Comparative Analysis and Structural Modeling of Elaeis oleifera FAD2, a Fatty Acid Desaturase Involved in Unsaturated Fatty Acid Composition of American Oil Palm
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. FAD2 Sequence Identification, Annotation, and Collection
2.2. Physico-Chemical Properties of the FAD2 Protein
2.3. Bioinformatics Analysis, Three-Dimensional (3D) Protein Structure Modeling, and Docking Analysis
2.4. Interspecific FAD2 SNPs Identification
2.5. Statistical Analysis
3. Results and Discussion
3.1. EoFAD2 Identification and Annotation
3.2. Comparative Analysis and Phylogenetic Study of fad2 Genes from Several Oilseed Species
- (a)
- The first group contained FAD2 members of six species: Glycine max, Arachis hypogaea, Arachis monticola, Helianthus annuum, and Sesamum indicum;
- (b)
- The second group corresponded to a single species: Brassica napus;
- (c)
- The third group included Zea mays with Elaeis oleifera.
3.3. Estimate of Evolutionary Divergence among the Sequences
3.4. Comparative Analysis between Interspecific FAD2 SNPs
3.4.1. In Silico SNP Prediction
3.4.2. Genotype/Phenotype Association Study of Predicted SNPs
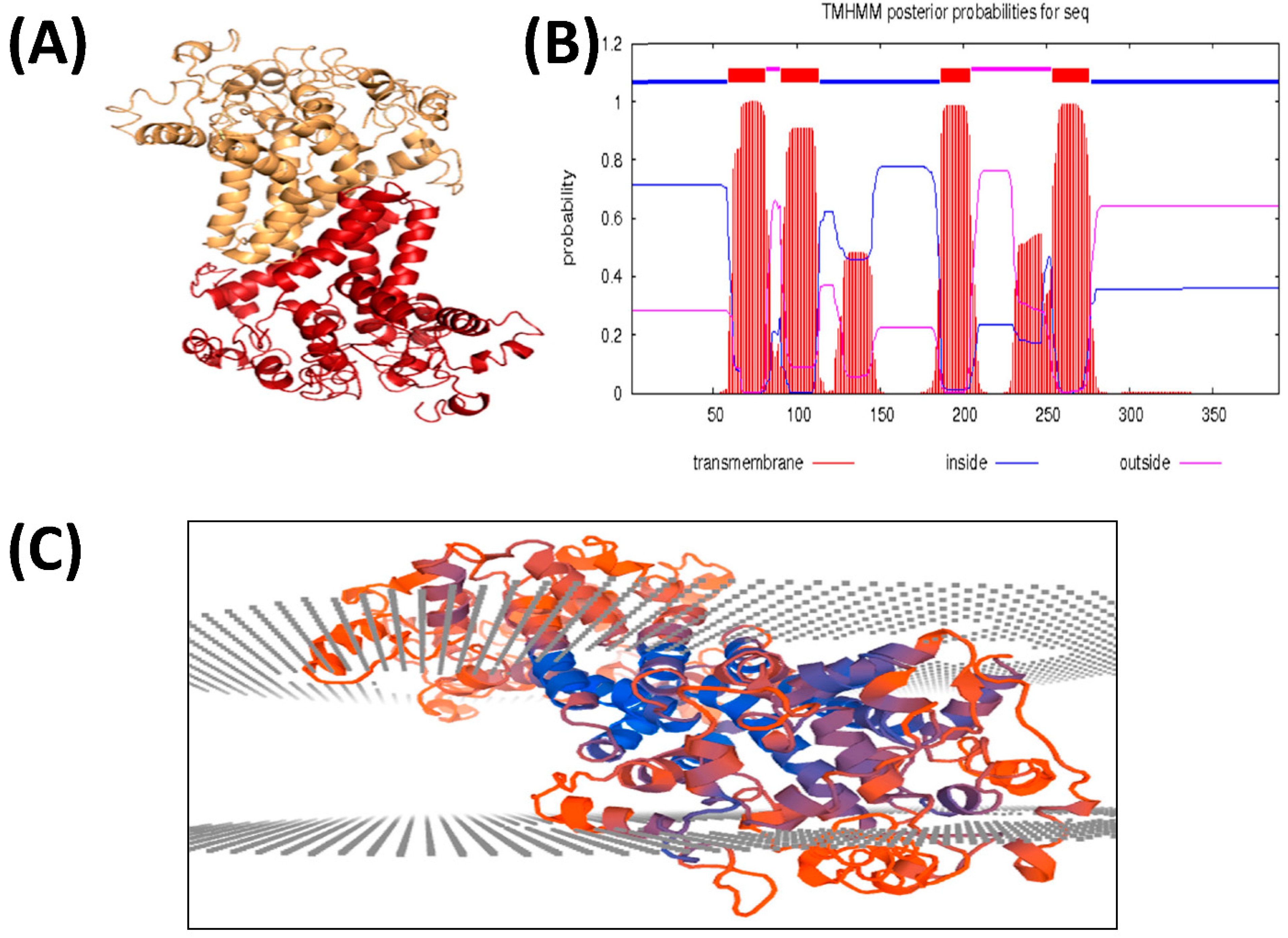
3.5. EoFAD2 Protein Features, 3D Structure Prediction, and Docking Analysis
3.5.1. Structure Prediction
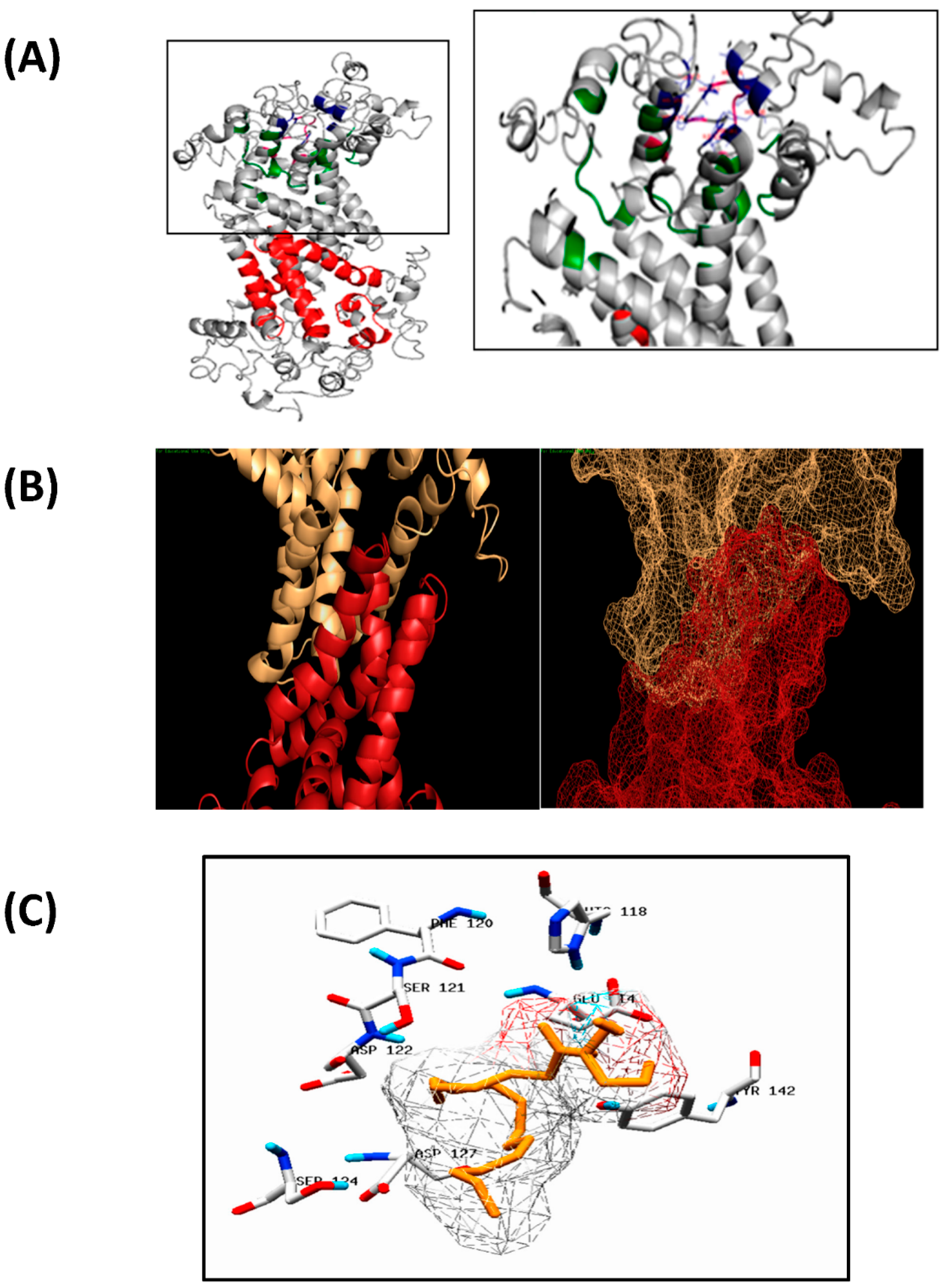
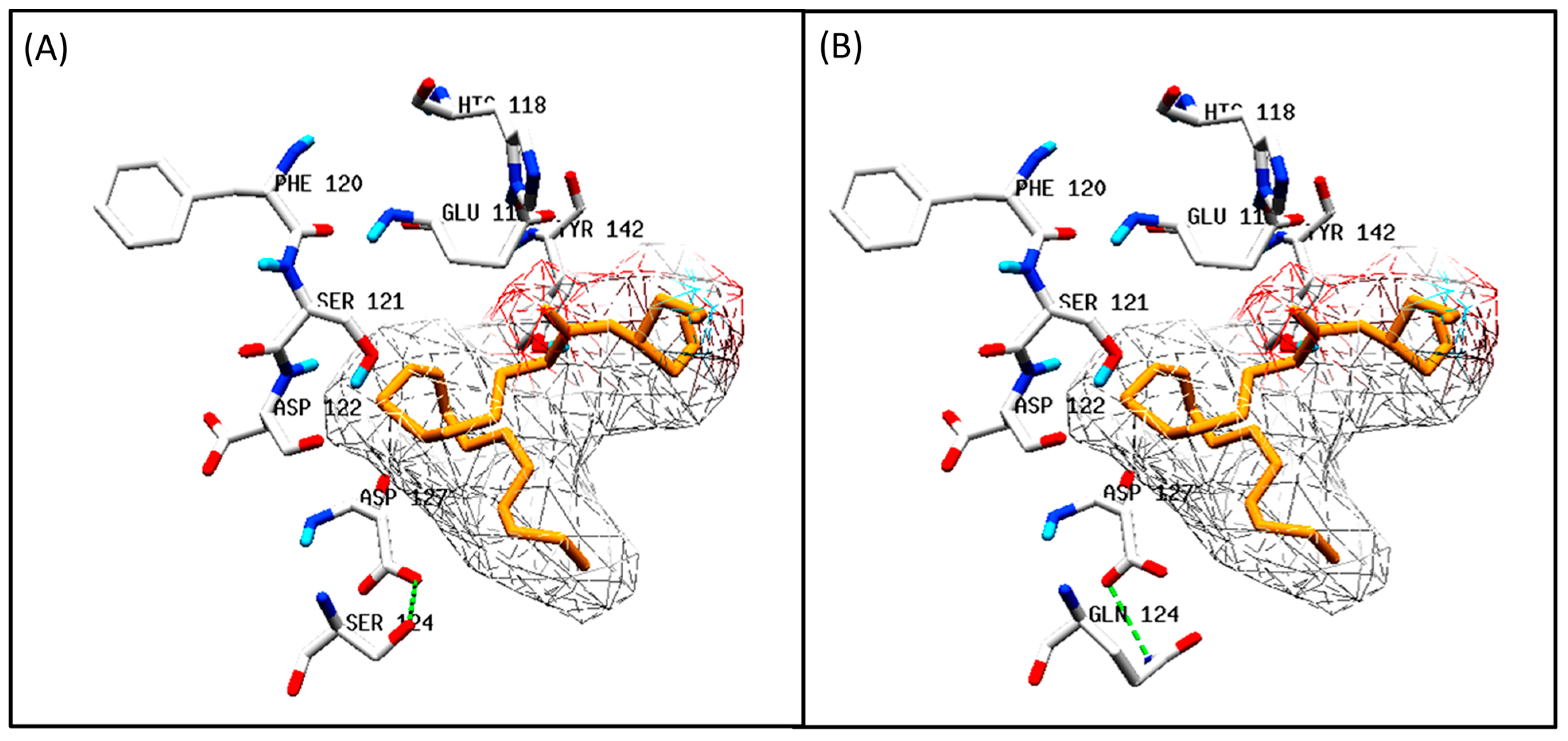
3.5.2. Docking Analysis between Oilseed Species
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Field, C.J. FATTY ACIDS|Dietary Importance. In Encyclopedia of Food Sciences and Nutrition, 2nd ed.; Caballero, B., Ed.; Academic Press: Oxford, UK, 2003; pp. 2317–2324. ISBN 978-0-12-227055-0. [Google Scholar]
- Kaur, N.; Chugh, V.; Gupta, A.K. Essential Fatty Acids as Functional Components of Foods—A Review. J. Food Sci. Technol. 2014, 51, 2289–2303. [Google Scholar] [CrossRef] [PubMed]
- Visioli, F.; Poli, A. Fatty Acids and Cardiovascular Risk. Evidence, Lack of Evidence, and Diligence. Nutrients 2020, 12, 3782. [Google Scholar] [CrossRef] [PubMed]
- Global Production Volume Palm Oil 2012–2021. Available online: https://www.statista.com/statistics/613471/palm-oil-production-volume-worldwide/ (accessed on 15 December 2021).
- Balić, A.; Vlašić, D.; Žužul, K.; Marinović, B.; Bukvić Mokos, Z. Omega-3 Versus Omega-6 Polyunsaturated Fatty Acids in the Prevention and Treatment of Inflammatory Skin Diseases. Int. J. Mol. Sci. 2020, 21, 741. [Google Scholar] [CrossRef]
- D’Angelo, S.; Motti, M.L.; Meccariello, R. ω-3 and ω-6 Polyunsaturated Fatty Acids, Obesity and Cancer. Nutrients 2020, 12, 2751. [Google Scholar] [CrossRef] [PubMed]
- Brondz, I. LIPIDS|Fatty Acids. In Encyclopedia of Analytical Science, 2nd ed.; Worsfold, P., Townshend, A., Poole, C., Eds.; Elsevier: Oxford, UK, 2005; pp. 76–88. ISBN 978-0-12-369397-6. [Google Scholar]
- de Carvalho, C.C.C.R.; Caramujo, M.J. The Various Roles of Fatty Acids. Molecules 2018, 23, 2583. [Google Scholar] [CrossRef]
- Aid, F. Plant Lipid Metabolism. In Advances in Lipid Metabolism; IntechOpen: London, UK, 2019. [Google Scholar] [CrossRef]
- Blanco, A.; Blanco, G. Chapter 5—Lipids. In Medical Biochemistry; Blanco, A., Blanco, G., Eds.; Academic Press: Cambridge, MA, USA, 2017; pp. 99–119. ISBN 978-0-12-803550-4. [Google Scholar]
- Alberts, B.; Johnson, A.; Lewis, J.; Raff, M.; Roberts, K.; Walter, P. The Lipid Bilayer. In Molecular Biology of the Cell, 4th ed.; Garland Science: New York, NY, USA, 2002. [Google Scholar]
- van Meer, G.; Voelker, D.R.; Feigenson, G.W. Membrane Lipids: Where They Are and How They Behave. Nat. Rev. Mol. Cell Biol. 2008, 9, 112–124. [Google Scholar] [CrossRef] [PubMed]
- Li-Beisson, Y.; Shorrosh, B.; Beisson, F.; Andersson, M.X.; Arondel, V.; Bates, P.D.; Baud, S.; Bird, D.; DeBono, A.; Durrett, T.P.; et al. Acyl-Lipid Metabolism. Arab. Book 2013, 11, e0161. [Google Scholar] [CrossRef]
- Brash, A.; Schneider, C.; Hamberg, M. Applications of Stereospecifically-Labeled Fatty Acids in Oxygenase and Desaturase Biochemistry. Lipids 2012, 47, 101–116. [Google Scholar] [CrossRef] [PubMed]
- He, M.; Qin, C.-X.; Wang, X.; Ding, N.-Z. Plant Unsaturated Fatty Acids: Biosynthesis and Regulation. Front. Plant Sci. 2020, 11, 390. [Google Scholar] [CrossRef]
- Review on Fatty Acid Desaturases and Their Roles in Temperature Acclimatisation. Available online: https://scialert.net/fulltext/?doi=jas.2017.282.295 (accessed on 27 January 2021).
- Dar, A.A.; Choudhury, A.R.; Kancharla, P.K.; Arumugam, N. The FAD2 Gene in Plants: Occurrence, Regulation, and Role. Front. Plant Sci. 2017, 8, 1789. [Google Scholar] [CrossRef] [PubMed]
- Schmid, K.M. Chapter 4—Lipid Metabolism in Plants. In Biochemistry of Lipids, Lipoproteins and Membranes, 7th ed.; Ridgway, N.D., McLeod, R.S., Eds.; Elsevier: Amsterdam, The Netherlands, 2021; pp. 121–159. ISBN 978-0-12-824048-9. [Google Scholar]
- Dong, C.-J.; Cao, N.; Zhang, Z.-G.; Shang, Q.-M. Characterization of the Fatty Acid Desaturase Genes in Cucumber: Structure, Phylogeny, and Expression Patterns. PLoS ONE 2016, 11, e0149917. [Google Scholar] [CrossRef] [PubMed]
- Hajiahmadi, Z.; Abedi, A.; Wei, H.; Sun, W.; Ruan, H.; Zhuge, Q.; Movahedi, A. Identification, Evolution, Expression, and Docking Studies of Fatty Acid Desaturase Genes in Wheat (Triticum aestivum L.). BMC Genom. 2020, 21, 778. [Google Scholar] [CrossRef] [PubMed]
- Raboanatahiry, N.; Yin, Y.; Chen, K.; He, J.; Yu, L.; Li, M. In Silico Analysis of Fatty Acid Desaturases Structures in Camelina Sativa, and Functional Evaluation of Csafad7 and Csafad8 on Seed Oil Formation and Seed Morphology. Int. J. Mol. Sci. 2021, 22, 10857. [Google Scholar] [CrossRef] [PubMed]
- Robin, J.; Gueroult, M.; Cheikhrouhou, R.; Guicherd, M.; Borsenberger, V.; Marty, A.; Bordes, F. Identification of a Crucial Amino Acid Implicated in the Hydroxylation/Desaturation Ratio of CpFAH12 Bifunctional Hydroxylase. Biotechnol. Bioeng. 2019, 116, 2451–2462. [Google Scholar] [CrossRef] [PubMed]
- Shanklin, J.; Guy, J.E.; Mishra, G.; Lindqvist, Y. Desaturases: Emerging Models for Understanding Functional Diversification of Diiron-Containing Enzymes. J. Biol. Chem. 2009, 284, 18559–18563. [Google Scholar] [CrossRef]
- Okuley, J.; Lightner, J.; Feldmann, K.; Yadav, N.; Lark, E.; Browse, J. Arabidopsis FAD2 Gene Encodes the Enzyme That Is Essential for Polyunsaturated Lipid Synthesis. Plant Cell 1994, 6, 147–158. [Google Scholar] [CrossRef]
- Elaeis oleifera American Oil Palm PFAF Plant Database. Available online: https://pfaf.org/user/Plant.aspx?LatinName=Elaeis+oleifera (accessed on 9 December 2021).
- Goh, K.J.; Wong, C.K.; Ng, P.H.C. Oil Palm. In Encyclopedia of Applied Plant Sciences, 2nd ed.; Thomas, B., Murray, B.G., Murphy, D.J., Eds.; Academic Press: Oxford, UK, 2017; pp. 382–390. ISBN 978-0-12-394808-3. [Google Scholar]
- Murphy, D.J. Using Modern Plant Breeding to Improve the Nutritional and Technological Qualities of Oil Crops. OCL 2014, 21, D607. [Google Scholar] [CrossRef][Green Version]
- Murphy, D.J. 12—Plant Breeding to Change Lipid Composition for Use in Food. In Modifying Lipids for Use in Food; Woodhead Publishing Series in Food Science, Technology and Nutrition; Gunstone, F.D., Ed.; Woodhead Publishing: Cambridge, UK, 2006; pp. 273–305. ISBN 978-1-85573-971-0. [Google Scholar]
- Borodovsky, M.; Rudd, K.E.; Koonin, E.V. Intrinsic and Extrinsic Approaches for Detecting Genes in a Bacterial Genome. Nucleic Acids Res. 1994, 22, 4756–4767. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Zheng, W.; Zhang, C.; Bell, E.W.; Zhang, Y. I-TASSER Gateway: A Protein Structure and Function Prediction Server Powered by XSEDE. Future Gener. Comput. Syst. 2019, 99, 73–85. [Google Scholar] [CrossRef]
- Zheng, W.; Zhang, C.; Wuyun, Q.; Pearce, R.; Li, Y.; Zhang, Y. LOMETS2: Improved Meta-Threading Server for Fold-Recognition and Structure-Based Function Annotation for Distant-Homology Proteins. Nucleic Acids Res. 2019, 47, W429–W436. [Google Scholar] [CrossRef] [PubMed]
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L. Predicting Transmembrane Protein Topology with a Hidden Markov Model: Application to Complete Genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, K.; Stoffel, W. Tmbase-A Database of Membrane Spanning Protein Segments. Biol. Chem. Hoppe-Seyler 1993, 346, 166. [Google Scholar]
- Kozakov, D.; Hall, D.R.; Xia, B.; Porter, K.A.; Padhorny, D.; Yueh, C.; Beglov, D.; Vajda, S. The ClusPro Web Server for Protein-Protein Docking. Nat. Protoc. 2017, 12, 255–278. [Google Scholar] [CrossRef] [PubMed]
- Studer, G.; Biasini, M.; Schwede, T. Assessing the Local Structural Quality of Transmembrane Protein Models Using Statistical Potentials (QMEANBrane). Bioinformatics 2014, 30, i505–i511. [Google Scholar] [CrossRef] [PubMed]
- Eberhardt, J.; Santos-Martins, D.; Tillack, A.F.; Forli, S. AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings. J. Chem. Inf. Model. 2021, 61, 3891–3898. [Google Scholar] [CrossRef]
- Thompson, J.D.; Gibson, T.J.; Higgins, D.G. Multiple Sequence Alignment Using ClustalW and ClustalX. Curr. Protoc. Bioinform. 2003, 1, 2.3.1–2.3.22. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, H.; Sun, J.; Li, B.; Zhu, Q.; Chen, S.; Zhang, H. Arabidopsis Fatty Acid Desaturase FAD2 Is Required for Salt Tolerance during Seed Germination and Early Seedling Growth. PLoS ONE 2012, 7, e30355. [Google Scholar] [CrossRef] [PubMed]
- Miquel, M.; James, D.; Dooner, H.; Browse, J. Arabidopsis Requires Polyunsaturated Lipids for Low-Temperature Survival. Proc. Natl. Acad. Sci. USA 1993, 90, 6208–6212. [Google Scholar] [CrossRef] [PubMed]
- Luisa Hernández, M.; Dolores Sicardo, M.; Arjona, P.M.; Martínez-Rivas, J.M. Specialized Functions of Olive FAD2 Gene Family Members Related to Fruit Development and the Abiotic Stress Response. Plant Cell Physiol. 2020, 61, 427–441. [Google Scholar] [CrossRef]
- Hernández, M.L.; Mancha, M.; Martínez-Rivas, J.M. Molecular Cloning and Characterization of Genes Encoding Two Microsomal Oleate Desaturases (FAD2) from Olive. Phytochemistry 2005, 66, 1417–1426. [Google Scholar] [CrossRef] [PubMed]
- Hernández, M.L.; Padilla, M.N.; Sicardo, M.D.; Mancha, M.; Martínez-Rivas, J.M. Effect of Different Environmental Stresses on the Expression of Oleate Desaturase Genes and Fatty Acid Composition in Olive Fruit. Phytochemistry 2011, 72, 178–187. [Google Scholar] [CrossRef] [PubMed]
- Hernández, M.L.; Sicardo, M.D.; Belaj, A.; Martínez-Rivas, J.M. The Oleic/Linoleic Acid Ratio in Olive (Olea europaea L.) Fruit Mesocarp Is Mainly Controlled by OeFAD2-2 and OeFAD2-5 Genes Together With the Different Specificity of Extraplastidial Acyltransferase Enzymes. Front. Plant Sci. 2021, 12, 345. [Google Scholar] [CrossRef] [PubMed]
- D’Angeli, S.; Altamura, M.M. Unsaturated Lipids Change in Olive Tree Drupe and Seed during Fruit Development and in Response to Cold-Stress and Acclimation. Int. J. Mol. Sci. 2016, 17, 1889. [Google Scholar] [CrossRef]
- Saeki, K.; Matsumoto, K.; Kinoshita, M.; Suzuki, I.; Tasaka, Y.; Kano, K.; Taguchi, Y.; Mikami, K.; Hirabayashi, M.; Kashiwazaki, N.; et al. Functional Expression of a Δ12 Fatty Acid Desaturase Gene from Spinach in Transgenic Pigs. Proc. Natl. Acad. Sci. USA 2004, 101, 6361–6366. [Google Scholar] [CrossRef] [PubMed]
- Cao, S.; Zhou, X.-R.; Wood, C.; Green, A.; Singh, S.; Liu, L.; Liu, Q. A Large and Functionally Diverse Family of Fad2 Genes in Safflower (Carthamus tinctorius L.). BMC Plant Biol. 2013, 12, 5. [Google Scholar] [CrossRef] [PubMed]
- Miao, X.; Zhang, L.; Hu, X.; Nan, S.; Chen, X.; Fu, H. Cloning and Functional Analysis of the FAD2 Gene Family from Desert Shrub Artemisia Sphaerocephala. BMC Plant Biol. 2019, 19, 481. [Google Scholar] [CrossRef]
- Yu, S.; Pan, L.; Yang, Q.; Min, P.; Ren, Z.; Zhang, H. Comparison of the Δ12 Fatty Acid Desaturase Gene between High-Oleic and Normal-Oleic Peanut Genotypes. J. Genet. Genom. 2008, 35, 679–685. [Google Scholar] [CrossRef]
- Janila, P.; Pandey, M.K.; Shasidhar, Y.; Variath, M.T.; Sriswathi, M.; Khera, P.; Manohar, S.S.; Nagesh, P.; Vishwakarma, M.K.; Mishra, G.P.; et al. Molecular Breeding for Introgression of Fatty Acid Desaturase Mutant Alleles (AhFAD2A and AhFAD2B) Enhances Oil Quality in High and Low Oil Containing Peanut Genotypes. Plant Sci. 2016, 242, 203–213. [Google Scholar] [CrossRef]
- Fan, R.; Li, L.; Cai, G.; Ye, J.; Liu, M.; Wang, S.; Li, Z. Molecular Cloning and Function Analysis of FAD2 Gene in Idesia Polycarpa. Phytochemistry 2019, 168, 112114. [Google Scholar] [CrossRef] [PubMed]
- Traore, S.M.; He, G. Soybean as a Model Crop to Study Plant Oil Genes: Mutations in FAD2 Gene Family; IntechOpen: London, UK, 2021; ISBN 978-1-83969-750-0. [Google Scholar]
- Lakhssassi, N.; Zhou, Z.; Liu, S.; Colantonio, V.; AbuGhazaleh, A.; Meksem, K. Characterization of the FAD2 Gene Family in Soybean Reveals the Limitations of Gel-Based TILLING in Genes with High Copy Number. Front. Plant Sci. 2017, 8, 324. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Salentijn, E.; Huang, B.; Krens, F.; Dechesne, A.; Visser, R.; Van Loo, E.N. Isolation and Characterization of the Omega-6 Fatty Acid Desaturase (FAD2) Gene Family in the Allohexaploid Oil Seed Crop Crambe Abyssinica Hochst. Mol. Breed. 2013, 32, 517–531. [Google Scholar] [CrossRef]
- Celik Altunoglu, Y.; Unel, N.M.; Baloglu, M.C.; Ulu, F.; Can, T.H.; Cetinkaya, R. Comparative Identification and Evolutionary Relationship of Fatty Acid Desaturase (FAD) Genes in Some Oil Crops: The Sunflower Model for Evaluation of Gene Expression Pattern under Drought Stress. Biotechnol. Biotechnol. Equip. 2018, 32, 846–857. [Google Scholar] [CrossRef]
- Dehghan Nayeri, F.; Yarizade, K. Bioinformatics Study of Delta-12 Fatty Acid Desaturase 2 (FAD2) Gene in Oilseeds. Mol. Biol. Rep. 2014, 41, 5077–5087. [Google Scholar] [CrossRef]
- Luo, J.; Liu, L.; Konik-Rose, C.; Tian, L.; Singh, S.; Howitt, C.A.; Li, Z.; Liu, Q. Down-Regulation of FAD2-1 Gene Expression Alters Lysophospholipid Composition in the Endosperm of Rice Grain and Influences Starch Properties. Foods 2021, 10, 1169. [Google Scholar] [CrossRef] [PubMed]
- Pirtle, I.L.; Kongcharoensuntorn, W.; Nampaisansuk, M.; Knesek, J.E.; Chapman, K.D.; Pirtle, R.M. Molecular Cloning and Functional Expression of the Gene for a Cotton Δ-12 Fatty Acid Desaturase (FAD2). Biochim. Biophys. Acta 2001, 1522, 122–129. [Google Scholar] [CrossRef]
- Liu, F.; Ma, L.; Wang, Y.; Li, Y.; Zhang, X.; Xue, F.; Nie, X.; Zhu, Q.; Sun, J. GhFAD2-3 Is Required for Anther Development in Gossypium Hirsutum. BMC Plant Biol. 2019, 19, 393. [Google Scholar] [CrossRef] [PubMed]
- Kargiotidou, A.; Deli, D.; Galanopoulou, D.; Tsaftaris, A.; Farmaki, T. Low Temperature and Light Regulate Delta 12 Fatty Acid Desaturases (FAD2) at a Transcriptional Level in Cotton (Gossypium hirsutum). J. Exp. Bot. 2008, 59, 2043–2056. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.-Y.; Hsieh, M.-J.; Tsai, Y.-T.; Chung, H.-H.; Shyur, L.-F.; Hsieh, C.-H.; To, K.-Y. Transformation and Characterization of Δ12-Fatty Acid Acetylenase and Δ12-Oleate Desaturase Potentially Involved in the Polyacetylene Biosynthetic Pathway from Bidens Pilosa. Plants 2020, 9, 1483. [Google Scholar] [CrossRef]
- Liu, K.; Zhao, S.; Wang, S.; Wang, H.; Zhang, Z. Identification and Analysis of the FAD Gene Family in Walnuts (Juglans regia L.) Based on Transcriptome Data. BMC Genom. 2020, 21, 299. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Liu, M.; Li, D.; Wu, J.; Li, X.; Yang, Y. Overexpression of FAD2 Promotes Seed Germination and Hypocotyl Elongation in Brassica Napus. Plant Cell Tissue Organ Cult. 2010, 102, 205–211. [Google Scholar] [CrossRef]
- Zhou, Z.; Wang, M.-J.; Zhao, S.-T.; Hu, J.-J.; Lu, M.-Z. Changes in Freezing Tolerance in Hybrid Poplar Caused by Up- and down-Regulation of PtFAD2 Gene Expression. Transgenic Res. 2010, 19, 647–654. [Google Scholar] [CrossRef] [PubMed]
- Zeng, F.; Roslinsky, V.; Cheng, B. Mutations in the Promoter, Intron and CDS of Two FAD2 Generate Multiple Alleles Modulating Linoleic Acid Level in Yellow Mustard. Sci. Rep. 2017, 7, 8284. [Google Scholar] [CrossRef] [PubMed]
- Sun, R.; Gao, L.; Yu, X.; Zheng, Y.; Li, D.; Wang, X. Identification of a Δ12 Fatty Acid Desaturase from Oil Palm (Elaeis guineensis Jacq.) Involved in the Biosynthesis of Linoleic Acid by Heterologous Expression in Saccharomyces Cerevisiae. Gene 2016, 591, 21–26. [Google Scholar] [CrossRef] [PubMed]
- Jadid, N.; Prasetyowati, I.; Rosidah, N.L.A.; Ermavitalini, D.; Nurhatika, S.; Nurhidayati, T.; Purnobasuki, H. In Silico Analysis of Partial Fatty Acid Desaturase 2 CDNA From Reutealis trisperma (Blanco) Airy Shaw. Bioinform. Biol. Insights 2021, 15, 11779322211005748. [Google Scholar] [CrossRef]
- Sequence Variations in the FAD2 Gene in Seeded Pumpkins. Available online: https://www.geneticsmr.com/articles/5754 (accessed on 25 February 2022).
- Kim, H.; Go, Y.S.; Kim, A.; Lee, S.; Kim, K.-N.; Lee, G.-J.; Kim, G.-J.; Suh, M. Isolation and Functional Analysis of Three Microsomal Delta-12 Fatty Acid Desaturase Genes from Camelina Sativa (L.) Cv. CAME. J. Plant Biotechnol. 2014, 41, 146–158. [Google Scholar] [CrossRef][Green Version]
- Kang, J.; Snapp, A.R.; Lu, C. Identification of Three Genes Encoding Microsomal Oleate Desaturases (FAD2) from the Oilseed Crop Camelina Sativa. Plant Physiol. Biochem. 2011, 49, 223–229. [Google Scholar] [CrossRef]
- Lei, N.; Peng, S.; Niu, B.; Chen, J.; Zhou, J.; Tang, L.; Xu, Y.; Wang, S.; Chen, F. Molecular Cloning and Characterization of a Novel Microsomal Oleate Desaturase Gene DiFAD2 from Davidia Involucrata Baill. Biol. Plant. 2010, 54, 41–46. [Google Scholar] [CrossRef]
- Suresha, G.S.; Santha, I. Molecular Cloning, Expression Analysis and Growth Temperature Dependent Regulation of a Novel Oleate Desaturase Gene (Fad2) Homologue from Brassica Juncea. Aust. J. Crop Sci. 2012, 6, 296–308. [Google Scholar]
- Suresha, G.S. Molecular Cloning and In Silico Analysis of Novel Oleate Desaturase Gene Homologues from Brassica Juncea through Sub-Genomic Library Approach. Plant Omics 2013, 6, 55–64. [Google Scholar]
- Mikkilineni, V.; Rocheford, T.R. Sequence Variation and Genomic Organization of Fatty Acid Desaturase-2 (Fad2) and Fatty Acid Desaturase-6 (Fad6) CDNAs in Maize. Theor. Appl. Genet. 2003, 106, 1326–1332. [Google Scholar] [CrossRef] [PubMed]
- Tao, F.; Zhu, S.-W.; Fan, J.; Cheng, B.-J. Cloning and Sequence Analysis of Maize FAD2 Gene. Zhi Wu Sheng Li Yu Fen Zi Sheng Wu Xue Xue Bao 2006, 32, 649–656. [Google Scholar] [PubMed]
- Utomo, C.; Subroto, A.P.; Darmawan, C.; Setyobudi, R.H.; Nugroho, Y.A.; Liwang, T. Construction for Δ-12 Fatty Acid Desaturase (FAD2) Silencing to Improve Oil Quality of Jatropha Curcas Linn. Energy Procedia 2015, 65, 36–41. [Google Scholar] [CrossRef]
- Zhang, Y.M.; Wang, C.C.; Hu, H.H.; Yang, L. Cloning and Expression of Three Fatty Acid Desaturase Genes from Cold-Sensitive Lima Bean (Phaseolus lunatus L.). Biotechnol. Lett. 2011, 33, 395–401. [Google Scholar] [CrossRef] [PubMed]
- Nordborg, M.; Hu, T.T.; Ishino, Y.; Jhaveri, J.; Toomajian, C.; Zheng, H.; Bakker, E.; Calabrese, P.; Gladstone, J.; Goyal, R.; et al. The Pattern of Polymorphism in Arabidopsis Thaliana. PLoS Biol. 2005, 3, e196. [Google Scholar] [CrossRef] [PubMed]
- Ben Ayed, R.; Moreau, F.; Ben Hlima, H.; Rebai, A.; Ercisli, S.; Kadoo, N.; Hanana, M.; Assouguem, A.; Ullah, R.; Ali, E.A. SNP Discovery and Structural Insights into OeFAD2 Unravelling High Oleic/Linoleic Ratio in Olive Oil. Comput. Struct. Biotechnol. J. 2022, 20, 1229–1243. [Google Scholar] [CrossRef]
- Lou, Y.; Schwender, J.; Shanklin, J. FAD2 and FAD3 Desaturases Form Heterodimers That Facilitate Metabolic Channeling in Vivo. J. Biol. Chem. 2014, 289, 17996–18007. [Google Scholar] [CrossRef]

| Species | Accession Number | Code |
|---|---|---|
| Elaeis oleifera | KY006847.1 | EoFAD2 |
| Glycine max | AY611472.1 | Gm1FAD2-1 |
| L43920.1 | Gm2FAD2-1 | |
| EU908061.1 | Gm1FAD2-1B | |
| EU908062.1 | Gm2FAD2-1B | |
| DQ532370.1 | Gm3FAD2-1B | |
| Brassica napus | FJ907397.1 | BnFAD2-1 |
| FJ907399.1 | BnFAD2-3 | |
| FJ907401.1 | BnFAD2-4 | |
| FJ907400.1 | BnFAD2-5 | |
| AF243045.1 | Bn1FAD2 | |
| DQ767949.1 | Bn2FAD2 | |
| AY577313.1 | Bn3FAD2 | |
| AY592975.1 | Bn4FAD2 | |
| Helianthus annuus | AY800245.1 | HaFAD2-1 |
| AY803008.1 | HaFAD2-3 | |
| AY802997.1 | Ha1FAD2-2 | |
| AY802995.1 | Ha2FAD2-2 | |
| Arachis hypogaea | AF030319.1 | AhFADs |
| DQ019933.1 | Ah1FAD2 | |
| AF248739.1 | Ah2FAD2 | |
| Arachis monticola | AY900663.1 | AmFAD2 |
| Zea mays | AB257309.1 | ZmFAD2 |
| Sesamum indicum | AF192486.1 | SiFAD2 |
| Gene ID/Name | Species | Function | Reference |
|---|---|---|---|
| L26296 | Arabidopsis thaliana | Polyunsaturated lipid synthesis, salt tolerance during seed germination and early seedling growth, and vegetative growth | [24,40,41] |
| AY733076, AY733077 | Olea europaea | Linoleic acid synthesis, wounding response of olive fruit mesocarp, cuticle formation, and cold-acclimation, and abiotic stresses (drought/cold) response | [42,43,44,45,46] |
| AB094415 | Spinacia oleracea | Linoleic acid synthesis | [47] |
| CtFAD2-1-11 | Carthamus tinctorius | Linoleic acid synthesis and crepenynic acid synthesis (with C16:1 as a substrate) | [48] |
| AsFAD2-1-24 | Artemisiasphaerocephala | Linoleic and palmitolinoleic acid biosynthesis | [49] |
| EF186911 | Arachis hypogaea | Enhancement of peanut oil quality | [50,51] |
| MF 693460 | Idesia polycarpa | Linoleic acid accumulation | [52] |
| GmFAD2–1a GmFAD2–1b | Glycine max | Increased oleic acid content in mutant lines | [53,54] |
| JX964741, JX964747 | Crambe abyssinica | Polyunsaturated fatty acids biosynthesis | [55] |
| HaFAD2-1-11 | Helianthus annuus | Linoleic acid synthesis and storage oil desaturation in seed | [56] |
| AAX11454, ACP39503, ACF49507, ABK59093, AAS19533, AEI60129, ACZ06072, AAN87573, AAF04094 | Sesamum indicum, Brassica napus, Linum usitatissinum, Ricinus communis, Cucurbita pepo, Vitis labrusca, Arachis hypogaea, Vernicia fordii, Vernonia galamensis | Linoleic acid synthesis and storage oil desaturation in seed | [57] |
| OsFAD2-1 RNAi | Orysa sativa | Alteration of lysophospholipid composition in the endosperm of rice grain and influence on starch properties | [58] |
| AF331163 | Gossypium hirsutum | Linoleic acid accumulation, anther development, and cold and light responsiveness | [59,60,61] |
| MF318524 | Bidens pilosa | Polyacetylene biosynthesis | [62] |
| XM_019004668.1, XM_019004667.1, XM_018993369.1, XM_018993367.1 | Juglans regia | Linoleic acid synthesis | [63] |
| AF243045 | Brassica napus | Promotion of seed germination and hypocotyl elongation | [64] |
| DQ31678 | Populus tomentosa | Freezing tolerance | [65] |
| SalFAD2.LIA1, SalFAD2.LIA2 | Sinapis alba | Linoleic acid accumulation | [66] |
| KT023602 | Elaeis guineensis | Linoleic acid synthesis | [67] |
| Partial RtFAD2 | Reutealis trisperma | Regulation of fatty acid desaturation | [68] |
| Csa3M808360, Csa4M286360 | Cucumis sativus | Temperature stress responsiveness | [19] |
| FAD2 (SNPs) | Cucurbita moschata, Cucurbita maxima, Cucurbita pepo, and Cucurbita ficifolia | Linoleic acid synthesis | [69] |
| CsFAD2 (SNPs) | Camelina sativa | Linoleic acid synthesis and storage oil desaturation in seed | [70,71] |
| EU275211 | Davidia involucrata | Linoleic acid synthesis | [72] |
| X91139, EF639848, FJ696650, FJ696651, FJ696652 | Brassica juncea | Biosynthesis of polyunsaturated fatty acids in seeds and cold responsiveness | [73,74] |
| DQ496227, ZmFAD2 (SNPs) | Zea mays | Ratio of oleic/linoleic acid | [75,76] |
| GU353167 | Jatropha curcas | Conversion of oleic acid to linoleic acid in the seed | [77] |
| HQ171179, HQ171180 | Phaseolus lunatus | Cold, drought, and salt stress responsiveness | [78] |
| Sequences | Region | n | s | π | D | θw |
|---|---|---|---|---|---|---|
| All Sequences | 1–1172 | 24 | 0.60 | 0.26 | 2.57 | 0.16 |
| C18:0 | C18:1 | C18:2 | C18:3 | |||||
|---|---|---|---|---|---|---|---|---|
| Mean ± SD | p-Value | Mean ± SD | p-Value | Mean ± SD | p-Value | Mean ± SD | p-Value | |
| SNP373 | ||||||||
| C | 2.972 ± 1.154 | 0.754 | 54.100 ± 11.948 | 0.006 | 23.747 ± 14.195 | 0.013 | 2.6150 ± 4.674 | 0.902 |
| T | 3.283 ± 1.338 | 21.333 ± 3.666 | 57.433 ± 5.749 | 3.053 ± 4.030 | ||||
| SNP718 | ||||||||
| G | 2.530 ± 0.907 | 0.282 | 58.700 ± 9.337 | 0.007 | 17.430 ± 7.925 | 0.002 | 3.486 ± 5.311 | 0.735 |
| A | 3.537 ± 1.205 | 26.075 ± 9.944 | 53.750 ± 8.735 | 2.29 ± 3.627 |
| Properties | Length (aa) | MW (kD) | pI | TM Regions | α-Helix Structure (%) | Extended Strand Structure (%) | Random Coil Structure (%) |
|---|---|---|---|---|---|---|---|
| EoFAD2 | 390 | 44.1 | 8.4 | 5 | 28 | 21 | 51 |
| Aa (%) | Leu | Ala | Val | Pro | Ser | Arg | Gly | Tyr | His |
|---|---|---|---|---|---|---|---|---|---|
| EoFAD2 | 10 | 9.7 | 7.7 | 7.4 | 6.4 | 6.2 | 6.2 | 5.4 | 5.1 |
| TM Position | Length | Orientation |
|---|---|---|
| 61–85 | 19 | o-i |
| 94–114 | 21 | i-o |
| 128–148 | 21 | o-i |
| 190–209 | 20 | i-o |
| 257–279 | 23 | o-i |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ben Ayed, R.; Chirmade, T.; Hanana, M.; Khamassi, K.; Ercisli, S.; Choudhary, R.; Kadoo, N.; Karunakaran, R. Comparative Analysis and Structural Modeling of Elaeis oleifera FAD2, a Fatty Acid Desaturase Involved in Unsaturated Fatty Acid Composition of American Oil Palm. Biology 2022, 11, 529. https://doi.org/10.3390/biology11040529
Ben Ayed R, Chirmade T, Hanana M, Khamassi K, Ercisli S, Choudhary R, Kadoo N, Karunakaran R. Comparative Analysis and Structural Modeling of Elaeis oleifera FAD2, a Fatty Acid Desaturase Involved in Unsaturated Fatty Acid Composition of American Oil Palm. Biology. 2022; 11(4):529. https://doi.org/10.3390/biology11040529
Chicago/Turabian StyleBen Ayed, Rayda, Tejas Chirmade, Mohsen Hanana, Khalil Khamassi, Sezai Ercisli, Ravish Choudhary, Narendra Kadoo, and Rohini Karunakaran. 2022. "Comparative Analysis and Structural Modeling of Elaeis oleifera FAD2, a Fatty Acid Desaturase Involved in Unsaturated Fatty Acid Composition of American Oil Palm" Biology 11, no. 4: 529. https://doi.org/10.3390/biology11040529
APA StyleBen Ayed, R., Chirmade, T., Hanana, M., Khamassi, K., Ercisli, S., Choudhary, R., Kadoo, N., & Karunakaran, R. (2022). Comparative Analysis and Structural Modeling of Elaeis oleifera FAD2, a Fatty Acid Desaturase Involved in Unsaturated Fatty Acid Composition of American Oil Palm. Biology, 11(4), 529. https://doi.org/10.3390/biology11040529









