Identification of Microorganisms Dwelling on the 19th Century Lanna Mural Paintings from Northern Thailand Using Culture-Dependent and -Independent Approaches
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
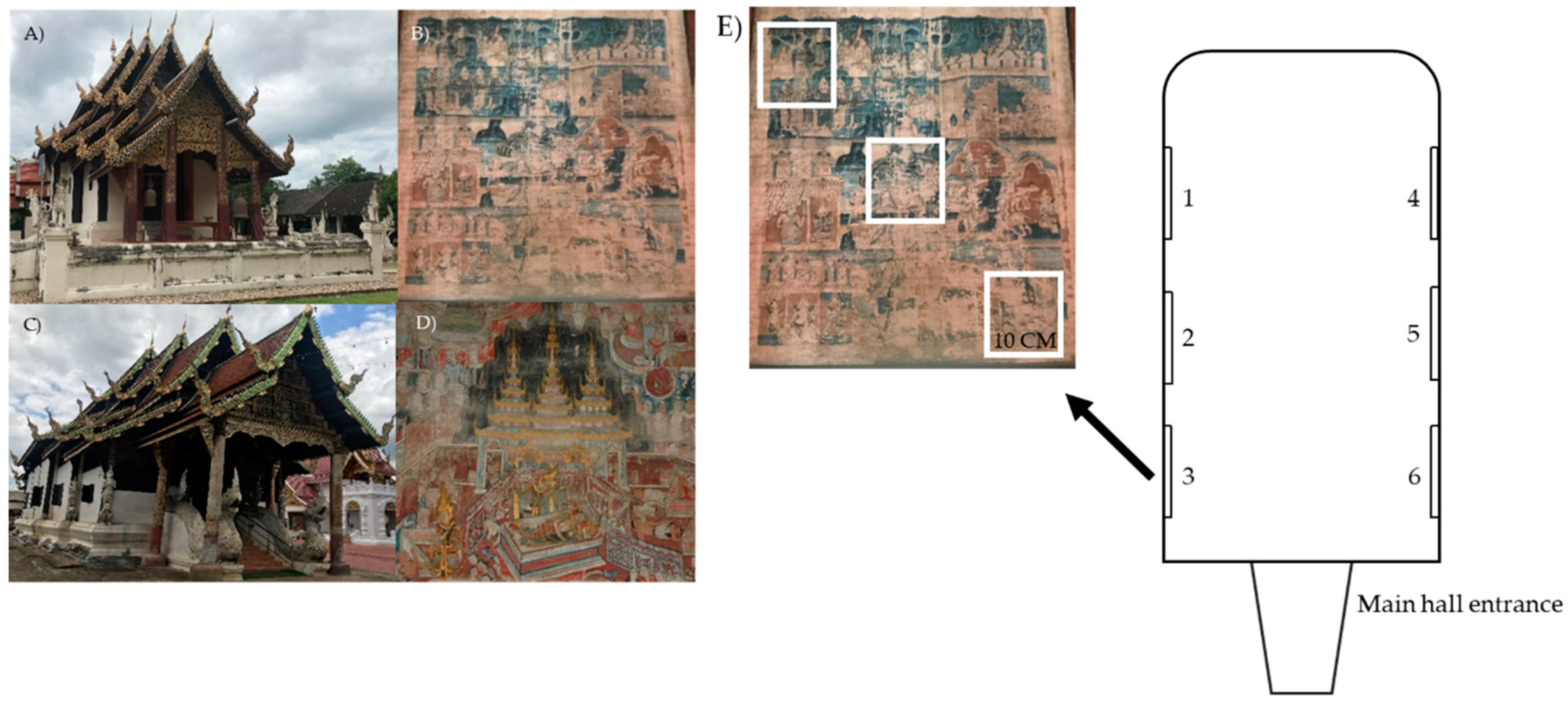
2.1. Sample Collections
2.1.1. Physical Properties Measurement and Visiting Frequency Estimation
2.1.2. Microorganism Collection
2.2. Culture-Dependent Study
2.2.1. Isolation and Identification of Microorganism
2.2.2. Biodeterioration Test of Microorganism and Acid Production Study of Microorganism
2.2.3. Mineralization and Assay
2.3. Culture-Independent Study
2.3.1. Genomic DNA Extraction, Sequencing, Bioinformatics and Data Processing for Fungi and Bacteria
2.3.2. Microbial Diversity and Function Study
2.3.3. Statistics Analysis
3. Results
3.1. Physical Property Measurements
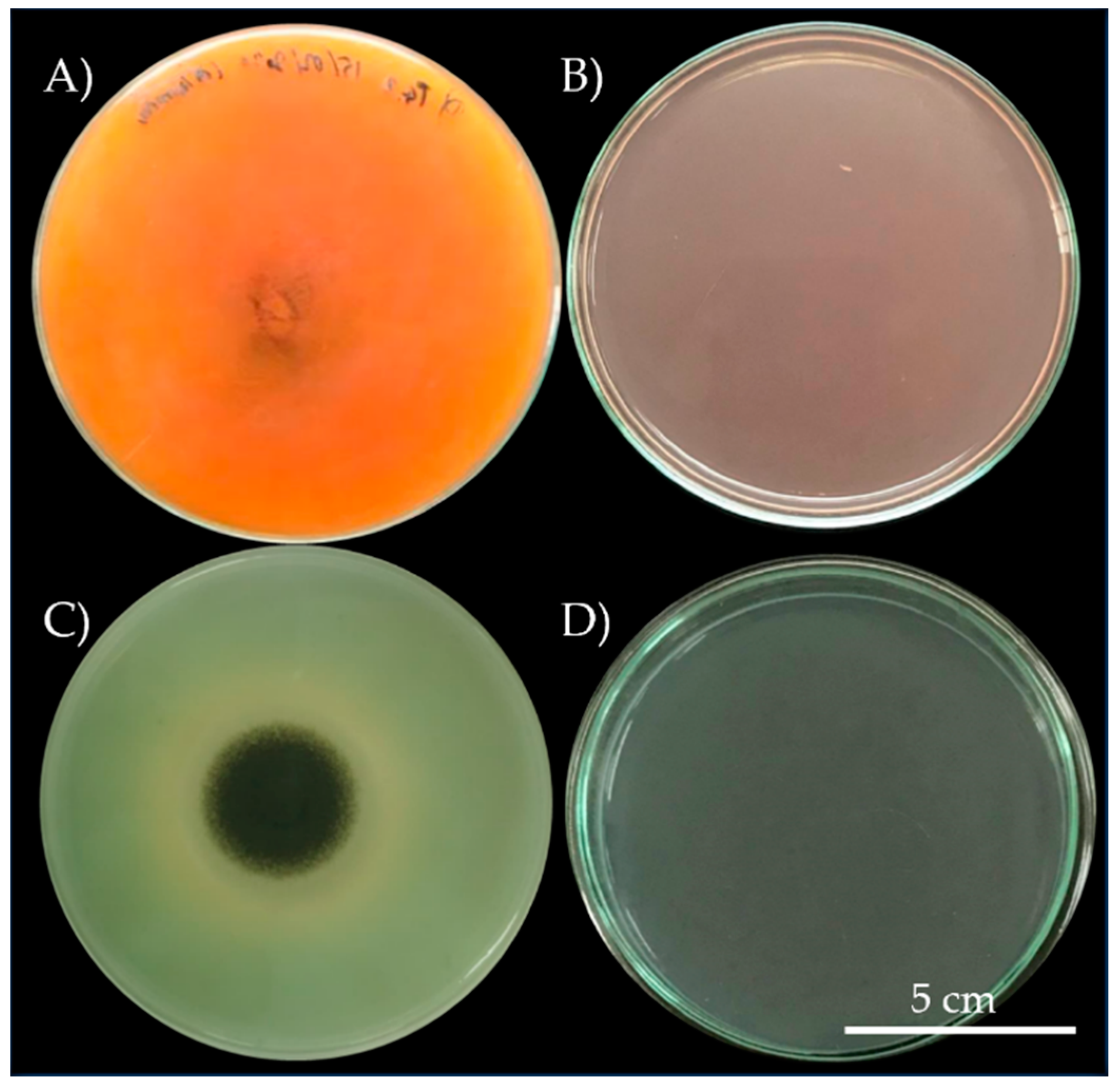
3.2. Microorganism Isolates and Their Bioterioration Activity
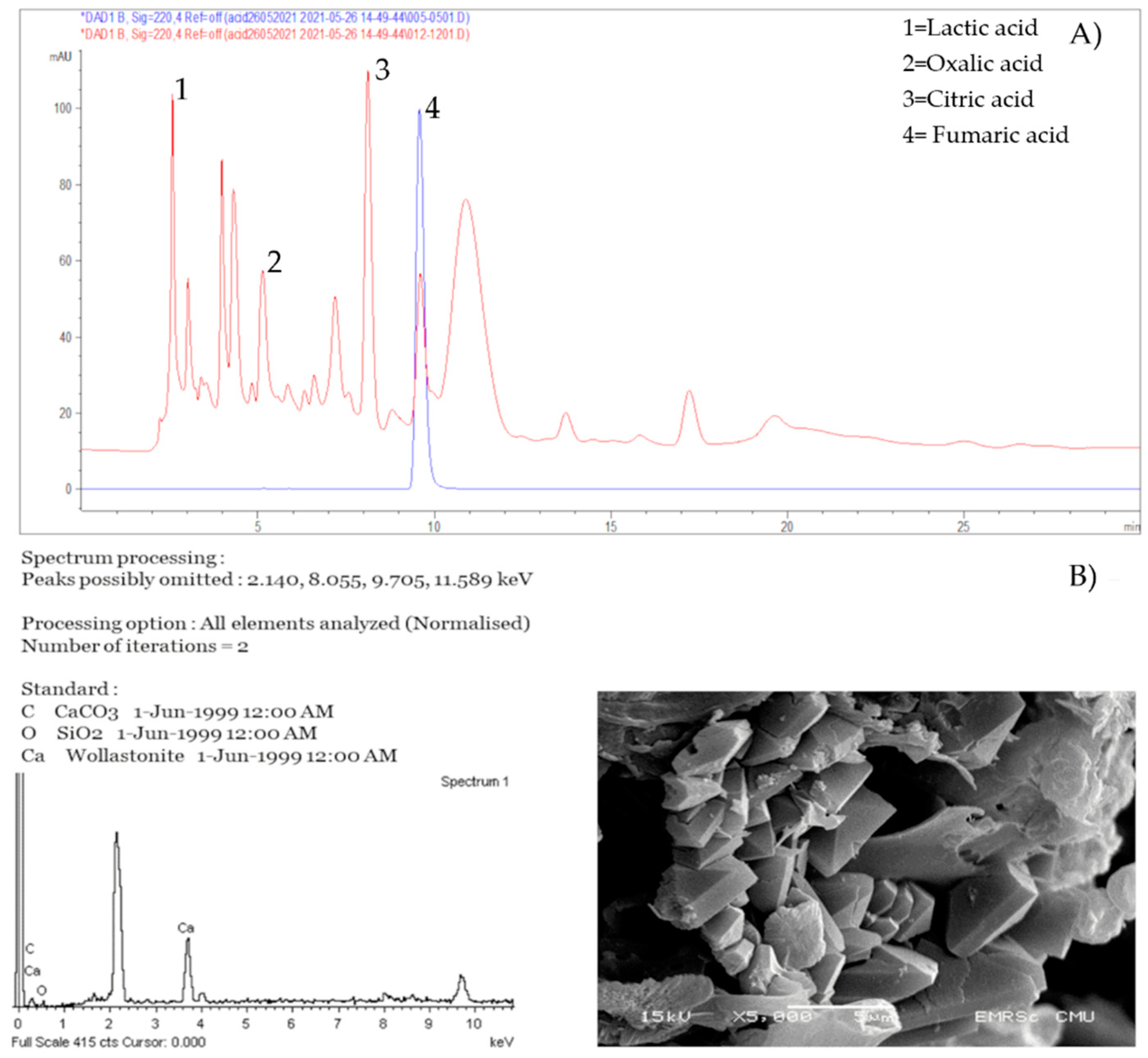
3.3. Organic Acid Production and Calcium Crystal Formation of Microorganism Isolates
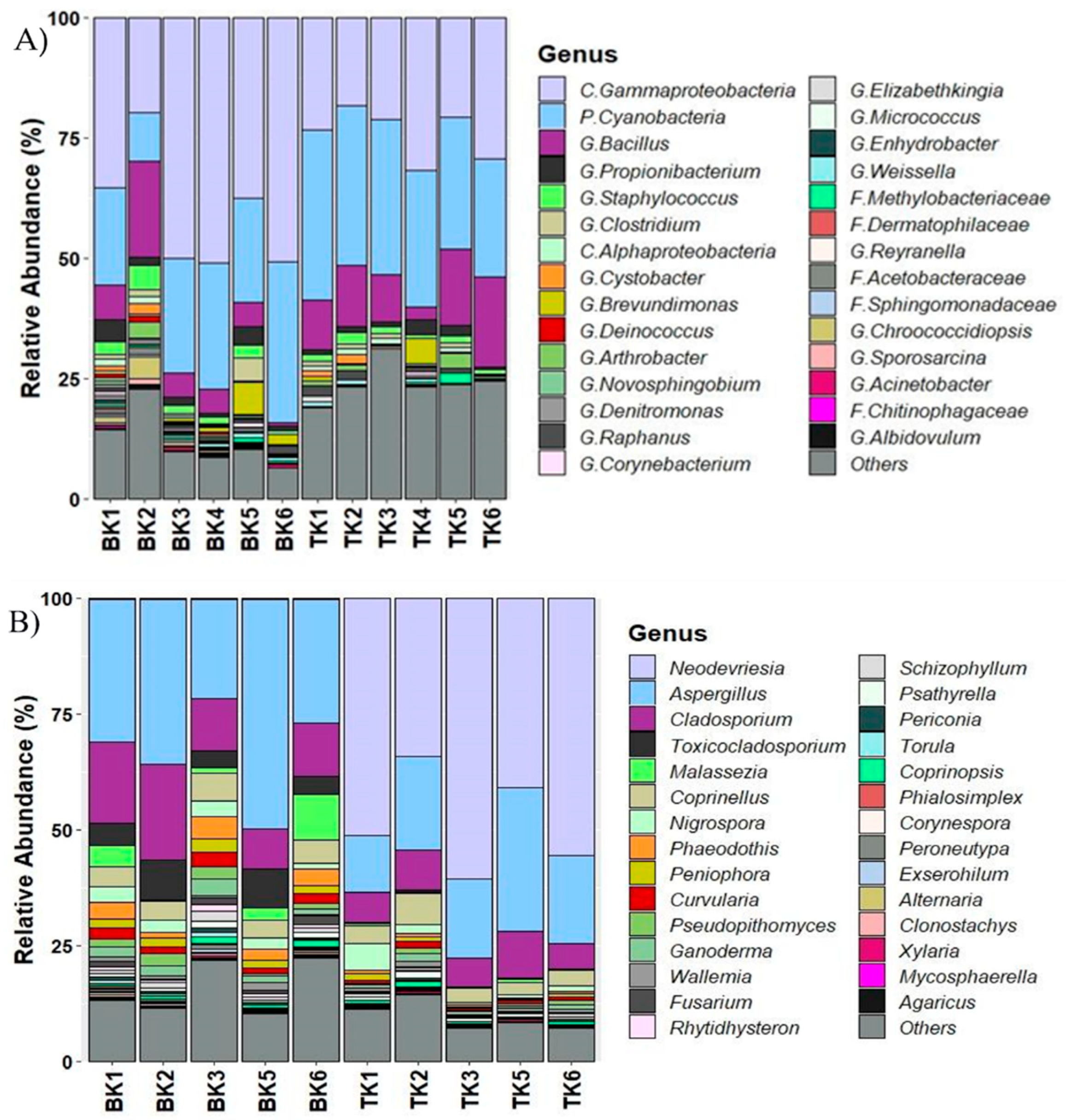
3.4. Microbial Community Structure in Mural Paintings Characterized by Culture-Independent Molecular Technique
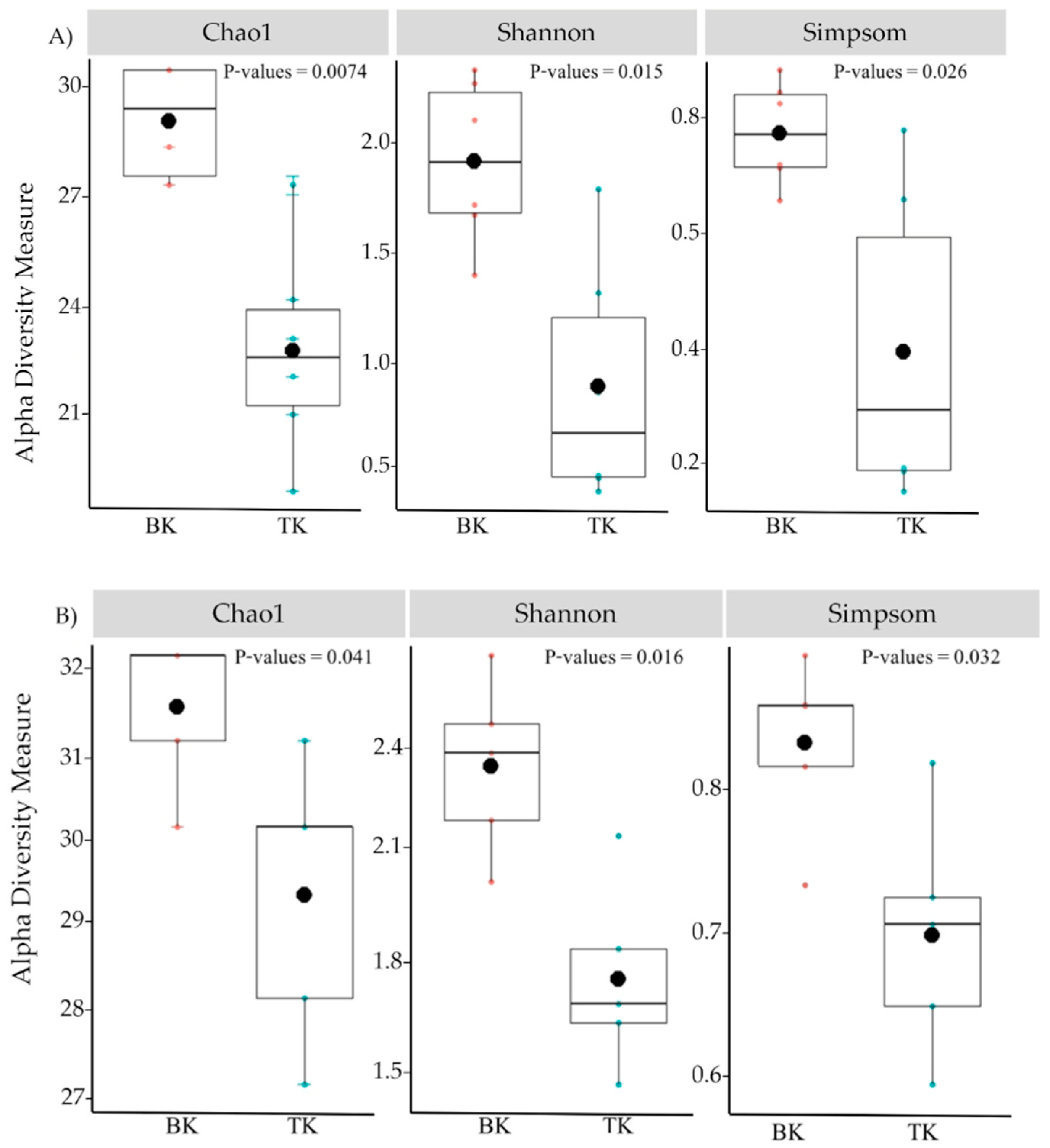
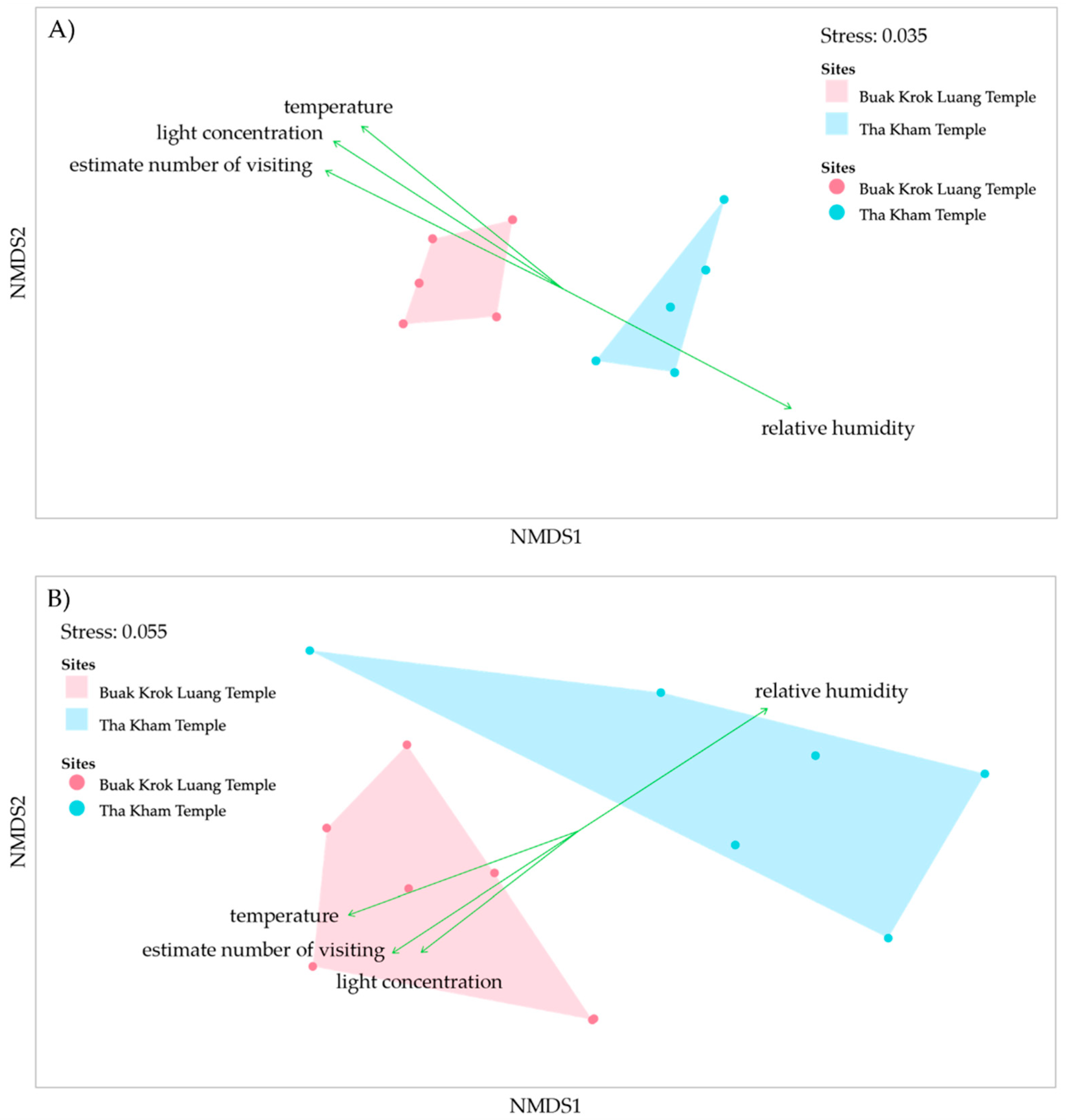
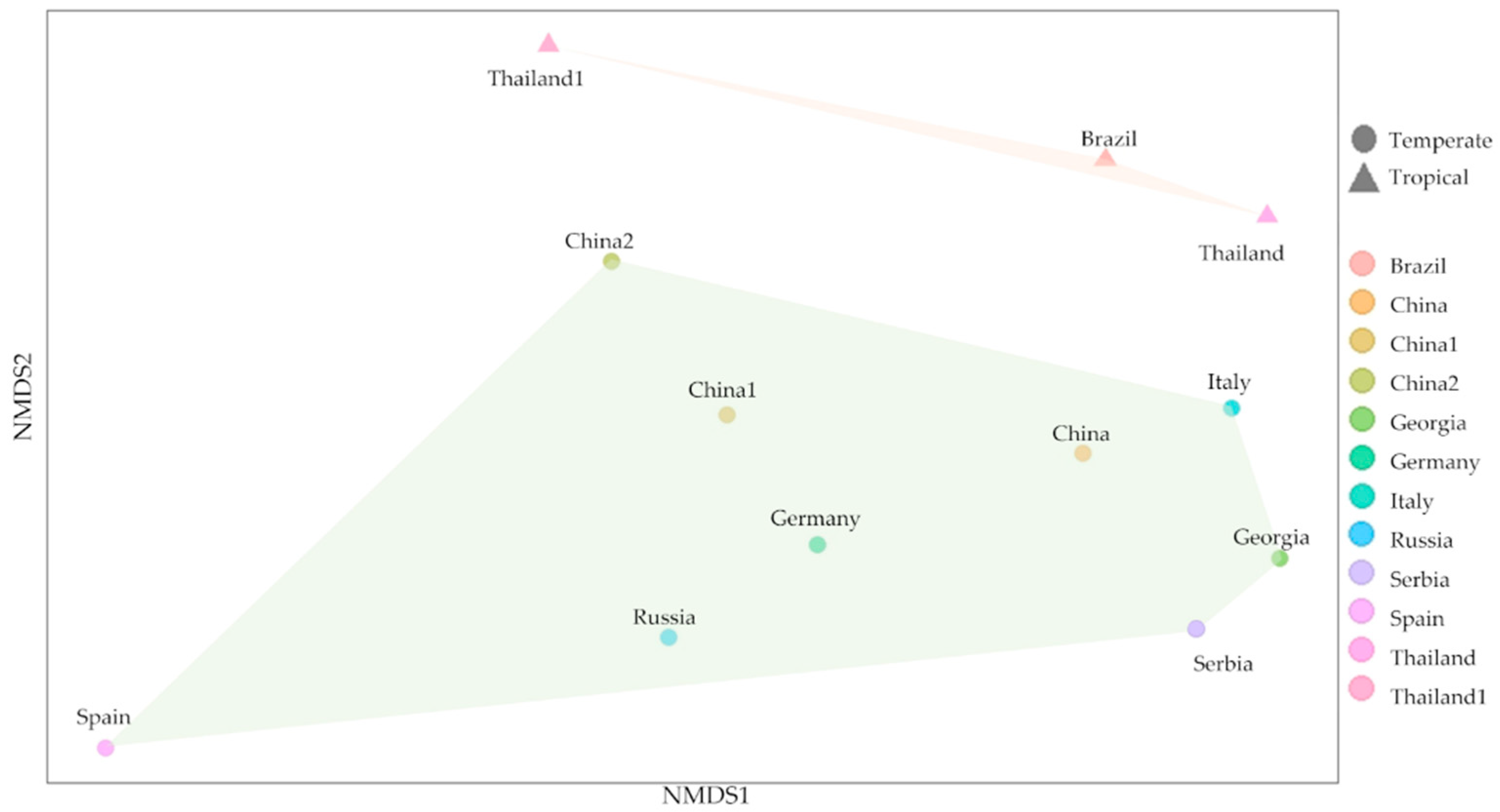
3.5. Comparison of Microbial Diversity and Community Composition Using NGS
3.6. Functional Roles of Microorganisms Detected from the Mural Paintings
3.7. Microbial Identification
3.8. Comparison between High-Throughput Sequencing and the Culture-Dependent Method
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Silverman, H.; Ruggles, D.F. Cultural Heritage and Human Rights; Springer: New York, NY, USA, 2007; pp. 3–29. [Google Scholar]
- Pinar, G.; Piombino-Mascali, D.; Maixner, F.; Zink, A.; Sterflinger, K. Microbial survey of the mummies from the Capuchin Catacombs of Palermo, Italy: Biodeterioration risk and contamination of the indoor air. FEMS Microbiol. Ecol. 2013, 86, 341–356. [Google Scholar] [CrossRef] [PubMed]
- Penth, H. A Brief History of Lan Na: Northern Thailand from Past to Present; Silkworm Books: Chiang Mai, Thailand, 2004. [Google Scholar]
- Thutongkinanon, P. The Ancient Colors of Mural Paintingwan Chapel: Re-Imaging of Faith in the Bhuddhaisa. J. Urban Cult. Res. 2011, 3, 122–129. [Google Scholar]
- Mydin, M.A.O.; Ramli, M.; Awang, H. Factors of deterioration in building and the principles of repair. Analele Universităţii" Eftimie Murgu 2012, 19, 345–352. [Google Scholar]
- Zhou, X.; Li, Y. Atlas of Oral Microbiology: From Healthy Microflora to Disease; Academic Press: Cambridge, MA, USA, 2015. [Google Scholar]
- Frindte, K.; Pape, R.; Werner, K.; Löffler, J.; Knief, C. Temperature and soil moisture control microbial community composition in an arctic–alpine ecosystem along elevational and micro-topographic gradients. ISME J. 2019, 13, 2031–2043. [Google Scholar] [CrossRef] [PubMed]
- Dahmani, J.; Benharbit, M.; Fassar, M.; Hajila, R.; Zidane, L.; Magri, N.; Belahbib, N. Vascular plants census linked to the biodeterioration process of the Portuguese city of Mazagan in El Jadida, Morocco. J. King Saud Univ. Sci. 2020, 32, 682–689. [Google Scholar] [CrossRef]
- Tiano, P. Biodegradation of cultural heritage: Decay mechanisms and control methods. In Seminar Article, New University of Lisbon, Department of Conservation and Restoration; University of Lisbon: Lisbon, Portugal, 2002; pp. 7–12. [Google Scholar]
- Sterflinger, K.; Piñar, G. Microbial deterioration of cultural heritage and works of art—Tilting at windmills? Appl. Microbiol. Biotechnol. 2013, 97, 9637–9646. [Google Scholar] [CrossRef] [PubMed]
- Sterflinger, K. Fungi as geologic agents. Geomicrobiol. J. 2000, 17, 97–124. [Google Scholar] [CrossRef]
- Warscheida, T.H.; Braamsb, J. Biodeterioration of stone: A review. Int. Biodeterior. Biodegrad. 2000, 46, 343–368. [Google Scholar] [CrossRef]
- Rosling, A.; Roose, T.; Herrmann, A.M.; Davidson, F.A.; Finlay, R.D.; Gadd, G.M. Approaches to modelling mineral weathering by fungi. Fungal Biol. Rev. 2009, 23, 138–144. [Google Scholar] [CrossRef]
- Lamprinou, V.; Skaraki, K.; Kotoulas, G.; Anagnostidis, K.; Economou-Amilli, A.; Pantazidou, A. A new species of Phormidium (Cyanobacteria, Oscillatoriales) from three Greek Caves: Morphological and molecular analysis. Fundam. Appl. Limnol. 2013, 182, 109–116. [Google Scholar] [CrossRef]
- Rosado, T.; Mirão, J.; Candeias, A.; Caldeira, A.T. Microbial communities analysis assessed by pyrosequencing—A new approach applied to conservation state studies of mural paintings. Anal. Bioanal. Chem. 2014, 406, 887–895. [Google Scholar] [CrossRef] [PubMed]
- Imperi, F.; Caneva, G.; Cancellieri, L.; Ricci, M.A.; Sodo, A.; Visca, P. The bacterial aetiology of rosy discoloration of ancient wall paintings. Environ. Microbiol. 2007, 9, 2894–2902. [Google Scholar] [CrossRef] [PubMed]
- Caneva, G.; Nugari, M.P.; Salvadori, O. Plant Biology for Cultural Heritage: Biodeterioration and Conservation; The Getty Conservation Institute: Los Angeles, CA, USA, 2008. [Google Scholar]
- Ranalli, G.; Zanardini, E.; Sorlini, C. Biodeterioration–including cultural heritage. In Encyclopedia of Microbiology, 3rd ed.; Schaechter, M., Ed.; Elsevier: Amsterdam, The Netherlands, 2009; pp. 191–205. [Google Scholar]
- Byrd, A.L.; Belkaid, Y.; Segre, J.A. The human skin microbiome. Nat. Rev. Microbiol. 2018, 16, 143. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Chen, H.; Yao, M.; Li, X. Bacterial pathogens were detected from human exhaled breath using a novel protocol. J. Aerosol Sci. 2018, 117, 224–234. [Google Scholar] [CrossRef]
- Rojas, T.I.; Aira, M.J.; Batista, A.; Cruz, I.L.; González, S. Fungal biodeterioration in historic buildings of Havana (Cuba). Grana 2012, 51, 44–51. [Google Scholar] [CrossRef]
- Senbua, W.; Wichitwechkarn, J. Molecular identification of fungi colonizing art objects in Thailand and their growth inhibition by local plant extracts. 3 Biotech 2019, 9, e356. [Google Scholar] [CrossRef]
- Ugbogu, O.C.; Awache, I.; Agwaranze, D.I.; Ogodo, A.C.; Ubandoma, A.; Yakubu, M.N. Microbial deterioration of painted wall surfaces in Wukari, Taraba State, Nigeria. Am. J. Microbiol. Biotechnol. 2017, 4, 31–34. [Google Scholar]
- Gorbushina, A.A.; Heyrman, J.; Dornieden, T.; Gonzalez-Delvalle, M.; Krumbein, W.E.; Laiz, L.; Swings, J. Bacterial and fungal diversity and biodeterioration problems in mural painting environments of St. Martins church (Greene–Kreiensen, Germany). Int. Biodeterior. Biodegrad. 2004, 53, 13–24. [Google Scholar] [CrossRef]
- Dissanayake, A.J.; Purahong, W.; Wubet, T.; Hyde, K.D.; Zhang, W.; Xu, H.; Yan, J. Direct comparison of culture-dependent and culture-independent molecular approaches reveal the diversity of fungal endophytic communities in stems of grapevine (Vitis vinifera). Fungal Divers. 2018, 90, 85–107. [Google Scholar] [CrossRef]
- Jayawardena, R.S.; Purahong, W.; Zhang, W.; Wubet, T.; Li, X.; Liu, M.; Yan, J. Biodiversity of fungi on Vitis vinifera L. revealed by traditional and high-resolution culture-independent approaches. Fungal Divers. 2018, 90, 1–84. [Google Scholar] [CrossRef]
- Sansupa, C.; Fareed Mohamed Wahdan, S.; Disayathanoowat, T.; Purahong, W. Identifying Hidden Viable Bacterial Taxa in Tropical Forest Soils Using Amplicon Sequencing of Enrichment Cultures. Biology 2021, 10, e569. [Google Scholar] [CrossRef] [PubMed]
- Sansupa, C.; Purahong, W.; Wubet, T.; Tiansawat, P.; Pathom-Aree, W.; Teaumroong, N.; Disayathanoowat, T. Soil bacterial communities and their associated functions for forest restoration on a limestone mine in northern Thailand. PLoS ONE 2021, 16, e0248806. [Google Scholar] [CrossRef] [PubMed]
- Ito, T.; Sekizuka, T.; Kishi, N.; Yamashita, A.; Kuroda, M. Conventional culture methods with commercially available media unveil the presence of novel culturable bacteria. Gut Microbes 2019, 10, 77–91. [Google Scholar] [CrossRef] [PubMed]
- Al-Awadhi, H.; Dashti, N.; Khanafer, M.; Al-Mailem, D.; Ali, N.; Radwan, S. Bias problems in culture—Independent analysis of environmental bacterial communities: A representative study on Hydrocarbonoclastic bacteria. SpringerPlus 2013, 2, e369. [Google Scholar] [CrossRef] [PubMed]
- Daniel, R. The metagenomics of soil. Nat. Rev. Microbiol. 2005, 3, 470–478. [Google Scholar] [CrossRef]
- Wu, B.; Hussain, M.; Zhang, W.; Stadler, M.; Liu, X.; Xiang, M. Current insights into fungal species diversity and perspective on naming the environmental DNA sequences of fungi. Mycology 2019, 10, 127–140. [Google Scholar] [CrossRef]
- Kaeberlein, T.; Lewis, K.; Epstein, S.S. Isolating “uncultivable” microorganisms in pure culture in a simulated natural environment. Science 2002, 296, 1127–1129. [Google Scholar] [CrossRef]
- Steen, A.D.; Crits-Christoph, A.; Carini, P.; DeAngelis, K.M.; Fierer, N.; Lloyd, K.G.; Thrash, J.C. High proportions of bacteria and archaea across most biomes remain uncultured. ISME J. 2019, 13, 3126–3130. [Google Scholar] [CrossRef]
- Stefani, F.O.P.; Bell, T.H.; Marchand, C.; de la Providencia, I.E.; Yassimi, A.E.; St-Arnaud, M.; Hijri, M. Culture-dependent and independent methods capture different microbial community fractions in hydrocarbon-contaminated soils. PLoS ONE 2015, 10, e0128272. [Google Scholar] [CrossRef]
- Nawaz, A.; Purahong, W.; Herrmann, M.; Küsel, K.; Buscot, F.; Wubet, T. DNA-and RNA-derived fungal communities in subsurface aquifers only partly overlap but react similarly to environmental factors. Microorganisms 2019, 7, e341. [Google Scholar] [CrossRef]
- Hongsanan, S.; Jeewon, R.; Purahong, W.; Xie, N.; Liu, J.K.; Jayawardena, R.S.; Peršoh, D. Can we use environmental DNA as holotypes? Fungal Divers. 2018, 92, 1–30. [Google Scholar] [CrossRef]
- Purahong, W.; Wubet, T.; Krüger, D.; Buscot, F. Application of next-generation sequencing technologies to conservation of wood-inhabiting fungi. Conserv. Biol. 2019, 33, 716–724. [Google Scholar] [CrossRef] [PubMed]
- Purahong, W.; Mapook, A.; Wu, Y.T.; Chen, C.T. Characterization of the Castanopsis carlesii deadwood mycobiome by Pacbio sequencing of the full-length fungal nuclear ribosomal internal transcribed spacer (ITS). Front. Microbiol. 2019, 10, e983. [Google Scholar] [CrossRef]
- Suphaphimol, N.; Attasopa, K.; Pakwan, C.; Chantawannakul, P.; Disayathanoowat, T. Cultured-dependent and cultured-independent study of bacteria associated with Thai commercial stingless bee Lepidotrigona terminata. J. Apic. Res. 2020, 60, 341–348. [Google Scholar] [CrossRef]
- Purahong, W.; Wahdan, S.F.M.; Heinz, D.; Jariyavidyanont, K.; Sungkapreecha, C.; Tanunchai, B.; Buscot, F. Back to the future: Decomposability of a biobased and biodegradable plastic in field soil environments and its microbiome under ambient and future climates. Environ. Sci. Technol. 2021, 55, 12337–12351. [Google Scholar] [CrossRef] [PubMed]
- Purahong, W.; Tanunchai, B.; Wahdan, S.F.M.; Buscot, F.; Schulze, E.D. Molecular Screening of Microorganisms Associated with Discolored Wood in Dead European Beech Trees Suffered from Extreme Drought Event Using Next Generation Sequencing. Plants 2021, 10, e2092. [Google Scholar] [CrossRef] [PubMed]
- Rocha-Martin, J.; Harrington, C.; Dobson, A.D.; O’Gara, F. Emerging strategies and integrated systems microbiology technologies for biodiscovery of marine bioactive compounds. Mar. Drugs 2014, 12, 3516–3559. [Google Scholar] [CrossRef] [PubMed]
- Duan, Y.; Wu, F.; Wang, W.; He, D.; Gu, J.D.; Feng, H.; An, L. The microbial community characteristics of ancient painted sculptures in Maijishan Grottoes, China. PLoS ONE 2017, 12, e0179718. [Google Scholar]
- Duan, Y.; Wu, F.; Wang, W.; Gu, J.D.; Li, Y.; Feng, H.; An, L. Differences of Microbial Community on the wall paintings preserved in situ and ex situ of the Tiantishan Grottoes, China. Int. Biodeterior. Biodegrad. 2018, 132, 102–113. [Google Scholar] [CrossRef]
- Ma, Y.; Zhang, H.; Du, Y.; Tian, T.; Xiang, T.; Liu, X.; Feng, H. The community distribution of bacteria and fungi on ancient wall paintings of the Mogao Grottoes. Sci. Rep. 2015, 5, e7752. [Google Scholar] [CrossRef]
- Torralba, M.G.; Kuelbs, C.; Moncera, K.J.; Roby, R.; Nelson, K.E. Characterizing Microbial Signatures on Sculptures and Paintings of Similar Provenance. Microb. Ecol. 2021, 81, 1098–1105. [Google Scholar] [CrossRef] [PubMed]
- Piñar, G.; Sclocchi, M.C.; Pinzari, F.; Colaizzi, P.; Graf, A.; Sebastiani, M.L.; Sterflinger, K. The microbiome of Leonardo da Vinci’s drawings: A bio-archive of their history. Front. Microbiol. 2020, 11, e2889. [Google Scholar] [CrossRef]
- Hall, T. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, e1547. [Google Scholar] [CrossRef] [PubMed]
- Ongsakul, S.; Jongjitngam, S.; Bussaban, B.; Pinngoen, C. The Sacread Area of Wat U-Mong Suan Bhuddha Dharma, History, Art and Science Chiang Mai Sun Lanna Suksa; Chiang Mai University: Chiang Mai, Thailand, 2018. [Google Scholar]
- Jang, J.Y.; Choi, Y.H.; Shin, T.S.; Kim, T.H.; Shin, K.S.; Park, H.W.; Kim, J.C. Biological control of Meloidogyne incognita by Aspergillus niger F22 producing oxalic acid. PLoS ONE 2016, 11, e0156230. [Google Scholar] [CrossRef] [PubMed]
- Kwak, A.M.; Lee, I.K.; Lee, S.Y.; Yun, B.S.; Kang, H.W. Oxalic acid from Lentinula edodes culture filtrate: Antimicrobial activity on phytopathogenic bacteria and qualitative and quantitative analyses. Mycobiology 2016, 44, 338–342. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.; Wang, Y.; Cao, H.; Zhao, Y.; Li, Z.; Wang, H.; Tang, Q. Simultaneous Determination of 13 Organic Acids in Liquid Culture Media of Edible Fungi Using High-Performance Liquid Chromatography. BioMed Res. Int. 2020, 2020, e2817979. [Google Scholar] [CrossRef]
- Ma, W.; Wu, F.; Tian, T.; He, D.; Zhang, Q.; Gu, J.D.; Feng, H. Fungal diversity and its contribution to the biodeterioration of mural paintings in two 1700-year-old tombs of China. Int. Biodeterior. Biodegrad. 2020, 152, e104972. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- DeSantis, T.Z.; Hugenholtz, P.; Larsen, N.; Rojas, M.; Brodie, E.L.; Keller, K.; Huber, T.; Dalevi, D.; Hu, P.; Andersen, G.L. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006, 72, 5069–5072. [Google Scholar] [CrossRef]
- Nilsson, R.H.; Larsson, K.H.; Taylor, A.F.S.; Bengtsson-Palme, J.; Jeppesen, T.S.; Schigel, D.; Kennedy, P.; Picard, K.; Glöckner, F.O.; Tedersoo, L.; et al. The UNITE database for molecular identification of fungi: Handling dark taxa and parallel taxonomic classifications. Nucleic acids Research 2019, 47, D259–D264. [Google Scholar] [CrossRef] [PubMed]
- Allaire, J. RStudio: Integrated Development Environment for R. Boston MA 2012, 770, 165–171. [Google Scholar]
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.R.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G.I. PICRUSt2 for Prediction of Metagenome Functions. Nat. Biotechnol. 2020, 38, 685–688. [Google Scholar] [CrossRef] [PubMed]
- Din, G.; Hassan, A.; Rafiq, M.; Hasan, F.; Badshah, M.; Khan, S.; Shah, A.A. Characterization of organic acid producing Asper-gillus tubingensis FMS1 and its role in metals leaching from soil. Geomicrobiol. J. 2020, 37, 336–344. [Google Scholar] [CrossRef]
- Li, Z.; Bai, T.; Dai, L.; Wang, F.; Tao, J.; Meng, S.; Hu, S. A study of organic acid production in contrasts between two phosphate solubilizing fungi: Penicillium oxalicum and Aspergillus niger. Sci. Rep. 2016, 6, 25313. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Alonso, M.; Castro, K.; Martinez-Arkarazo, I.; Angulo, M.; Olazabal, M.A.; Madariaga, J.M. Analysis of bulk and inorganic degradation products of stones, mortars and wall paintings by portable Raman microprobe spectroscopy. Anal. Bioanal. Chem. 2004, 379, 42–50. [Google Scholar] [CrossRef]
- Nevin, A.; Melia, J.L.; Osticioli, I.; Gautier, G.; Colombini, M.P. The identification of copper oxalates in a 16th century Cypriot exterior wall painting using micro FTIR, micro Raman spectroscopy and Gas Chromatography-Mass Spectrometry. J. Cult. Herit. 2008, 9, 154–161. [Google Scholar] [CrossRef]
- Sarmiento, A.; Maguregui, M.; Martinez-Arkarazo, I.; Angulo, M.; Castro, K.; Olazábal, M.A.; Madariaga, J.M. Raman spectroscopy as a tool to diagnose the impacts of combustion and greenhouse acid gases on properties of Built Heritage. J. Raman Spectrosc. Int. J. Orig. Work. All Asp. Raman Spectrosc. Incl. High. Order Processes Also Brillouin Rayleigh Scatt. 2008, 39, 1042–1049. [Google Scholar] [CrossRef]
- Unković, N.; Erić, S.; Šarić, K.; Stupar, M.; Savkoviša, S.; Stanković, S.; Stanojević, O.; Dimkić, I.; Vukojević, J.; Grbić, M.L. Biogenesis of secondary mycogenic minerals related to wall paintings deterioration process. Micron. 2017, 100, 1–9. [Google Scholar] [CrossRef]
- Herrera, L.K.; Videla, H.A. Surface analysis and materials characterization for the study of biodeterioration and weathering effects on cultural property. Int. Biodeterior. Biodegrad. 2009, 63, 813–822. [Google Scholar] [CrossRef]
- Torre, M.A.D.L.; Gomezalarcon, G.; Melgarejo, P.; Saizjimenez, C. Fungi in weathered sandstone from salamanca cathedral, Spain. Sci. Total Environ. 1991, 107, 159–168. [Google Scholar] [CrossRef]
- Torre, M.A.D.L.; Gomezalarcon, G.; Viscano, C.; Garca, T.M. Biochemical mechanisms of stone alteration carried out by filamentous fungi living in monuments. Biogeochemistry 1993, 19, 129–147. [Google Scholar] [CrossRef]
- Vasanthakumar, A.; Dearaujo, A.; Mazurek, J.; Schilling, M.; Mitchell, R. Microbiological survey for analysis of the brown spots on the walls of the tomb of King Tutankhamun. Int. Biodeterior. Biodegrad. 2013, 79, 56–63. [Google Scholar] [CrossRef]
- Sayer, J.A.; Gadd, G.M. Solubilization and transformation of insoluble inorganic metal compounds to insoluble metal oxalates by Aspergillus niger. Mycol. Res. 1997, 101, 653–661. [Google Scholar] [CrossRef]
- Hueck, H.J. The biodeterioration of materials as part of hylobiology. Mater. Org. 1965, 1, 5–34. [Google Scholar]
- Saiz-Jimenez, C. Weathering and colonization of limestones in an urban environment. In Soil Biology and Conservation of the Biosphere; Szegi, J., Ed.; Akademiai Kiado: Budapest, Hungary, 1984; Volume 2, pp. 757–767. [Google Scholar]
- Viles, H.A. Blue-green algae and terrestrial limestone weathering on Aldabra Atoll: An SEM and light microscope study. Earth Surf. Processes Landf. 1987, 12, 319–330. [Google Scholar] [CrossRef]
- El-Gorj, F.M.; Aisha, M.A.; Maznah, W.O. Isolation, Definition and Chemical Control some of the Bacteria that Cause Contamination of Wall Paintings in Caves. Asian J. Pharm. Res. Dev. 2019, 7, 6–11. [Google Scholar] [CrossRef]
- Nugari, M.P.; Pietrini, A.M.; Caneva, G.; Imperi, F.; Visca, P. Biodeterioration of mural paintings in a rocky habitat: The Crypt of the Original Sin (Matera, Italy). Int. Biodeterior. Biodegrad. 2009, 63, 705–711. [Google Scholar] [CrossRef]
- Tescari, M.; Frangipani, E.; Caneva, G.; Municchia, A.C.; Sodo, A.; Visca, P. Arthrobacter agilis and rosy discoloration in “Terme del Foro” (Pompeii, Italy). Int. Biodeterior. Biodegrad. 2018, 130, 48–54. [Google Scholar] [CrossRef]
- Heyrman, J.; Verbeeren, J.; Schumann, P.; Swings, J.; De Vos, P. Six novel Arthrobacter species isolated from deteriorated mural paintings. Int. J. Syst. Evol. Microbiol. 2005, 55, 1457–1464. [Google Scholar] [CrossRef] [PubMed]
- Altenburgera, P.; Kämpferb, P.; Makristathisc, A.; Lubitza, W.; Bussea, H.J. Classification of bacteria isolated from a medieval wall painting. J. Biotechnol. 1996, 47, 39–52. [Google Scholar] [CrossRef]
- Unković, N.; Dimkić, I.; Stupar, M.; Stanković, S.; Vukojević, J.; Ljaljević Grbić, M. Biodegradative potential of fungal isolates from sacral ambient: In vitro study as risk assessment implication for the conservation of wall paintings. PLoS ONE 2018, 13, e0190922. [Google Scholar]
- Jurado, V.; Gonzalez-Pimentel, J.L.; Hermosin, B.; Saiz-Jimenez, C. Biodeterioration of Salón de Reinos, Museo Nacional del Prado, Madrid, Spain. Appl. Sci. 2021, 11, e8858. [Google Scholar] [CrossRef]
- Vukojevi, J.; Grbi, M.L. Moulds on paintings in Serbian fine art museums. Afr. J. Microbiol. Res. 2010, 4, 1453–1456. [Google Scholar]
- Rosado, T.; Martins, M.R.; Pires, M.; Mirão, J.; Candeias, A.; Caldeira, A.T. Enzymatic monitorization of mural paintings biodegradation and biodeterioration. Int. J. Conserv. Sci. 2013, 4, 603–612. [Google Scholar]
- Ortega-Morales, B.O.; Narváez-Zapata, J.; Reyes-Estebanez, M.; Quintana, P.; de la Rosa-García, S.C.; Bullen, H.; Chan-Bacab, M.J. Bioweathering potential of cultivable fungi associated with semi-arid surface microhabitats of Mayan buildings. Front. Microbiol. 2016, 7, e201. [Google Scholar] [CrossRef]
- Ljaljević-Grbić, M.V.; Vukojević, J.B. Role of fungi in biodeterioration process of stone in historic buildings. Proc. Nat. Sci. 2009, 116, 245–251. [Google Scholar] [CrossRef]
- Albertano, P.; Urzì, C. Structural interactions among epilithic cyanobacteria and heterotrophic microorganisms in Roman Hypogea. Microb. Ecol. 1999, 38, 244–252. [Google Scholar] [CrossRef]
- Ciferri, O. Microbial degradation of paintings. Appl. Environ. Microbiol. 1999, 65, 879–885. [Google Scholar] [CrossRef]
- Bock, E.; Sand, W. The microbiology of masonry biodeterioration. J. Appl. Bacteriol. 1993, 74, 503–514. [Google Scholar]
- Kraková, L.; Chovanová, K.; Selim, S.A.; Šimonovičová, A.; Puškarová, A.; Maková, A.; Pangallo, D. A multiphasic approach for investigation of the microbial diversity and its biodegradative abilities in historical paper and parchment documents. Int. Biodeterior. Biodegrad. 2012, 70, 117–125. [Google Scholar] [CrossRef]
- Coutinho, M.L.; Miller, A.Z.; Gutierrez-Patricio, S.; Hernandez-Marine, M.; Gomez-Bolea, A.; Rogerio-Candelera, M.A.; Macedo, M.F. Microbial communities on deteriorated artistic tiles from Pena National Palace (Sintra, Portugal). Int. Biodeterior. Biodegrad. 2013, 84, 322–332. [Google Scholar] [CrossRef]
- Hu, H.; Ding, S.; Katayama, Y.; Kusumi, A.; Li, S.X.; De Vries, R.P.; Gu, J.D. Occurrence of Aspergillus allahabadii on sandstone at Bayon temple, Angkor Thom, Cambodia. Int. Biodeterior. Biodegrad. 2013, 76, 112–117. [Google Scholar] [CrossRef]
- Abdollahzadeh, J.; Groenewald, J.Z.; Coetzee, M.P.A.; Wingfield, M.J.; Crous, P.W. Evolution of lifestyles in Capnodiales. Stud. Mycol. 2020, 95, 381–414. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.M.; Shenoy, B.D.; Li, W.; Cai, L. Molecular phylogeny of Neodevriesia, with two new species and several new combinations. Mycologia 2017, 109, 965–974. [Google Scholar] [CrossRef]
- Schubert, K. Morphotaxonomic Revision of Foliicolous Cladosporium Species (Hyphomycetes); Martin-Luther-Universität Halle-Wittenberg: Halle, Germany, 2005. [Google Scholar]
- Strong, C.L.; Bullard, J.E.; Burford, M.A.; McTainsh, G.H. Response of cyanobacterial soil crusts to moisture and nutrient availability. Catena 2013, 109, 195–202. [Google Scholar] [CrossRef]
- Cuezva, S.; Fernandez-Cortes, A.; Porca, E.; Pašić, L.; Jurado, V.; Hernandez-Marine, M.; Saiz-Jimenez, C. The biogeochemical role of Actinobacteria in Altamira cave, Spain. FEMS Microbiol. Ecol. 2012, 81, 281–290. [Google Scholar] [CrossRef]
- de Leo, F.; Iero, A.; Zammit, G.; Urzi, C.E. Chemoorganotrophic bacteria isolated from biodeteriorated surfaces in cave and catacombs. Int. J. Speleol. 2012, 41, 125–136. [Google Scholar] [CrossRef]
- Hsieh, P.; Pedersen, J.Z.; Bruno, L. Photoinhibition of cyanobacteria and its application in cultural heritage conservation. Photochem. Photobiol. 2014, 90, 533–543. [Google Scholar] [CrossRef]
- Grice, E.A.; Kong, H.H.; Conlan, S.; Deming, C.B.; Davis, J.; Young, A.C.; Segre, J.A. Topographical and temporal diversity of the human skin microbiome. Science 2019, 324, 1190–1192. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Tseng, C.H.; Pei, Z.; Blaser, M.J. Molecular analysis of human forearm superficial skin bacterial biota. Proc. Natl. Acad. Sci. USA 2007, 104, 2927–2932. [Google Scholar] [CrossRef] [PubMed]
- Świzdor, A.; Panek, A.; Milecka-Tronina, N.; Kołek, T. Biotransformations utilizing β-oxidation cycle reactions in the synthesis of natural compounds and medicines. Int. J. Mol. Sci. 2012, 13, 16514–16543. [Google Scholar] [CrossRef] [PubMed]
- Guerin, M.E.; Korduláková, J.; Alzari, P.M.; Brennan, P.J.; Jackson, M. Molecular basis of phosphatidyl-myo-inositol mannoside biosynthesis and regulation in mycobacteria. J. Biol. Chem. 2010, 285, 33577–33583. [Google Scholar] [CrossRef]
- Dobos, K.M.; Khoo, K.H.; Swiderek, K.M.; Brennan, P.J.; Belisle, J.T. Definition of the full extent of glycosylation of the 45-kilodalton glycoprotein of Mycobacterium tuberculosis. J. Bacteriol. 1996, 178, 2498–2506. [Google Scholar] [CrossRef]
- Unden, G. Transcriptional regulation and energetics of alternative respiratory pathways in facultatively anaerobic bacteria. Biochim. Biophys. Acta (BBA) Bioenerg. 1998, 1365, 220–224. [Google Scholar] [CrossRef][Green Version]
- Boniek, D.; Bonadio, L.; Santos de Abreu, C.; Dos Santos, A.F.B.; de Resende Stoianoff, M.A. Fungal bioprospecting and antifungal treatment on a deteriorated Brazilian contemporary painting. Lett. Appl. Microbiol. 2018, 67, 337–342. [Google Scholar] [CrossRef]
- Coronado-Ruiz, C.; Avendaño, R.; Escudero-Leyva, E.; Conejo-Barboza, G.; Chaverri, P.; Chavarría, M. Two new cellulolytic fungal species isolated from a 19 th-century art collection. Sci. Rep. 2018, 8, e7492. [Google Scholar] [CrossRef]
- De Jesús Cortés Sánchez, A.; Ramirez, M.D.; Barrientos, R.G.; Sharma, A. The genus Staphylococcus: Harmful and Beneficial Microorganisms in the Environment. Pakistan. Pakistan. J. Life Soc. Sci. 2017, 15, 72–83. [Google Scholar]
- Caselli, E.; Pancaldi, S.; Baldisserotto, C.; Petrucci, F.; Impallaria, A.; Volpe, L.; Mazzacane, S. Characterization of biodegradation in a 17th century easel painting and potential for a biological approach. PLoS ONE 2018, 13, e0207630. [Google Scholar] [CrossRef]
- López-Miras, M.; Piñar, G.; Romero-Noguera, J.; Bolivar-Galiano, F.C.; Ettenauer, J.; Sterflinger, K.; Martin-Sanchez, I. Microbial communities adhering to the obverse and reverse sides of an oil painting on canvas: Identification and evaluation of their biodegradative potential. Aerobiologia 2013, 29, 301–314. [Google Scholar] [CrossRef] [PubMed]
- Pavić, A.; Ilić-Tomić, T.; Pačevski, A.; Nedeljković, T.; Vasiljević, B.; Morić, I. Diversity and biodeteriorative potential of bacterial isolates from deteriorated modern combined-technique canvas painting. Int. Biodeterior. Biodegrad. 2015, 97, 40–50. [Google Scholar] [CrossRef]
- Voronina, L.I. Nekotorye svedeniya o gribakh, razrushayushchikh proiz-vedeniya zhivopisi. Soobshcheniya VCNILKR 1966, 17–18, 117–124. [Google Scholar]
- Kuritsyna, D.S. Mould fungi destroying old Russian mural painting and the struggle with them (in Russian). Vestnik Moskovskogo Universiteta 1968, 4, 31–41. [Google Scholar]
- Shirakawa, M.A.; Loh, K.; John, V.M.; Silva, M.E.S.; Gaylarde, C.C. Biodeterioration of painted mortar surfaces in tropical urban and coastal situations: Comparison of four paint formulations. Int. Biodeterior. Biodegrad. 2011, 65, 669–674. [Google Scholar] [CrossRef]
- Tonolo, A.; Giacobini, C. Microbiological change of frescoes. In Recent Advances in Conservation; Thomson, G., Ed.; Butterworths: London, UK, 1961; pp. 62–67. [Google Scholar]

| Isolates | Code | Acid Production | Calcium Formation | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Malic | Acetic | Citric | Lactic | Fumaric | Succinic | Oxalic | |||
| Trichoderma aethiopicum | BK1 | ||||||||
| Trichoderma longibrachiatum | BK2 | ||||||||
| Aspergillus niger | BTK1 | ||||||||
| Fusarium solani | BTK2 | ||||||||
| Aspergillus fumigatus | BK3 | ||||||||
| Penicillium citrinum | TK2 | ||||||||
| Penicillium hetheringtonii | TK3 | ||||||||
| Penicillium oxalicum | TK4 | ||||||||
| Aspergillus aculeatinus | TK5 | ||||||||
| Trichoderma harzianum | TK6 | ||||||||
| Aspergillus piperis | TK8 | ||||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Suphaphimol, N.; Suwannarach, N.; Purahong, W.; Jaikang, C.; Pengpat, K.; Semakul, N.; Yimklan, S.; Jongjitngam, S.; Jindasu, S.; Thiangtham, S.; et al. Identification of Microorganisms Dwelling on the 19th Century Lanna Mural Paintings from Northern Thailand Using Culture-Dependent and -Independent Approaches. Biology 2022, 11, 228. https://doi.org/10.3390/biology11020228
Suphaphimol N, Suwannarach N, Purahong W, Jaikang C, Pengpat K, Semakul N, Yimklan S, Jongjitngam S, Jindasu S, Thiangtham S, et al. Identification of Microorganisms Dwelling on the 19th Century Lanna Mural Paintings from Northern Thailand Using Culture-Dependent and -Independent Approaches. Biology. 2022; 11(2):228. https://doi.org/10.3390/biology11020228
Chicago/Turabian StyleSuphaphimol, Nattaphon, Nakarin Suwannarach, Witoon Purahong, Churdsak Jaikang, Kamonpan Pengpat, Natthawat Semakul, Saranphong Yimklan, Surachai Jongjitngam, Saiklang Jindasu, Sathaporn Thiangtham, and et al. 2022. "Identification of Microorganisms Dwelling on the 19th Century Lanna Mural Paintings from Northern Thailand Using Culture-Dependent and -Independent Approaches" Biology 11, no. 2: 228. https://doi.org/10.3390/biology11020228
APA StyleSuphaphimol, N., Suwannarach, N., Purahong, W., Jaikang, C., Pengpat, K., Semakul, N., Yimklan, S., Jongjitngam, S., Jindasu, S., Thiangtham, S., Chantawannakul, P., & Disayathanoowat, T. (2022). Identification of Microorganisms Dwelling on the 19th Century Lanna Mural Paintings from Northern Thailand Using Culture-Dependent and -Independent Approaches. Biology, 11(2), 228. https://doi.org/10.3390/biology11020228








