High-Throughput Chemical Screening and Structure-Based Models to Predict hERG Inhibition
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Thallium Flux Assay
2.3. Active Chemical Identification
2.4. Data Preparation for Molecular Modeling
2.5. Chemical Category Assignments
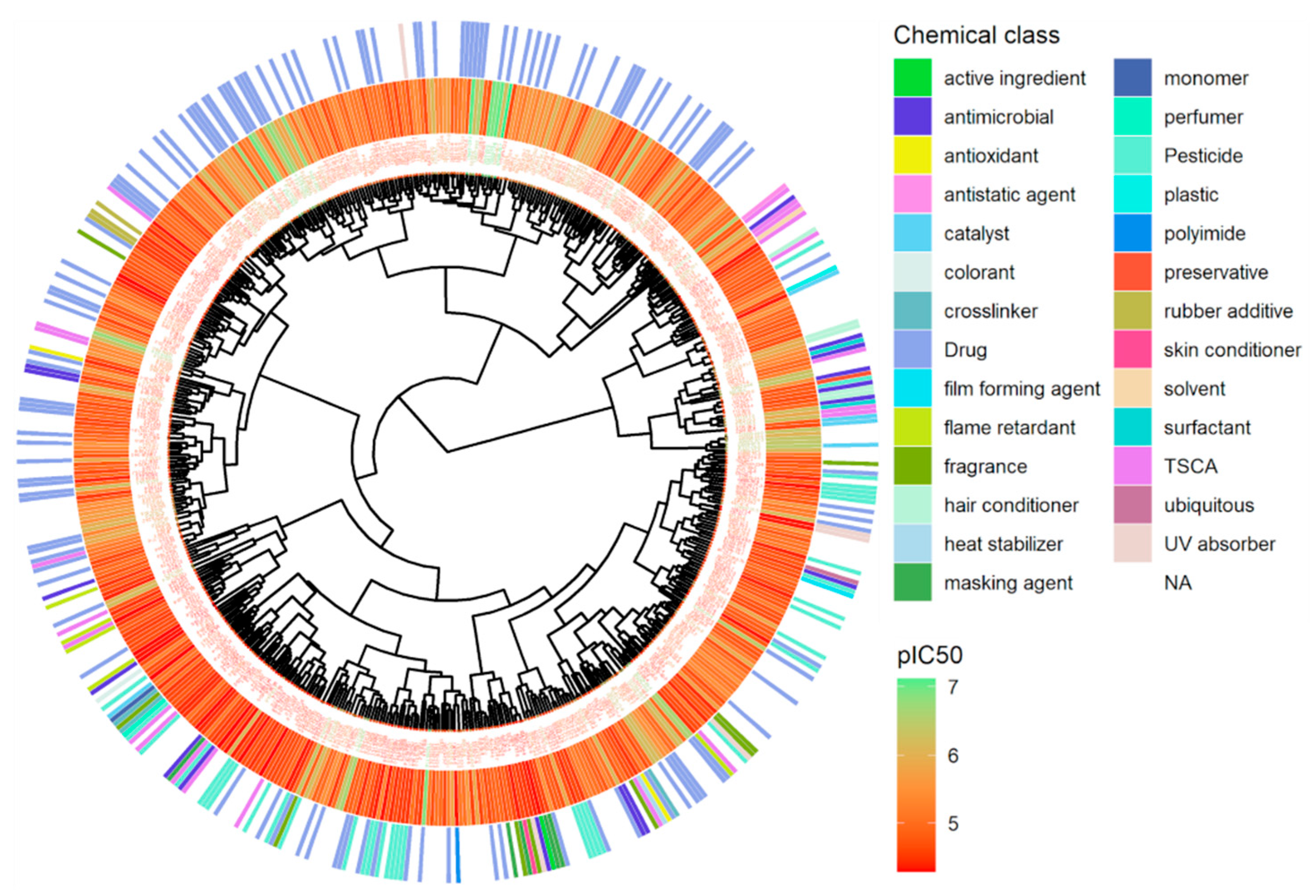
2.6. Structural Clustering
2.7. Chemotype Enrichment Analysis
2.8. QSAR Modeling
2.9. Machine Learning
2.10. Under-Sampling Protocol
2.11. Evaluation of the Classification Model Performance
2.12. Dataset Enrichment
2.13. Validation Sets
2.14. Applicability Domain (AD)
3. Results
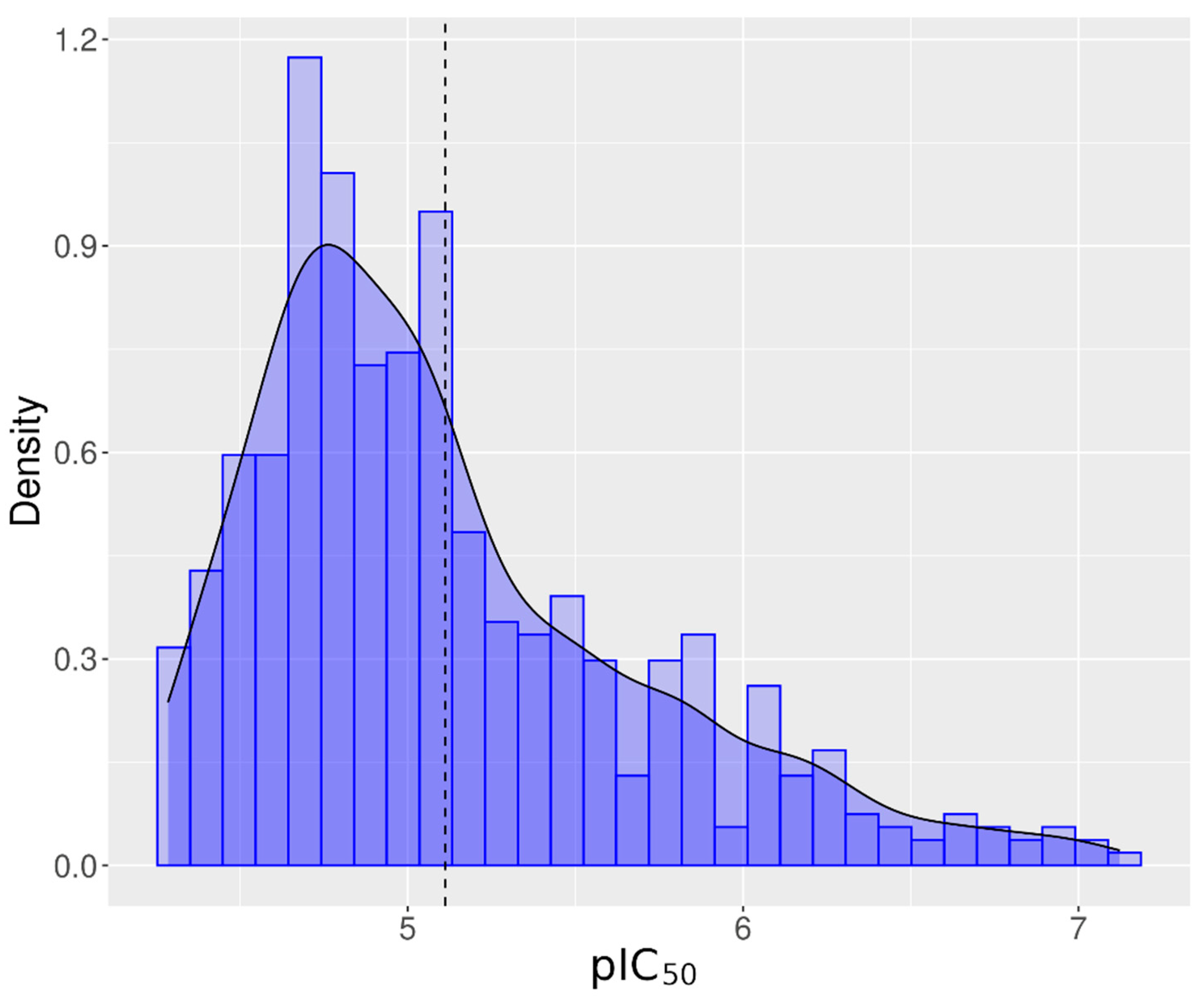
3.1. Chemical Activity for hERG Inhibition
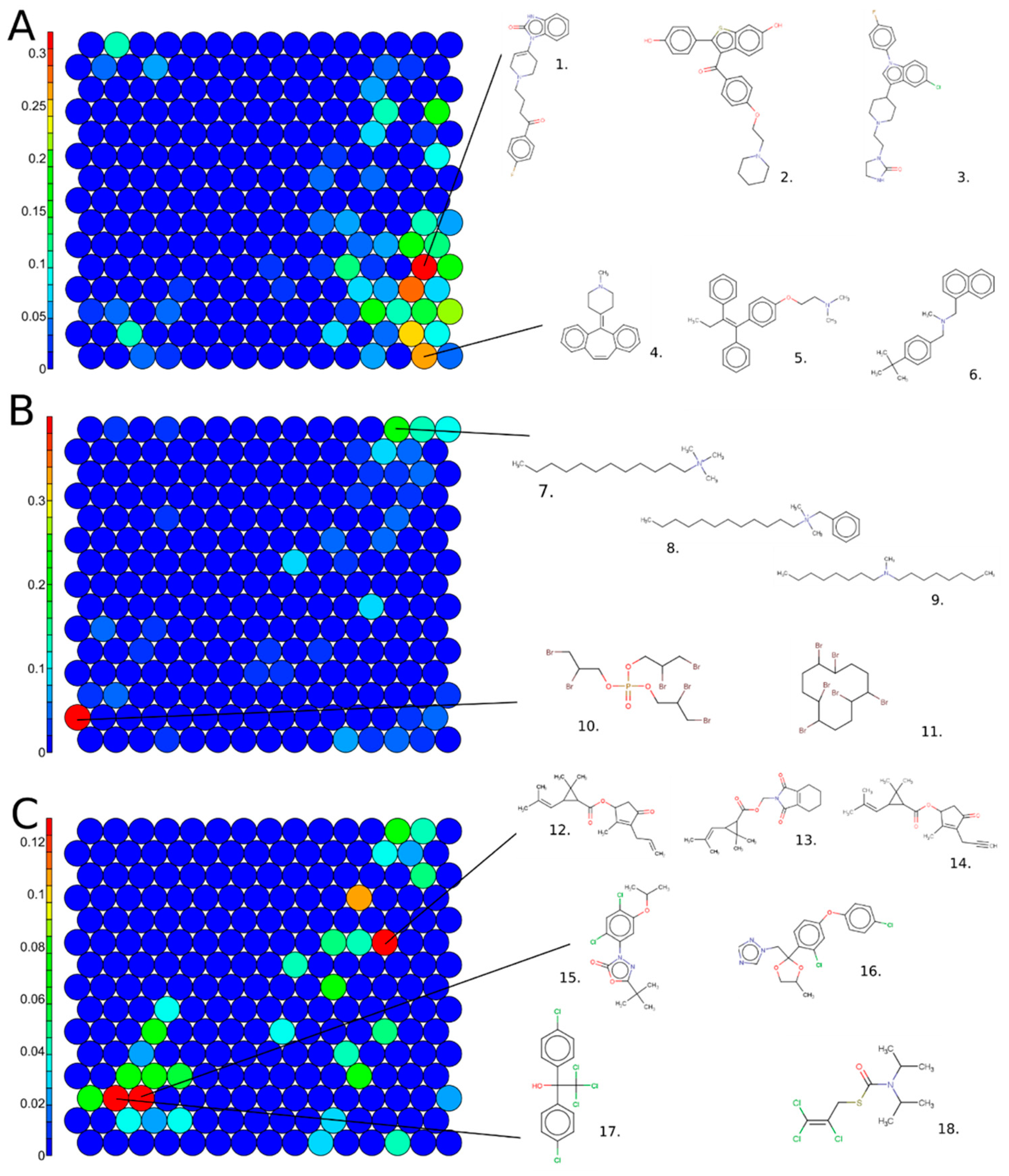
3.2. Active Chemical Categories
3.3. Most Active Chemicals
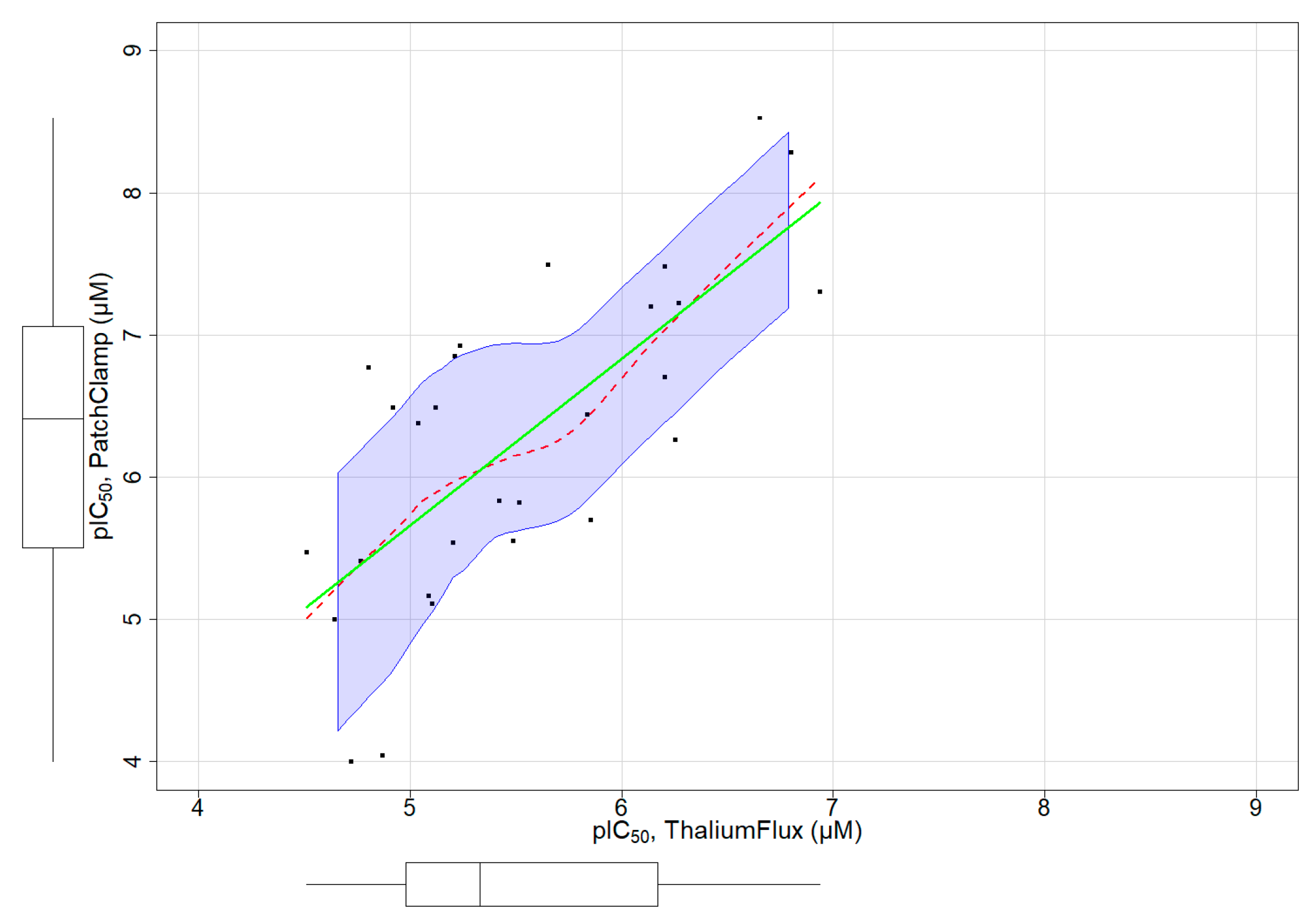
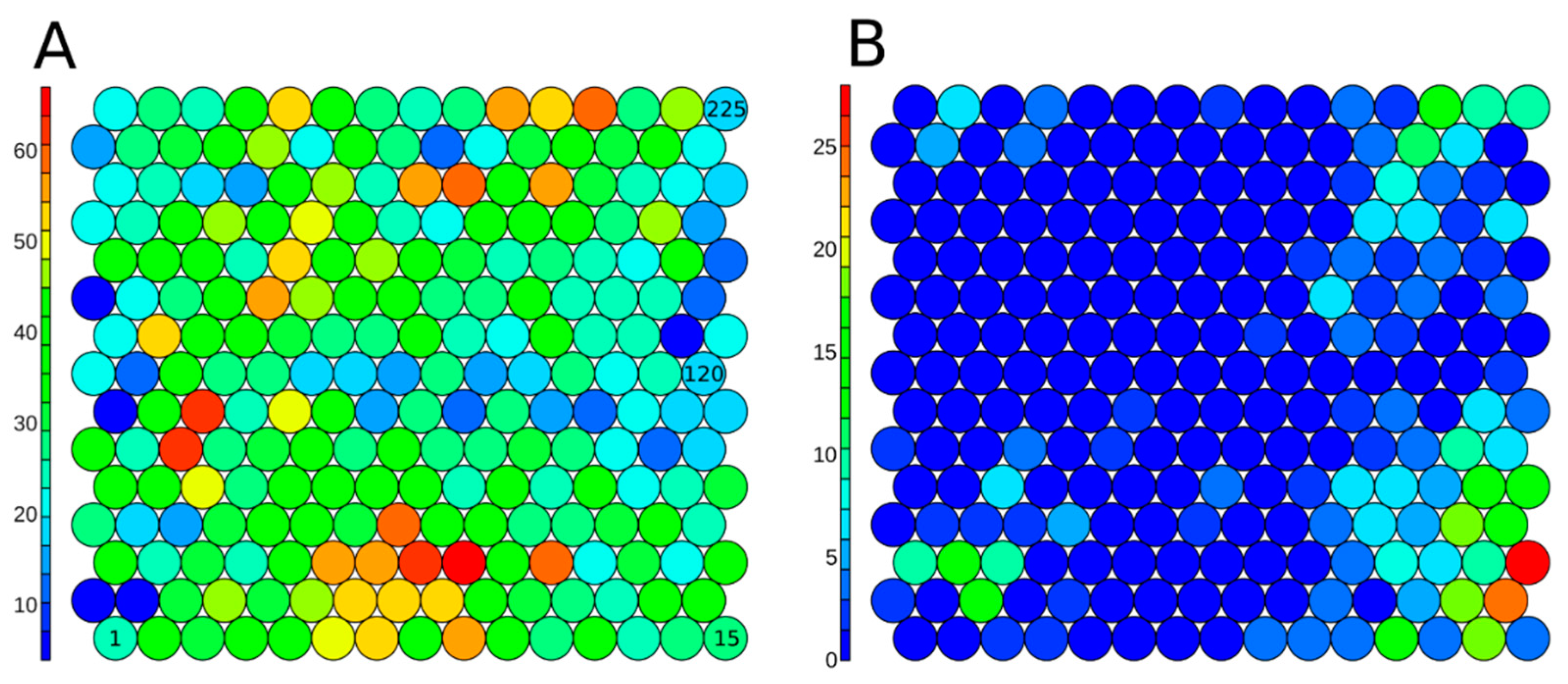
3.4. Assay Dependent Potency Shift
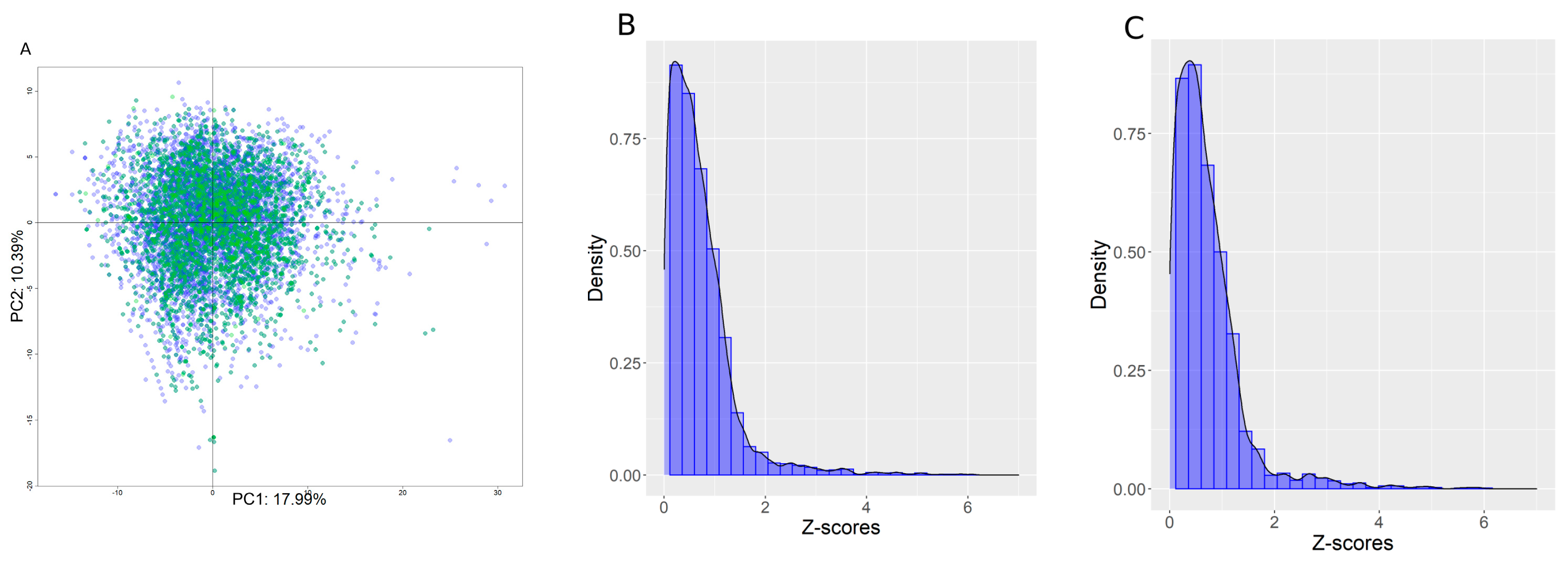
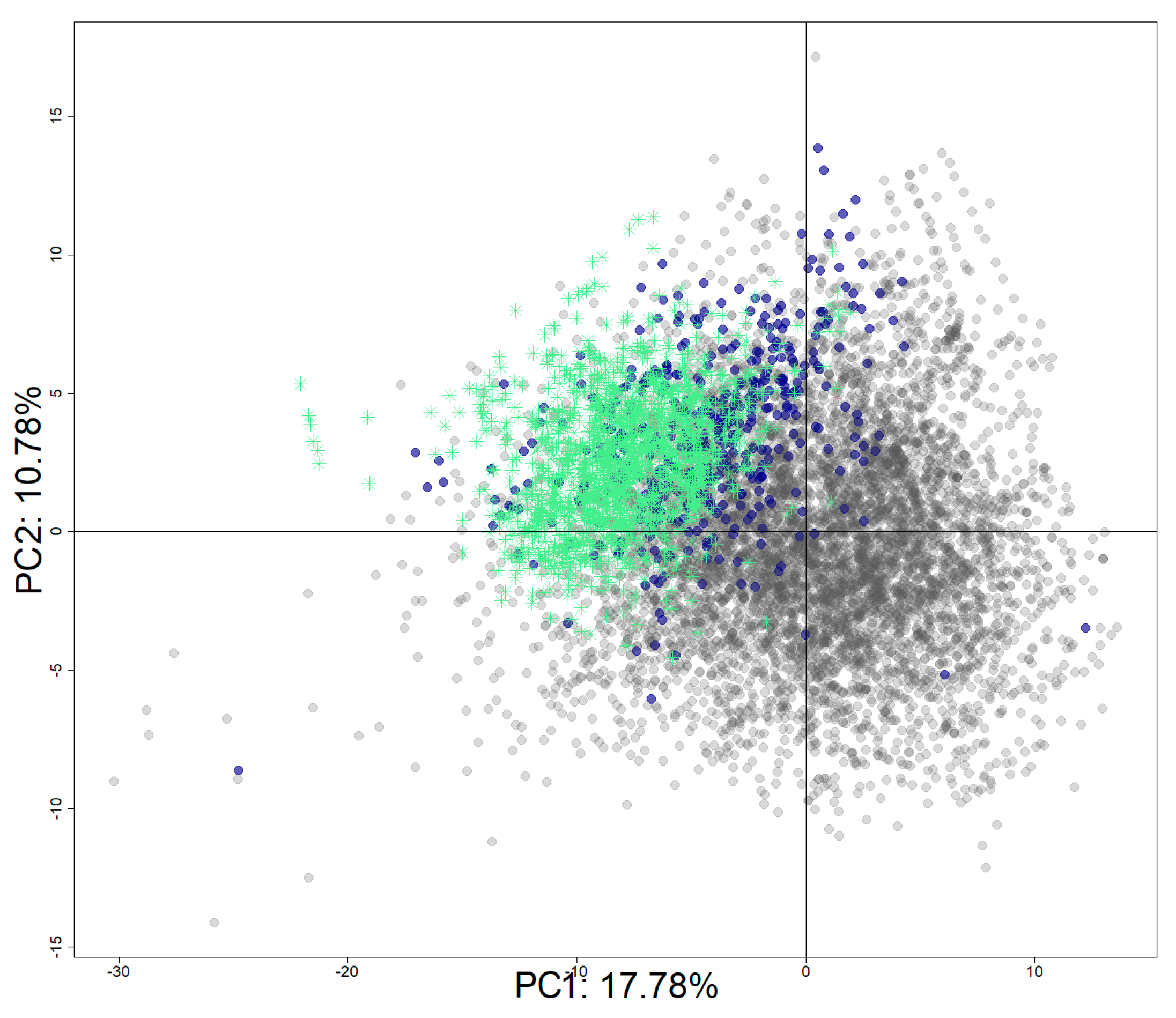
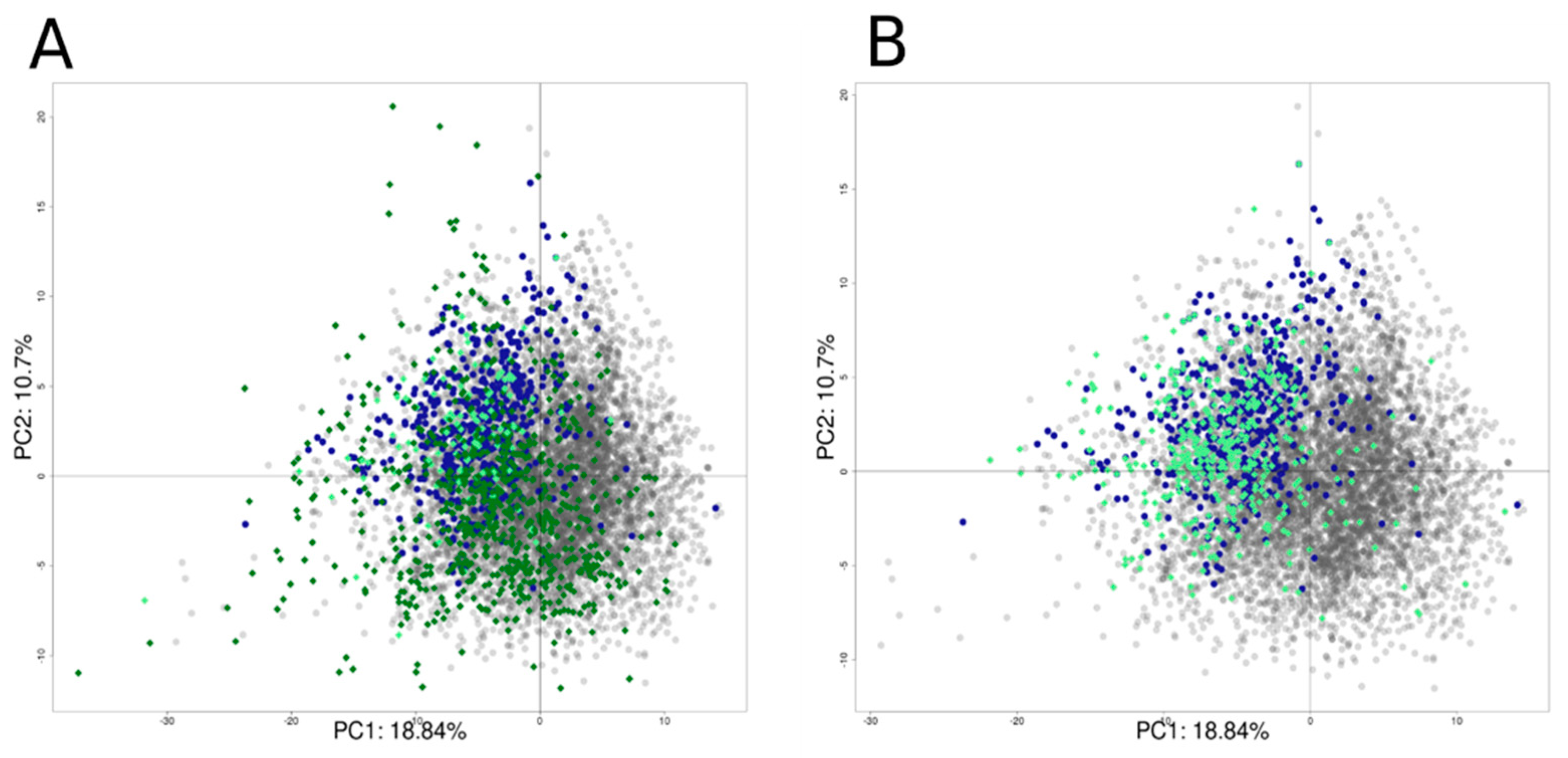
3.5. Structural Activity Patterns
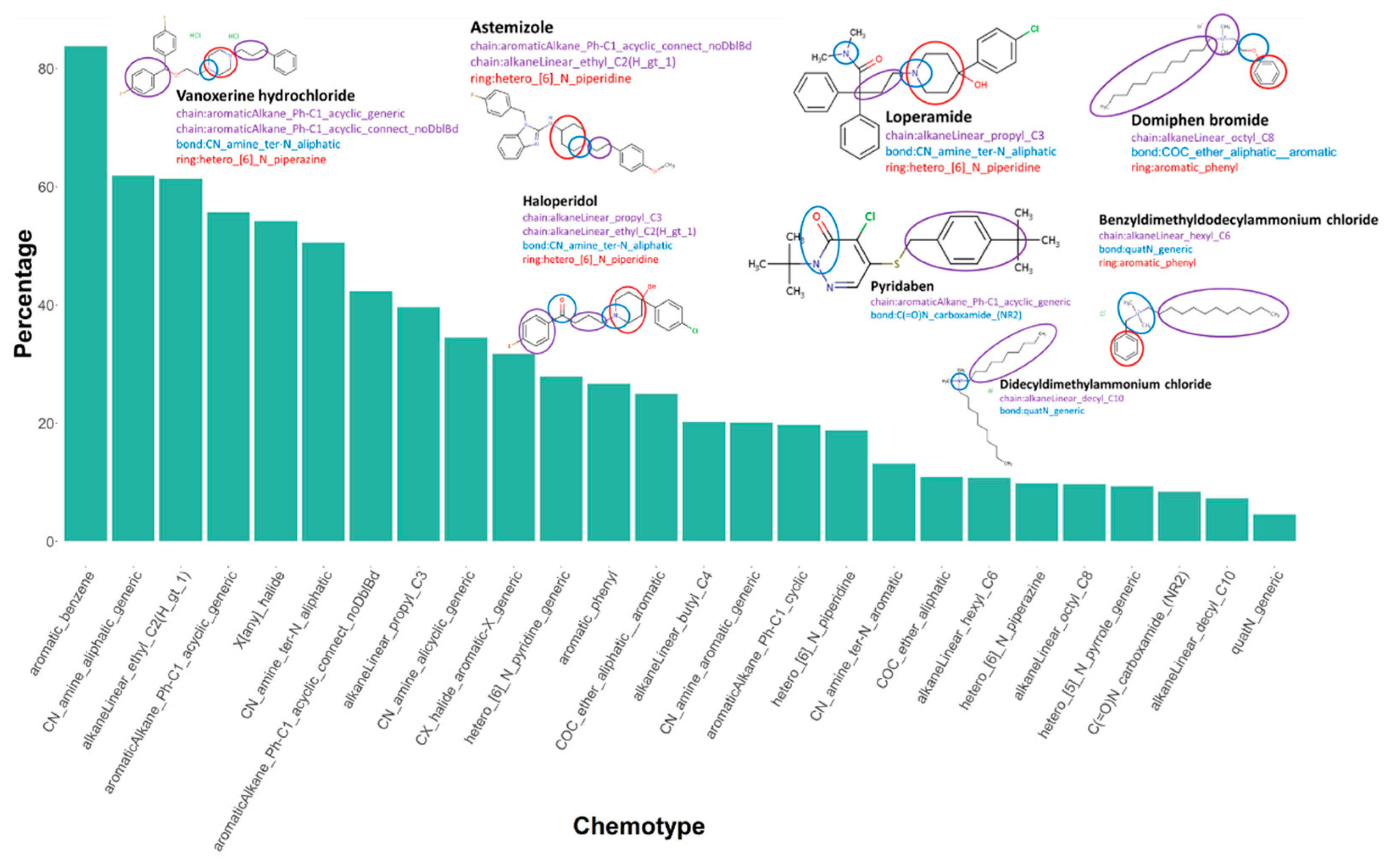
3.6. Chemotype Enrichment
3.7. QSAR Classification Models for hERG Inhibition Using the Tox21 FluxOR Thallium Influx Assay Dataset
3.8. QSAR Classification Models for hERG Inhibition Using the Enriched Dataset
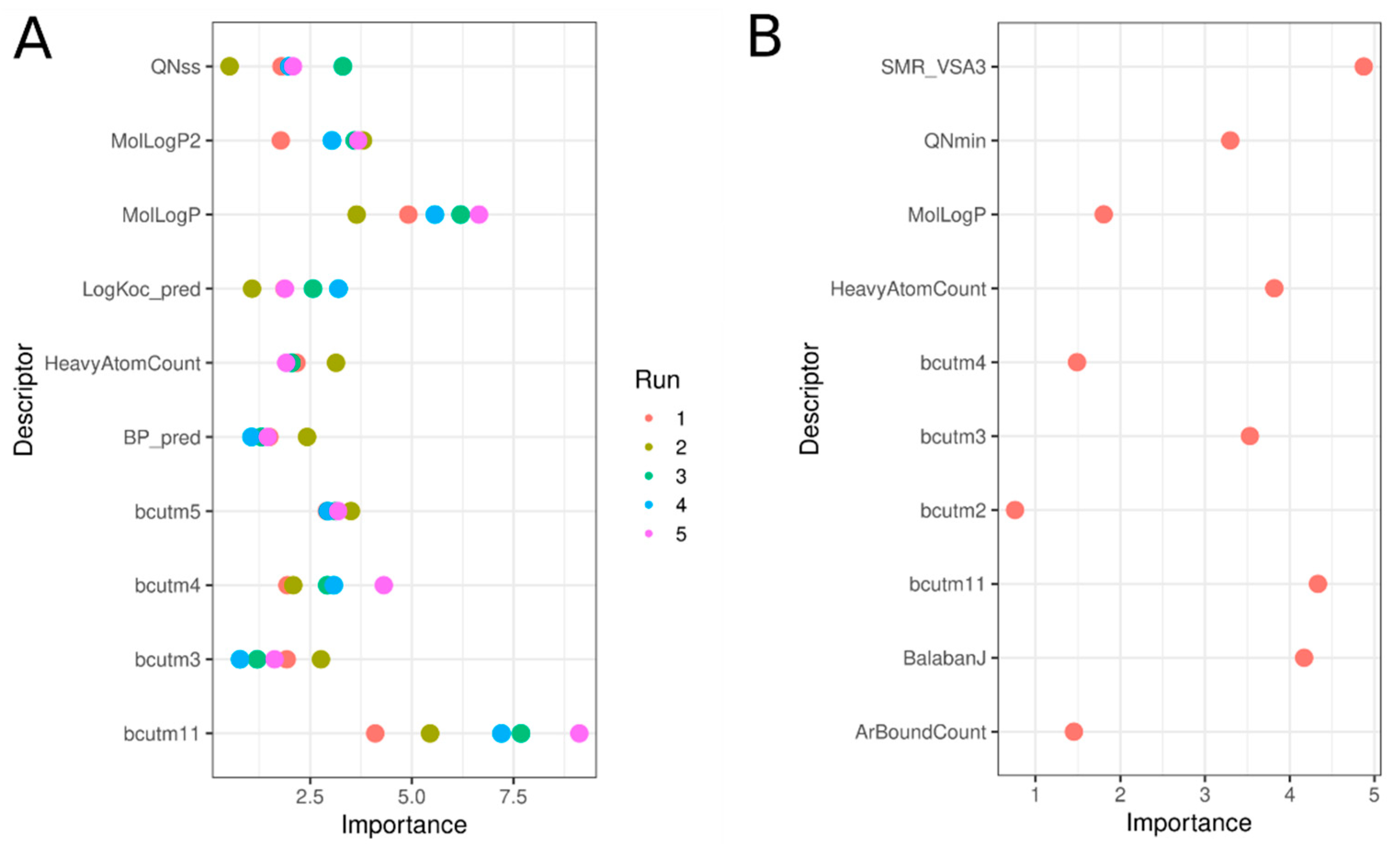
3.9. Significant Molecular Descriptors
3.10. External Validation of the QSAR Models
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Brown, A.M. Drugs, hERG and sudden death. Cell Calcium 2004, 35, 543–547. [Google Scholar] [CrossRef] [PubMed]
- Curran, M.E.; Splawski, I.; Timothy, K.W.; Vincen, G.M.; Green, E.D.; Keating, M.T. A molecular basis for cardiac arrhythmia: HERG mutations cause long QT syndrome. Cell 1995, 80, 795–803. [Google Scholar] [CrossRef] [Green Version]
- Sanguinetti, M.C.; Jiang, C.; Curran, M.E.; Keating, M.T. A mechanistic link between an inherited and an acquired cardiac arrhythmia: HERG encodes the IKr potassium channel. Cell 1995, 81, 299–307. [Google Scholar] [CrossRef] [Green Version]
- Kalyaanamoorthy, S.; Barakat, K.H. Development of Safe Drugs: The hERG Challenge. Med. Res. Rev. 2018, 38, 525–555. [Google Scholar] [CrossRef] [PubMed]
- Priest, B.T.; Bell, I.M.; Garcia, M.L. Role of hERG potassium channel assays in drug development. Channels 2008, 2, 87–93. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siramshetty, V.B.; Nickel, J.; Omieczynski, C.; Gohlke, B.O.; Drwal, M.N.; Preissner, R. WITHDRAWN—a resource for withdrawn and discontinued drugs. Nucleic Acids Res. 2016, 44, D1080–D1086. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- International Council for Harmonisation of Technical Requirements for Pharmaceuticals for Human Use. ICH Harmonized Tripartite Guideline “Safety Pharmacology Studies for Human Pharmaceuticals S7A”. In Proceedings of the International Conference on Harmonisation of Technical Requirements for Registration of Pharmaceuticals for Human Use, Geneva, Switzerland, 8 November 2000. [Google Scholar]
- Committee for Medicinal Products. ICH E14 Note for Guidance on the Clinical Evaluation of QT/Qtc Interval Prolongation and Proarrhythmic Potential for Non-Antiarrhythmic Drugs (CHMP/ICH/2/04), in Committee for Medicinal Products for Human Use (CHMP); European Medicines Agency: London, UK, 2005. [Google Scholar]
- Kratz, J.M.; Grienke, U.; Scheel, O.; Mann, S.A.; Rollinger, J.M. Natural products modulating the hERG channel: Heartaches and hope. Nat. Prod. Rep. 2017, 34, 957–980. [Google Scholar] [CrossRef] [Green Version]
- Witchel, H.J. Drug-induced hERG block and long QT syndrome. Cardiovasc. Ther. 2011, 29, 251–259. [Google Scholar] [CrossRef]
- Garrido, A.; Lepailleur, A.; Mignani, S.M.; Dallemagne, P.; Rochais, C. hERG toxicity assessment: Useful guidelines for drug design. Eur. J. Med. Chem. 2020, 195, 112290. [Google Scholar] [CrossRef]
- Villoutreix, B.O.; Taboureau, O. Computational investigations of hERG channel blockers: New insights and current predictive models. Adv. Drug Deliv. Rev. 2015, 86, 72–82. [Google Scholar] [CrossRef]
- Strauss, D.G.; Gintant, G.; Li, Z.; Wu, W.; Blinova, K.; Vicente, J.; Turner, J.R.; Sager, P.T. Comprehensive In Vitro Proarrhythmia Assay (CiPA) Update from a Cardiac Safety Research Consortium / Health and Environmental Sciences Institute / FDA Meeting. Innov. Regul. Sci. 2019, 53, 519–525. [Google Scholar] [CrossRef] [PubMed]
- Xia, M.; Shahane, S.A.; Huang, R.; Titus, S.A.; Shum, E.; Zhao, Y.; Southall, N.; Zheng, W.; Witt, K.L.; Tice, R.R.; et al. Identification of quaternary ammonium compounds as potent inhibitors of hERG potassium channels. Toxicol. Appl. Pharm. 2011, 252, 250–258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krishna, S.; Berridge, B.; Kleinstreuer, N. High-Throughput Screening to Identify Chemical Cardiotoxic Potential. Chem. Res. Toxicol. 2021, 34, 566–583. [Google Scholar] [CrossRef] [PubMed]
- Belpomme, D.; Irigaray, P.; Hardell, L.; Clapp, R.; Montagnier, L.; Epstein, S.; Sasco, A.J. The multitude and diversity of environmental carcinogens. Environ. Res. 2007, 105, 414–429. [Google Scholar] [CrossRef]
- Collins, F.S.; Gray, G.M.; Bucher, J.R. Toxicology. Transforming environmental health protection. Science 2008, 319, 906–907. [Google Scholar] [CrossRef] [Green Version]
- Thomas, R.S.; Paules, R.S.; Simeonov, A.; Fitzpatrick, S.C.; Crofton, K.M.; Casey, W.M.; Mendrick, D.L. The US Federal Tox21 Program: A strategic and operational plan for continued leadership. ALTEX 2018, 35, 163–168. [Google Scholar] [CrossRef]
- Richard, A.M.; Huang, R.; Waidyanatha, S.; Shinn, P.; Collins, B.J.; Thillainadarajah, I.; Grulke, C.M.; Williams, A.J.; Lougee, R.R.; Judson, R.S.; et al. The Tox21 10K Compound Library: Collaborative Chemistry Advancing Toxicology. Chem. Res. Toxicol. 2021, 34, 189–216. [Google Scholar] [CrossRef]
- Zhao, J.; Xia, M. Cell-based hERG Channel Inhibition Assay in High-throughput Format. In High-Throughput Screening Assay in Toxicology, 2nd ed.; Unpublished Work; 2022; ISBN 978-1-0716-2212-4. [Google Scholar]
- Huang, R. A Quantitative High-Throughput Screening Data Analysis Pipeline for Activity Profiling. Methods Mol. Biol. 2016, 1473, 111–122. [Google Scholar]
- Inglese, J.; Auld, D.S.; Jadhav, A.; Johnson, R.L.; Simeonov, A.; Yasgar, A.; Zheng, W.; Austin, C.P. Quantitative high-throughput screening: A titration-based approach that efficiently identifies biological activities in large chemical libraries. Proc. Natl. Acad. Sci. USA 2006, 103, 11473–11478. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Huang, R. Correction of Microplate Data from High-Throughput Screening. Methods Mol. Biol. 2016, 1473, 123–134. [Google Scholar]
- Wang, Y.; Jadhav, A.; Southal, N.; Huang, R.; Nguyen, D.T. A grid algorithm for high throughput fitting of dose-response curve data. Curr. Chem. Genom. 2010, 4, 57–66. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.; Xia, M.; Cho, M.H.; Sakamuru, S.; Shinn, P.; Houck, K.A.; Dix, D.J.; Judson, R.S.; Witt, K.L.; Kavlock, R.J.; et al. Chemical genomics profiling of environmental chemical modulation of human nuclear receptors. Env. Health Perspect. 2011, 119, 1142–1148. [Google Scholar] [CrossRef] [PubMed]
- Huang, R. A Quantitative High-Throughput Screening Data Analysis Pipeline for Activity Profiling. In High-Throughput Screening Assays in Toxicology; Zhu, H., Xia, M., Eds.; Humana Press: Totova, NJ, USA, 2016. [Google Scholar]
- Fourches, D.; Muratov, E.; Tropsha, A. Trust, but verify: On the importance of chemical structure curation in cheminformatics and QSAR modeling research. J. Chem. Inf. Model. 2010, 50, 1189–1204. [Google Scholar] [CrossRef] [PubMed]
- Fourches, D.; Muratov, E.; Tropsha, A. Trust, but Verify II: A Practical Guide to Chemogenomics Data Curation. J. Chem. Inf. Model. 2016, 56, 1243–1252. [Google Scholar] [CrossRef] [Green Version]
- Mansouri, K.; Grulke, C.M.; Judson, R.S.; Williams, A.J. OPERA models for predicting physicochemical properties and environmental fate endpoints. J. Cheminform 2018, 10, 10. [Google Scholar] [CrossRef] [Green Version]
- Dionisio, K.L.; Phillips, K.; Price, P.S.; Grulke, C.M.; Williams, A.; Biryol, D.; Hong, T.; Isaacs, K.K. The Chemical and Products Database, a resource for exposure-relevant data on chemicals in consumer products. Sci. Data 2018, 5, 180125. [Google Scholar] [CrossRef] [Green Version]
- Kohonen, T. Essentials of the self-organizing map. Neural Netw. 2013, 37, 52–65. [Google Scholar] [CrossRef]
- Borrel, A.; Huang, R.; Sakamuru, S.; Xia, M.; Simeonov, A.; Mansouri, K.; Houck, K.A.; Judson, R.S.; Kleinstreuer, N.C. High-Throughput Screening to Predict Chemical-Assay Interference. Sci. Rep. 2020, 10, 3986. [Google Scholar] [CrossRef] [Green Version]
- Lynch, C.; Mackowiak, B.; Huang, R.; Li, L.; Heyward, S.; Sakamuru, S.; Wang, H.; Xia, M. Identification of Modulators That Activate the Constitutive Androstane Receptor From the Tox21 10K Compound Library. Toxicol. Sci. 2019, 167, 282–292. [Google Scholar] [CrossRef]
- Yang, C.; Tarkhov, A.; Marusczyk, J.; Bienfait, B.; Gasteiger, J.; Kleinoeder, T.; Magdziarz, T.; Sacher, O.; Schwab, C.H.; Schwoebel, J.; et al. New publicly available chemical query language, CSRML, to support chemotype representations for application to data mining and modeling. J. Chem. Inf. Model. 2015, 55, 510–528. [Google Scholar] [CrossRef]
- Ashby, J.; Tennant, R.W. Definitive relationships among chemical structure, carcinogenicity and mutagenicity for 301 chemicals tested by the U.S. NTP. Mutat. Res. 1991, 257, 229–306. [Google Scholar] [CrossRef]
- Kroes, R.; Renwick, A.G.; Cheeseman, M.; Kleiner, J.; Mangelsdorf, I.; Piersma, A.; Schilter, B.; Schlatter, J.; Van Schothorst, F.; Vos, J.G.; et al. Structure-based thresholds of toxicological concern (TTC): Guidance for application to substances present at low levels in the diet. Food Chem. Toxicol. 2004, 42, 65–83. [Google Scholar] [CrossRef] [PubMed]
- Cherkasov, A.; Muratov, E.N.; Fourches, D.; Varnek, A.; Baskin, I.I.; Cronin, M.; Dearden, J.; Gramatica, P.; Martin, Y.C.; Todeschini, R.; et al. QSAR modeling: Where have you been? Where are you going to? J. Med. Chem. 2014, 57, 4977–5010. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Golbraikh, A.; Muratov, E.; Fourches, D.; Tropsha, A. Data set modelability by QSAR. J. Chem. Inf. Model. 2014, 54, 1–4. [Google Scholar] [CrossRef] [Green Version]
- Tropsha, A.; Golbraikh, A. Predictive QSAR modeling workflow, model applicability domains, and virtual screening. Curr. Pharm. Des. 2007, 13, 3494–3504. [Google Scholar] [CrossRef]
- Breiman, L.; Friedman, J.H.; Olshen, R.A.; Stone, C.J. Classification and Regression Trees; Monterey, C., Ed.; Brooks/Cole Publishing: Monterey, CA, USA.
- Ripley, B. (Ed.) Pattern Recognition and Neural Networks; Cambridge University Press: Cambridge, UK, 1996. [Google Scholar]
- Vapnik, C. Support-vector networks. Mach. Learn. 1995, 3, 273–297. [Google Scholar]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef] [Green Version]
- Fisher, R. The Use of Multiple Measurements in Taxonomic Problems. Ann. Eugen. 1936, 7, 179–188. [Google Scholar] [CrossRef]
- Basheer, I.A.; Hajmeer, M. Artificial neural networks: Fundamentals, computing, design, and application. J. Microbiol. Methods 2000, 43, 3–31. [Google Scholar] [CrossRef]
- Mendez, D.; Gaulton, A.; Bento, A.P.; Chambers, J.; De Veij, M.; Félix, E.; Magariños, M.P.; Mosquera, J.F.; Mutowo, P.; Nowotka, M.; et al. ChEMBL: Towards direct deposition of bioassay data. Nucleic Acids Res. 2019, 47, D930–D940. [Google Scholar] [CrossRef]
- Siramshetty, V.B.; Nguyen, D.T.; Martinez, N.J.; Southall, N.T.; Simeonov, A.; Zakharov, A.V. Critical Assessment of Artificial Intelligence Methods for Prediction of hERG Channel Inhibition in the "Big Data" Era. J. Chem. Inf. Model. 2020, 60, 6007–6019. [Google Scholar] [CrossRef] [PubMed]
- Kiss, L.; Bennett, P.B.; Uebele, V.N.; Koblan, K.S.; Kane, S.A.; Neagle, B.; Schroeder, K. High throughput ion-channel pharmacology: Planar-array-based voltage clamp. Assay Drug Dev. Technol. 2003, 1 (Suppl. 2), 127–135. [Google Scholar] [CrossRef]
- Polonchuk, L. Toward a New Gold Standard for Early Safety: Automated Temperature-Controlled hERG Test on the PatchLiner. Front. Pharm. 2012, 3, 3. [Google Scholar] [CrossRef] [Green Version]
- Siramshetty, V.B.; Chen, Q.; Devarakonda, P.; Preissner, R. The Catch-22 of Predicting hERG Blockade Using Publicly Accessible Bioactivity Data. J. Chem. Inf. Model. 2018, 58, 1224–1233. [Google Scholar] [CrossRef] [PubMed]
- Dubin, A.E.; Nasser, N.; Rohrbacher, J.; Hermans, A.N.; Marrannes, R.; Grantham, C.; Van Rossem, K.; Cik, M.; Chaplan, S.R.; Gallacher, D.; et al. Identifying modulators of hERG channel activity using the PatchXpress planar patch clamp. J. Biomol. Screen. 2005, 10, 168–181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bridal, T.R.; Margulis, M.; Wang, X.; Donio, M.; Sorota, S. Comparison of human Ether-a-go-go related gene screening assays based on IonWorks Quattro and thallium flux. Assay Drug Dev. Technol. 2010, 8, 755–765. [Google Scholar] [CrossRef]
- Rezazadeh, S.; Hesketh, J.C.; Fedida, D. Rb+ flux through hERG channels affects the potency of channel blocking drugs: Correlation with data obtained using a high-throughput Rb+ efflux assay. J. Biomol. Screen. 2004, 9, 588–597. [Google Scholar] [CrossRef]
- Weaver, C.D. Thallium Flux Assay for Measuring the Activity of Monovalent Cation Channels and Transporters. Methods Mol. Biol. 2018, 1684, 105–114. [Google Scholar]
- Lacerda, A.E.; Kuryshev, Y.A.; Yan, G.X.; Waldo, A.L.; Brown, A.M. Vanoxerine: Cellular mechanism of a new antiarrhythmic. J. Cardiovasc. Electrophysiol. 2010, 21, 301–310. [Google Scholar] [CrossRef] [Green Version]
- Olsen, R.E.; Kroken, R.A.; Bjorhovde, S.; Aanesen, K.; Jorgensen, H.A.; Loberg, E.M.; Johnsen, E. Influence of different second generation antipsychotics on the QTc interval: A pragmatic study. World J. Psychiatry 2016, 6, 442–448. [Google Scholar] [CrossRef]
- Polcwiartek, C.; Kragholm, K.; Schjerning, O.; Graff, C.; Nielsen, J. Cardiovascular safety of antipsychotics: A clinical overview. Expert Opin. Drug Saf. 2016, 15, 679–688. [Google Scholar] [CrossRef] [PubMed]
- Silke, B.; Campbell, C.; King, D.J. The potential cardiotoxicity of antipsychotic drugs as assessed by heart rate variability. J. Psychopharmacol. 2002, 16, 355–360. [Google Scholar] [CrossRef] [PubMed]
- Suessbrich, H.; Schonherr, R.; Heinemann, S.H.; Attali, B.; Lang, F.; Busch, A.E. The inhibitory effect of the antipsychotic drug haloperidol on HERG potassium channels expressed in Xenopus oocytes. Br. J. Pharmacol. 1997, 120, 968–974. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mitcheson, J.S.; Chen, J.; Lin, M.; Culberson, C.; Sanguinetti, M.C. A structural basis for drug-induced long QT syndrome. Proc. Natl. Acad. Sci. USA 2000, 97, 12329–12333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Redfern, W.S.; Carlsson, L.; Davis, A.S.; Lynch, W.G.; MacKenzie, I.; Palethorpe, S.; Siegl, P.K.S.; Strang, I.; Sullivan, A.T.; Wallis, R.; et al. Relationships between preclinical cardiac electrophysiology, clinical QT interval prolongation and torsade de pointes for a broad range of drugs: Evidence for a provisional safety margin in drug development. Cardiovasc. Res. 2003, 58, 32–45. [Google Scholar] [CrossRef]
- Ridley, J.M.; Milnes, J.T.; Hancox, J.C.; Witchel, H.J. Clemastine, a conventional antihistamine, is a high potency inhibitor of the HERG K+ channel. J. Mol. Cell. Cardiol. 2006, 40, 107–118. [Google Scholar] [CrossRef]
- Rossi, M.; Giorgi, G. Domperidone and long QT syndrome. Curr. Drug Saf. 2010, 5, 257–262. [Google Scholar] [CrossRef]
- Suessbrich, H.; Schonherr, R.; Heinemann, S.H.; Attali, B.; Lang, F.; Busch, A.E. Blockade of HERG channels expressed in Xenopus oocytes by the histamine receptor antagonists terfenadine and astemizole. FEBS Lett. 1996, 385, 77–80. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, H.; Takahashi, Y.; Hamaguchi, S.; Iida-Tanaka, N.; Oka, T.; Nishio, M.; Ohtsuki, A.; Namekata, I. Effect of terfenadine and pentamidine on the HERG channel and its intracellular trafficking: Combined analysis with automated voltage clamp and confocal microscopy. Biol. Pharm. Bull. 2014, 37, 1826–1830. [Google Scholar] [CrossRef] [Green Version]
- Walker, B.D.; Singleton, C.B.; Bursill, J.A.; Wyse, K.R.; Valenzuela, S.M.; Qiu, M.R.; Breit, S.N.; Campbell, T.J. Inhibition of the human ether-a-go-go-related gene (HERG) potassium channel by cisapride: Affinity for open and inactivated states. Br. J. Pharm. 1999, 128, 444–450. [Google Scholar] [CrossRef]
- Coppola, C.; Rienzo, A.; Piscopo, G.; Barbieri, A.; Arra, C.; Maurea, N. Management of QT prolongation induced by anti-cancer drugs: Target therapy and old agents. Different algorithms for different drugs. Cancer Treat. Rev. 2018, 63, 135–143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duan, J.; Tao, J.; Zhai, M.; Li, C.; Zhou, N.; Lv, J.; Wang, L.; Lin, L.; Bai, R. Anticancer drugs-related QTc prolongation, torsade de pointes and sudden death: Current evidence and future research perspectives. Oncotarget 2018, 9, 25738–25749. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sara, J.D.; Kaur, J.; Khodadadi, R.; Rehman, M.; Lobo, R.; Chakrabarti, S.; Herrmann, J.; Lerman, A.; Grothey, A. 5-fluorouracil and cardiotoxicity: A review. Adv. Med. Oncol. 2018, 10, 1758835918780140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.H.; Dempsey, C.E.; Hancox, J.C. The Basis for Low-affinity hERG Potassium Channel Block by Sotalol. J. Pharm. Pharm. 2017, 8, 130–131. [Google Scholar]
- Heir, E.; Sundheim, G.; Holck, A.L. Identification and characterization of quaternary ammonium compound resistant staphylococci from the food industry. Int. J. Food Microbiol. 1999, 48, 211–219. [Google Scholar] [CrossRef]
- McDonnell, G.; Russell, A.D. Antiseptics and disinfectants: Activity, action, and resistance. Clin. Microbiol. Rev. 1999, 12, 147–179. [Google Scholar] [CrossRef] [Green Version]
- Long, Y.; Chen, W.; Lin, Z.; Sun, H.; Xia, M.; Zheng, W.; Li, Z. Inhibition of HERG potassium channels by domiphen bromide and didecyl dimethylammonium bromide. Eur. J. Pharm. 2014, 737, 202–209. [Google Scholar] [CrossRef] [Green Version]
- Long, Y.; Lin, Z.; Xia, M.; Zheng, W.; Li, Z. Mechanism of HERG potassium channel inhibition by tetra-n-octylammonium bromide and benzethonium chloride. Toxicol. Appl. Pharm. 2013, 267, 155–166. [Google Scholar] [CrossRef] [Green Version]
- Baker, N.; Williams, A.J.; Tropsha, A.; Ekins, S. Repurposing Quaternary Ammonium Compounds as Potential Treatments for COVID-19. Pharm. Res. 2020, 37, 104. [Google Scholar] [CrossRef]
- Ogilvie, B.H.; Solis-Leal, A.; Lopez, J.B.; Poole, B.D.; Robison, R.A.; Berges, B.K. Alcohol-free hand sanitizer and other quaternary ammonium disinfectants quickly and effectively inactivate SARS-CoV-2. J. Hosp. Infect 2021, 108, 142–145. [Google Scholar] [CrossRef]
- Schrank, C.L.; Minbiole, K.P.C.; Wuest, W.M. Are Quaternary Ammonium Compounds, the Workhorse Disinfectants, Effective against Severe Acute Respiratory Syndrome-Coronavirus-2? ACS Infect Dis. 2020, 6, 1553–1557. [Google Scholar] [CrossRef] [PubMed]
- Konda, L.S.K.; Praba, S.K.; Kristam, R. hERG liability classification models using machine learning techniques. Comput. Toxicol. 2019, 12, 100089. [Google Scholar] [CrossRef]
- Liu, M.; Zhang, L.; Li, S.; Yang, T.; Liu, L.; Zhao, J.; Liu, H. Prediction of hERG potassium channel blockage using ensemble learning methods and molecular fingerprints. Toxicol. Lett. 2020, 332, 88–96. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Zhou, Y.; Gu, S.; Wu, Z.; Wu, W.; Liu, C.; Wang, K.; Liu, G.; Li, W.; Lee, P.W.; et al. In silico prediction of hERG potassium channel blockage by chemical category approaches. Toxicol. Res. 2016, 5, 570–582. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, H.; Huang, R.; Xia, M.; Shahane, S.; Southall, N.; Wang, Y. Prediction of hERG Liability—Using SVM Classification, Bootstrapping and Jackknifing. Mol. Inf. 2017, 36, 1600126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
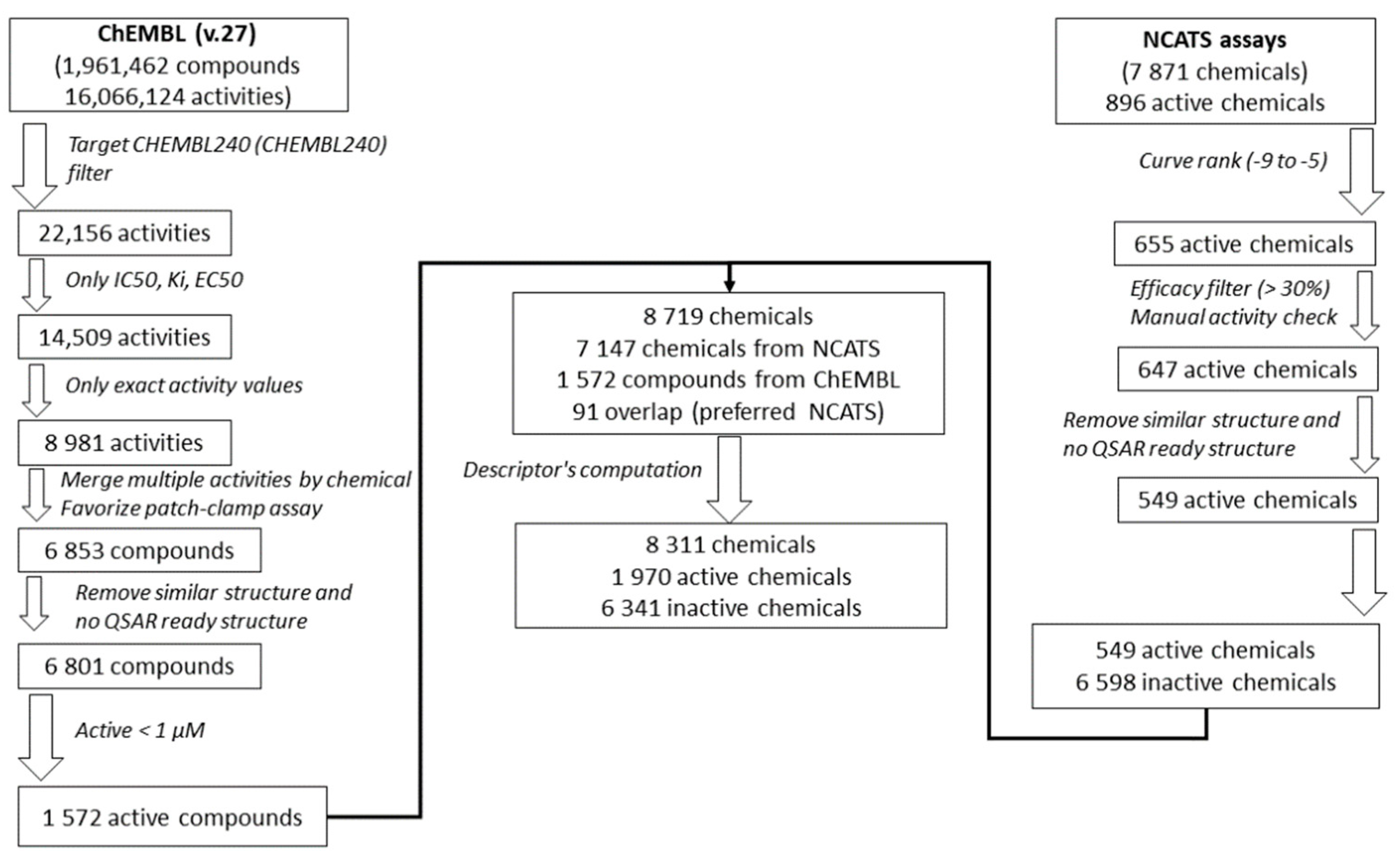
| Filtering Step | % of Active Chemicals | |||||
|---|---|---|---|---|---|---|
| Filtering Step Applied Sequentially | Initial Outcome | Curve Rank (−9 to −5) | Efficacy (>30%) | Manual Activity Curve Check | Chemical Standardization | |
| Count of active chemicals | 896 | 655 | 655 | 647 | 549 | 7.78% |
| 10-Fold Cross-Validation (for five Undersampled Training Set, n = 2023) | |||||
| Q | Qb | Sp | Se | MCC | |
| CART | 0.88 (+/−0.005) | 0.757 (+/−0.011) | 0.948 (+/−0.003) | 0.566 (+/−0.018) | 0.561 (+/−0.019) |
| NN | 0.868 (+/−0.008) | 0.75 (+/−0.058) | 0.933 (+/−0.013) | 0.566 (+/−0.103) | 0.526 (+/−0.057) |
| DNN | 0.88 (+/−0.005) | 0.824 (+/−0.017) | 0.927 (+/−0.007) | 0.721 (+/−0.039) | 0.659 (+/−0.021) |
| SVM-linear | 0.896 (+/−0.004) | 0.803 (+/−0.008) | 0.947 (+/−0.004) | 0.658 (+/−0.011) | 0.629 (+/−0.013) |
| SVM-radial | 0.913 (+/−0.004) | 0.82 (+/−0.009) | 0.964 (+/−0.002) | 0.676 (+/−0.016) | 0.685 (+/−0.016) |
| SVM-sigmoid | 0.89 (+/−0.002) | 0.758 (+/−0.007) | 0.962 (+/−0.002) | 0.553 (+/−0.012) | 0.587 (+/−0.009) |
| RF | 0.907 (+/−0.004) | 0.795 (+/−0.006) | 0.969 (+/−0.003) | 0.621 (+/−0.009) | 0.657 (+/−0.014) |
| LDA | 0.895 (+/−0.002) | 0.805 (+/−0.004) | 0.944 (+/−0.002) | 0.666 (+/−0.005) | 0.629 (+/−0.008) |
| Fitting (for five undersampled training set, n = 2023) | |||||
| Q | Qb | Sp | Se | MCC | |
| CART | 0.914 (+/−0.002) | 0.86 (+/−0.011) | 0.961 (+/−0.005) | 0.759 (+/−0.017) | 0.751 (+/−0.005) |
| NN | 0.895 (+/−0.01) | 0.852 (+/−0.029) | 0.931 (+/−0.016) | 0.773 (+/−0.041) | 0.704 (+/−0.028) |
| DNN | 0.983 (+/−0.005) | 0.975 (+/−0.005) | 0.927 (+/−0.007) | 0.962 (+/−0.014) | 0.951 (+/−0.014) |
| SVM-linear | 0.921 (+/−0.004) | 0.881 (+/−0.008) | 0.955 (+/−0.004) | 0.806 (+/−0.012) | 0.773 (+/−0.011) |
| SVM-radial | 0.972 (+/−0.008) | 0.958 (+/−0.015) | 0.983 (+/−0.002) | 0.933 (+/−0.028) | 0.92 (+/−0.023) |
| SVM-sigmoid | 0.884 (+/−0.004) | 0.805 (+/−0.008) | 0.952 (+/−0.005) | 0.658 (+/−0.011) | 0.657 (+/−0.011) |
| RF | 0.997(+/−0.002) | 0.996 (+/−0.004) | 0.998 (+/−0.001) | 0.993 (+/−0.006) | 0.99 (+/−0.005) |
| LDA | 0.902 (+/−0.005) | 0.849 (+/−0.006) | 0.948 (+/−0.005) | 0.749 (+/−0.006) | 0.718 (+/−0.013) |
| External validation (test set, n = 1072) | |||||
| Q | Qb | Sp | Se | MCC | |
| CART | 0.898 (+/−0.005) | 0.775 (+/−0.016) | 0.92 (+/−0.006) | 0.629 (+/−0.026) | 0.447 (+/−0.016) |
| NN | 0.893 (+/−0.008) | 0.791 (+/−0.019) | 0.911 (+/−0.009) | 0.671 (+/−0.028) | 0.456 (+/−0.021) |
| DNN | 0.913 (+/−0.007) | 0.812 (+/−0.027) | 0.931 (+/−0.006) | 0.693 (+/−0.052) | 0.517 (+/−0.041) |
| SVM-linear | 0.906 (+/−0.005) | 0.802 (+/−0.012) | 0.925 (+/−0.006) | 0.678 (+/−0.017) | 0.492 (+/−0.014) |
| SVM-radial | 0.929 (+/−0.004) | 0.818 (+/−0.011) | 0.948 (+/−0.003) | 0.688 (+/−0.018) | 0.563 (+/−0.02) |
| SVM-sigmoid | 0.92 (+/−0.004) | 0.784 (+/−0.005) | 0.944 (+/−0.004) | 0.624 (+/−0.005) | 0.505 (+/−0.012) |
| RF | 0.928 (+/−0.004) | 0.814 (+/−0.021) | 0.948 (+/−0.005) | 0.68 (+/−0.037) | 0.557 (+/−0.019) |
| LDA | 0.909 (+/−0.007) | 0.795 (+/−0.015) | 0.93 (+/−0.006) | 0.659 (+/−0.024) | 0.491 (+/−0.028) |
| 10-Fold Cross-Validation (Full Training Set, n = 7064) | |||||
| Q | Qb | Sp | Se | MCC | |
| CART | 0.921 | 0.887 | 0.822 | 0.951 | 0.779 |
| NN | 0.890 | 0.826 | 0.707 | 0.947 | 0.684 |
| DNN | 0.941 | 0.917 | 0.962 | 0.873 | 0.836 |
| SVM-linear | 0.945 | 0.924 | 0.885 | 0.963 | 0.847 |
| SVM-radial | 0.953 | 0.932 | 0.893 | 0.972 | 0.870 |
| SVM-sigmoid | 0.939 | 0.916 | 0.873 | 0.959 | 0.831 |
| RF | 0.951 | 0.925 | 0.875 | 0.975 | 0.863 |
| LDA | 0.938 | 0.911 | 0.861 | 0.962 | 0.828 |
| Fitting (training set, n = 7064) | |||||
| Q | Qb | Sp | Se | MCC | |
| CART | 0.930 | 0.900 | 0.845 | 0.956 | 0.805 |
| NN | 0.935 | 0.922 | 0.897 | 0.947 | 0.826 |
| DNN | 0.930 | 0.984 | 0.994 | 0.974 | 0.971 |
| SVM-linear | 0.952 | 0.932 | 0.895 | 0.970 | 0.867 |
| SVM-radial | 0.981 | 0.970 | 0.951 | 0.990 | 0.946 |
| SVM-sigmoid | 0.941 | 0.920 | 0.881 | 0.960 | 0.838 |
| RF | 0.999 | 0.998 | 0.996 | 0.999 | 0.997 |
| LDA | 0.939 | 0.913 | 0.863 | 0.963 | 0.831 |
| External validation (test set, n = 1247) | |||||
| Q | Qb | Sp | Se | MCC | |
| CART | 0.929 | 0.904 | 0.858 | 0.951 | 0.804 |
| NN | 0.929 | 0.917 | 0.895 | 0.939 | 0.810 |
| DNN | 0.933 | 0.909 | 0.956 | 0.861 | 0.816 |
| SVM-linear | 0.949 | 0.926 | 0.882 | 0.970 | 0.857 |
| SVM-radial | 0.958 | 0.937 | 0.895 | 0.978 | 0.884 |
| SVM-sigmoid | 0.942 | 0.924 | 0.889 | 0.959 | 0.842 |
| RF | 0.949 | 0.926 | 0.882 | 0.970 | 0.857 |
| LDA | 0.937 | 0.906 | 0.848 | 0.964 | 0.823 |
| Descriptor | Description | M Active | M Inactive | p-Value |
|---|---|---|---|---|
| Physicochemical descriptors | ||||
| BP_pred | Boiling point prediction | 358.27 | 279.15 | *** |
| LogKoc_pred | Log of soil adsorption coefficient of organic compounds. The ratio of the amount of chemical adsorbed per unit weight of organic carbon in the soil or sediment to the concentration of the chemical in solution at equilibrium. | 3.56 | 2.53 | *** |
| MolLogP2 | Crippen method to estimate log(P)2 | 22.12 | 8.76 | *** |
| MolLogP | Crippen method to estimate log(P) | 4.41 | 2.19 | *** |
| MOE type | ||||
| SMR_VSA3 | MOE-type descriptors using molecular refractivity contributions and surface area contributions | 13.05 | 3.29 | *** |
| Topological | ||||
| BalabanJ | Balaban’s J index (J) | 1.52 | 2.65 | *** |
| Charge descriptor | ||||
| QNss | Sum of squares of charges on N atoms | 0.16 | 0.1 | *** |
| QNmin | Most negative charge on N atoms | −0.32 | −0.16 | *** |
| Burden descriptors | ||||
| bcutm2 | Highest eigenvalue 2 for burden matrix/weighted by atomic masses | 3.89 | 3.59 | *** |
| bcutm3 | Highest eigenvalue 3 for burden matrix/weighted by atomic masses | 1.73 | 1.37 | *** |
| bcutm5 | Highest eigenvalue 5 for burden matrix/weighted by atomic masses | 3.15 | 2.52 | *** |
| bcutm4 | Highest eigenvalue 4 for burden matrix/weighted by atomic masses | 3.36 | 2.85 | *** |
| bcutm11 | Highest eigenvalue 11 for burden matrix/weighted by atomic masses | 1.73 | 1.37 | *** |
| Composition descriptor | ||||
| HeavyAtomCount | Count of heavy atom | 25.14 | 16.18 | *** |
| ArBoundCount | Count of aromatic bonds | 15.74 | 5.84 | *** |
| PubChem (ID: AID588834) (135 Actives and 876 Inactives) | |||||
| TOX21 Model | Q | Qb | Sp | Se | MCC |
| DNN | 0.665 | 0.676 | 0.661 | 0.690 | 0.245 |
| RF | 0.920 | 0.793 | 0.966 | 0.619 | 0.631 |
| Consensus | 0.873 | 0.788 | 0.904 | 0.673 | 0.518 |
| TOX21-ChEMBL | |||||
| DNN | 0.693 | 0.703 | 0.690 | 0.717 | 0.286 |
| RF | 0.926 | 0.766 | 0.984 | 0.549 | 0.641 |
| Combined (RF + DNN) | 0.900 | 0.791 | 0.945 | 0.637 | 0.582 |
| Lit-based hERG inhibitors (393 actives) | |||||
| TOX21 model | Q | TP | FN | ||
| DNN | 0.41 | 161 | 231 | ||
| RF | 0.40 | 157 | 235 | ||
| Combined (RF + DNN) | 0.40 | 157 | 235 | ||
| TOX21-ChEMBL | |||||
| DNN | 0.389 | 153 | 249 | ||
| RF | 0.341 | 134 | 258 | ||
| Combined (RF + DNN) | 0.341 | 134 | 258 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Krishna, S.; Borrel, A.; Huang, R.; Zhao, J.; Xia, M.; Kleinstreuer, N. High-Throughput Chemical Screening and Structure-Based Models to Predict hERG Inhibition. Biology 2022, 11, 209. https://doi.org/10.3390/biology11020209
Krishna S, Borrel A, Huang R, Zhao J, Xia M, Kleinstreuer N. High-Throughput Chemical Screening and Structure-Based Models to Predict hERG Inhibition. Biology. 2022; 11(2):209. https://doi.org/10.3390/biology11020209
Chicago/Turabian StyleKrishna, Shagun, Alexandre Borrel, Ruili Huang, Jinghua Zhao, Menghang Xia, and Nicole Kleinstreuer. 2022. "High-Throughput Chemical Screening and Structure-Based Models to Predict hERG Inhibition" Biology 11, no. 2: 209. https://doi.org/10.3390/biology11020209
APA StyleKrishna, S., Borrel, A., Huang, R., Zhao, J., Xia, M., & Kleinstreuer, N. (2022). High-Throughput Chemical Screening and Structure-Based Models to Predict hERG Inhibition. Biology, 11(2), 209. https://doi.org/10.3390/biology11020209






