Defense Strategies: The Role of Transcription Factors in Tomato–Pathogen Interaction
Abstract
:Simple Summary
Abstract
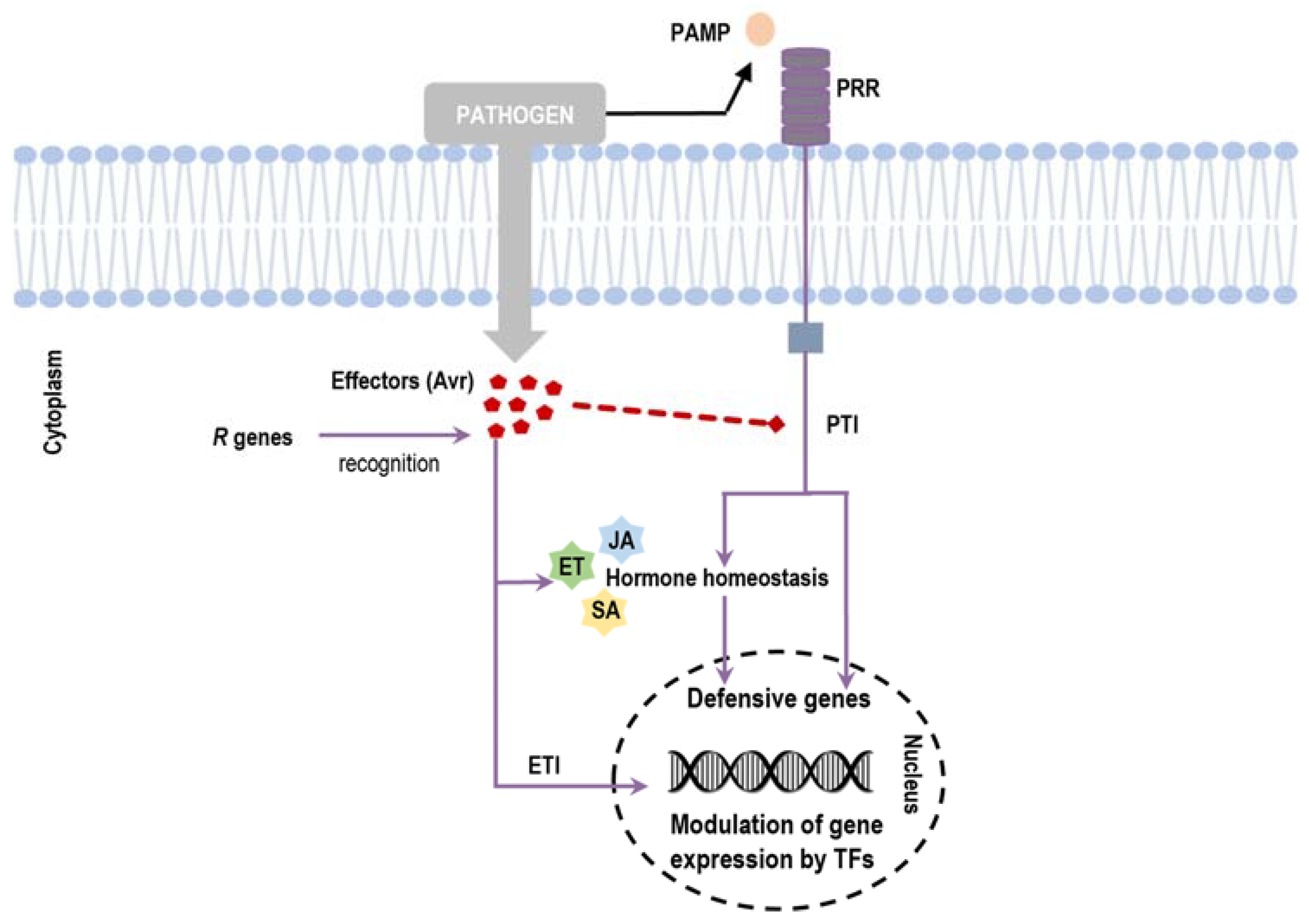
1. Introduction
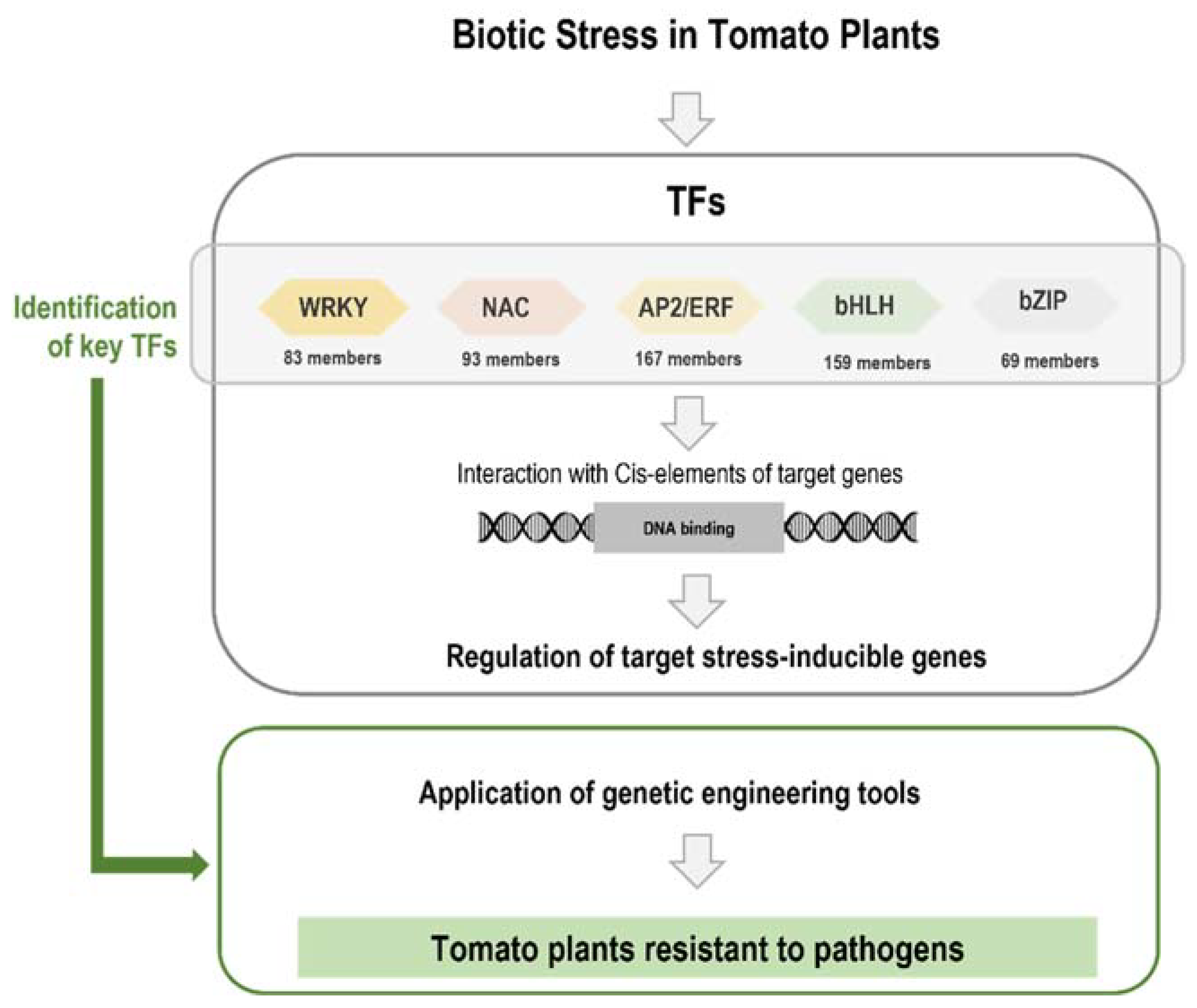
2. Genome Editing to Increase Tomato Resistance to Biotic Stress
3. Transcription Factors Are Involved in Plant Defense Response
3.1. WRKY
3.2. NAC
3.3. AP2/ERF
3.4. bHLH
3.5. bZIP
4. Transcription Factors Are Involved in Tomato Resistance to Biotic Stresses
5. Final Considerations
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Campos, M.D.; Patanita, M.; Varanda, C.; Materatski, P.; Félix, M.R. Plant-Pathogen Interaction. Biology 2021, 10, 444. [Google Scholar] [CrossRef] [PubMed]
- Burdon, J.J.; Zhan, J. Climate change and disease in plant communities. PLoS Biol. 2020, 18, e3000949. [Google Scholar] [CrossRef] [PubMed]
- Campos, M.D.; Patanita, M.; Campos, C.; Materatski, P.; Varanda, C.M.R.; Brito, I.; Félix, M.R. Detection and quantification of Fusarium spp. (F. oxysporum, F. verticillioides, F. graminearum) and Magnaporthiopsis maydis in maize using real-time PCR targeting the ITS region. Agronomy 2019, 9, 45. [Google Scholar] [CrossRef] [Green Version]
- Varanda, C.M.R.; Materatski, P.; Landum, M.; Campos, M.D.; Félix, M.R. Fungal communities associated with peacock and cercospora leaf spots in olive. Plants 2019, 8, 169. [Google Scholar] [CrossRef] [Green Version]
- Campos, M.D.; Zellama, M.S.; Varanda, C.; Materatski, P.; Peixe, A.; Chaouachi, M.; Félix, M.R. Establishment of a sensitive qPCR methodology for detection of the olive-infecting viruses in portuguese and tunisian orchards. Front. Plant Sci. 2019, 10, 694. [Google Scholar] [CrossRef] [PubMed]
- Buscaill, P.; Rivas, S. Transcriptional control of plant defence responses. Curr. Opin. Plant Biol. 2014, 20, 35–46. [Google Scholar] [CrossRef]
- Dodds, P.N.; Rathjen, J.P. Plant immunity: Towards an integrated view of plant–pathogen interactions. Nat. Rev. Genet. 2010, 11, 539–548. [Google Scholar] [CrossRef]
- El Hadrami, A.; Adam, L.R.; Daayf, F. Biocontrol treatments confer protection against Verticillium dahliae infection of potato by inducing antimicrobial metabolites. Mol. Plant-Microbe Interact. 2011, 24, 328–335. [Google Scholar] [CrossRef] [Green Version]
- Guttman, D.S.; McHardy, A.C.; Schulze-Lefert, P. Microbial genome-enabled insights into plant-microorganism interactions. Nat. Rev. Genet. 2014, 15, 797–813. [Google Scholar] [CrossRef]
- Amorim, L.; Santos, R.; Neto, J.; Guida-Santos, M.; Crovella, S.; Benko-Iseppon, A. Transcription factors involved in plant resistance to pathogens. Curr. Protein Pept. Sci. 2016, 18, 335–351. [Google Scholar] [CrossRef]
- Chisholm, S.T.; Coaker, G.; Day, B.; Staskawicz, B.J. Host-microbe interactions: Shaping the evolution of the plant immune response. Cell 2006, 124, 803–814. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, J.D.G.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jalali, B.L.; Bhargava, S.; Kamble, A. Signal transduction and transcriptional regulation of plant defence responses. J. Phytopathol. 2006, 154, 65–74. [Google Scholar] [CrossRef]
- van Verk, M.C.; Gatz, C.; Linthorst, H.J.M. Chapter 10 Transcriptional regulation of plant defense responses. Adv. Bot. Res. 2009, 51, 397–438. [Google Scholar] [CrossRef]
- Rezzonico, F.; Rupp, O.; Fahrentrapp, J. Pathogen recognition in compatible plant-microbe interactions. Sci. Rep. 2017, 7, 6383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Inukai, S.; Kock, K.H.; Bulyk, M.L. Transcription factor–DNA binding: Beyond binding site motifs. Curr. Opin. Genet. Dev. 2017, 43, 110–119. [Google Scholar] [CrossRef] [Green Version]
- Seo, E.; Choi, D. Functional studies of transcription factors involved in plant defenses in the genomics era. Brief. Funct. Genomics 2015, 14, 260–267. [Google Scholar] [CrossRef] [Green Version]
- Javed, T.; Shabbir, R.; Ali, A.; Afzal, I.; Zaheer, U.; Gao, S.J. Transcription factors in plant stress responses: Challenges and potential for sugarcane improvement. Plants 2020, 9, 491. [Google Scholar] [CrossRef]
- Campos, M.D.; Félix, M.R.; Patanita, M.; Materatski, P.; Varanda, C. High throughput sequencing unravels tomato-pathogen interactions towards a sustainable plant breeding. Hortic. Res. 2021, 8, 171. [Google Scholar] [CrossRef]
- Zhao, M.; Ji, H.M.; Gao, Y.; Cao, X.X.; Mao, H.Y.; Ouyang, S.Q.; Liu, P. An integrated analysis of mRNA and srna transcriptional profiles in tomato root: Insights on tomato wilt disease. PLoS ONE 2018, 13, e0206765. [Google Scholar] [CrossRef]
- Gerszberg, A.; Hnatuszko-Konka, K.; Kowalczyk, T.; Kononowicz, A.K. Tomato (Solanum lycopersicum L.) in the service of biotechnology. Plant Cell. Tissue Organ Cult. 2015, 120, 881–902. [Google Scholar] [CrossRef] [Green Version]
- Panno, S.; Davino, S.; Caruso, A.G.; Bertacca, S.; Crnogorac, A.; Mandi, A. A review of the most common and economically important diseases that undermine the cultivation of tomato crop in the mediterranean basin. Agronomy 2021, 11, 2188. [Google Scholar] [CrossRef]
- Simko, I.; Jia, M.; Venkatesh, J.; Kang, B.C.; Weng, Y.; Barcaccia, G.; Lanteri, S.; Bhattarai, G.; Foolad, M.R. Genomics and marker-assisted improvement of vegetable crops. CRC. Crit. Rev. Plant Sci. 2021, 40, 303–365. [Google Scholar] [CrossRef]
- Salava, H.; Thula, S.; Mohan, V.; Kumar, R.; Maghuly, F. Application of genome editing in tomato breeding: Mechanisms, advances, and prospects. Int. J. Mol. Sci. 2021, 22, 682. [Google Scholar] [CrossRef] [PubMed]
- Singh, V.K.; Singh, A.K.; Kumar, A. Disease management of tomato through PGPB: Current trends and future perspective. 3 Biotech 2017, 7, 255. [Google Scholar] [CrossRef] [PubMed]
- Nekrasov, V.; Wang, C.; Win, J.; Lanz, C.; Weigel, D.; Kamoun, S. Rapid generation of a transgene-free powdery mildew resistant tomato by genome deletion. Sci. Rep. 2017, 7, 482. [Google Scholar] [CrossRef] [Green Version]
- Parmar, N.; Singh, K.H.; Sharma, D.; Singh, L.; Kumar, P.; Nanjundan, J.; Khan, Y.J.; Chauhan, D.K.; Thakur, A.K. Genetic engineering strategies for biotic and abiotic stress tolerance and quality enhancement in horticultural crops: A comprehensive review. 3 Biotech 2017, 7, 239. [Google Scholar] [CrossRef]
- Capriotti, L.; Baraldi, E.; Mezzetti, B.; Limera, C.; Sabbadini, S. Biotechnological approaches: Gene overexpression, gene silencing, and genome editing to control fungal and oomycete diseases in grapevine. Int. J. Mol. Sci. 2020, 21, 5701. [Google Scholar] [CrossRef]
- Louwaars, N.; Jochemsen, H. An ethical and societal analysis for biotechnological methods in plant breeding. Agronomy 2021, 11, 1183. [Google Scholar] [CrossRef]
- Xia, X.; Cheng, X.; Li, R.; Yao, J.; Li, Z.; Cheng, Y. Advances in application of genome editing in tomato and recent development of genome editing technology. Theor. Appl. Genet. 2021, 134, 2727–2747. [Google Scholar] [CrossRef]
- Pathak, K.; Gogoi, B. RNA interference (RNAi): Application in crop improvement: A review. Agric. Rev. 2016, 37. [Google Scholar] [CrossRef]
- Lange, M.; Yellina, A.L.; Orashakova, S.; Becker, A. Virus-induced gene silencing (VIGS) in plants: An overview of target species and the virus-derived vector systems. Methods Mol. Biol. 2013, 975, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Varanda, C.M.; Félix, M.R.; Campos, M.D.; Patanita, M.; Materatski, P. Plant viruses: From targets to tools for CRISPR. Viruses 2021, 13, 141. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Zhang, H.; Zhu, H. CRISPR technology is revolutionizing the improvement of tomato and other fruit crops. Hortic. Res. 2019, 6, 77. [Google Scholar] [CrossRef] [Green Version]
- Chaudhary, R.; Atamian, H.S. Resistance-gene-mediated defense responses against biotic stresses in the crop model plant tomato. J. Plant Pathol. Microbiol. 2017, 8. [Google Scholar] [CrossRef]
- Huibers, R.P.; Loonen, A.E.H.M.; Gao, D.; Van den Ackerveken, G.; Visser, R.G.F.; Bai, Y. Powdery mildew resistance in tomato by impairment of SlPMR4 and SlDMR1. PLoS ONE 2013, 8, e67467. [Google Scholar] [CrossRef] [Green Version]
- Rushton, P.J.; Somssich, I.E.; Ringler, P.; Shen, Q.J. WRKY transcription factors. Trends Plant Sci. 2010, 15, 247–258. [Google Scholar] [CrossRef]
- Chen, X.; Li, C.; Wang, H.; Guo, Z. WRKY transcription factors: Evolution, binding, and action. Phytopathol. Res. 2019, 1, 13. [Google Scholar] [CrossRef]
- Baillo, E.H.; Kimotho, R.N.; Zhang, Z.; Xu, P. Transcription factors associated with abiotic and biotic stress tolerance and their potential for crops improvement. Genes 2019, 10, 771. [Google Scholar] [CrossRef] [Green Version]
- Bai, Y.; Sunarti, S.; Kissoudis, C.; Visser, R.G.F.; van der Linden, C.G. The role of tomato WRKY genes in plant responses to combined abiotic and biotic stresses. Front. Plant Sci. 2018, 9, 801. [Google Scholar] [CrossRef] [Green Version]
- Xie, Zhen, Zhong-Lin Zhang, Xiaolu Zou, Jie Huang, Paul Ruas, Daniel Thompson, and Q. J.S. Annotations and functional analyses of the rice WRKY gene superfamily reveal positive and negative regulators of abscisic acid signaling in. 2005, 137, 176–189. [CrossRef]
- Dong, J.; Chen, C.; Chen, Z. Expression profiles of the Arabidopsis WRKY gene superfamily during plant defense response. Plant Mol. Biol. 2003, 51, 21–37. [Google Scholar] [CrossRef] [PubMed]
- Li, M.Y.; Xu, Z.S.; Tian, C.; Huang, Y.; Wang, F.; Xiong, A.S. Genomic identification of WRKY transcription factors in carrot (Daucus carota) and analysis of evolution and homologous groups for plants. Sci. Rep. 2016, 6, 23101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bakshi, M.; Oelmüller, R. Wrky transcription factors jack of many trades in plants. Plant Signal. Behav. 2014, 9, e27700. [Google Scholar] [CrossRef] [Green Version]
- Riechmann, J.L.; Heard, J.; Martin, G.; Reuber, L.; Jiang, C.; Keddie, J.; Adam, L.; Pineda, O.; Ratcliffe, O.J.; Samaha, R.R.; et al. Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science 2000, 290, 2105–2110. [Google Scholar] [CrossRef]
- Zhang, H.; Kang, H.; Su, C.; Qi, Y.; Liu, X.; Pu, J. Genome-wide identification and expression profile analysis of the NAC transcription factor family during abiotic and biotic stress in woodland strawberry. PLoS ONE 2018, 13, e0197892. [Google Scholar] [CrossRef]
- Shen, H.; Yin, Y.; Chen, F.; Xu, Y.; Dixon, R.A. A bioinformatic analysis of NAC genes for plant cell wall development in relation to lignocellulosic bioenergy production. Bioenergy Res. 2009, 2, 217–232. [Google Scholar] [CrossRef]
- Fang, Y.; You, J.; Xie, K.; Xie, W.; Xiong, L. Systematic sequence analysis and identification of tissue-specific or stress-responsive genes of NAC transcription factor family in rice. Mol. Genet. Genom. 2008, 280, 547–563. [Google Scholar] [CrossRef]
- Jin, J.F.; Wang, Z.Q.; He, Q.Y.; Wang, J.Y.; Li, P.F.; Xu, J.M.; Zheng, S.J.; Fan, W.; Yang, J.L. Genome-wide identification and expression analysis of the NAC transcription factor family in tomato (Solanum lycopersicum) during aluminum stress. BMC Genomics 2020, 21, 288. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Li, X.; Chao, J.; Zhang, Z.; Wang, W.; Guo, Y. NAC Family Transcription factors in tobacco and their potential role in regulating leaf senescence. Front. Plant Sci. 2018, 9, 1900. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Wang, T.; Bartholomew, E.; Black, K.; Dong, M.; Zhang, Y.; Yang, S.; Cai, Y.; Xue, S.; Weng, Y.; et al. Comprehensive analysis of NAC transcription factors and their expression during fruit spine development in cucumber (Cucumis sativus L.). Hortic. Res. 2018, 5, 31. [Google Scholar] [CrossRef] [PubMed]
- Yuan, X.; Wang, H.; Cai, J.; Li, D.; Song, F. NAC transcription factors in plant immunity. Phytopathol. Res. 2019, 1, 3. [Google Scholar] [CrossRef]
- Thirumalaikumar, V.P.; Devkar, V.; Mehterov, N.; Ali, S.; Ozgur, R.; Turkan, I.; Mueller-Roeber, B.; Balazadeh, S. NAC transcription factor JUNGBRUNNEN1 enhances drought tolerance in tomato. Plant Biotechnol. J. 2018, 16, 354–366. [Google Scholar] [CrossRef] [PubMed]
- Nuruzzaman, M.; Sharoni, A.M.; Kikuchi, S. Roles of NAC transcription factors in the regulation of biotic and abiotic stress responses in plants. Front. Microbiol. 2013, 4, 248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Licausi, F.; Ohme-Takagi, M.; Perata, P. APETALA2/Ethylene Responsive Factor (AP2/ERF) transcription factors: Mediators of stress responses and developmental programs. New Phytol. 2013, 199, 639–649. [Google Scholar] [CrossRef]
- Akbudak, M.A.; Filiz, E.; Kontbay, K. DREB2 (dehydration-responsive element-binding protein 2) type transcription factor in sorghum (Sorghum bicolor): Genome-wide identification, characterization and expression profiles under cadmium and salt stresses. 3 Biotech 2018, 8, 426. [Google Scholar] [CrossRef]
- Chen, C.Y.; Lin, P.H.; Chen, K.H.; Cheng, Y.S. Structural insights into Arabidopsis ethylene response factor 96 with an extended N-terminal binding to GCC box. Plant Mol. Biol. 2020, 104, 483–498. [Google Scholar] [CrossRef]
- Gutterson, N.; Reuber, T.L. Regulation of disease resistance pathways by AP2/ERF transcription factors. Curr. Opin. Plant Biol. 2004, 7, 465–471. [Google Scholar] [CrossRef]
- Chopra, R.; Burow, G.; Hayes, C.; Emendack, Y.; Xin, Z.; Burke, J. Transcriptome profiling and validation of gene based single nucleotide polymorphisms (SNPs) in sorghum genotypes with contrasting responses to cold stress. BMC Genom. 2015, 16, 1040. [Google Scholar] [CrossRef] [Green Version]
- Bihani, P.; Char, B.; Bhargava, S. Transgenic expression of sorghum DREB2 in rice improves tolerance and yield under water limitation. J. Agric. Sci. 2011, 149, 95–101. [Google Scholar] [CrossRef]
- Guo, B.; Wei, Y.; Xu, R.; Lin, S.; Luan, H.; Lv, C.; Zhang, X.; Song, X.; Xu, R. Genome-wide analysis of APETALA2/ethylene-responsive factor (AP2/ERF) gene family in barley (Hordeum vulgare L.). PLoS ONE 2016, 11, e0161322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gu, C.; Guo, Z.H.; Hao, P.P.; Wang, G.M.; Jin, Z.M.; Zhang, S.L. Multiple regulatory roles of AP2/ERF transcription factor in angiosperm. Bot. Stud. 2017, 58, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, R.; Song, J.; Liu, S.; Chen, C.; Zhang, S.; Wang, J.; Xiao, Y.; Cao, B.; Lei, J.; Zhu, Z. Genome-wide identification of the capsicum bHLH transcription factor family: Discovery of a candidate regulator involved in the regulation of species-specific bioactive metabolites. BMC Plant Biol. 2021, 21, 262. [Google Scholar] [CrossRef] [PubMed]
- Bai, G.; Yang, D.H.; Chao, P.; Yao, H.; Fei, M.L.; Zhang, Y.; Chen, X.; Xiao, B.; Li, F.; Wang, Z.Y.; et al. Genome-wide identification and expression analysis of NtbHLH gene family in tobacco (Nicotiana tabacum L.) and the role of NtbHLH86 in drought adaptation. Plant Divers. 2021, 43, 510–522. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Zhao, P.; Kong, N.; Lu, R.; Pei, Y.; Huang, C.; Ma, H.; Chen, Q. Genome-wide identification and characterization of the potato bHLH transcription factor family. Genes 2018, 9, 54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, H.; Fan, H.J.; Ling, H.Q. Genome-wide identification and characterization of the bHLH gene family in tomato. BMC Genom. 2015, 16, 9. [Google Scholar] [CrossRef] [Green Version]
- Toledo-Ortiz, G.; Huq, E.; Quail, P.H. The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 2003, 15, 1749–1770. [Google Scholar] [CrossRef] [Green Version]
- Atchley, W.R.; Terhalle, W.; Dress, A. Positional dependence, cliques, and predictive motifs in the bHLH protein domain. J. Mol. Evol. 1999, 48, 501–516. [Google Scholar] [CrossRef] [PubMed]
- Fan, M.; Bai, M.Y.; Kim, J.G.; Wang, T.; Oh, E.; Chen, L.; Park, C.H.; Son, S.H.; Kim, S.K.; Mudgett, M.B.; et al. The bHLH transcription factor HBI1 mediates the trade-off between growth and pathogen-associated molecular pattern-triggered immunity in Arabidopsis. Plant Cell 2014, 26, 828–841. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.H.; Oikawa, T.; Kyozuka, J.; Wong, H.L.; Umemura, K.; Kishi-Kaboshi, M.; Takahashi, A.; Kawano, Y.; Kawasaki, T.; Shimamoto, K. The bHLH Rac immunity1 (RAI1) is activated by OsRac1 via OsMAPK3 and OsMAPK6 in rice immunity. Plant Cell Physiol. 2012, 53, 740–754. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.G.; Mudgett, M.B. Tomato bHLH132 transcription factor controls growth and defense and is activated by Xanthomonas euvesicatoria effector XopD during pathogenesis. Mol. Plant-Microbe Interact. 2019, 32, 1614–1622. [Google Scholar] [CrossRef] [PubMed]
- Jakoby, M.; Weisshaar, B.; Dröge-Laser, W.; Vicente-Carbajosa, J.; Tiedemann, J.; Kroj, T.; Parcy, F. bZIP transcription factors in Arabidopsis. Trends Plant Sci. 2002, 7, 106–111. [Google Scholar] [CrossRef]
- Wei, K.; Chen, J.; Wang, Y.; Chen, Y.; Chen, S.; Lin, Y.; Pan, S.; Zhong, X.; Xie, D. Genome-wide analysis of bZIP-encoding genes in maize. DNA Res. 2012, 19, 463–476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gai, W.X.; Ma, X.; Qiao, Y.M.; Shi, B.H.; ul Haq, S.; Li, Q.H.; Wei, A.M.; Liu, K.K.; Gong, Z.H. Characterization of the bZIP transcription factor family in pepper (Capsicum annuum L.): CabZIP25 positively modulates the salt Tolerance. Front. Plant Sci. 2020, 11, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Fu, F.; Zhang, H.; Song, F. Genome-wide systematic characterization of the bZIP transcriptional factor family in tomato (Solanum lycopersicum L.). BMC Genom. 2015, 16, 771. [Google Scholar] [CrossRef] [Green Version]
- Kesarwani, M.; Yoo, J.; Dong, X. Genetic interactions of TGA transcription factors in the regulation of pathogenesis-related genes and disease resistance in Arabidopsis. Plant Physiol. 2007, 144, 336–346. [Google Scholar] [CrossRef] [Green Version]
- Lim, C.W.; Baek, W.; Lim, S.; Han, S.W.; Lee, S.C. Expression and functional roles of the pepper pathogen-induced bZIP transcription factor CabZIP2 in enhanced disease resistance to bacterial pathogen infection. Mol. Plant-Microbe Interact. 2015, 28, 825–833. [Google Scholar] [CrossRef] [Green Version]
- Sato, S.; Tabata, S.; Hirakawa, H.; Asamizu, E.; Shirasawa, K.; Isobe, S.; Kaneko, T.; Nakamura, Y.; Shibata, D.; Aoki, K.; et al. The tomato genome sequence provides insights into fleshy fruit evolution. Nature 2012, 485, 635–641. [Google Scholar] [CrossRef] [Green Version]
- Liu, B.; Hong, Y.; Zhang, Y.; Li, X.; Huang, L.; Zhang, H. Tomato WRKY transcriptional factor SlDRW1 is required for disease resistance against Botrytis cinerea and tolerance to oxidative stress. Plant Sci. 2014, 227, 145–156. [Google Scholar] [CrossRef]
- Li, J.; Luan, Y.; Liu, Z. SpWRKY1 mediates resistance to Phytophthora infestans and tolerance to salt and drought stress by modulating reactive oxygen species homeostasis and expression of defense-related genes in tomato. Plant Cell. Tissue Organ Cult. 2015, 123, 67–81. [Google Scholar] [CrossRef]
- Cui, J.; Xu, P.; Meng, J.; Li, J.; Jiang, N.; Luan, Y. Transcriptome signatures of tomato leaf induced by Phytophthora infestans and functional identification of transcription factor SpWRKY3. Theor. Appl. Genet. 2018, 131, 787–800. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Gao, Y.; Liu, J.; Peng, X.; Niu, X.; Fei, Z.; Cao, S.; Liu, Y. Genome-wide analysis of WRKY transcription factors in Solanum lycopersicum. Mol. Genet. Genom. 2012, 287, 495–513. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Gao, Y.; Li, H.; Yang, S.; Liu, Y. sheng Over-expression of SlWRKY39 leads to enhanced resistance to multiple stress factors in tomato. J. Plant Biol. 2015, 58, 52–60. [Google Scholar] [CrossRef]
- Bhattarai, K.K.; Atamian, H.S.; Kaloshian, I.; Eulgem, T. WRKY72-type transcription factors contribute to basal immunity in tomato and Arabidopsis as well as gene-for-gene resistance mediated by the tomato R gene Mi-1. Plant J. 2010, 63, 229–240. [Google Scholar] [CrossRef]
- Chinnapandi, B.; Bucki, P.; Braun Miyara, S. SlWRKY45, nematode-responsive tomato WRKY gene, enhances susceptibility to the root knot nematode; M. javanica infection. Plant Signal. Behav. 2017, 12. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.; Li, M.Y.; Wu, P.; Xu, Z.S.; Que, F.; Wang, F.; Xiong, A.S. Members of WRKY Group III transcription factors are important in TYLCV defense signaling pathway in tomato (Solanum lycopersicum). BMC Genom. 2016, 17, 788. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Zheng, C.; Shao, X.; Hu, Z.; Li, J.; Wang, P.; Wang, A.; Yu, J.; Shi, K. Transcriptomic and genetic approaches reveal an essential role of the NAC transcription factor SlNAP1 in the growth and defense response of tomato. Hortic. Res. 2020, 7. [Google Scholar] [CrossRef]
- Liu, B.; Ouyang, Z.; Zhang, Y.; Li, X.; Hong, Y.; Huang, L.; Liu, S.; Zhang, H.; Li, D.; Song, F. Tomato NAC transcription factor SlSRN1 positively regulates defense response against biotic stress but negatively regulates abiotic stress response. PLoS ONE 2014, 9, e102067. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Miao, M.; Kud, J.; Niu, X.; Ouyang, B.; Zhang, J.; Ye, Z.; Kuhl, J.C.; Liu, Y.; Xiao, F. SlNAC1, a stress-related transcription factor, is fine-tuned on both the transcriptional and the post-translational level. New Phytol. 2013, 197, 1214–1224. [Google Scholar] [CrossRef]
- Selth, L.A.; Dogra, S.C.; Rasheed, M.S.; Healy, H.; Randles, J.W.; Rezaian, M.A. A NAC domain protein interacts with tomato leaf curl virus replication accessory protein and enhances viral replication. Plant Cell 2005, 17, 311–325. [Google Scholar] [CrossRef] [Green Version]
- Upadhyay, P.; Ganie, S.H.; Rai, A.; Singh, M.; Sinha, B. Identification of transcription factors in tomato, potentially related to early blight resistance at invasion in host tissue using, microarray expression profiling. S. Afr. J. Bot. 2016, 106, 165–173. [Google Scholar] [CrossRef]
- Yang, H.; Sun, Y.; Wang, H.; Zhao, T.; Xu, X.; Jiang, J.; Li, J. Genome-wide identification and functional analysis of the ERF2 gene family in response to disease resistance against Stemphylium lycopersici in tomato. BMC Plant Biol. 2021, 21, 72. [Google Scholar] [CrossRef] [PubMed]
- Buxdorf, K.; Rubinsky, G.; Barda, O.; Burdman, S.; Aharoni, A.; Levy, M. The transcription factor SlSHINE3 modulates defense responses in tomato plants. Plant Mol. Biol. 2014, 84, 37–47. [Google Scholar] [CrossRef] [PubMed]
- Shu, P.; Li, Z.; Min, D.; Zhang, X.; Ai, W.; Li, J.; Zhou, J.; Li, Z.; Li, F.; Li, X. CRISPR/Cas9-mediated SlMYC2 mutagenesis adverse to tomato plant growth and MeJA-induced fruit resistance to Botrytis cinerea. J. Agric. Food Chem. 2020, 68, 5529–5538. [Google Scholar] [CrossRef] [PubMed]
- Orellana, S.; Yañez, M.; Espinoza, A.; Verdugo, I.; González, E.; Ruiz-Lara, S.; Casaretto, J.A. The transcription factor SlAREB1 confers drought, salt stress tolerance and regulates biotic and abiotic stress-related genes in tomato. Plant Cell Environ. 2010, 33, 2191–2208. [Google Scholar] [CrossRef]
- Karkute, S.G.; Gujjar, R.S.; Rai, A.; Akhtar, M.; Singh, M.; Singh, B. Genome wide expression analysis of WRKY genes in tomato (Solanum lycopersicum) under drought stress. Plant Gene 2018, 13, 8–17. [Google Scholar] [CrossRef]
- Atamian, H.S.; Eulgem, T.; Kaloshian, I. SlWRKY70 is required for Mi-1-mediated resistance to aphids and nematodes in tomato. Planta 2012, 235, 299–309. [Google Scholar] [CrossRef]
- Gao, Y.; Wei, W.; Zhao, X.; Tan, X.; Fan, Z.; Zhang, Y.; Jing, Y.; Meng, L.; Zhu, B.; Zhu, H.; et al. A NAC transcription factor, NOR-like1, is a new positive regulator of tomato fruit ripening. Hortic. Res. 2018, 5, 75. [Google Scholar] [CrossRef] [Green Version]
- Zhu, M.; Chen, G.; Zhou, S.; Tu, Y.; Wang, Y.; Dong, T.; Hu, Z. A new tomato NAC (NAM ATAF1/2/CUC2) transcription factor, SlNAC4, functions as a positive regulator of fruit ripening and carotenoid accumulation. Plant Cell Physiol. 2014, 55, 119–135. [Google Scholar] [CrossRef] [Green Version]
- Du, M.; Zhai, Q.; Deng, L.; Li, S.; Li, H.; Yan, L.; Huang, Z.; Wang, B.; Jiang, H.; Huang, T.; et al. Closely related NAC transcription factors of tomato differentially regulate stomatal closure and reopening during pathogen attack. Plant Cell 2014, 26, 3167–3184. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.; Zhang, B.; Sun, S.; Xing, G.; Wang, F.; Li, M.; Tian, Y.; Xiong, A. AP2/ERF transcription actors involved in response to tomato yellow leaf curly virus in tomato. Plant Genome 2016, 9, plantgenome2015-09. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.; Wildermuth, M.C.; Chakravarthy, S.; Loh, Y.; Yang, C.; He, X.; Han, Y.; Martin, G.B. Tomato transcription factors Pti4, Pti5, and Pti6 activate defense responses when expressed in Arabidopsis. Plant Cell 2002, 14, 817–831. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kazan, K.; Manners, J.M. MYC2: The master in action. Mol. Plant 2013, 6, 686–703. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| TF Family | TF Target Gene | Effect | Method | Ref. |
|---|---|---|---|---|
| WRKY | SlDRW1 | - Gene expression significantly induced by Botrytis cinerea. - Silencing increase severity of disease caused by B. cinerea. | VIGS | [79] |
| SpWRKY1 | - Overexpression increase resistance to Phytophthora infestans. | Expression vector | [80] | |
| SpWRKY3 | - Gene expression significantly induced by Phytophthora infestans. - Silencing impaired the resistance to P. infestans. - Overexpression increase resistance to P. infestans. | VIGS and expression vector | [81] | |
| SlWRKY8, SlWRKY23, SlWRKY39, SlWRKY53, SlWRKY80, SlWRKY81 | - Up-regulated genes expression in response to Pseudomonas syringae (Pst) pv. tomato DC3000 infection. | - | [82] | |
| SlWRKY39 | - Overexpressing increased resistance to P. syringae. | Expression vector | [83] | |
| SlWRKY72a, SlWRKY72b | - Up-regulated during root-knot nematodes (RKN) disease resistance mediated by the R gene Mi-1. - Silencing resulted in a reduction of Mi-1-mediated resistance and basal defense against RKN. | VIGS | [84] | |
| SlWRKY45 | - Overexpression enhanced tomato susceptibility to RKN | Expression vector | [85] | |
| SolyWRKY41, SolyWRKY42, SolyWRKY53, SolyWRKY54, SolyWRKY80, SolyWRKY8 | - Genes responsive to Tomato yellow leaf curly virus (TYLCV) infection. - Silencing of SolyWRKY41 and SolyWRKY54 decrease accumulation of TYLCV DNA. | VIGS | [86] | |
| NAC | SlNAP1 | - Overexpressing enhanced defense against Pst DC3000 and Ralstonia solanacearum. | Expression vector | [87] |
| SlSRN1 | - Gene expression induced by infection with B. cinerea and Pst DC3000. - Silencing increased severity of disease caused by B. cinerea. | VIGS | [88] | |
| SlNAC1 | - Upregulated gene expression during Pst DC3000 infection. | - | [89] | |
| SlNAC1 | - Overexpression enhanced the accumulation of Tomato leaf curl virus (TLCV) DNA. | Expression vector | [90] | |
| AP2/ERF | ERF-3 | - Upregulated gene expression to Alternaria solani infection, using a resistant genotype. | - | [91] |
| ERF-2 | - Silencing revealed aggravated diseases symptoms caused by Stemphylium lycopersici. | VIGS | [92] | |
| SlSHINE3 | - Silencing revealed higher sensitivity to B. cinerea. - Overexpression revealed resistance to B. cinerea and to Xanthomonas campestris pv. vesicatoria infection. | Not referred | [93] | |
| bHLH | bHLH132 | - Transcriptionally highly induced by Xanthomonas euvesicatoria. - Silencing enhanced susceptibility to X. euvesicatoria. | Expression vector | [71] |
| MYC2 | - Knockout aggravated the B. cinerea disease symptoms. | CRISPR/Cas9 | [94] | |
| bZIP | SlAREB1 | - Overexpression up-regulate several defense genes associated with biotic stress. | Expression vector | [95] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Campos, M.D.; Félix, M.d.R.; Patanita, M.; Materatski, P.; Albuquerque, A.; Ribeiro, J.A.; Varanda, C. Defense Strategies: The Role of Transcription Factors in Tomato–Pathogen Interaction. Biology 2022, 11, 235. https://doi.org/10.3390/biology11020235
Campos MD, Félix MdR, Patanita M, Materatski P, Albuquerque A, Ribeiro JA, Varanda C. Defense Strategies: The Role of Transcription Factors in Tomato–Pathogen Interaction. Biology. 2022; 11(2):235. https://doi.org/10.3390/biology11020235
Chicago/Turabian StyleCampos, Maria Doroteia, Maria do Rosário Félix, Mariana Patanita, Patrick Materatski, André Albuquerque, Joana A. Ribeiro, and Carla Varanda. 2022. "Defense Strategies: The Role of Transcription Factors in Tomato–Pathogen Interaction" Biology 11, no. 2: 235. https://doi.org/10.3390/biology11020235
APA StyleCampos, M. D., Félix, M. d. R., Patanita, M., Materatski, P., Albuquerque, A., Ribeiro, J. A., & Varanda, C. (2022). Defense Strategies: The Role of Transcription Factors in Tomato–Pathogen Interaction. Biology, 11(2), 235. https://doi.org/10.3390/biology11020235









