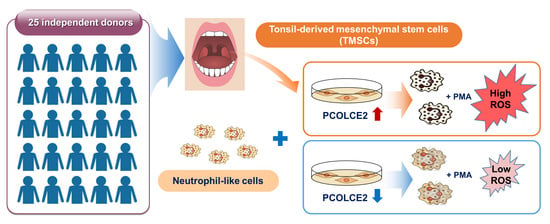
Procollagen C-Endopeptidase Enhancer 2 Secreted by Tonsil-Derived Mesenchymal Stem Cells Increases the Oxidative Burst of Promyelocytic HL-60 Cells
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Differentiation of HL-60 Cells into Granulocyte-Like Cells
2.3. Coculture of MSCs and dHL-60 Cells
2.4. Measurement of ROS
2.5. Collection of Conditioned Medium (CM)
2.6. Western Blotting
2.7. Real-Time Polymerase Chain Reaction (PCR)
2.8. Transfection of siRNA
2.9. Measurement of Viability of HL-60 Cells
2.10. RNA Sequencing (RNA-seq)
2.11. Statistical Analysis of Gene Expression Levels in RNA-seq
2.12. Statistical Analysis
3. Results
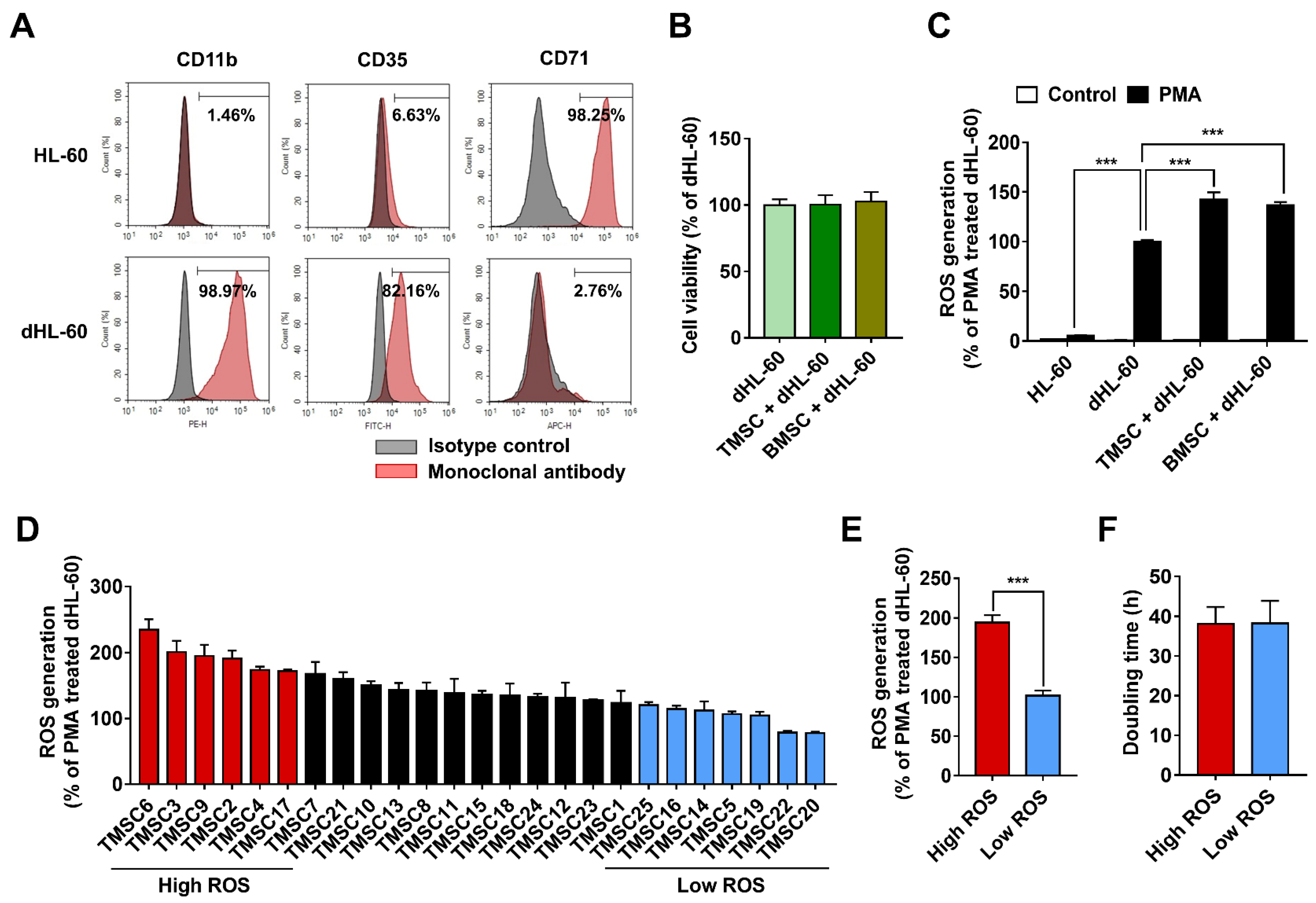
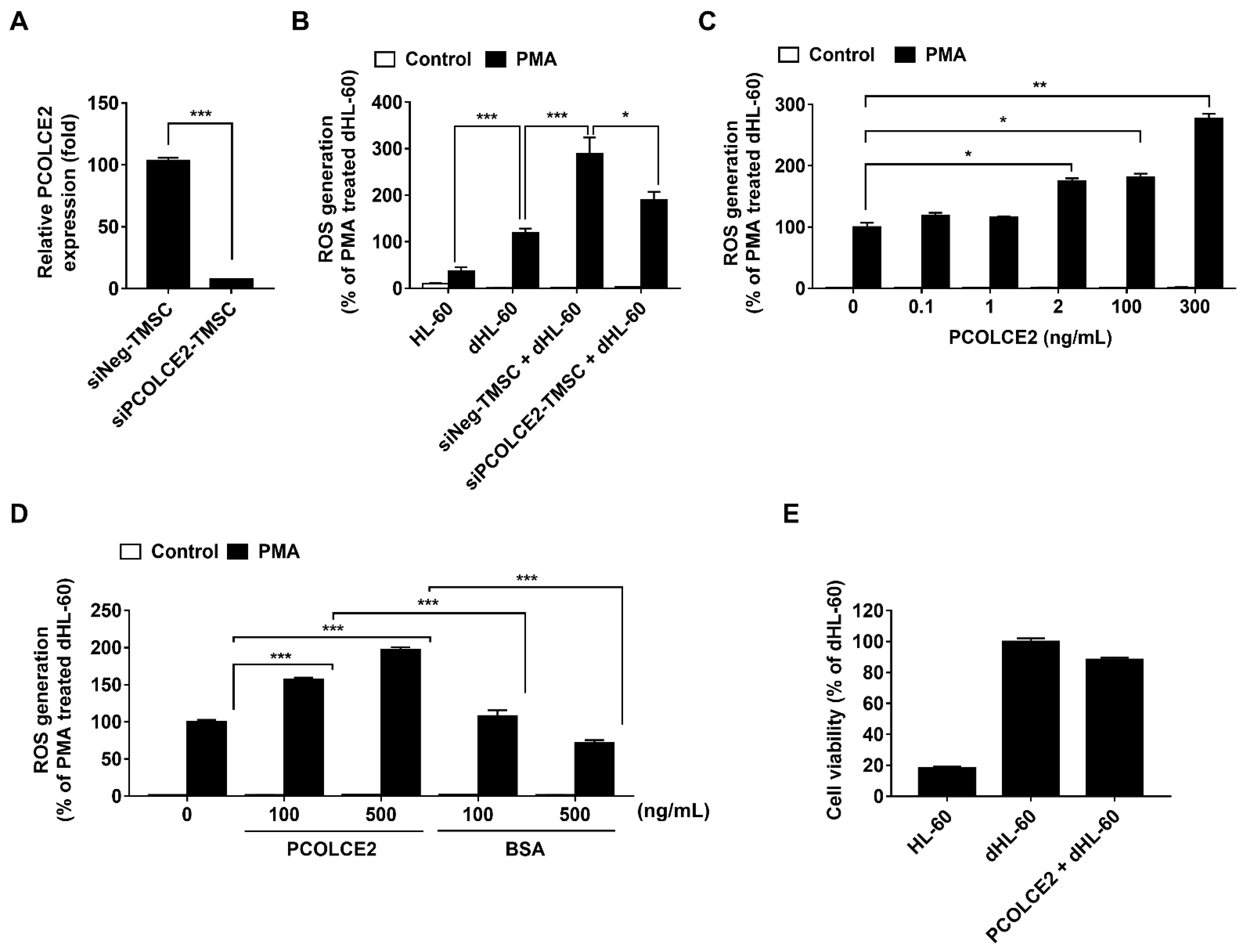
3.1. TMSCs Augment ROS Generation from Differentiated HL-60
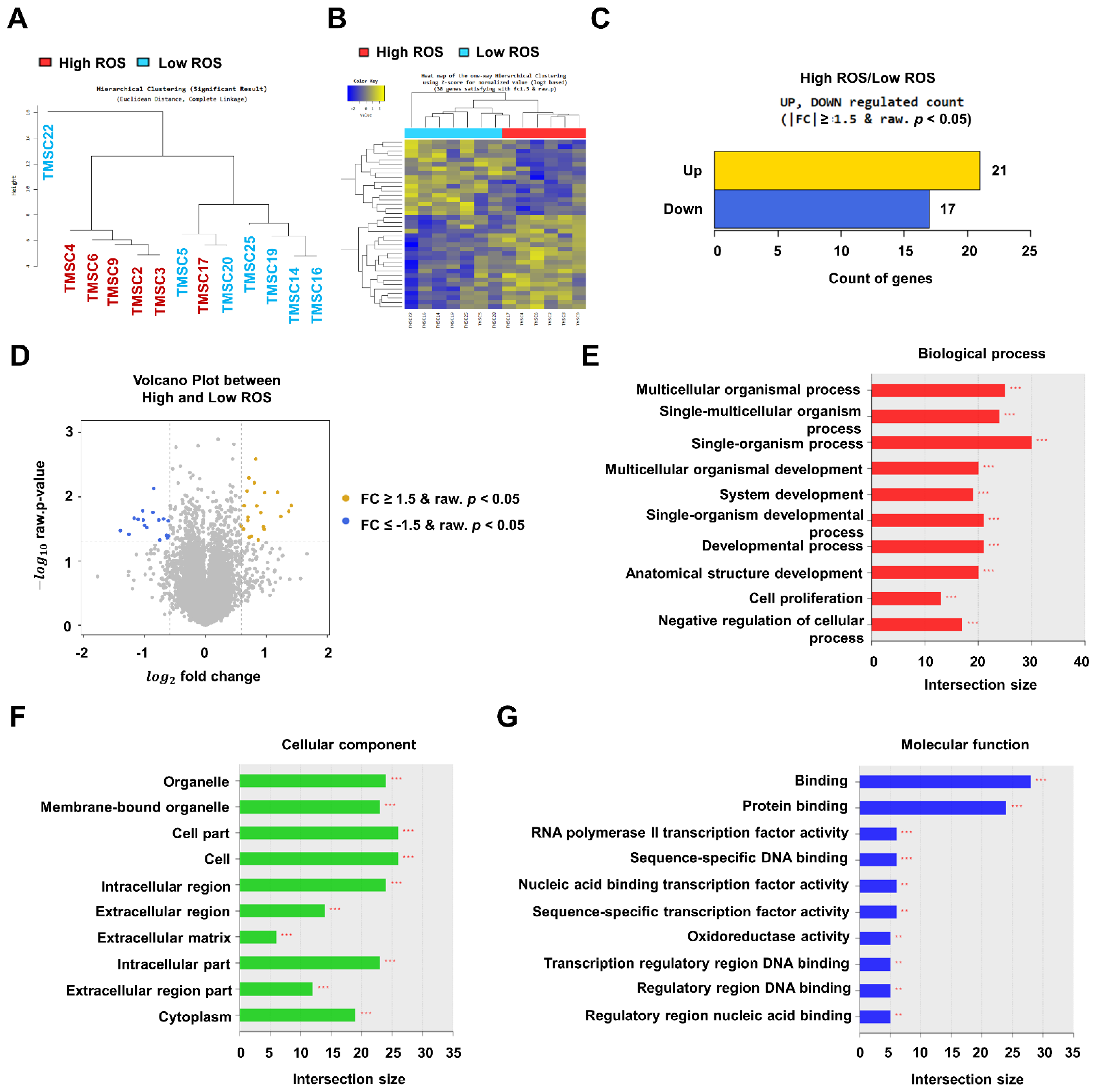
3.2. RNA-Sequencing Data Analysis of TMSCs
3.3. PCOLCE2 mRNA and Protein Levels Are Elevated in the High ROS Group when Compared with the Low ROS Group
3.4. Neutrophil ROS Production Is Enhanced by PCOLCE2
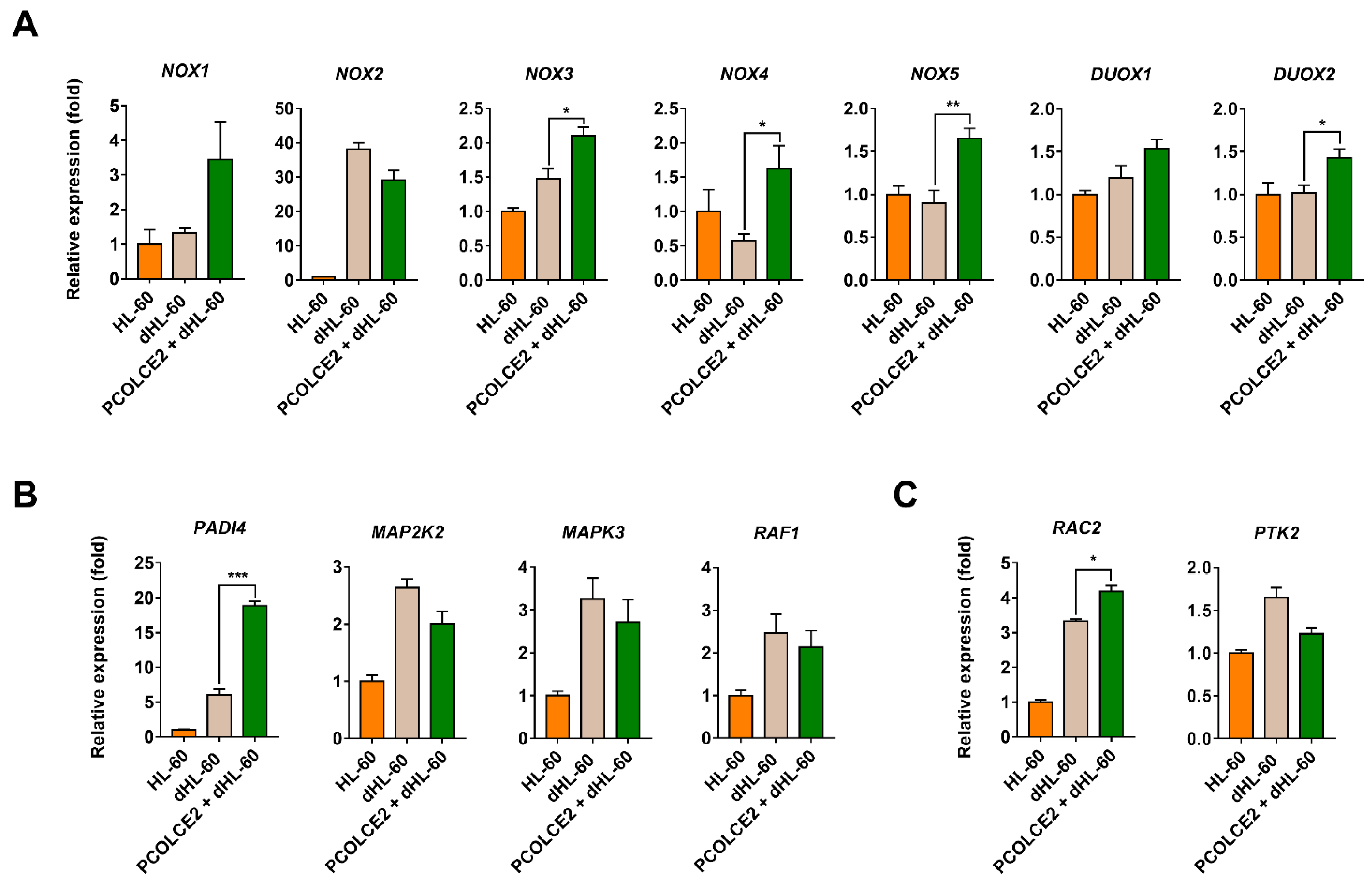
3.5. PCOLCE2 Alters the Levels of Genes Involved in Neutrophil Function
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
List of Abbreviations
| AMSC | Adipose tissue-derived mesenchymal stem cell |
| APCDD1 | Adenomatosis polyposis downregulated 1 |
| BMP | Bone morphogenetic protein |
| BMSC | Bone marrow-derived mesenchymal stem cell |
| BSA | bovine serum albumin |
| CD | Clusters of differentiation |
| CM | Conditioned medium |
| COL8A1 | Collagen type VIII alpha 1 chai |
| dHL-60 | Differentiated HL-60 |
| DUOX | Dual oxidase |
| DEG | Differentially expressed genes |
| DMF, MAPK3 | Mitogen-activated protein kinase 3 |
| FKRM | Fragments Per Kilobase of exon per Million fragments mapped |
| GO | Gene ontology |
| MAP2K2 | Mitogen-activated protein kinase kinase 2 |
| MEDAG | Mesenteric estrogen-dependent adipogenesis |
| MFAP5 | Microfibrillar-associated protein 5 |
| MSC | Mesenchymal stem cell |
| NET | Neutrophil extracellular trap |
| NOX | NADPH oxidase |
| PADI4 | Peptidyl arginine deiminase 4 |
| PCOLCE2 | Procollagen C-endopeptidase enhancer 2 |
| PCR | polymerase chain reaction |
| PDPN | Podoplanin |
| PMA | phorbol 12-myristate 13-acetate |
| PSG1 | Pregnancy-specific beta-1-glycoprotein 1 |
| PTK2 | Protein tyrosine kinase 2 |
| RAC2 | Rac family small GTPase 2 |
| RAF1 | Raf-1 proto-oncogene serine/threonine- kinase |
| RNA-seq | RNA sequencing |
| ROS | Reactive oxygen species |
| RUNX3 | Runt related transcription factor 3 |
| SFRP4 | Secreted frizzled-related protein 4 |
| SNAP25 | Synaptosome-associated protein 25 |
| TMSC | Tonsil-derived mesenchymal stem cell |
References
- Nguyen, G.T.; Green, E.R.; Mecsas, J. Neutrophils to the ROScue: Mechanisms of NADPH Oxidase Activation and Bacterial Resistance. Front. Cell Infect. Microbiol. 2017, 7, 373. [Google Scholar] [CrossRef] [PubMed]
- Rosales, C. Neutrophil: A Cell with Many Roles in Inflammation or Several Cell Types? Front. Physiol. 2018, 9, 113. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, M.E.; Borthwick, A.; Rigg, A.; Leary, A.; Assersohn, L.; Last, K.; Tan, S.; Milan, S.; Tait, D.; Smith, I.E. Mortality within 30 days of chemotherapy: A clinical governance benchmarking issue for oncology patients. Br. J. Cancer 2006, 95, 1632–1636. [Google Scholar] [CrossRef] [PubMed]
- Kong, S.G.; Jeong, S.; Lee, S.; Jeong, J.Y.; Kim, D.J.; Lee, H.S. Early transplantation-related mortality after allogeneic hematopoietic cell transplantation in patients with acute leukemia. BMC Cancer 2021, 21, 177. [Google Scholar] [CrossRef] [PubMed]
- Gan, Y.H. Host susceptibility factors to bacterial infections in type 2 diabetes. PLoS Pathog. 2013, 9, e1003794. [Google Scholar] [CrossRef]
- Ullah, I.; Subbarao, R.B.; Rho, G.J. Human mesenchymal stem cells—Current trends and future prospective. Biosci. Rep. 2015, 35, e00191. [Google Scholar] [CrossRef]
- Salami, F.; Tavassoli, A.; Mehrzad, J.; Parham, A. Immunomodulatory effects of mesenchymal stem cells on leukocytes with emphasis on neutrophils. Immunobiology 2018, 223, 786–791. [Google Scholar] [CrossRef]
- Joel, M.D.M.; Yuan, J.; Wang, J.; Yan, Y.; Qian, H.; Zhang, X.; Xu, W.; Mao, F. MSC: Immunoregulatory effects, roles on neutrophils and evolving clinical potentials. Am. J. Transl. Res. 2019, 11, 3890–3904. [Google Scholar]
- Taghavi-Farahabadi, M.; Mahmoudi, M.; Hashemi, S.M.; Rezaei, N. Evaluation of the effects of mesenchymal stem cells on neutrophils isolated from severe congenital neutropenia patients. Int. Immunopharmacol. 2020, 83, 106463. [Google Scholar] [CrossRef]
- Hall, S.R.; Tsoyi, K.; Ith, B.; Padera, R.F., Jr.; Lederer, J.A.; Wang, Z.; Liu, X.; Perrella, M.A. Mesenchymal stromal cells improve survival during sepsis in the absence of heme oxygenase-1: The importance of neutrophils. Stem. Cells. 2013, 31, 397–407. [Google Scholar] [CrossRef]
- Wang, L.T.; Wang, H.H.; Chiang, H.C.; Huang, L.Y.; Chiu, S.K.; Siu, L.K.; Liu, K.J.; Yen, M.L.; Yen, B.L. Human Placental MSC-Secreted IL-1beta Enhances Neutrophil Bactericidal Functions during Hypervirulent Klebsiella Infection. Cell. Rep. 2020, 32, 108188. [Google Scholar] [CrossRef]
- Park, Y.S.; Lim, G.W.; Cho, K.A.; Woo, S.Y.; Shin, M.; Yoo, E.S.; Chan Ra, J.; Ryu, K.H. Improved viability and activity of neutrophils differentiated from HL-60 cells by co-culture with adipose tissue-derived mesenchymal stem cells. Biochem. Biophys. Res. Commun. 2012, 423, 19–25. [Google Scholar] [CrossRef] [PubMed]
- Mumaw, J.L.; Schmiedt, C.W.; Breidling, S.; Sigmund, A.; Norton, N.A.; Thoreson, M.; Peroni, J.F.; Hurley, D.J. Feline mesenchymal stem cells and supernatant inhibit reactive oxygen species production in cultured feline neutrophils. Res. Vet. Sci. 2015, 103, 60–69. [Google Scholar] [CrossRef] [PubMed]
- Jiang, D.; Muschhammer, J.; Qi, Y.; Kugler, A.; de Vries, J.C.; Saffarzadeh, M.; Sindrilaru, A.; Beken, S.V.; Wlaschek, M.; Kluth, M.A.; et al. Suppression of Neutrophil-Mediated Tissue Damage—A Novel Skill of Mesenchymal Stem Cells. Stem. Cells 2016, 34, 2393–2406. [Google Scholar] [CrossRef] [PubMed]
- Ryu, K.H.; Cho, K.A.; Park, H.S.; Kim, J.Y.; Woo, S.Y.; Jo, I.; Choi, Y.H.; Park, Y.M.; Jung, S.C.; Chung, S.M.; et al. Tonsil-derived mesenchymal stromal cells: Evaluation of biologic, immunologic and genetic factors for successful banking. Cytotherapy 2012, 14, 1193–1202. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.Y.; Kim, Y.R.; Park, W.J.; Kim, H.S.; Jung, S.C.; Woo, S.Y.; Jo, I.; Ryu, K.H.; Park, J.W. Characterisation of insulin-producing cells differentiated from tonsil derived mesenchymal stem cells. Differentiation 2015, 90, 27–39. [Google Scholar] [CrossRef]
- Oh, S.Y.; Choi, Y.M.; Kim, H.Y.; Park, Y.S.; Jung, S.C.; Park, J.W.; Woo, S.Y.; Ryu, K.H.; Kim, H.S.; Jo, I. Application of Tonsil-Derived Mesenchymal Stem Cells in Tissue Regeneration: Concise Review. Stem Cells 2019, 37, 1252–1260. [Google Scholar] [CrossRef]
- Park, S.; Jung, N.; Myung, S.; Choi, Y.; Chung, K.W.; Choi, B.O.; Jung, S.C. Differentiation of Human Tonsil-Derived Mesenchymal Stem Cells into Schwann-Like Cells Improves Neuromuscular Function in a Mouse Model of Charcot-Marie-Tooth Disease Type 1A. Int. J. Mol. Sci. 2018, 19, 2393. [Google Scholar] [CrossRef]
- Kim, H.Y.; Lee, Y.; Yoon, H.S.; Kim, Y.H.; Cho, K.A.; Woo, S.Y.; Kim, H.S.; Park, B.Y.; Jung, S.C.; Jo, I.; et al. A Novel Method to Differentiate Tonsil-Derived Mesenchymal Stem Cells In Vitro into Estrogen-Secreting Cells. Tissue Eng. Regen. Med. 2021, 18, 253–264. [Google Scholar] [CrossRef]
- Ryu, K.H.; Kim, S.Y.; Kim, Y.R.; Woo, S.Y.; Sung, S.H.; Kim, H.S.; Jung, S.C.; Jo, I.; Park, J.W. Tonsil-derived mesenchymal stem cells alleviate concanavalin A-induced acute liver injury. Exp. Cell Res. 2014, 326, 143–154. [Google Scholar] [CrossRef]
- Kim, J.Y.; Park, M.; Kim, Y.H.; Ryu, K.H.; Lee, K.H.; Cho, K.A.; Woo, S.Y. Tonsil-derived mesenchymal stem cells (T-MSCs) prevent Th17-mediated autoimmune response via regulation of the programmed death-1/programmed death ligand-1 (PD-1/PD-L1) pathway. J. Tissue Eng. Regen. Med. 2018, 12, e1022–e1033. [Google Scholar] [CrossRef]
- Cho, K.A.; Kim, Y.H.; Park, M.; Kim, H.J.; Woo, S.Y.; Park, J.W.; Ryu, K.H. Conditioned medium from human palatine tonsil mesenchymal stem cells attenuates acute graftvs.host disease in mice. Mol. Med. Rep. 2019, 19, 609–616. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Learoyd, J.; Duan, Y.; Leff, A.R.; Zhu, X. Hematopoietic Pyk2 regulates migration of differentiated HL-60 cells. J. Inflamm. 2010, 7, 26. [Google Scholar] [CrossRef] [PubMed]
- Heo, S.H.; Jang, S.I.; Kim, S.Y.; Choi, B.; Lee, D.K.; Lee, H.K.; Chang, E.J. Characterization of Circulating IL-7R Positive Cell Populations for Early Detection of Pancreatic Ductal Adenocarcinoma. J. Clin. Med. 2021, 10, 4157. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Raudvere, U.; Kolberg, L.; Kuzmin, I.; Arak, T.; Adler, P.; Peterson, H.; Vilo, J.G. Profiler: A web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 2019, 47, W191–W198. [Google Scholar] [CrossRef]
- Swamydas, M.; Lionakis, M.S. Isolation, purification and labeling of mouse bone marrow neutrophils for functional studies and adoptive transfer experiments. J. Vis. Exp. 2013, 10, e50586. [Google Scholar] [CrossRef]
- Blanter, M.; Gouwy, M.; Struyf, S. Studying Neutrophil Function in vitro: Cell Models and Environmental Factors. J. Inflamm. Res. 2021, 14, 141–162. [Google Scholar] [CrossRef]
- Carrigan, S.O.; Weppler, A.L.; Issekutz, A.C.; Stadnyk, A.W. Neutrophil differentiated HL-60 cells model Mac-1 (CD11b/CD18)-independent neutrophil transepithelial migration. Immunology 2005, 115, 108–117. [Google Scholar] [CrossRef]
- Manda-Handzlik, A.; Bystrzycka, W.; Wachowska, M.; Sieczkowska, S.; Stelmaszczyk-Emmel, A.; Demkow, U.; Ciepiela, O. The influence of agents differentiating HL-60 cells toward granulocyte-like cells on their ability to release neutrophil extracellular traps. Immunol Cell Biol. 2018, 96, 413–425. [Google Scholar] [CrossRef]
- Marsee, D.K.; Pinkus, G.S.; Yu, H. CD71 (transferrin receptor): An effective marker for erythroid precursors in bone marrow biopsy specimens. Am. J. Clin. Pathol. 2010, 134, 429–435. [Google Scholar] [CrossRef]
- Kuwabara, W.M.; Zhang, L.; Schuiki, I.; Curi, R.; Volchuk, A.; Alba-Loureiro, T.C. NADPH oxidase-dependent production of reactive oxygen species induces endoplasmatic reticulum stress in neutrophil-like HL60 cells. PLoS ONE 2015, 10, e0116410. [Google Scholar] [CrossRef] [PubMed]
- Park, J.W.; Kuehn, H.S.; Kim, S.Y.; Chung, K.M.; Choi, H.; Kim, M.; Kim, J.; Lee, S.Y.; Bae, D.S.; Jin, D.K.; et al. Downregulation of Wnt-mediated ROS generation is causally implicated in leprechaunism. Mol. Cells 2010, 29, 63–69. [Google Scholar] [CrossRef] [PubMed]
- Warrier, S.; Balu, S.K.; Kumar, A.P.; Millward, M.; Dharmarajan, A. Wnt antagonist, secreted frizzled-related protein 4 (sFRP4), increases chemotherapeutic response of glioma stem-like cells. Oncol. Res. 2013, 21, 93–102. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Park, Y.S.; Kim, H.S.; Kim, H.Y.; Jin, Y.M.; Jung, S.C.; Ryu, K.H.; Jo, I. Characterization of long-term in vitro culture-related alterations of human tonsil-derived mesenchymal stem cells: Role for CCN1 in replicative senescence-associated increase in osteogenic differentiation. J. Anat. 2014, 225, 510–518. [Google Scholar] [CrossRef]
- Lee, H.J.; Kim, Y.H.; Choi, D.W.; Cho, K.A.; Park, J.W.; Shin, S.J.; Jo, I.; Woo, S.Y.; Ryu, K.H. Tonsil-derived mesenchymal stem cells enhance allogeneic bone marrow engraftment via collagen IV degradation. Stem. Cell Res. Ther. 2021, 12, 329. [Google Scholar] [CrossRef] [PubMed]
- Choi, D.W.; Cho, K.A.; Lee, H.J.; Kim, Y.H.; Woo, K.J.; Park, J.W.; Ryu, K.H.; Woo, S.Y. Cotransplantation of tonsilderived mesenchymal stromal cells in bone marrow transplantation promotes thymus regeneration and T cell diversity following cytotoxic conditioning. Int. J. Mol. Med. 2020, 46, 1166–1174. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Zhou, L.; Wang, Z.; Gao, W.; Chen, W.; Zhang, H.; Jing, B.; Zhu, X.; Chen, L.; Zheng, C.; et al. Eradication of specific donor-dependent variations of mesenchymal stem cells in immunomodulation to enhance therapeutic values. Cell Death Dis. 2021, 12, 357. [Google Scholar] [CrossRef]
- Trivedi, A.; Miyazawa, B.; Gibb, S.; Valanoski, K.; Vivona, L.; Lin, M.; Potter, D.; Stone, M.; Norris, P.J.; Murphy, J.; et al. Bone marrow donor selection and characterization of MSCs is critical for pre-clinical and clinical cell dose production. J. Transl. Med. 2019, 17, 128. [Google Scholar] [CrossRef]
- Haraguchi, R.; Kitazawa, R.; Mori, K.; Tachibana, R.; Kiyonari, H.; Imai, Y.; Abe, T.; Kitazawa, S. sFRP4-dependent Wnt signal modulation is critical for bone remodeling during postnatal development and age-related bone loss. Sci. Rep. 2016, 6, 25198. [Google Scholar] [CrossRef] [PubMed]
- Steiglitz, B.M.; Keene, D.R.; Greenspan, D.S. PCOLCE2 encodes a functional procollagen C-proteinase enhancer (PCPE2) that is a collagen-binding protein differing in distribution of expression and post-translational modification from the previously described PCPE1. J. Biol. Chem. 2002, 277, 49820–49830. [Google Scholar] [CrossRef] [PubMed]
- Touyz, R.M.; Anagnostopoulou, A.; Rios, F.; Montezano, A.C.; Camargo, L.L. NOX5: Molecular biology and pathophysiology. Exp. Physiol. 2019, 104, 605–616. [Google Scholar] [CrossRef] [PubMed]
- Guichard, C.; Pedruzzi, E.; Fay, M.; Ben Mkaddem, S.; Coant, N.; Daniel, F.; Ogier-Denis, E. The Nox/Duox family of ROS-generating NADPH oxidases. Med. Sci. 2006, 22, 953–959. [Google Scholar] [CrossRef]
- Rohrbach, A.S.; Slade, D.J.; Thompson, P.R.; Mowen, K.A. Activation of PAD4 in NET formation. Front. Immunol. 2012, 3, 360. [Google Scholar] [CrossRef]
- Liu, X.; Arfman, T.; Wichapong, K.; Reutelingsperger, C.P.M.; Voorberg, J.; Nicolaes, G.A.F. PAD4 takes charge during neutrophil activation: Impact of PAD4 mediated NET formation on immune-mediated disease. J. Thromb. Haemost. 2021, 19, 1607–1617. [Google Scholar] [CrossRef]
- Vonica, A.; Bhat, N.; Phan, K.; Guo, J.; Iancu, L.; Weber, J.A.; Karger, A.; Cain, J.W.; Wang, E.C.E.; DeStefano, G.M.; et al. Apcdd1 is a dual BMP/Wnt inhibitor in the developing nervous system and skin. Dev. Biol. 2020, 464, 71–87. [Google Scholar] [CrossRef]
- Ma, Z.H.; Ma, J.H.; Jia, L.; Zhao, Y.F. Effect of enhanced expression of COL8A1 on lymphatic metastasis of hepatocellular carcinoma in mice. Exp. Ther. Med. 2012, 4, 621–626. [Google Scholar] [CrossRef][Green Version]
- Blois, S.M.; Sulkowski, G.; Tirado-Gonzalez, I.; Warren, J.; Freitag, N.; Klapp, B.F.; Rifkin, D.; Fuss, I.; Strober, W.; Dveksler, G.S. Pregnancy-specific glycoprotein 1 (PSG1) activates TGF-beta and prevents dextran sodium sulfate (DSS)-induced colitis in mice. Mucosal. Immunol. 2014, 7, 348–358. [Google Scholar] [CrossRef] [PubMed]
- Astarita, J.L.; Acton, S.E.; Turley, S.J. Podoplanin: Emerging functions in development, the immune system, and cancer. Front. Immunol. 2012, 3, 283. [Google Scholar] [CrossRef]
- Antonucci, F.; Corradini, I.; Fossati, G.; Tomasoni, R.; Menna, E.; Matteoli, M. SNAP-25, a Known Presynaptic Protein with Emerging Postsynaptic Functions. Front. Synaptic. Neurosci. 2016, 8, 7. [Google Scholar] [CrossRef] [PubMed]
- Taghavi-Farahabadi, M.; Mahmoudi, M.; Rezaei, N.; Hashemi, S.M. Wharton’s Jelly Mesenchymal Stem Cells Exosomes and Conditioned Media Increased Neutrophil Lifespan and Phagocytosis Capacity. Immunol. Investig. 2021, 50, 1042–1057. [Google Scholar] [CrossRef] [PubMed]
- Raffaghello, L.; Bianchi, G.; Bertolotto, M.; Montecucco, F.; Busca, A.; Dallegri, F.; Ottonello, L.; Pistoia, V. Human mesenchymal stem cells inhibit neutrophil apoptosis: A model for neutrophil preservation in the bone marrow niche. Stem. Cells 2008, 26, 151–162. [Google Scholar] [CrossRef] [PubMed]
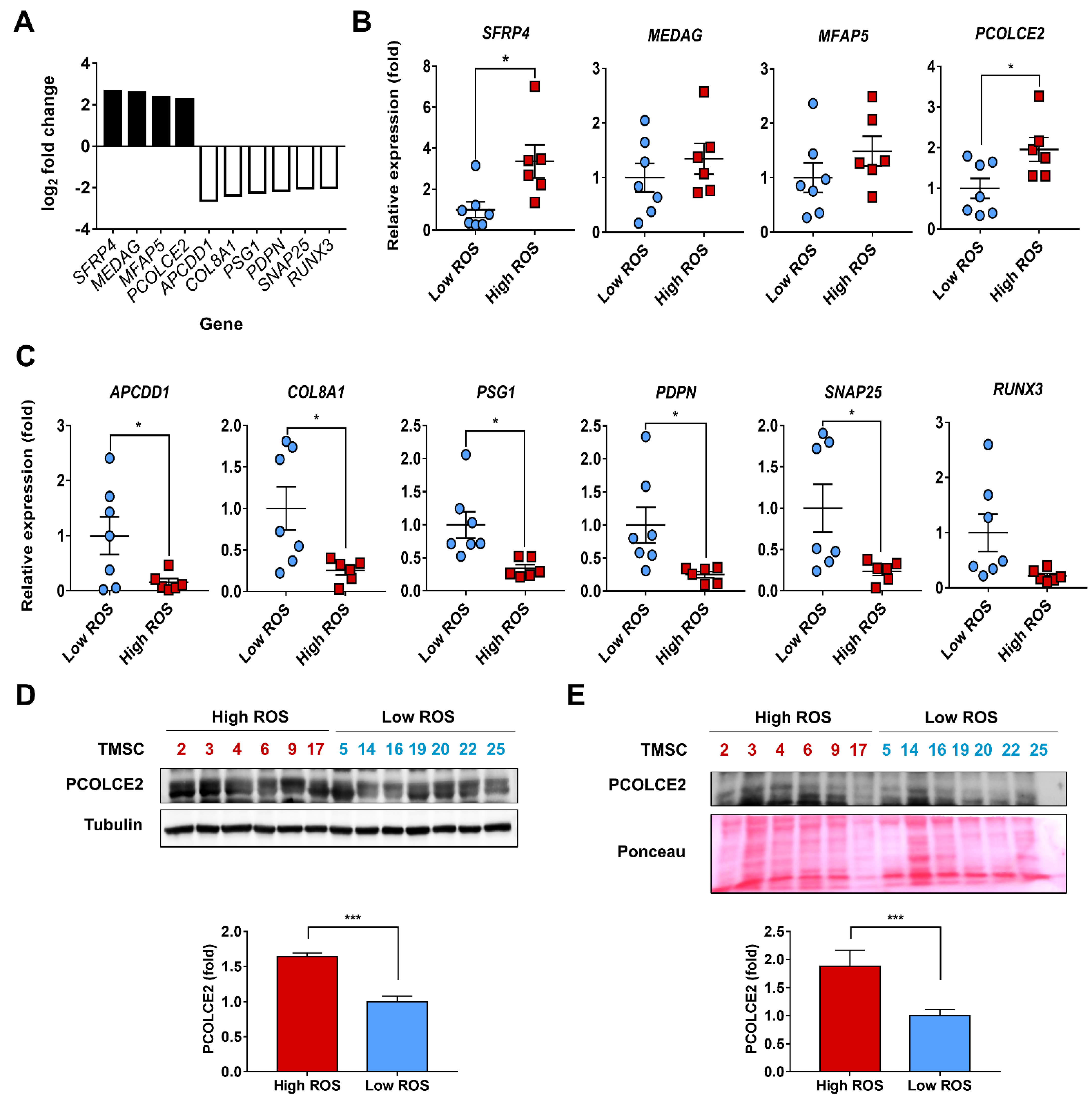
| Gene Number | Gene Symbol | Gene Full Name | High/Low Group (Fold) |
|---|---|---|---|
| NM_003014 | SFRP4 | Secreted frizzled-related protein 4 | 2.66 |
| NM_032849 | MEDAG | Mesenteric estrogen-dependent adipogenesis | 2.58 |
| NM_001297709 | MFAP5 | Microfibrillar-associated protein 5 | 2.36 |
| NM_013363 | PCOLCE2 | Procollagen C-endopeptidase enhancer 2 | 2.27 |
| NM_001162371 | LOC728392 | Uncharacterised LOC728392 | 1.96 |
| NM_000908 | NPR3 | Natriuretic peptide receptor 3 | 1.95 |
| NM_001309443 | SPARC | Secreted protein acidic and cysteine-rich | 1.93 |
| NR_001591 | TPTEP1 | Transmembrane phosphatase with tensin homology pseudogene 1 | 1.88 |
| NM_001039667 | ANGPTL4 | Angiopoietin-like 4 | 1.82 |
| NM_001134851 | TCF7 | Transcription factor 7 (T-cell specific, HMG-box) | 1.79 |
| NM_001307952 | HAPLN3 | Hyaluronan and proteoglycan link protein 3 | 1.77 |
| NM_006332 | IFI30 | IFI30, lysosomal thiol reductase | 1.75 |
| NM_000398 | CYB5R3 | Cytochrome b5 reductase 3 | 1.69 |
| NM_001190737 | NFIB | Nuclear factor I B | 1.64 |
| NM_178507 | OAF | Out at first homolog | 1.64 |
| NM_006617 | NES | Nestin | 1.62 |
| NM_000710 | BDKRB1 | Bradykinin receptor B1 | 1.62 |
| NM_005251 | FOXC2 | Forkhead box C2 (MFH-1, mesenchyme forkhead 1) | 1.60 |
| NM_144617 | HSPB6 | Heat shock protein family B (small) member 6 | 1.56 |
| NM_016932 | SIX2 | SIX homeobox 2 | 1.54 |
| NM_173554 | C10orf107 | Chromosome 10 open reading frame 107 | 1.50 |
| Gene Number | Gene Symbol | Gene Full Name | High/Low Group (Fold) |
|---|---|---|---|
| NM_153000 | APCDD1 | APC downregulated 1 | −2.63 |
| NM_001850 | COL8A1 | Collagen type VIII alpha 1 chain | −2.38 |
| NM_001184825 | PSG1 | Pregnancy-specific beta-1-glycoprotein 1 | −2.25 |
| NM_001006624 | PDPN | Podoplanin | −2.15 |
| NM_001322902 | SNAP25 | Synaptosome associated protein 25 | −2.04 |
| NM_001031680 | RUNX3 | Runt-related transcription factor 3 | −2.02 |
| NM_001242702 | MKX | Mohawk homeobox | −1.99 |
| NM_000902 | MME | Membrane metalloendopeptidase | −1.95 |
| NM_001287426 | UPP1 | Uridine phosphorylase 1 | −1.82 |
| NM_016246 | HSD17B14 | Hydroxysteroid 17-beta dehydrogenase 14 | −1.80 |
| NM_133448 | TMEM132D | Transmembrane protein 132D | −1.70 |
| NM_001128166 | PAK3 | p21 (RAC1) activated kinase 3 | −1.68 |
| NM_001253835 | IGFBP7 | Insulin-like growth factor-binding protein 7 | −1.61 |
| NM_207510 | LCNL1 | Lipocalin like 1 | −1.56 |
| NM_002153 | HSD17B2 | Hydroxysteroid 17-beta dehydrogenase 2 | −1.54 |
| NM_000827 | GRIA1 | Glutamate ionotropic receptor AMPA type subunit 1 | −1.52 |
| NM_001002294 | FMO3 | Glutamate ionotropic receptor AMPA type subunit 1 | −1.52 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yoon, H.-S.; Kim, H.-Y.; Cho, K.-A.; Kim, Y.-H.; Woo, S.-Y.; Kim, H.-S.; Kang, J.-L.; Ryu, K.-H.; Park, J.-W. Procollagen C-Endopeptidase Enhancer 2 Secreted by Tonsil-Derived Mesenchymal Stem Cells Increases the Oxidative Burst of Promyelocytic HL-60 Cells. Biology 2022, 11, 255. https://doi.org/10.3390/biology11020255
Yoon H-S, Kim H-Y, Cho K-A, Kim Y-H, Woo S-Y, Kim H-S, Kang J-L, Ryu K-H, Park J-W. Procollagen C-Endopeptidase Enhancer 2 Secreted by Tonsil-Derived Mesenchymal Stem Cells Increases the Oxidative Burst of Promyelocytic HL-60 Cells. Biology. 2022; 11(2):255. https://doi.org/10.3390/biology11020255
Chicago/Turabian StyleYoon, Hee-Soo, Hee-Yeon Kim, Kyung-Ah Cho, Yu-Hee Kim, So-Youn Woo, Han-Su Kim, Jihee-Lee Kang, Kyung-Ha Ryu, and Joo-Won Park. 2022. "Procollagen C-Endopeptidase Enhancer 2 Secreted by Tonsil-Derived Mesenchymal Stem Cells Increases the Oxidative Burst of Promyelocytic HL-60 Cells" Biology 11, no. 2: 255. https://doi.org/10.3390/biology11020255
APA StyleYoon, H.-S., Kim, H.-Y., Cho, K.-A., Kim, Y.-H., Woo, S.-Y., Kim, H.-S., Kang, J.-L., Ryu, K.-H., & Park, J.-W. (2022). Procollagen C-Endopeptidase Enhancer 2 Secreted by Tonsil-Derived Mesenchymal Stem Cells Increases the Oxidative Burst of Promyelocytic HL-60 Cells. Biology, 11(2), 255. https://doi.org/10.3390/biology11020255