merritt, B.B.; Rieger, L.; Frenger, M.S.; Marschall, A.; Franke, R.B.; Pattathil, S.; et al. Cell wall composition and penetration resistance against the fungal pathogen Colletotrichum higginsianum are affected by impaired starch turnover in Arabidopsis mutants. J. Exp. Bot. 2017, 68, 701–713. [Google Scholar] [CrossRef] [PubMed]
- Serrano, M.; Coluccia, F.; Torres, M.; L’Haridon, F.; Métraux, J.P. The cuticle and plant defense to pathogens. Front. Plant Sci. 2014, 5, 274. [Google Scholar] [CrossRef] [PubMed]
- Wu, P.; Wu, Y.; Liu, C.C.; Liu, L.W.; Ma, F.F.; Wu, X.Y.; Wu, M.; Hang, Y.Y.; Chen, J.Q.; Shao, Z.Q.; et al. Identification of arbuscular mycorrhiza (AM)-responsive microRNAs in Tomato. Front. Plant Sci. 2016, 7, 429. [Google Scholar] [CrossRef] [PubMed]
- Rai, K.M.; Thu, S.W.; Balasubramanian, V.K.; Cobos, C.J.; Disasa, T.; Mendu, V. Identification, characterization, and expression analysis of cell wall related genes in Sorghum bicolor (L.) moench, a food, fodder, and biofuel crop. Front. Plant Sci. 2016, 7, 1287. [Google Scholar] [CrossRef]
- Song, Y.; Chen, D.; Lu, K.; Sun, Z.; Zeng, R. Enhanced tomato disease resistance primed by arbuscular mycorrhizal fungus. Front. Plant Sci. 2015, 6, 786. [Google Scholar] [CrossRef]
- Mustafa, G.; Randoux, B.; Tisserant, B.; Fontaine, J.; Magnin-Robert, M.; Lounès-Hadj Sahraoui, A.; Reignault, P. Phosphorus supply, arbuscular mycorrhizal fungal species, and plant genotype impact on the protective efficacy of mycorrhizal inoculation against wheat powdery mildew. Mycorrhiza 2016, 26, 685–697. [Google Scholar] [CrossRef]
- Fiorilli, V.; Vannini, C.; Ortolani, F.; Garcia-Seco, D.; Chiapello, M.; Novero, M.; Domingo, G.; Terzi, V.; Morcia, C.; Bagnaresi, P.; et al. Omics approaches revealed how arbuscular mycorrhizal symbiosis enhances yield and resistance to leaf pathogen in wheat. Sci. Rep. 2018, 8, 9625. [Google Scholar] [CrossRef]
- Underwood, W. The plant cell wall: A dynamic barrier against pathogen invasion. Front. Plant Sci. 2012, 3, 85. [Google Scholar] [CrossRef]
- Carpita, N.C.; McCann, M. The cell wall. In Biochemistry and Molecular Biology of Plants; Rockville, M.D., American Society of Plant Physiologists, Buchanan, B., Gruissem, W., Jones, R.L., Eds.; Wiley: New York, NY, USA, 2000; pp. 52–108. [Google Scholar]
- Chylińska, M.; Szymańska-Chargot, M.; Zdunek, A. Imaging of polysaccharides in the tomato cell wall with Raman microspectroscopy. Plant Methods 2014, 10, 14. [Google Scholar] [CrossRef]
- Lunn, D.; Phan, T.D.; Tucker, G.A.; Lycett, G.W. Cell wall composition of tomato fruit changes during development and inhibition of vesicle trafficking is associated with reduced pectin levels and reduced softening. Plant Physiol. Biochem. 2013, 66, 91–97. [Google Scholar] [CrossRef]
- Zheng, W.; Ma, L.; Zhao, J.; Li, Z.; Sun, F.; Lu, X. Comparative transcriptome analysis of two rice varieties in response to rice stripe virus and small brown planthoppers during early interaction. PLoS ONE 2013, 8, e82126. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, M.T.; Stein, M.; Hou, B.-H.; Vogel, J.P.; Edwards, H.; Somerville, S.C. Loss of a callose synthase results in salicylic acid-dependent disease resistance. Science 2003, 301, 969–972. [Google Scholar] [CrossRef] [PubMed]
- Zeyen, R.J.; Carver, T.L.W.; Lyngkjaer, M.F. Epidermal cell papillae. In The Powdery Mildews: A Comprehensive Treatise; Belanger, R.R., Buschnell, W.R., Dik, A.J., Carver, T.L.W., Eds.; APS Press: Saint Paul, MN, USA, 2002; pp. 107–125. [Google Scholar]
- Vega-Sánchez, M.E.; Verhertbruggen, Y.; Christensen, U.; Chen, X.; Sharma, V.; Varanasi, P.; Jobling, S.A.; Talbot, M.; White, R.G.; Joo, M.; et al. Loss of Cellulose synthase-like F6 function affects mixed-linkage glucan deposition, cell wall mechanical properties, and defense responses in vegetative tissues of rice. Plant Physiol. 2012, 159, 56–69. [Google Scholar] [CrossRef] [PubMed]
- Kesten, C.; Menna, A.; Sanchez-Rodriguez, C. Regulation of cellulose synthesis in response to stress. Curr. Opin. Plant Biol. 2017, 40, 106–113. [Google Scholar] [CrossRef]
- Wiethölter, N.; Graessner, B.; Mierau, M.; Mort, A.J.; Moerschbacher, B.M. Differences in the methyl ester distribution of homogalacturonans from near-isogenic wheat lines resistant and susceptible to the wheat stem rust fungus. Mol. Plant Microbe Interact. 2003, 16, 945–952. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.H.; Citovsky, V. Systemic movement of a tobamovirus requires host cell pectin methylesterase. Plant J. 2003, 35, 386–392. [Google Scholar] [CrossRef]
- Liu, N.; Sun, Y.; Pei, Y.; Zhang, X.; Wang, P.; Li, X.; Li, F.; Hou, Y. A pectin methylesterase inhibitor enhances resistance to Verticillium wilt. Plant Physiol. 2018, 176, 2202–2220. [Google Scholar] [CrossRef]
- An, S.H.; Sohn, K.H.; Choi, H.W.; Hwang, I.S.; Lee, S.C.; Hwang, B.K. Pepper pectin methylesterase inhibitor protein CaPMEI1 is required for antifungal activity, basal disease resistance and abiotic stress tolerance. Planta 2008, 228, 61–78. [Google Scholar] [CrossRef]
- Badmi, R.; Payyavula, R.S.; Bali, G.; Guo, H.B.; Jawdy, S.S.; Gunter, L.E.; Yang, X.; Winkeler, K.A.; Collins, C.; Rottmann, W.H.; et al. A New Calmodulin-Binding Protein Expresses in the Context of Secondary Cell Wall Biosynthesis and Impacts Biomass Properties in Populus. Front. Plant Sci. 2018, 9, 1669. [Google Scholar] [CrossRef]
- Lecourieux, D.; Mazars, C.; Pauly, N.; Ranjeva, R.; Pugin, A. Analysis and effects of cytosolic free calcium increases in response to elicitors in Nicotiana plumbaginifolia cells. Plant Cell 2002, 14, 2627–2641. [Google Scholar] [CrossRef]
- Lv, T.; Li, X.; Fan, T.; Luo, H.; Xie, C.; Zhou, Y.; Tian, C.E. The calmodulin-binding protein IQM1 interacts with CATALASE2 to affect pathogen defense. Plant Physiol. 2019, 181, 1314–1327. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.C.; Lee, S.H.; Kim, J.K.; Chun, H.J.; Choi, M.S.; Chung, W.S.; Moon, B.C.; Kang, C.H.; Park, C.Y.; Yoo, J.H.; et al. Mlo, a modulator of plant defense and cell death, is a novel calmodulin-binding protein. Isolation and characterization of a rice Mlo homologue. J. Biol. Chem. 2002, 277, 19304–19314. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Van Houten, J.; Gonzalez, G.; Xiao, H.; van der Knaap, E. Genome-wide identification, phylogeny and expression analysis of SUN, OFP and YABBY gene family in tomato. Mol. Genet. Genomics. 2013, 288, 111–129. [Google Scholar] [CrossRef] [PubMed]
- Criqui, M.C.; Parmentier, Y.; Derevier, A.; Shen, W.H.; Dong, A.; Genschik, P. Cell cycle-dependent proteolysis and ectopic overexpression of cyclin B1 in tobacco BY2 cells. Plant J. 2000, 24, 763–773. [Google Scholar] [CrossRef]
- Weimer, A.K.; Biedermann, S.; Harashima, H.; Roodbarkelari, F.; Takahashi, N.; Foreman, J.; Guan, Y.; Pochon, G.; Heese, M.; Van Damme, D.; et al. The plant-specific CDKB1-CYCB1 complex mediates homologous recombination repair in Arabidopsis. EMBO J. 2016, 35, 2068–2086. [Google Scholar] [CrossRef]
- Ambastha, V.; Leshem, Y. Cyclin B1;1 activity is observed in lateral roots but not in the primary root during lethal salinity and salt stress recovery. Plant Signal Behav. 2020, 15, 1776026. [Google Scholar] [CrossRef]
- Berger, Y.; Harpaz-Saad, S.; Brand, A.; Melnik, H.; Sirding, N.; Alvarez, J.P.; Zinder, M.; Samach, A.; Eshed, Y.; Ori, N. The NAC-domain transcription factor GOBLET specifies leaflet boundaries in compound tomato leaves. Development 2009, 136, 823–832. [Google Scholar] [CrossRef]
- Guo, Y.; Gan, S. AtNAP, a NAC family transcription factor, has an important role in leaf senescence. Plant J. 2006, 46, 601–612. [Google Scholar] [CrossRef]
- Mitsuda, N.; Seki, M.; Shinozaki, K.; Ohme-Takagi, M. The NAC transcription factors NST1 and NST2 of Arabidopsis regulate secondary wall thickenings and are required for anther dehiscence. Plant Cell 2005, 17, 2993–3006. [Google Scholar] [CrossRef]
- Nuruzzaman, M.; Sharoni, M.; Kikuchi, S. Roles of NAC transcription factors in the regulation of biotic and abiotic stress responses in plants. Front. Microbiol. 2013, 4, 248. [Google Scholar] [CrossRef]
- Yokotani, N.; Ichikawa, T.; Kondou, Y.; Matsui, M.; Hirochika, H.; Iwabuchi, M.; Oda, K. Tolerance to various environmental stresses conferred by the salt-responsive rice gene ONAC063 in transgenic Arabidopsis. Planta 2009, 229, 1065–1075. [Google Scholar] [CrossRef] [PubMed]
- Shahnejat-Bushehri, S.; Mueller-Roeber, B.; Balazadeh, S. Arabidopsis NAC transcription factor JUNGBRUNNEN1 affects thermomemory-associated genes and enhances heat stress tolerance in primed and unprimed conditions. Plant Signal Behav. 2012, 7, 1518–1521. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Wang, W.; Zeng, J.; Zhang, J.; Grierson, D.; Li, X.; Yin, X.; Chen, K. A NAC transcription factor, EjNAC1, affects lignification of loquat fruit by regulating lignin. Postharvest Biol. Technol. 2015, 102, 25–31. [Google Scholar] [CrossRef]
- Zhong, R.; Richardson, E.A.; Ye, Z.H. Two NAC domain transcription factors, SND1 and NST1, function redundantly in regulation of secondary wall synthesis in fibers of Arabidopsis. Planta 2007, 225, 1603–1611. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Jiang, C.; Jiang, R.; Wang, F.; Zhang, Z.; Zeng, J. A Novel NAC Transcription Factor from Eucalyptus, EgNAC141, Positively Regulates Lignin Biosynthesis and Increases Lignin Deposition. Front. Plant Sci. 2021, 12, 642090. [Google Scholar] [CrossRef]
- Hu, H.; Dai, M.; Yao, J.; Xiao, B.; Li, X.; Zhang, Q.; Xiong, L. Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc. Natl. Acad. Sci. USA 2006, 103, 12987–12992. [Google Scholar] [CrossRef]
- Nakashima, K.; Tran, L.S.; Van Nguyen, D.; Fujita, M.; Maruyama, K.; Todaka, D.; Ito, Y.; Hayashi, N.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice. Plant J. 2007, 51, 617–630. [Google Scholar] [CrossRef]
- Kaur, P.; Shukla, N.; Joshi, G.; VijayaKumar, C.; Jagannath, A.; Agarwal, M.; Goel, S.; Kumar, A. Genome-wide identification and characterization of miRNAome from tomato (Solanum lycopersicum) roots and root-knot nematode (Meloidogyne incognita) during susceptible interaction. PLoS ONE 2017, 12, e0175178. [Google Scholar] [CrossRef]
- Zheng, H.Y.; York, W.S.; Darvill, A.G. Important new players in secondary wall synthesis. Trends Plant Sci. 2006, 11, 162–164. [Google Scholar]
- Pan, Y.X.; Wang, X.F.; Liu, H.W.; Zhang, G.Y.; Ma, Z.Y. Molecular cloning of three UDP-glucuronate decarboxylase genes that are preferentially expressed in gossypium fibers from elongation to secondary cell wall synthesis. J. Plant Biol. 2010, 53, 367–373. [Google Scholar] [CrossRef]
- Crowe, J.D.; Hao, P.; Pattathil, S.; Pan, H.; Ding, S.Y.; Hodge, D.B.; Jensen, J.K. Xylan Is Critical for Proper Bundling and Alignment of Cellulose Microfibrils in Plant Secondary Cell Walls. Front. Plant Sci. 2021, 12, 737690. [Google Scholar] [CrossRef] [PubMed]
- Simmons, T.J.; Mortimer, J.C.; Bernardinelli, O.D.; Pöppler, A.-C.; Brown, S.P.; de Azevedo, E.R.; Dupree, R.; Dupree, P. Folding of xylan onto cellulose fibrils in plant cell walls revealed by solid-state NMR. Nat. Commun. 2016, 7, 13902. [Google Scholar] [CrossRef]
- Kang, X.; Kirui, A.; Dickwella Widanage, M.C.; Mentink-Vigier, F.; Cosgrove, D.J.; Wang, T. Lignin-polysaccharide interactions in plant secondary cell walls revealed by solid-state NMR. Nat. Commun. 2019, 10, 347. [Google Scholar] [CrossRef] [PubMed]
- Ludueña, R.F. Multiple forms of tubulin: Different gene products and covalent modifications. Int. Rev. Cytol. 1998, 178, 207–275. [Google Scholar] [CrossRef]
- Nogales, E. Structural insights into microtubule function. Annu. Rev. Biochem. 2000, 69, 277–302. [Google Scholar] [CrossRef]
- Spokevicius, A.V.; Southerton, S.G.; MacMillan, C.P.; Qiu, D.; Gan, S.; Tibbits, J.F.; Moran, G.F.; Bossinger, G. beta-tubulin affects cellulose microfibril orientation in plant secondary fibre cell walls. Plant J. 2007, 51, 717–726. [Google Scholar] [CrossRef] [PubMed]
- Gavnholt, B.; Larsen, K. Molecular biology of plant laccases in relation to lignin formation. Physiol. Plant. 2002, 116, 273–280. [Google Scholar] [CrossRef]
- Lee, B.R.; Muneer, S.; Jung, W.J.; Avice, J.C.; Ourry, A.; Kim, T.H. Mycorrhizal colonization alleviates drought-induced oxidative damage and lignification in the leaves of drought-stressed perennial ryegrass (Lolium perenne). Physiol. Plant. 2012, 145, 440–449. [Google Scholar] [CrossRef]
- Baslam, M.; Antolín, M.C.; Gogorcena, Y.; Muñoz, F.; Goicoechea, N. Changes in alfalfa forage quality and stem carbohydrates induced by arbuscular mycorrhizal fungi and elevated atmospheric CO2. Ann. Appl. Biol. 2013, 164, 190–199. [Google Scholar] [CrossRef]
- Pan, L.; Zhao, H.; Yu, Q.; Bai, L.; Dong, L. miR397/Laccase Gene Mediated Network Improves Tolerance to Fenoxprop-P-ethyl in Beckmannia syzigachne and Oryza sativa. Front. Plant Sci. 2017, 8, 879. [Google Scholar] [CrossRef]
- Chen, J.; Zhang, H.; Zhang, X.; Tang, M. Arbuscular mycorrhizal symbiosis mitigates oxidative injury in black locust under salt stress through modulating antioxidant defence of the plant. Environ. Exp. Bot. 2020, 175, 104034. [Google Scholar] [CrossRef]
- Cui, J.; Jiang, N.; Hou, X.; Wu, S.; Zhang, Q.; Meng, J.; Luan, Y. Genome-Wide Identification of lncRNAs and Analysis of ceRNA Networks During Tomato Resistance to Phytophthora infestans. Phytopathology 2020, 110, 456–464. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, D.; Maji, R.K.; Dey, S.; Sarkar, A.; Ghosh, Z.; Kundu, P. Integrated miRNA and mRNA expression profiling reveals the response regulators of a susceptible tomato cultivar to early blight disease. DNA Res. 2017, 24, 235–250. [Google Scholar] [CrossRef] [PubMed]


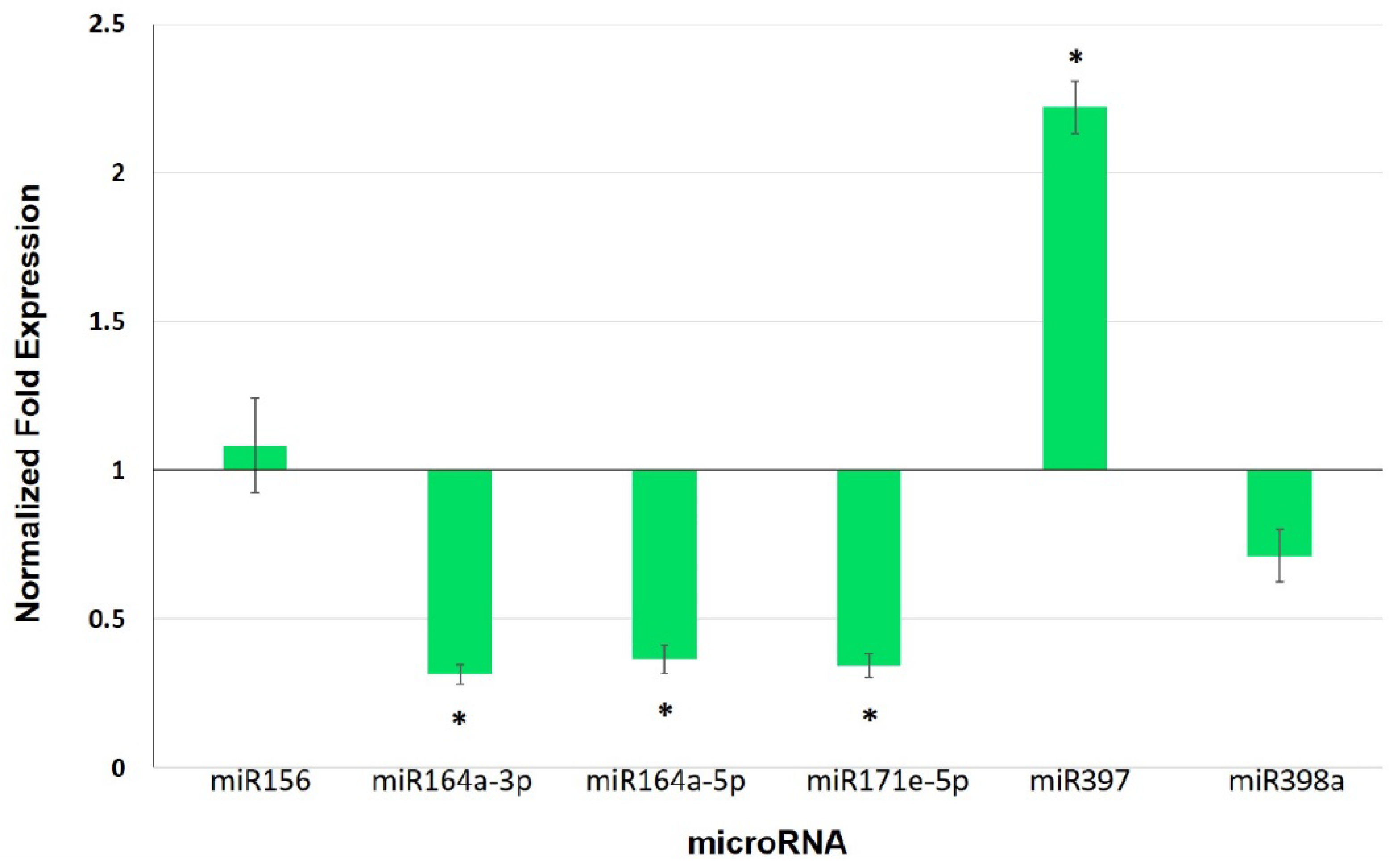
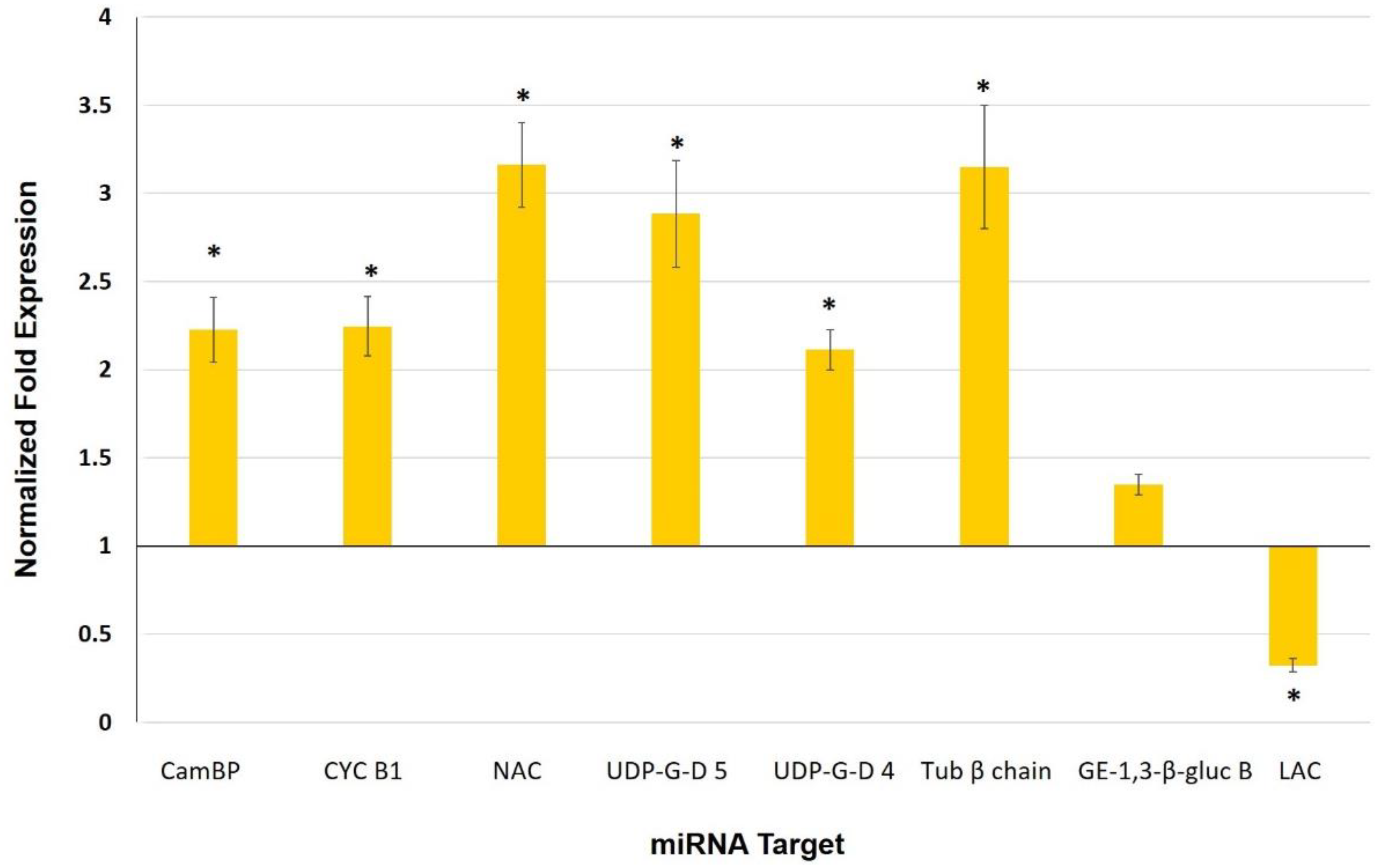
| miRNA | Target | Gene | Expectation |
|---|---|---|---|
| sly-miR156 | Acyl-CoA dehydrogenase family | Solyc05g054370.2.1 | 1.5 |
| sly-miR164a-3p | Calmodulin-binding protein | Solyc09g082560.2.1 | 2.0 |
| Cyclin B1 | Solyc01g009040.2.1 | 3.0 | |
| sly-miR164a-5p | NAC domain protein | Solyc07g066330.2.1 | 1.0 |
| UDP-glucuronic acid decarboxylase 5 | Solyc11g066150.1.1 | 1.5 | |
| UDP-glucuronic acid decarboxylase 4 | Solyc01g066710.2.1 | 3.0 | |
| sly-miR171e-5p | Tubulin β chain | Solyc03g025730.2.1 | 2.5 |
| Glucan endo-1,3-β-glucosidase B | Solyc01g059980.2.1 | 3.0 | |
| sly-miR397 | Laccase | Solyc07g049460.2.1 | 0.5 |
| sly-miR398a | Endoglucanase 1 | Solyc08g083210.2.1 | 3.0 |
| Endoglucanase 1 | Solyc09g075360.2.1 | 3.5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mendoza-Soto, A.B.; Rodríguez-Corral, A.Z.; Bojórquez-López, A.; Cervantes-Rojo, M.; Castro-Martínez, C.; Lopez-Meyer, M. Arbuscular Mycorrhizal Symbiosis Leads to Differential Regulation of Genes and miRNAs Associated with the Cell Wall in Tomato Leaves. Biology 2022, 11, 854. https://doi.org/10.3390/biology11060854
Mendoza-Soto AB, Rodríguez-Corral AZ, Bojórquez-López A, Cervantes-Rojo M, Castro-Martínez C, Lopez-Meyer M. Arbuscular Mycorrhizal Symbiosis Leads to Differential Regulation of Genes and miRNAs Associated with the Cell Wall in Tomato Leaves. Biology. 2022; 11(6):854. https://doi.org/10.3390/biology11060854
Chicago/Turabian StyleMendoza-Soto, Ana Belén, Amada Zulé Rodríguez-Corral, Adriana Bojórquez-López, Maylin Cervantes-Rojo, Claudia Castro-Martínez, and Melina Lopez-Meyer. 2022. "Arbuscular Mycorrhizal Symbiosis Leads to Differential Regulation of Genes and miRNAs Associated with the Cell Wall in Tomato Leaves" Biology 11, no. 6: 854. https://doi.org/10.3390/biology11060854
APA StyleMendoza-Soto, A. B., Rodríguez-Corral, A. Z., Bojórquez-López, A., Cervantes-Rojo, M., Castro-Martínez, C., & Lopez-Meyer, M. (2022). Arbuscular Mycorrhizal Symbiosis Leads to Differential Regulation of Genes and miRNAs Associated with the Cell Wall in Tomato Leaves. Biology, 11(6), 854. https://doi.org/10.3390/biology11060854






