Simple Summary
During embryonic development, cells proliferate and move to form structures essential to each organ. As these dynamic processes occur cells must form adhesions with their neighbors. These adhesions serve as communication centers to amplify the behavior of individual cells into collective cellular events. Here, we review cell-to-cell adhesions and focus on their role in the development of the face and palate.
Abstract
Morphogenesis requires a tight coordination between mechanical forces and biochemical signals to inform individual cellular behavior. For these developmental processes to happen correctly the organism requires precise spatial and temporal coordination of the adhesion, migration, growth, differentiation, and apoptosis of cells originating from the three key embryonic layers, namely the ectoderm, mesoderm, and endoderm. The cytoskeleton and its remodeling are essential to organize and amplify many of the signaling pathways required for proper morphogenesis. In particular, the interaction of the cell junctions with the cytoskeleton functions to amplify the behavior of individual cells into collective events that are critical for development. In this review we summarize the key morphogenic events that occur during the formation of the face and the palate, as well as the protein complexes required for cell-to-cell adhesions. We then integrate the current knowledge into a comprehensive review of how mutations in cell-to-cell adhesion genes lead to abnormal craniofacial development, with a particular focus on cleft lip with or without cleft palate.
1. Introduction
Morphogenesis is a complex process in which cells divide, move, and interact with one another and extracellular matrices. These events are highly coordinated across a range of timescales and physical spaces, resulting in the formation of tissues and organs that are essential to a functional body. Morphogenesis is highly dynamic; it integrates mechanical forces with biochemical signals, both sensed and produced by cells, to inform individual cellular behavior. At the cross-roads of force signaling and cell fate is the internal cytoskeleton and its remodeling, which, through the intermediary of cell junctions, amplify individual cell behavior into collective movement, which is critical for the development of organisms.
Amongst all developmental processes, the morphogenesis of the face is one of the most fascinating. It transforms the cranial part of the neural tube by way of distinct embryological processes that form the eyes, nose, chin, and mouth, each defining the identity of the organism. Not only is this morphogenic event extremely dynamic, but it also has the unique characteristic that most of the events are visible from the outside, allowing for a direct appreciation of the cellular movements and interactions. When the coordinated interplay of cellular migration, adhesion, and proliferation is disrupted it can lead to structural birth defects, such as orofacial clefts.
In this review we focus on craniofacial morphogenesis, with a particular emphasis on the formation of the lip and palate, and examine how the structures interacting with the cytoskeleton—namely cell–cell junctions—contribute to the morphogenesis of these tissues in humans and mice. We discuss the need to study the cytoskeletal rearrangements that regulate biomechanical processes to understand how cells move and adhere during embryonic development.
2. Craniofacial Morphogenesis
The development of the cranium and face is a highly complex process. It involves the precise spatial and temporal coordination of the migration, adhesion, growth, differentiation, and apoptosis of cells originating from the ectoderm, mesoderm, and endoderm. The result of these morphogenic events is a constellation of structures that is distinctly recognizable as human and critical to our interaction with the world.
2.1. Early Embryogenesis
One of the critical early events of embryogenesis is the transition from a bilaminar to a trilaminar embryo. The addition of the third embryological layer starts at an estimated gestational age (EGA) of 14 days, with the formation of the primitive streak [1]. This streak is the result of the differentiation of ectodermal cells into rapidly dividing mesenchymal cells, starting at the caudal end of the embryo and moving cranially. These cells proliferate and migrate to fill the space between the endoderm and the ectoderm, with the exception of the oropharyngeal and cloacal membranes. Further differentiation of the primitive streak leads to the formation of the notochord, which serves as the embryonic axial skeleton and as a signaling center to the overlying ectoderm to differentiate into the neural plate (Figure 1A,B,B′).
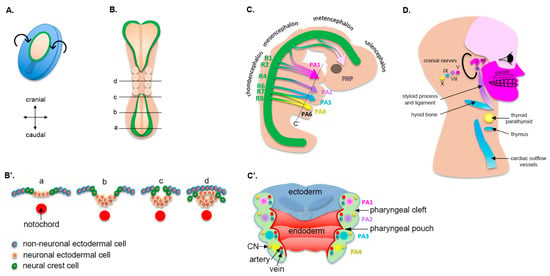
Figure 1.
Morphogenesis of pharyngeal arches and their derivatives. (A) Dorsal view of a human embryo at approximately 16 d EGA. The neural plate is being specified (pale area) and will undergo dorsal folding. (B) Dorsal view of a human embryo at about 22 d EGA. Note that the neural folds (green color) start to fuse in the midline. Transverse views at different levels of the embryo are shown in (B′). (C) Lateral view of a human embryo at approximately 32 d EGA. The neural tube (green) is specified in different rhombomeres (R1 to R8), all of which (except R3 and R5) will contribute to pharyngeal arches. (C′) Longitudinal view of the pharyngeal arches. Each pharyngeal arch is composed of a core mesenchyme derived from the neural crest, which includes a unique cranial nerve, an artery, and a vein (see Table 1 for details). The narrowing between two pharyngeal arches is called a cleft (on the outside of the embryo) or a pouch (on the inside of the embryo). (D) Derivatives of pharyngeal arches in a human adult. CN = cranial nerve; i = incus; m = maleus; and s = stapes.
The specification of the neural plate from the ectoderm occurs at 17 d EGA. At 22 d EGA the borders of the neural plate, also called neural folds, converge at the dorsal midline and begin to fuse, forming the neural tube (Figure 1B). First, the fusion occurs in the cranial region between the 3rd and 5th somite, which corresponds to what will eventually become the occipital region. Then, the fusion of the remaining neural folds proceeds caudally to complete the formation of the neural tube along the body axis (Figure 1B,B′). The failure of fusion in either direction results in the pathologies broadly termed as neural tube defects (for example, anencephaly from failure in the cranial direction and spina bifida caudally), some of the most common birth defects. In the context of this review, it is notable that there is an increased incidence of neural tube defects in individuals with orofacial clefts, indicating an overlap in risk factors for these two structural birth defects [2].
2.2. Cranial Neural Crest Cells
Cells that reside at the dorsal edge of the neural tube (i.e., crest of the neural tube), where the neural folds fused, are called neural crest cells (NCCs, Figure 1B,B′) [3]. These cells are specified at the border of the neural plate and the non-neural ectoderm via signaling gradients, which include WNT, BMP, and FGF, along with a large gene regulatory network of transcription factors. Following induction, NCCs undergo an epithelial-to-mesenchymal transition (EMT). They lose their cell-to-cell contact by decreasing E-cadherin levels, activate mechanosensitive molecules, including the small Rho GTPases and YAP, and migrate following the stimulation of growth factors such as WNT, BMP, and FGF [4]. NCCs ultimately differentiate into a variety of connective, muscular, nervous, endocrine, and pigmentary tissue, and induce the differentiation of overlying tissues that they invade. The reader is invited to read reviews on NCCs for extensive details [3,5].
Neural crest cells are grouped based on their anatomical position along the body axis, i.e., cranial, cardiac, or trunk. In the context of craniofacial development, cranial neural crest cells (CNCCs) are the key determinant of facial morphogenesis (Figure 1C). The migration of CNCCs from the cranial neural tube to ventral swellings and the first pharyngeal arch is considered the initial event of craniofacial development [4]. These cells determine the structures that will develop in each arch (Figure 1C,C′). Eventually, they form the facial cartilage, bone, dentin, muscles and tendons, and dermis, along with the forebrain, midbrain, hindbrain, and sensory neurons (Figure 1D, Table 1). Consequently, the embryological origin of all these structures in the face (CNCCs) is different than that of the rest of the body (often the mesoderm).

Table 1.
Derivatives of pharyngeal arches.
2.3. Pharyngeal Arches
The pharyngeal arches (or branchial arches, from the Greek branchia for gill) are bilateral pairs of swellings (or outpouchings) of the mesoderm that surround the developing pharynx of the embryo. They project forward from the dorsal aspect of the embryo and fuse in the ventral central midline (Figure 1C). Pharyngeal arches develop in a cranial-to-caudal sequence, with the first pharyngeal arch being the most cranial and initiating its morphogenesis around 20 d EGA, followed by the second and the third. By the time that the last arches develop the first two are no longer visible externally, as they progressed through morphogenesis. Consequently, diagrams, including ours, which show distinct arches at the same stage of development are, in fact, not an accurate representation of morphogenesis.
The first arch, as the first to form, separates the mouth pit (or stomodeum) from the externally developing pericardium. By differential growth, the neck elongates and new arches form, such that the embryo ultimately has six arches (Figure 1C). The first four are the most prominent and clinically relevant. The fifth pharyngeal arch involutes quickly after it is formed and is not relevant for subsequent development. The central core of each pharyngeal arch is composed of mesoderm-derived tissue, in which CNCCs migrate as they delaminate from the neural crest (Figure 1C′). As such, this tissue has a mixed embryological origin and is often referred to as the ecto-mesenchyme. Surrounding the ecto-mesenchyme are the endoderm-derived (medially) and ectoderm-derived (laterally) epithelia. At the junction of each arch the ectoderm and endoderm layers come closer to each other, but remain always separated by an ecto-mesenchymal layer. They form recesses called pouches on the endodermal side in addition to grooves or clefts on the lateral ectodermal surface (Figure 1C′) [6,7].
Each arch develops its own neurovascular structures that supply a distinct muscle group and skeletal tissue [8]. Derivatives of each arch (its core and associated pouch and cleft) are unique and listed in detail in Table 1. The first two arches are the most relevant for this review, as they give rise to the major bony structures of the middle ear, upper neck, and face, including the mandible (via Meckel’s cartilage), the maxilla, and the muscles of mastication and facial expression that overlie the bony structures. The last three pairs of arches give rise to the bones, muscles, and glands of the neck as well as the outflow tract of the heart, including the aortic arch and pulmonary arteries.
2.4. Formation of the Upper Lip, Nose, and Primary Palate
At the end of the fourth week of gestation, as embryogenesis progresses, the unpaired frontonasal prominence becomes distinct on the ventral side of the forebrain. In addition, the first and second pharyngeal arches continue to swell ventrally and give rise to two pairs of processes: the maxillary and mandibular, both surrounding the primitive mouth (stomodeum) (Figure 2). Collectively, it is the well-orchestrated proliferation and migration of CNCCs in these five prominences as well as the specific tissue fusions between them that will lead to the formation of the face [7]. On both sides of the frontonasal prominence, local thickening of the ectoderm leads to the formation of nasal placodes (Figure 2). During the fifth week of gestation, these nasal placodes invaginate and become depressed nasal pits. At the same time, mesenchymal cells proliferate around the placodes, and the sides of these swellings form the medial and lateral nasal prominences. As the maxillary prominences continue growing they merge laterally with the mandibular prominences to form the cheeks. Medially, the growth of the maxillary prominences compresses the medial nasal prominences and causes them to fuse around the 10th week of development (Figure 2). Hence, the upper lip is formed by the two medial nasal prominences and the two maxillary prominences. This fusion creates the intermaxillary segment, composed of a labial component that forms the philtrum of the upper lip, an upper jaw component with the four incisors, and a palatal component that forms the triangular primary palate. The lateral nasal prominences give rise to the nasal alae, and their interaction with the maxillary prominences forms the nasolacrimal groove and nasolacrimal duct system.
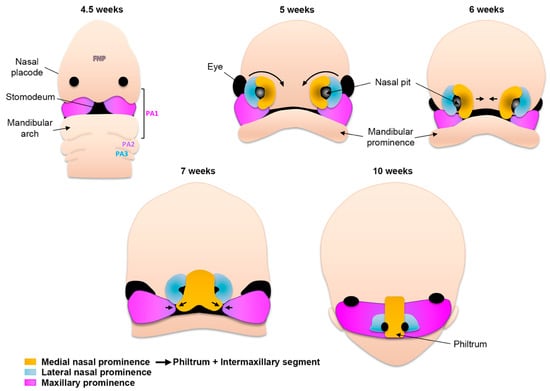
Figure 2.
Schematic of human facial morphogenesis (frontal views). At 4.5 weeks the frontonasal prominence (FNP) constitutes the most rostral part of the embryo. Derivatives of the first pharyngeal arch (PA) contribute structures below the nasal placodes, including the mandibular arch. Note the presence of the stomodeum (primitive mouth). At 5 weeks the thickening of nasal placodes gives rise to medial and lateral prominences. Medial rotation (6 weeks) and fusion (7 weeks) of the medial prominences give rise to the philtrum of the lip and the intermaxillary segment (10 weeks). The lateral prominences contribute to the nasal ala.
2.5. Formation of the Secondary Palate
The secondary palate is formed by pair-wise outgrowths from the maxillary prominences (Figure 3A). The reader is invited to consult complementary reviews related to human [9] and murine [10] palatogenesis. At around the sixth week of gestation in humans (E11.5 in mice, see Figure 4 for timeline correlation between mice and humans) these outgrowths, namely the palatine shelves, appear on each side of the tongue and begin to grow downward (Figure 3B,C). In the seventh week (E13.5–E14 in mice) these shelves reorient themselves and acquire a horizontal position as the tongue depresses inferiorly. At this stage the palatine shelf is composed of an ecto-mesenchyme (CNNC-derived) core surrounded by an epithelium (ectodermal-derived) with at least two layers of oral keratinocytes: a basal layer and a superficial squamous layer called the periderm (Figure 3C, see below for details). As the two palatine shelves come into closer proximity the palatal epithelia make contact in the midline. For adhesion, rather than just contact, to occur the periderm must slough off from the oppositional surfaces, and epithelial cells from each of the shelves intercalate to form a single-layered epithelial seam (termed medial edge epithelium or medial edge seam, MEE or MES) (Figure 3B,C) [11]. From a transverse view, this initial adhesion occurs in the middle of the secondary palate and proceeds anteriorly and posteriorly, similar to a “zipper” closing in both directions, ending at the incisive foramen anteriorly and the uvula posteriorly (Figure 3A). Between nine and twelve weeks of gestation (E15–E16 in mice), the epithelial cells of the MEE disappear, leading to mesenchymal confluence of the palate and the completion of palatogenesis. The mechanisms and cellular dynamics of this disappearance have been debated between three primary mechanisms (which may be involved in combination): epithelial-to-mesenchymal transition (EMT), apoptosis/non-apoptotic cell death, and migration to the nasal and oral surfaces [10]. Live imaging studies further support orthogonal cell displacement and epithelial cell extrusion driven by activation of the Rho and myosin light chain kinases, and ultimately the activation of the non-muscle myosin IIA [11]. The completion of palatogenesis by 12 weeks of gestation (E17 in mice) results in the division of the oral cavity into the oral cavity proper and the nasal cavity (Figure 3).
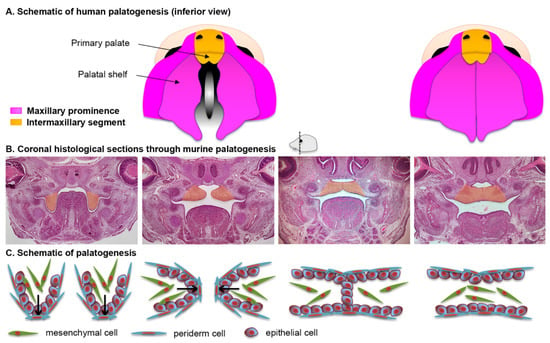
Figure 3.
Palatal morphogenesis. (A) Schematic of human palatogenesis from an inferior view. (B) Coronal sections of murine embryonic heads stained with hematoxylin and eosin. The palatal shelves, highlighted in orange, change from a vertical position (approximately E13.5) to a horizontal position (approximately E14); their epithelial cells adhere to eventually leave a confluent bridge of mesenchymal cells (approximately E15). (C) Schematic representation of the histological views in (B).
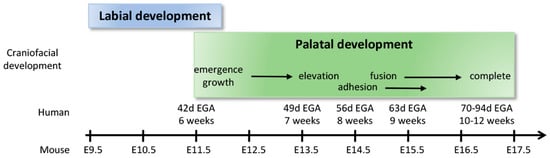
Figure 4.
Timeline correlation between human and murine labial and palatal development.
2.6. The Periderm
The periderm is a single layer of squamous cells that is considered to be the first stratification event of the epidermis around 55 d EGA. It is distinct from—and precedes—the later-developed process of normal epidermal differentiation, which involves distinct spinous, granular, and cornified layers. In mice, the formation of the periderm begins on the tips of the tail and distal limbs at E9 [12]. Periderm cells are distinct from basal cells from many perspectives: they exhibit an elongated cytoplasmic and nuclear shape, they are unable to retain [3H]thymidine for long periods of time (actively cycling), and they express keratin 6, 8, 17, 18, and 19, all characteristics of simple and glandular epithelia [13]. They are thought to originate from the basal layer by delamination as they lose contact with the underlying basement membrane [14]. These cells proceed to cover the facial structures by E10.5 and the intraoral structures soon after [12].
In the context of craniofacial development, the periderm is essential for palatogenesis at two critical developmental stages: First, as the palatine shelves initiate their vertical-to-horizontal transition, this single-cell layer acts as a nonstick barrier, preventing the developing palatine shelves from becoming adherent to the tongue, maxillary epithelium, or mandibular epithelium. Later, once the palatine shelves reorient to the horizontal position and make contact in the midline, the periderm needs to slough off for basal keratinocytes to adhere and fuse [15]. The failure of the periderm to form or function leads to craniofacial defects, discussed later.
3. Cellular Adhesions
3.1. Overview
Cellular adhesions are the main cellular structures that mediate the integrity of epithelial tissue and confer its barrier function. These cellular structures are present in most tissues, and may be classified according to (1) their ability to interact with the actin cytoskeleton or the intermediate filaments, or (2) whether the adhesions are formed between two neighboring cells or between a cell and the extracellular matrix. Cellular adhesions are highly dynamic and need to be modified and rearranged during morphogenic events such as embryonic development and tissue repair. Mutations impairing the assembly of adhesions or adhesion dynamics result in severe defects, frequently causing early embryonic death.
This section focuses on cell–cell adhesions. There are four main types of cell–cell adhesions (Figure 5). Tight junctions (TJs, also called zonula occludens) and adherens junctions (AJs, also called zonula adherens) link the actin cytoskeleton of neighboring cells. Desmosomes (also called macula adherens), however, link the intermediate filaments of neighboring cells. Finally, gap junctions are clusters of channels that form tunnels between two neighboring cells.
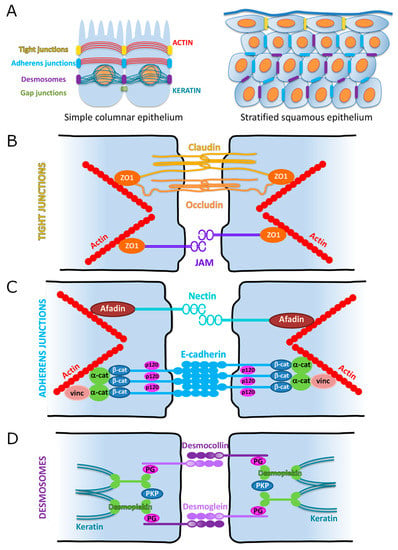
Figure 5.
Schematic representation of cell–cell adhesion junctions in simple columnar and stratified epithelia. (A) Classic organization of cell–cell adhesions between two epithelial cells of a simple columnar epithelium (left). Note the apical localization of the tight junctions, superficial to the adherens junctions. Both of these junctions connect to the actin cytoskeleton. Desmosomes are more baso-lateral and connect to the keratin intermediate filaments. This organization is preserved in a stratified epithelium (right), with desmosomes and adherens junctions mainly in the basal and suprabasal layers, while the tight junctions are exclusively found in the uppermost layers. (B) Schematic of tight junctions. (C) Schematic of adherens junctions. α-cat = alpha-catenin; β-cat = beta-catenin; p-120 = p120-catenin; and vinc = vinculin. (D) Schematic of desmosomes. PG = plakoglobin; PKP = plakophilin.
Where are these adhesion complexes located in epithelial cells? The classic model uses intestinal or renal cells of a simple columnar epithelium as the standard. In this model the TJs are always apico-lateral and the AJs alternate with desmosomes in a more baso-lateral pattern. However, this organization only applies to simple epithelia. In stratified epithelia, such as the lining of the oral cavity or the epidermis, the organization of these junctions has to take into account the presence of multilayered epithelial cells. With this in mind, one can think of the entire stratified epithelium as a simple intestinal cell with TJs stitching upper suprabasal cells together, in addition to AJs and desmosomes connecting basal and lower suprabasal cells (Figure 5A).
3.2. Adherens Junctions (AJs)
AJs are one of the best characterized types of cell–cell adhesions. They are comprised of two transmembrane proteins: cadherins and nectins (Figure 5C). Through the extracellular regions of these proteins, they link the actin cytoskeleton of neighboring cells. As such, they confer to cells their ability to sense, generate forces, and become de facto mechanotransducers [16]. Through their intracellular regions they interact with numerous proteins to regulate signaling events, ultimately leading to the regulation of gene expression.
3.2.1. Cadherin-Based Adhesions
Cadherins are transmembrane glycoproteins of 750–900 amino acids that mediate cell–cell adhesion in a homotypic and calcium-dependent manner. The family of classical cadherins includes more than 20 different types, of which epithelial cadherin (E-cadherin) is the most studied, as well as being the focus of this review. As with all classical cadherins its structure comprises extracellular, transmembrane, and intracellular domains [16,17,18,19,20,21]. The large extracellular domain contains five subdomains of approximately 100 residues each. Biochemical studies with Xenopus cadherin indicate that cadherins first form lateral cis-dimers with cadherins of the same cell [22]. Upon binding to three ions of calcium, these dimers then form homodimers (trans) with cadherins of a neighboring cell [23]. The region responsible for this homophilic recognition is located at the carboxyl terminal end of the first extracellular subdomain [24]. The intracellular N-terminus domain contains approximately 150 residues and is necessary for the adhesion function of E-cadherin [25,26]. The molecules recruited to interact with the cytoplasmic tail are β-catenin and p120-catenin, two molecules belonging to the family of armadillo proteins. The interaction of E-cadherin and β-catenin maintains β-catenin in the cytoplasm, where it activates its non-canonical Wnt-mediated pathway. The interaction of E-cadherin and p120-catenin, however, maintains E-cadherin at the cell membrane by preventing the phosphorylation of key tyrosine residues on the cytoplasmic tail of E-cadherin, which would otherwise send it to recycling endosomes [27,28]. E-cadherin does not directly bind the actin cytoskeleton. Instead, actin filaments bind directly to α-catenin (which can bind to β-catenin) and interact with β-catenin-associated proteins, such as vinculin, to reorganize both the cytoskeleton and the AJ complex (Figure 5C).
E-cadherin is an essential molecule in many morphogenetic processes that take place during embryonic development. It is the first cadherin to be expressed during development, where it participates in the compaction of the eight-cell-stage embryo [29]. Murine embryos lacking the Cdh1 gene (encoding the E-cadherin protein) do not develop past embryonic day four due to their inability to form trophectodermal epithelium [30]. The transcriptional regulation of E-cadherin is multifactorial, with transcription factors such as RB, c-Myc, and AP-2 promoting its transcription, and Smad interacting protein 1 (SIP1), E12/E47, and members of the Snail family inhibiting its transcription [31,32]. This tight regulation is necessary to control the formation of adhesion complexes [33].
In addition to transcriptional regulation of E-cadherin, its post-translational modifications also affect the function of AJs. Much of the existing data suggest that these modifications occur primarily via the phosphorylation and dephosphorylation of tyrosine residues [34]. The phosphorylation of tyrosine residues results in the loss of cell contacts [35], while the phosphorylation of serine residues by casein kinase II increases the interaction with β-catenin thus maintaining cell contacts [36]. Additional post-translational modifications of β-catenin also affect AJ function: phosphorylation by the activation of Src kinase or the EGF receptor prevents adhesions [37,38,39], whereas its dephosphorylation by leukocyte common antigen-related protein (LAR) or protein tyrosine phosphatase (PTP) increases cell–cell adhesion and prevents cell migration [40,41]. Lastly, cadherin clustering stimulates Rho GTPases [42] to promote AJ dynamics.
3.2.2. Nectin-Based Adhesions
The nectin family contains four members, nectin-1–4 [43,44,45,46], each with cell- and tissue-specific alternative splicing. Nectin-based adhesions, similar to cadherin-based adhesions, are formed by cis- and trans-interactions with dimers of the same (homo) or different (hetero) member of the family [45,47,48]. Nectins contain immunoglobulin-like extracellular loops that are necessary for their dimerization. Their cytoplasmic C-terminal domain contains a PDZ binding motif that is required to bind afadin, an actin-binding protein that links nectins to the actin cytoskeleton [45,46,48].
Contrary to cadherin-based adhesions, nectins do not require calcium to engage in adhesions [45,48]. As such, nectins are nascent adhesions (likely via the proximity of neighboring cells) that subsequently facilitate the recruitment of proteins required for the assembly of other types of cell–cell adhesions, i.e., cadherin-based adhesions.
3.3. Tight Junctions (TJs)
Tight junctions are the most intimate contacts formed between cells (Figure 5A,B). They prevent the diffusion of molecules between adjacent cells and the lateral migration of membrane proteins and lipids. The first protein identified as being associated with TJs was Zonula Occludens 1 (ZO-1) [49,50]. Since then, more than 30 proteins have been characterized in association with these junctions [51]. Typically, a tight junction is composed of a transmembrane protein (i.e., claudin, occludin, or a junctional adhesion molecule) and a cytoplasmic protein linked to the actin cytoskeleton (i.e., ZO-1). These proteins interact in the intercellular space with peer proteins from adjacent cells in a homophilic or heterophilic fashion (Figure 5B) [52,53,54].
3.3.1. Claudins
Claudins are the most important component of the tight junctions. They have four hydrophobic transmembrane domains, two extracellular loops, and two cytoplasmic domains that correspond to their amino- and carboxy-terminal ends (Figure 5B). Extracellular loops are critical for homophilic and heterophilic interactions, as well as for the formation of selective ion channels. The C-terminal domain is necessary for the localization of the claudins to the TJs. It contains a PDZ motif through which it binds scaffolding proteins containing PDZ binding domains (i.e., ZO-1) [55,56,57]. Distinct claudins and associated PDZ proteins form different tight junctions, which determine their permeability [58]. In fact, up to 24 claudin isoforms have been identified in humans [59], with claudin-1 and -2 constituting the main components of the TJs [56].
3.3.2. Occludin
Occludin is an enzyme (EC1.6) that contributes to the function of the epithelial barrier. Together with claudins, it is a major component of TJs (Figure 5B). Contrary to claudins, however, occludin is dispensable for the assembly of TJs. Occludin has four transmembrane domains, two extracellular loops, and two cytoplasmic ends. This molecule regulates selective paracellular permeability. The intracellular C-terminus interacts with ZO-1 via the PDZ binding domain and links occludin to the actin cytoskeleton. As a result of alternative splicing several isoforms have been identified, each of which has a unique cellular distribution and interaction with other molecules [60].
3.3.3. Zonula Occludens
Many molecules localize to the intracellular region of tight junctions to modulate and stabilize this type of adhesion. Amongst them, Zonula Occludens (ZO) 1, ZO2, and ZO3 are the main components of this intracellular network. These three proteins co-immunoprecipitate with each other and share a similar structure and functional domains. However, they are distinct due to their unique C-terminal regions, which define their specific functions [33,61,62]. ZO1 is usually considered the central molecule responsible for scaffolding and organizing the cytoplasmic events associated with TJs. ZO1 is a 220 kDa protein containing multiple functional domains, among which are PSD95, DlgA, ZO1 homology (PDZ) domains, SRC homology 3 (SH3) domains, and a guanylate kinase homology (GUK) domain. These domains define the specific functions of ZO1. For example, the PDZ, GUK, and C-terminal domains mediate the interaction with claudin, JAMs, occluding, and F-actin, respectively [63,64]. Alternative splicing confers tissue specificity of these components [61].
3.3.4. Junctional Adhesion Molecules (JAMs)
Junctional adhesion molecules (JAMs) are proteins that belong to the superfamily of immunoglobulins. They are subdivided into four types: JAM-A, JAM-B, JAM-C, and JAM-L/JAM4 [65]. From a structural perspective, JAMs have an extracellular domain, a single transmembrane domain, and a cytoplasmic domain. The extracellular domain plays an important regulatory role by being able to bind multiple ligands, which have been proposed to regulate cellular functions and paracellular permeability [66]. The C-terminal region contains a PDZ binding domain (except for JAM-L/JAM4). This domain serves as an anchor to the actin cytoskeleton and to many TJ-associated scaffolding molecules, including ZO1 and the polarity protein Par3. These interactions are necessary for the proper function of the TJs [67,68]. JAM-A is necessary for the formation and assembly of TJs in epithelial cells in a homophilic pattern [69]; however, other studies have shown heterophilic interactions among the JAM family members as well as with other components of adhesion complexes [70,71].
3.4. Desmosomes
In 1920 Josef Schaffer coined the term “Desmosome”. It has its origins in the Greek words for bond (desmo) and body (soma). Also called macula adherens (Latin for “adherent spot”), a desmosome is a “binding body”, or a cell structure specialized for cell-to-cell adhesion randomly arranged along the plasma membrane. Desmosomes are one of the strongest cell adhesion types and are found in tissue that experience high mechanical stress, such as epithelia and cardiac muscle. Desmosomes share some structural organization with AJs and TJs: a transmembrane region formed by desmosomal cadherins (desmoglein and desmocollin) that interact in the intercellular space with neighboring cells, and a series of cytoplasmic adaptor proteins that link the cadherins to the cytoskeleton. Desmosomes are also quite distinct from other adhesion complexes. First, desmosomes link the cytoskeletal intermediate filaments (i.e., keratins in epithelia) of adjacent cells, as opposed to the actin filaments. Second, their cytoplasmic proteins are organized in two plaques: the outer dense plaque, where the N-terminus domain of desmoplakin binds the desmosomal cadherins, and the inner dense plaque, where the C-terminus domain of desmoplakin binds the intermediate filaments (inner dense; Figure 5D) [72,73,74]. The presence of desmoplakin as an intermediate between the cadherins and the cytoskeleton is likely to be responsible for the increased strength of the desmosomal adhesion.
The molecular composition of desmosomes is variable, showing tissue- and cell-specific isoforms, conferring functional specificity based on the physiological context [75]. As with AJs, extracellular calcium promotes the assembly of desmosomal proteins [76,77], a characteristic that has been extensively used in keratinocyte cultures to study their dynamics [78].
3.5. Desmosomal Cadherins (Desmogleins, Desmocollins)
Desmosomal cadherins belong to the superfamily of cadherins. They contain five extracellular domains and have calcium binding motifs. In the presence of calcium the extracellular domains become rigid and promote adhesion [79,80]. The cytoplasmic tail of these cadherins bind to plakoglobin and plakophilin, members of the armadillo protein family [81]. To date, four desmoglein isoforms and three desmocollin isoforms have been reported. In stratified epithelia, some of these isoforms are ubiquitously detected throughout the tissue (desmoglein 1, desmocollin 2), whereas others show a more restricted localization to specific cell layers (desmoglein 2–4, desmocollin 1, 3, and 4) [82,83]
3.6. Desmosomal Armadillo Proteins (Plakophilin, Plakoglobin)
Armadillo proteins are characterized by a central domain containing α-helix-forming repeated units of ~42 amino acids, initially characterized in the Drosophila segment polarity protein armadillo [84,85,86]. Armadillo repeat units fold together as a superhelix, thus creating a functionally rich region able to interact with many protein partners that contribute to the diverse range of functions associated with this family of proteins. In the desmosomes, plakophilin-1–3 and plakoglobin (also named γ-catenin) anchor the cytoplasmic tail of desmosomal cadherins to desmoplakin in the outer dense plaque. In contrast to cadherins, their tissue distribution is uniform throughout stratified epithelia.
3.7. Desmoplakin
Desmoplakin is the most abundant and essential component of desmosomes [87]. It belongs to the family of plakins and binds to intermediate filaments in the inner dense plaque [88,89]. Its N-terminal globular head domain is required for both the localization to the desmosome and the interaction with armadillo proteins and desmosomal cadherins [88]. The C-terminal region is composed of three plakin repeats essential for binding to the intermediate filaments [88]. Serine phosphorylation of the C-terminal region inhibits interaction with the intermediate filaments, ultimately disassembling the adhesion complex [90,91].
4. Contribution of Cellular Adhesion to Craniofacial Morphogenesis
Because of their critical role in the assembly of individual cells into three-dimensional tissues, cellular adhesions contribute to every step of craniofacial morphogenesis. This includes the formation and delamination of neural crest cells and their migration into pharyngeal arches, the formation and expansion of facial processes, and the fusion of these processes to form the face. We will limit our review to the later stages of facial morphogenesis with an emphasis on the formation of the lip and the palate. We will first review the contribution of the cytoskeleton and then take a more systemic approach by evaluating the role of adhesion molecules in facial morphogenesis. We will learn from human genetic studies of individuals with cleft lip with or without cleft palate (CL/P) and then from animal models to highlight the contribution of cellular adhesion to facial morphogenesis. A summary of these intersecting genes is presented in Figure 6 and Table 2.
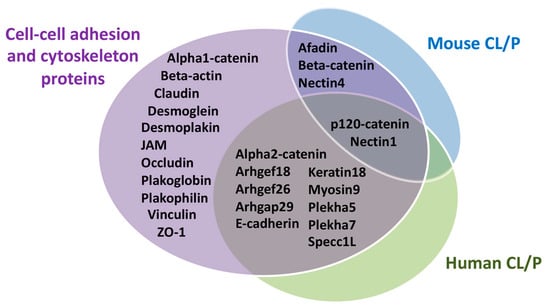
Figure 6.
Summary of cell–cell adhesion and the cytoskeletal proteins involved in human and mouse cleft lip with or without palate. Adhesion molecules discussed in this review were distributed on the basis of whether they played a role in human cleft lip with or without palate (CL/P), mouse CL/P, both, or had no identified role in craniofacial morphogenesis.
4.1. Cytoskeleton in Craniofacial Morphogenesis
The remodeling of the cytoskeleton is essential during morphogenesis. During the formation of the lip and the palate, two main cell types undergo dynamic remodeling: ectodermal-derived epithelial cells and neural-crest-derived mesenchymal cells. Amongst the filaments constituting their cytoskeleton, both populations contain actin and microtubules. However, their intermediate filaments are distinct, with epithelial cells characterized by keratin filaments while mesenchymal cells contain vimentin. Although genetic variants in the ACTIN gene are not associated with orofacial clefting, early studies demonstrated the active production of actin in palatal shelves as they transition from a vertical to a horizontal position [92], supporting the dynamic remodeling of the cytoskeleton. During that process mesenchymal cells align their actin cytoskeleton, further demonstrating an actin-dependent cell contractility that drives palatal shelf elevation [93]. Actomyosin contractility is also required during the fusion of the secondary palatal shelves, as cells from the medial epithelial seam intercalate to form a single-layered epithelium concomitantly with epithelial cell extrusion [11]. Not surprisingly, genetic variants in the MYOSIN9 gene have been reported in the study of families with orofacial clefts and found to be associated with an increased risk for the defect [94,95,96]. Novel variants in the KRT18 gene have also been shown to contribute to non-syndromic CL/P (NSCL/P) [97]. Although keratins can remodel the cytoskeleton via desmosomal adhesions, the association of KRT18 to NSCL/P may be more related to its putative function in the periderm [98] than to a role in remodeling the cytoskeleton.

Table 2.
Summary comparison of cell adhesion molecules, cleft lip with or without palate in human, and murine knockout phenotypes.
Table 2.
Summary comparison of cell adhesion molecules, cleft lip with or without palate in human, and murine knockout phenotypes.
| Gene (Protein) | Type of Human Craniofacial Clefts | Human Variants Associated with the CL/P Phenotype | Mouse Knockouts Craniofacial Phenotype |
|---|---|---|---|
| AFDN (Afadin) | No CL/P | KO embryonic lethal [99] K14-Cre cKO no CP [100] In utero cKO CP and oral adhesions [100] | |
| CTNNA2 (Alpha2-catenin) | NSCL/P [101] | g.82025185 | None |
| ARHGEF18 (Arhgef18) | NSCL/P [102] | c.1484G>A p.Arg495Gln | None |
| ARHGEF26 (Arhgef26) | NSCL/P [103] | g.153840512:A>T g.153943770:C>G | None |
| ARHGAP29 (Arhgap29) | NSCL/P [104,105,106,107] | c.62_63delCT p.Ser21Tyrfs*20 c.91C>T p.Leu31Phe c.698-1G>C c.888G>C p.Arg296Ser c.976A>T p.Lys326* c.1252G>A p.Val418Ile c.1475C>A p.Ser492* c.1576+1G>A c.1654T>C p.Ser552Pro c.1847G>A p.Arg616His c.1865C>T p.Thr622Met c.2017T>G p.Phe673Val c.2109+1G>A c.2393G>A p.Arg798Gln c.2533A>G p.Ile845Val c.2617C>T p.Arg873Cys c.3023G>A. p.(Arg1008Lys) c.3326_3328delCAA p.Thr1109del c.3339T>G p.Ile1113Met g.94545160:T>C g.94547883:C>G g.94547889:G>A | KO embryonic lethal [108] Heterozygote no CP but oral adhesion [108] |
| CTNNB1 (Beta-catenin) | No CL/P [109] | cKO CP [110] | |
| CTNND1 (p120-catenin) | Syndromic CL/P [111] NSCL/P [112,113] | c.606_627del p.Pro203Leufs*25 c.1093C>T p.Gln365* c.2098C>T p.Arg700* g.57559005:C>G p.Gln19Glu g.57564445_5756446del p.Asp313Profs*9 g.57569255:G>A p.Trp336* g.57571168:A>G p.Asp499Gly g.57572202:C>T p.Leu558Phe g.57573381:C>T p.Arg584Trp g57575761:G>T p.Try690Cys g.57576939:G>T c.2417+1G>T g.57578892:C>T p.Arg852* c.1381C>T p.Arg461* c1481_1485del p.Leu494Argfs*5 c.2598_2601dupTGAT p.Ser868* c.2737dupC p.His913Profs*3 | CreCT cKO 47% CP [112] |
| CDH1 (E-cadherin) | Syndromic CL/P [111] NSCL/P [112,114,115,116,117,118] | c.760G>T p.Asp254Tyr c.770A>T p.Asp257Val c.1320G>T p. ? c.1320+1G>C p. ? c.1361_1363del p.Val454del c.387+5G>A p.? c.468G>C p.Trp156Cys c.752C>T p.Thr251Met c.760G>A p.Asp254Asn c.768T>A p.Asn256Lys c.1023T>G p.Tyr341* c.1489G>A p.Glu497Lys c.1766A>T p.Asn589Ile c.2351G>A p.Arg784His c.2426_2427del p.Asn809Ilefs*3 | KO embryonic lethal [119] Wnt1-Cre cKO no cleft [120] K14-Cre cKO no cleft [121] |
| KRT18 (Keratin18) | NSCL/P [97] | g.53344318:G>T | None |
| MYOSIN9 (Myosin9) | NSCL/P [94,95,96] | g.35044605:C>T g.35048804:C>T g.35007860:T>C | None |
| PVRL1 (Nectin1) | Syndromic CL/P [122] | p.Tryp185X | KO no CL/P [123,124] In utero cKO CP [100] |
| PVRL4 (Nectin4) | No CL/P [125] | KO 11–40% CP [100] | |
| PLEKHA5 (Plekha5) | NSCL/P [112] | g.19440414:A>G p.Tyr590Cys | None |
| PLEKHA7 (Plekha7) | NSCL/P [112] | g.16838582:C>T p.Gly544Asp g.16838676:G>A p.Arg513Trp g.16834682:T>C p.Asp662Gly | None |
| SPECC1L (Specc1L) | Syndromic CL/P [126] NSCL/P [127] | c.569C>T p.Thr190Met c.1244A>C p.Gln415Pro c.273G>A p.Met91Iso c.256G>A p.Ala86Thr c.895A>G p.Thr299Ala c.1637G>A p.Arg546Gln | KO embryonic lethal [127] In-frame deletion CP [128] |
4.2. Role of Adherens Junctions in Facial Morphogenesis
4.2.1. Cadherin-Based Adhesions: Lessons from Patients
One of the most important components of AJs is E-cadherin, encoded by the CDH1 gene. Missense mutations and rare variants in CDH1 were identified in patients with non-syndromic CL/P [112,114,115,116,117], patients with hereditary diffuse gastric cancer and CL/P [118], as well as in one of the syndromic forms of CL/P (blepharocheilodontic syndrome, BCDS [111]). Interestingly, most pathogenic CL/P variants cluster in the linker regions between the extracellular domains of E-cadherin [118], likely affecting its chelation to calcium and altering its function.
In the cytoplasm, E-cadherin interacts with beta-catenin (encoded by the CTNNB1 gene), which is also a major player in the Wnt signaling pathway. De novo nonsense and frameshift mutations in CTNNB1 were reported in patients with craniofacial phenotypes, but they did not present with CL/P [109], suggesting that in humans mutations in CTNNB1 are not directly associated with CL/P. However, many other members of the Wnt signaling pathway have been associated with CL/P, including numerous Wnt ligands, Wnt receptors (Frizzled), and co-receptor ROR2 (for a full review of the Wnt signaling contribution to orofacial clefting, see [129]).
Two other catenins are part of AJs: p120-catenin (encoded by the CTNND1 gene), which anchors E-cadherin at the plasma membrane, and α-catenin (encoded by the CTNNA1 gene), which mediates forces and interacts with the actin cytoskeleton. Genetic variants in CTNND1 cause blepharocheilodontic syndrome [111] and increase the risk for non-syndromic CL/P [112,113]. Novel protein-truncating variants and de novo variants were also identified in families with craniofacial dysmorphisms and cardiac, limb, and neurodevelopmental anomalies [113]. Some of these variants affected the binding of p120-catenin to E-cadherin [112,113], suggesting an alteration in the AJ complex and cellular adhesions in these individuals. To the best of our knowledge, no genetic variants in CTNNA1 have so far been reported in association with CL/P. However, variants in CTNNA2 were identified in consanguineous families with NSCL/P [101]. This isoform is typically detected in the brain, reinforcing previous evidence of a correlation between neuronal and craniofacial defects [130,131].
4.2.2. Cadherin-Based Adhesions: Lessons from Murine Models
Because of its prominent role in cellular adhesion, animals deficient in E-cadherin do not undergo morphogenesis and are embryonic lethal [119]. However, animals with a conditional knockout of E-cadherin in neural crest cells survived embryonic development and exhibited craniofacial defects related to skeletal development, but no cleft palate [120]. Similarly, cleft palate was not observed when E-cadherin was ablated in epithelial cells using a keratin 14-cre driver [121]. Conditional ablation of p120-catenin using an ectoderm-specific Cre driver (CreCT) resulted in 47% of animals with overt clefts, while the remaining animals exhibited delays in medial growth of the maxillary labial processes [112]. Conditional knockout and conditional gain of function of β-catenin in palatal epithelial cells, however, led to cleft palate [110]. The higher penetrance of a cleft palate phenotype in animals with altered levels of β-catenin may be due not to its role as a component of the AJ complex (non-canonical) but as a transcription factor (canonical) with many downstream effectors, a property not associated with p120-catenin or E-cadherin.
4.2.3. Nectin-Based Adhesions: Lessons from Patients
Although nectin-based adhesions have not been extensively studied compared to AJs or TJs, mutations in both NECTIN1 (encoded by the PVRL1 gene) and NECTIN4 (encoded by the PVRL4 gene) cause syndromes involving orofacial clefts (NECTIN1) or phenotypes previously associated with clefting (NECTIN4), supporting their critical role in facial morphogenesis. PVRL1 was identified as the gene responsible for the autosomal recessive CLP-ectodermal dysplasia (ED) syndrome (CLPED1, previously ED4), characterized by CL/P, hidrotic ED, developmental defects in the hands, and occasional mental retardation [122]. Homozygous nonsense (W185X) and frameshift mutations identified in patients truncate the nectin-1 protein and abolish its interaction with afadin, likely abolishing the calcium-independent cell–cell adhesion complex [122]. Mutations in PVRL4 cause ED syndactyly syndrome 1 (EDSS1, previously ED1), a syndrome similar to CLPED1 yet distinct because of a lack of orofacial defects [125]. These mutations affect different protein interaction domains, including the afadin binding site [125]. To date, no human genetic variant in the AFADIN gene has been reported to be associated with orofacial clefting; however, only 25% of clefting heritability is currently known [132].
4.2.4. Nectin-Based Adhesions: Lessons from Murine Models
Despite the clear evidence that mutations in PVRL1 cause a syndromic form of CL/P in humans, murine models have failed to recapitulate this phenotype. In fact, no CL/P was observed in mice homozygous null for Nectin-1 or in compound mutants where Nectin-1 and -3 were deleted [123,124]. Recently, Lough et al. revisited the loss of nectin-1 and nectin-4 in mice. Rather than deleting these alleles using homologous recombination or tissue-specific Cre drivers, the investigators took advantage of an in utero lentiviral-mediated genetic approach to knockdown these proteins [100]. The loss of Nectin-1 and the loss of Nectin-4 led to cleft palate with a penetrance of 11–40%. Interestingly, synergistic dual loss of Nectin-1 and Nectin-4 caused highly penetrant cleft palate, with oral adhesions between the palatal shelves and the tongue due to the partial loss of periderm cells [100]. Because afadin is an obligate binding partner of nectin function [133], one would expect orofacial clefting in an afadin loss-of-function model. Germline homozygous null mice were early embryonic lethal [99], preventing the evaluation of afadin’s role in facial morphogenesis, while the epithelial-specific loss of afadin (using a K14-Cre driver) showed proper palatogenesis [100]. However, lentiviral-mediated delivery of the Cre recombinase a few days earlier than the onset of the K14-Cre driver led to the failure of palatal shelf elevation and fusion due to intraoral adhesions [100]. Collectively, these studies are important for two reasons: First, they highlight the importance of the timing of the delivery of the Cre recombinase in tissue-specific knockout and warrant the potential re-examination of mutants where the phenotype of the murine model did not phenocopy the human phenotype. Second, they reinforce the role of the periderm in palatogenesis. Amongst the different nectins, nectin-4 is specific to the periderm. Its loss, or the loss of its binding partner, afadin, showed strong intraoral adhesions, further supporting the theory of a role for nectin-based adhesions in facial morphogenesis.
4.3. Role of Other Adhesion Components in Facial Morphogenesis
4.3.1. Lessons from Patients
Despite the fact that TJs anchor to the actin cytoskeleton and that actin cytoskeleton remodeling is necessary for facial morphogenesis, to the best of our knowledge no human genetic variants in any of the proteins comprising the TJ complex have been associated with CL/P. Similarly, none of the desmosomal proteins, critical for remodeling the keratin of epithelial cells, have been associated with CL/P. The reason for this lack of association in not clear, but one could speculate that the role of TJs in barrier formation is so critical that a small defect would be incompatible with life. As for a lack of desmosomal contribution to facial morphogenesis, we may want to consider an evolutionary perspective. Desmosomes are sophisticated structures that appeared with the appearance of stratified epithelia [83]. Palatogenesis occurs at a time when the oral epithelium resembles a simple epithelial sheet, in which the main form of cell-to-cell junctions is AJs. The appearance of desmosomes would occur after the palate has formed, potentially diminishing the role of desmosomes in this morphogenic event.
4.3.2. Lessons from Murine Models
We investigated the Mouse Genome Informatics database for “craniofacial phenotype”, “abnormal craniofacial development”, and “abnormal palate development”. From the 192 genotypes corresponding to these phenotypes, none were deficient in desmosomal or tight junction proteins.
4.4. Other Molecules Modulating Cell–Cell Junctions Critical for Facial Morphogenesis
4.4.1. SPECC1L and Pleckstrin
Amongst the numerous proteins that interact with actin and microtubules, three showed an association with human CL/P. The first is SPECC1L (sperm antigen with calponin homology and coiled-coil domains 1 like). Mutations in this gene are responsible for oblique facial clefts [126], a rare form of orofacial clefts [134], and are clustered in the second coiled-coil and calponin homology domains. Human genetic variants outside of the coiled-coil domain were also reported in individuals with non-syndromic CL/P and resulted in milder functional defects than those of variants associated with syndromic clefts [127]. The loss of SPECC1L in mice resulted in early embryonic lethality. However, mice homozygous for a truncation allele showed an altered palatal rugae phenotype with oral adhesions and a reduction in interferon regulatory factor 6 (IRF6) in these structures, but no cleft palate [127]. Homozygotes for in-frame deletions in the second coiled-coil domain resulted in exencephaly, cleft palate, and ventral body wall closure defects [128], demonstrating the essential functional role of the coiled-coil domain. In vitro, functional studies demonstrated that SPECC1L colocalized with microtubules and filamentous actin in palatal mesenchymal cells [126]. Deletions or mutations in the coiled-coil domain severely affected its ability to associate with microtubules, reducing its ability to traffic in the cells and resulting in a perinuclear accumulation. This uneven distribution of SPECC1L in the cells resulted in increased actin and non-muscle myosin II bundles at the cell periphery [128]. Ultimately, the disrupted actomyosin cytoskeleton would affect cell alignment and coordinated movement required for elevation and fusion of the palatal shelves.
The other two cleft- and microtubule-associated proteins are pleckstrin-homology-domain-containing protein 5 and 7 (PLEKHA5 and PLEKHA7). These molecules are cytoplasmic accessory members of AJs. Although very little is known about the function of PLEKHA5, studies show that PLEKHA7 contributes to the integrity of AJs by linking the E-cadherin/p120 catenin complex to the minus end of microtubules [135]. It also stabilizes nectin-based junctions by recruiting the PDZD11 protein to the adhesion complex. Although mice deficient in Plekha7 exhibited normal gross morphology, including craniofacial appearance, genetic variants in both PLEKHA5 and PLEKHA7 were associated with CL/P in multiple populations [112]. Interestingly, genetic variants in PLEKHG3, another family member related to PLEKHA5 and PLEKHA7, were also reported in association with CL/P [103], further supporting the theory of a role for these cytoplasmic proteins in the cell–cell adhesion stability required for proper facial morphogenesis.
4.4.2. IRF6 and the Rho Pathway
In addition to molecules that directly participate in the formation of an adhesion complex, some transcription factors and growth factors are essential modulators of cellular adhesion and are involved in syndromic and non-syndromic orofacial clefts. One such transcription factor is interferon regulatory factor 6 (IRF6). Mutations in IRF6 cause Van der Woude syndrome, the most common syndromic form of CL/P, in which the vast majority of patients present with a cleft lip with or without cleft palate and lip pits in their lower lips [136]. Genetic variants in IRF6 also contribute to an increased risk for non-syndromic CL/P [137], making IRF6 one of the prominent contributors to CL/P. No patients have been reported with homozygous IRF6 mutations, likely because of the fact that the loss of IRF6 would be incompatible with life, as demonstrated by animal models. Indeed, IRF6-deficient mice die postnatally from dehydration and an inability to suckle, due to the absence of an epidermal barrier and a fused oral cavity [138]. Further investigation into the cause of oral fusion demonstrated that IRF6-deficient embryos lack an oral periderm, allowing abnormal tissue adhesions between the tongue and palatal shelves or between two opposing maxillary and mandibular oral epithelia [12,138]. In the absence of IRF6, E-cadherin, which is normally restricted to the lateral and basal sides of the most superficial layers of oral epithelial cells, was detected on the apical side. This ectopic localization was due in part to altered tight junctions, and promoted the formation of adhesion complexes with opposing epithelial cells [12]. In vitro, our recent studies indicate that IRF6 regulates the delivery of E-cadherin to the plasma membrane [139]. How IRF6 participates in E-cadherin turnover is not fully elucidated, although its association with NME1/2 could contribute to adhesion protein trafficking or recycling to the plasma membrane [112]. Other adhesion molecules were reduced at the plasma membrane of IRF6-deficient keratinocytes, supporting a more global effect of IRF6 on cellular adhesions.
In addition to its role in tissue and cellular adhesions, IRF6 inhibits the small GTPase RhoA [140], a major signaling node that influences cytoskeleton changes downstream of cell adhesions [141,142]. RhoA cycles between an active-GTP bound (activated by guanine nucleotide exchange factors, GEFs) and an inactive-GDP bound form (inactivated by RhoA GTPase-activating proteins, GAPs). In the absence of IRF6, keratinocytes exhibited increased stress fibers and reduced migration [140]. During this process we found that one of the GAPs, namely ARHGAP29, was downregulated [140]. This is significant because human genetic variants in ARHGAP29 have been associated with CL/P in numerous populations [104,105,106,107]. In addition, although ARHGAP29 homozygous null embryos die early during embryogenesis before facial morphogenesis, mice heterozygous for a patient-derived mutation knock-in allele presented with abnormal oral epithelial adhesions [108]. ARHGAP29 also plays a role in the migration of endothelial cells by binding afadin to regulate Rho-associated kinase activity [143]. Collectively, these studies highlight a critical role for IRF6 in regulating the actin cytoskeleton by modulating RhoA activity during craniofacial morphogenesis. Other GAPs and GEFs, in addition to ARHGAP29, have been associated with CL/P in humans. Variants reported in ARHGEF18 [102] and ARHGEF26 [103] reinforce the fundamental role of actin cytoskeletal remodeling during craniofacial development.
5. Conclusions
Cells are the base units of all organisms. They touch, adhere, and engage their cytoskeletons in response to each other’s shape. Cells also move, both alone and collectively, following cues to ultimately form structures. These cues vary in their nature, ranging from a leader cell who is secreting growth factors to the stiffness of extracellular matrices. They also include forces exerted by neighboring cells and sensed by target cells via adhesion receptors. By reviewing the evidence linking gene regulatory networks involved in cellular adhesions with orofacial morphogenesis, it becomes clear that a more integrated approach that includes biomechanical processes—mediated in part by cellular contacts—will be essential to further our understanding of in vivo embryonic development. This will require the generation of new tools to measure forces in vivo as embryos develop in addition to improved live imaging to track cells as they move within increasingly complex structures.
Author Contributions
Conceptualization: A.A., B.J.P. and M.D. Project Administration: M.D. Writing—Original Draft Preparation, Review and Editing: A.A., B.J.P. and M.D. Visualization: M.D. Funding Acquisition: M.D. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by grants from the American Association for Anatomy (FGAP) and the National Institute of Health (R01-AR067739 to M.D. and F31-AR078659 to A.A.).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
The authors would like to thank all the current and past members of the Dunnwald lab for critically reviewing the manuscript. A special thank you to Lindsey Rhea and Darren Hoffmann for illustration suggestions.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Chuai, M.; Weijer, C.J. The mechanisms underlying primitive streak formation in the chick embryo. Curr. Top. Dev. Biol. 2008, 81, 135–156. [Google Scholar]
- Kousa, Y.A.; Mansour, T.A.; Seada, H.; Matoo, S.; Schutte, B.C. Shared molecular networks in orofacial and neural tube development. Birth Defects Res. 2017, 109, 169–179. [Google Scholar] [CrossRef]
- Munoz, W.A.; Trainor, P.A. Neural crest cell evolution: How and when did a neural crest cell become a neural crest cell. Curr. Top. Dev. Biol. 2015, 111, 3–26. [Google Scholar]
- Minoux, M.; Rijli, F.M. Molecular mechanisms of cranial neural crest cell migration and patterning in craniofacial development. Development 2010, 137, 2605–2621. [Google Scholar] [CrossRef]
- Simoes-Costa, M.; Bronner, M.E. Establishing neural crest identity: A gene regulatory recipe. Development 2015, 142, 242–257. [Google Scholar] [CrossRef]
- Santagati, F.; Rijli, F.M. Cranial neural crest and the building of the vertebrate head. Nat. Rev. Neurosci. 2003, 4, 806–818. [Google Scholar] [CrossRef]
- Frisdal, A.; Trainor, P.A. Development and evolution of the pharyngeal apparatus. Wiley Interdiscip. Rev. Dev. Biol. 2014, 3, 403–418. [Google Scholar] [CrossRef]
- Guthrie, S. Patterning and axon guidance of cranial motor neurons. Nat. Rev. Neurosci. 2007, 8, 859–871. [Google Scholar] [CrossRef]
- Diewert, V.M. A morphometric analysis of craniofacial growth and changes in spatial relations during secondary palatal development in human embryos and fetuses. Am. J. Anat. 1983, 167, 495–522. [Google Scholar] [CrossRef]
- Bush, J.O.; Jiang, R. Palatogenesis: Morphogenetic and molecular mechanisms of secondary palate development. Development 2012, 139, 231–243. [Google Scholar] [CrossRef]
- Kim, S.; Lewis, A.E.; Singh, V.; Ma, X.; Adelstein, R.; Bush, J.O. Convergence and extrusion are required for normal fusion of the mammalian secondary palate. PLoS Biol. 2015, 13, e1002122. [Google Scholar] [CrossRef] [PubMed]
- Richardson, R.J.; Ammond, N.L.; Coulombe, P.A.; Saloranta, C.; Nousiainen, H.O.; Salonen, R.; Berry, A.; Hanley, N.; Headon, D.; Karikoski, R.; et al. Periderm prevents pathological epithelial adhesions during embryogenesis. J. Clin. Investig. 2014, 124, 3891–3900. [Google Scholar] [CrossRef] [PubMed]
- Holbrook, K.A. Structure and Function of the Developing Human Skin. In Physiology, Biochemistry, and Molecular Biology of the Skin; Goldsmith, L.A., Ed.; Oxford University Press: New York, NY, USA, 1991; pp. 63–110. [Google Scholar]
- McGowan, K.M.; Coulombe, P.A. Onset of keratin 17 expression coincides with the definition of major epithelial lineages during skin development. J. Cell Biol. 1998, 143, 469–486. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Liu, J.; Li, Z.; Ozturk, F.; Gurumurthy, C.; Romano, R.A.; Sinha, S.; Nawshad, A. TGFbeta3 regulates periderm removal through DeltaNp63 in the developing palate. J. Cell. Physiol. 2015, 230, 1212–1225. [Google Scholar] [CrossRef] [PubMed]
- Nagafuchi, A.; Takeichi, M. Cell binding function of E-cadherin is regulated by the cytoplasmic domain. EMBO J. 1988, 7, 3679–3684. [Google Scholar] [CrossRef]
- Danjo, Y.; Gipson, I.K. Actin ‘purse string’ filaments are anchored by E-cadherin-mediated adherens junctions at the leading edge of the epithelial wound, providing coordinated cell movement. J. Cell Sci. 1998, 111, 3323–3332. [Google Scholar] [CrossRef]
- Yulis, M.; Kusters, D.H.M.; Nusrat, A. Cadherins: Cellular adhesive molecules serving as signalling mediators. J. Physiol. 2018, 596, 3883–3898. [Google Scholar] [CrossRef]
- McCrea, P.D.; Gumbiner, B.M. Purification of a 92-kDa cytoplasmic protein tightly associated with the cell-cell adhesion molecule E-cadherin (uvomorulin). Characterization and extractability of the protein complex from the cell cytostructure. J. Biol. Chem. 1991, 266, 4514–4520. [Google Scholar] [CrossRef]
- Stappert, J.; Kemler, R. A short core region of E-cadherin is essential for catenin binding and is highly phosphorylated. Cell Adhes. Commun. 1994, 2, 319–327. [Google Scholar] [CrossRef]
- Su, W.; Kowalczyk, A.P. The VE-cadherin cytoplasmic domain undergoes proteolytic processing during endocytosis. Mol. Biol. Cell 2017, 28, 76–84. [Google Scholar] [CrossRef]
- Brieher, W.M.; Yap, A.S.; Gumbiner, B.M. Lateral dimerization is required for the homophilic binding activity of C-cadherin. J. Cell Biol. 1996, 135, 487–496. [Google Scholar] [CrossRef] [PubMed]
- Nagar, B.; Overduin, M.; Ikura, M.; Rini, J.M. Structural basis of calcium-induced E-cadherin rigidification and dimerization. Nature 1996, 380, 360–364. [Google Scholar] [CrossRef]
- Shapiro, L.; Kwong, P.D.; Fannon, A.M.; Colman, D.R.; Hendrickson, W.A. Considerations on the folding topology and evolutionary origin of cadherin domains. Proc. Natl. Acad. Sci. USA 1995, 92, 6793–6797. [Google Scholar] [CrossRef]
- Drees, F.; Pokutta, S.; Yamada, S.; Nelson, W.J.; Weis, W.I. Alpha-catenin is a molecular switch that binds E-cadherin-beta-catenin and regulates actin-filament assembly. Cell 2005, 123, 903–915. [Google Scholar] [CrossRef]
- Yamada, S.; Pokutta, S.; Drees, F.; Weis, W.I.; Nelson, W.J. Deconstructing the cadherin-catenin-actin complex. Cell 2005, 123, 889–901. [Google Scholar] [CrossRef] [PubMed]
- Davis, M.A.; Ireton, R.C.; Reynolds, A.B. A core function for p120-catenin in cadherin turnover. J. Cell Biol. 2003, 163, 525–534. [Google Scholar] [CrossRef]
- Shibamoto, S.; Hayakawa, M.; Takeuchi, K.; Hori, T.; Miyazawa, K.; Kitamura, N.; Johnson, K.R.; Wheelock, M.J.; Matsuyoshi, N.; Takeichi, M.; et al. Association of p120, a tyrosine kinase substrate, with E-cadherin/catenin complexes. J. Cell Biol. 1995, 128, 949–957. [Google Scholar] [CrossRef] [PubMed]
- Fleming, T.P.; Sheth, B.; Fesenko, I. Cell adhesion in the preimplantation mammalian embryo and its role in trophectoderm differentiation and blastocyst morphogenesis. Front. Biosci. 2001, 6, 1000–1007. [Google Scholar] [CrossRef]
- Larue, L.; Ohsugi, M.; Hirchenhain, J.; Kemler, R. E-cadherin null mutant embryos fail to form a trophectoderm epithelium. Proc. Natl. Acad. Sci. USA 1994, 91, 8263–8267. [Google Scholar] [CrossRef]
- Perez-Moreno, M.; Davis, M.A.; Wong, E.; Pasolli, H.A.; Reynolds, A.B. p120-catenin mediates inflammatory responses in the skin. Cell 2006, 124, 631–644. [Google Scholar] [CrossRef]
- Batsche, E.; Cremisi, C. Opposite transcriptional activity between the wild type c-myc gene coding for c-Myc1 and c-Myc2 proteins and c-Myc1 and c-Myc2 separately. Oncogene 1999, 18, 5662–5671. [Google Scholar] [CrossRef][Green Version]
- Gumbiner, B.M. Regulation of cadherin adhesive activity. J. Cell Biol. 2000, 148, 399–403. [Google Scholar] [CrossRef] [PubMed]
- Daniel, J.M.; Reynolds, A.B. Tyrosine phosphorylation and cadherin/catenin function. Bioessays 1997, 19, 883–891. [Google Scholar] [CrossRef] [PubMed]
- Fujita, Y.; Krause, G.; Scheffner, M.; Zechner, D.; Leddy, H.E.; Behrens, J.; Sommer, T.; Birchmeier, W. Hakai, a c-Cbl-like protein, ubiquitinates and induces endocytosis of the E-cadherin complex. Nat. Cell Biol. 2002, 4, 222–231. [Google Scholar] [CrossRef]
- Lickert, H.; Bauer, A.; Kemler, R.; Stappert, J. Casein kinase II phosphorylation of E-cadherin increases E-cadherin/beta-catenin interaction and strengthens cell-cell adhesion. J. Biol. Chem. 2000, 275, 5090–5095. [Google Scholar] [CrossRef]
- Thomas, S.M.; Brugge, J.S. Cellular functions regulated by Src family kinases. Annu. Rev. Cell Dev. Biol. 1997, 13, 513–609. [Google Scholar] [CrossRef]
- Behrens, J.; Vakaet, L.; Friis, R.; Winterhager, E.; Van Roy, F.; Mareel, M.M.; Birchmeier, W. Loss of epithelial differentiation and gain of invasiveness correlates with tyrosine phosphorylation of the E-cadherin/beta-catenin complex in cells transformed with a temperature-sensitive v-SRC gene. J. Cell Biol. 1993, 120, 757–766. [Google Scholar] [CrossRef]
- Hazan, R.B.; Norton, L. The epidermal growth factor receptor modulates the interaction of E-cadherin with the actin cytoskeleton. J. Biol. Chem. 1998, 273, 9078–9084. [Google Scholar] [CrossRef] [PubMed]
- Kypta, R.M.; Su, H.; Reichardt, L.F. Association between a transmembrane protein tyrosine phosphatase and the cadherin-catenin complex. J. Cell Biol. 1996, 134, 1519–1529. [Google Scholar] [CrossRef]
- Muslin, A.J.; Tanner, J.W.; Allen, P.M.; Shaw, A.S. Interaction of 14-3-3 with signaling proteins is mediated by the recognition of phosphoserine. Cell 1996, 84, 889–897. [Google Scholar] [CrossRef]
- Braga, V.M.; Machesky, L.M.; Hall, A.; Hotchin, N.A. The small GTPases Rho and Rac are required for the establishment of cadherin-dependent cell-cell contacts. J. Cell Biol. 1997, 137, 1421–1431. [Google Scholar] [CrossRef]
- Morrison, M.E.; Racaniello, V.R. Molecular cloning and expression of a murine homolog of the human poliovirus receptor gene. J. Virol. 1992, 66, 2807–2813. [Google Scholar] [CrossRef] [PubMed]
- Cocchi, F.; Menotti, L.; Mirandola, P.; Lopez, M.; Campadelli-Fiume, G. The ectodomain of a novel member of the immunoglobulin subfamily related to the poliovirus receptor has the attributes of a bona fide receptor for herpes simplex virus types 1 and 2 in human cells. J. Virol. 1998, 72, 9992–10002. [Google Scholar] [CrossRef]
- Satoh-Horikawa, K.; Nakanishi, H.; Takahashi, K.; Miyahara, M.; Nishimura, M.; Tachibana, K.; Mizoguchi, A.; Takai, Y. Nectin-3, a new member of immunoglobulin-like cell adhesion molecules that shows homophilic and heterophilic cell-cell adhesion activities. J. Biol. Chem. 2000, 275, 10291–10299. [Google Scholar] [CrossRef] [PubMed]
- Reymond, N.; Fabre, S.; Lecocq, E.; Adelaïde, J.; Dubreuil, P.; Lopez, M. Nectin4/PRR4, a new afadin-associated member of the nectin family that trans-interacts with nectin1/PRR1 through V domain interaction. J. Biol. Chem. 2001, 276, 43205–43215. [Google Scholar] [CrossRef] [PubMed]
- Sakisaka, T.; Nakanishi, H.; Takahashi, K.; Mandai, K.; Miyahara, M.; Satoh, A.; Takaishi, K.; Takai, Y. Different behavior of l-afadin and neurabin-II during the formation and destruction of cell-cell adherens junction. Oncogene 1999, 18, 1609–1617. [Google Scholar] [CrossRef][Green Version]
- Takahashi, K.; Nakanishi, H.; Miyahara, M.; Mandai, K.; Satoh, K.; Satoh, A.; Nishioka, H.; Aoki, J.; Nomoto, A.; Mizoguchi, A. Nectin/PRR: An immunoglobulin-like cell adhesion molecule recruited to cadherin-based adherens junctions through interaction with Afadin, a PDZ domain-containing protein. J. Cell Biol. 1999, 145, 539–549. [Google Scholar] [CrossRef]
- Rajasekaran, A.K.; Hojo, M.; Huima, T.; Rodriguez-Boulan, E. Catenins and zonula occludens-1 form a complex during early stages in the assembly of tight junctions. J. Cell Biol. 1996, 132, 451–463. [Google Scholar] [CrossRef]
- Morita, K.; Itoh, M.; Saitou, M.; Ando-Akatsuka, Y.; Furuse, M.; Yoneda, K.; Imamura, S.; Fujimoto, K.; Tsukita, S. Subcellular distribution of tight junction-associated proteins (occludin, ZO-1, ZO-2) in rodent skin. J. Investig. Derm. 1998, 110, 862–866. [Google Scholar] [CrossRef]
- Ooshio, T.; Kobayashi, R.; Ikeda, W.; Miyata, M.; Fukumoto, Y.; Matsuzawa, N.; Ogita, H.; Takai, Y. Involvement of the interaction of afadin with ZO-1 in the formation of tight junctions in Madin-Darby canine kidney cells. J. Biol. Chem. 2010, 285, 5003–5012. [Google Scholar] [CrossRef]
- Anderson, J.M. Molecular structure of tight junctions and their role in epithelial transport. Physiology 2001, 16, 126–130. [Google Scholar] [CrossRef]
- Fanning, A.S.; Anderson, J.M. Zonula occludens-1 and -2 are cytosolic scaffolds that regulate the assembly of cellular junctions. Ann. N. Y. Acad. Sci. 2009, 1165, 113–120. [Google Scholar] [CrossRef] [PubMed]
- Fanning, A.S.; Van Itallie, C.M.; Anderson, J.M. Zonula occludens-1 and -2 regulate apical cell structure and the zonula adherens cytoskeleton in polarized epithelia. Mol. Biol. Cell 2012, 23, 577–590. [Google Scholar] [CrossRef]
- Furuse, M.; Fujita, K.; Hiiragi, T.; Fujimoto, K.; Tsukita, S. Claudin-1 and -2: Novel integral membrane proteins localizing at tight junctions with no sequence similarity to occludin. J. Cell Biol. 1998, 141, 1539–1550. [Google Scholar] [CrossRef]
- Furuse, M.; Hata, M.; Furuse, K.; Yoshida, Y.; Haratake, A.; Sugitani, Y.; Noda, T.; Kubo, A.; Tsukita, S. Claudin-based tight junctions are crucial for the mammalian epidermal barrier: A lesson from claudin-1-deficient mice. J. Cell Biol. 2002, 156, 1099–1111. [Google Scholar] [CrossRef]
- Itoh, M.; Nagafuchi, A.; Moroi, S.; Tsukita, S. Involvement of ZO-1 in cadherin-based cell adhesion through its direct binding to alpha catenin and actin filaments. J. Cell Biol. 1997, 138, 181–192. [Google Scholar] [CrossRef] [PubMed]
- Poliak, S.; Matlis, S.; Ullmer, C.; Scherer, S.S.; Peles, E. Distinct claudins and associated PDZ proteins form different autotypic tight junctions in myelinating Schwann cells. J. Cell Biol. 2002, 159, 361–372. [Google Scholar] [CrossRef]
- Mineta, K.; Yamamoto, Y.; Yamazaki, Y.; Tanaka, H.; Tada, Y.; Saito, K.; Tamura, A.; Igarashi, M.; Endo, T.; Takeuchi, K.; et al. Predicted expansion of the claudin multigene family. FEBS Lett. 2011, 585, 606–612. [Google Scholar] [CrossRef]
- Groschwitz, K.R.; Hogan, S.P. Intestinal barrier function: Molecular regulation and disease pathogenesis. J. Allergy Clin. Immunol. 2009, 124, 3–20, quiz 21–2. [Google Scholar] [CrossRef] [PubMed]
- Balda, M.S.; Anderson, J.M. Two classes of tight junctions are revealed by ZO-1 isoforms. Am. J. Physiol. 1993, 264, C918–C924. [Google Scholar] [CrossRef]
- Haskins, J.; Gu, L.; Wittchen, E.S.; Hibbard, J.; Stevenson, B.R. ZO-3, a novel member of the MAGUK protein family found at the tight junction, interacts with ZO-1 and occludin. J. Cell Biol. 1998, 141, 199–208. [Google Scholar] [CrossRef]
- Rodgers, L.S.; Beam, M.T.; Anderson, J.M.; Fanning, A.S. Epithelial barrier assembly requires coordinated activity of multiple domains of the tight junction protein ZO-1. J. Cell Sci. 2013, 126, 1565–1575. [Google Scholar] [CrossRef]
- Stevenson, B.R.; Siliciano, J.D.; Mooseker, M.S.; Goodenough, D.A. Identification of ZO-1: A high molecular weight polypeptide associated with the tight junction (zonula occludens) in a variety of epithelia. J. Cell Biol. 1986, 103, 755–766. [Google Scholar] [CrossRef] [PubMed]
- Mandell, K.J.; Babbin, B.A.; Nusrat, A.; Parkos, C.A. Junctional adhesion molecule 1 regulates epithelial cell morphology through effects on beta1 integrins and Rap1 activity. J. Biol. Chem. 2005, 280, 11665–11674. [Google Scholar] [CrossRef]
- Kostrewa, D.; Brockhaus, M.; D’Arcy, A.; Dale, G.E.; Nelboeck, P.; Schmid, G.; Mueller, F.; Bazzoni, G.; Dejana, E.; Bartfai, T.; et al. X-ray structure of junctional adhesion molecule: Structural basis for homophilic adhesion via a novel dimerization motif. EMBO J. 2001, 20, 4391–4398. [Google Scholar] [CrossRef] [PubMed]
- Ebnet, K.; Suzuki, A.; Horikoshi, Y.; Hirose, T.; Meyer Zu Brickwedde, M.K.; Ohno, S.; Vestweber, D. The cell polarity protein ASIP/PAR-3 directly associates with junctional adhesion molecule (JAM). EMBO J. 2001, 20, 3738–3748. [Google Scholar] [CrossRef] [PubMed]
- Tsukita, S.; Furuse, M.; Itoh, M. Multifunctional strands in tight junctions. Nat. Rev. Mol. Cell Biol. 2001, 2, 285–293. [Google Scholar] [CrossRef]
- Bazzoni, G.; Martinez-Estrada, O.M.; Orsenigo, F.; Cordenonsi, M.; Citi, S.; Dejana, E. Interaction of junctional adhesion molecule with the tight junction components ZO-1, cingulin, and occludin. J. Biol. Chem. 2000, 275, 20520–20526. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, S.A.; Rodriguez, J.M.; Arrate, M.P.; Tran, T.M.; Brock, T.A. JAM2 interacts with alpha4beta1. Facilitation by JAM3. J. Biol. Chem. 2002, 277, 27589–27592. [Google Scholar] [CrossRef]
- Liang, T.W.; Chiu, H.H.; Gurney, A.; Sidle, A.; Tumas, D.B.; Schow, P.; Foster, J.; Klassen, T.; Dennis, K.; DeMarco, R.A.; et al. Vascular endothelial-junctional adhesion molecule (VE-JAM)/JAM 2 interacts with T, NK, and dendritic cells through JAM 3. J. Immunol. 2002, 168, 1618–1626. [Google Scholar] [CrossRef]
- Franke, W.W.; Schmid, E.; Winter, S.; Osborn, M.; Weber, K. Widespread occurrence of intermediate-sized filaments of the vimentin-type in cultured cells from diverse vertebrates. Exp. Cell Res. 1979, 123, 25–46. [Google Scholar] [CrossRef]
- Troyanovsky, S.M.; Eshkind, L.G.; Troyanovsky, R.B.; Leube, R.E.; Franke, W.W. Contributions of cytoplasmic domains of desmosomal cadherins to desmosome assembly and intermediate filament anchorage. Cell 1993, 72, 561–574. [Google Scholar] [CrossRef]
- Kowalczyk, A.P.; Bornslaeger, E.A.; Norvell, S.M.; Palka, H.L.; Green, K.J. Desmosomes: Intercellular adhesive junctions specialized for attachment of intermediate filaments. Int. Rev. Cytol. 1999, 185, 237–302. [Google Scholar]
- Garrod, D.; Chidgey, M. Desmosome structure, composition and function. Biochim. Biophys. Acta 2008, 1778, 572–587. [Google Scholar] [CrossRef] [PubMed]
- Watt, F.M.; Mattey, D.L.; Garrod, D.R. Calcium-induced reorganization of desmosomal components in cultured human keratinocytes. J. Cell Biol. 1984, 99, 2211–2215. [Google Scholar] [CrossRef] [PubMed]
- Stuart, R.O.; Sun, A.; Bush, K.T.; Nigam, S.K. Dependence of epithelial intercellular junction biogenesis on thapsigargin-sensitive intracellular calcium stores. J. Biol. Chem. 1996, 271, 13636–13641. [Google Scholar] [CrossRef]
- Kimura, T.E.; Merritt, A.J.; Garrod, D.R. Calcium-independent desmosomes of keratinocytes are hyper-adhesive. J. Investig. Dermatol. 2007, 127, 775–781. [Google Scholar] [CrossRef]
- Boggon, T.J.; Murray, J.; Chappuis-Flament, S.; Wong, E.; Gumbiner, B.M.; Shapiro, L. C-cadherin ectodomain structure and implications for cell adhesion mechanisms. Science 2002, 296, 1308–1313. [Google Scholar] [CrossRef] [PubMed]
- Hulpiau, P.; van Roy, F. Molecular evolution of the cadherin superfamily. Int. J. Biochem. Cell Biol. 2009, 41, 349–369. [Google Scholar] [CrossRef]
- Hatzfeld, M. Plakophilins: Multifunctional proteins or just regulators of desmosomal adhesion? Biochim. Biophys. Acta 2007, 1773, 69–77. [Google Scholar] [CrossRef]
- Mahoney, M.G.; Hu, Y.; Brennan, D.; Bazzi, H.; Christiano, A.M.; Wahl, J.K., 3rd. Delineation of diversified desmoglein distribution in stratified squamous epithelia: Implications in diseases. Exp. Derm. 2006, 15, 101–109. [Google Scholar] [CrossRef] [PubMed]
- Rubsam, M.; Broussard, J.A.; Wickström, S.A.; Nekrasova, O.; Green, K.J.; Niessen, C.M. Adherens Junctions and Desmosomes Coordinate Mechanics and Signaling to Orchestrate Tissue Morphogenesis and Function: An Evolutionary Perspective. Cold Spring Harb. Perspect. Biol. 2018, 10, a029207. [Google Scholar] [CrossRef] [PubMed]
- Peifer, M.; Sweeton, D.; Casey, M.; Wieschaus, E. Wingless signal and Zeste-white 3 kinase trigger opposing changes in the intracellular distribution of Armadillo. Development 1994, 120, 369–380. [Google Scholar] [CrossRef] [PubMed]
- Hatzfeld, M.; Nachtsheim, C. Cloning and characterization of a new armadillo family member, p0071, associated with the junctional plaque: Evidence for a subfamily of closely related proteins. J. Cell Sci. 1996, 109, 2767–2778. [Google Scholar] [CrossRef]
- Choi, H.J.; Weis, W.I. Structure of the armadillo repeat domain of plakophilin 1. J. Mol. Biol. 2005, 346, 367–376. [Google Scholar] [CrossRef] [PubMed]
- Mueller, H.; Franke, W.W. Biochemical and immunological characterization of desmoplakins I and II, the major polypeptides of the desmosomal plaque. J. Mol. Biol. 1983, 163, 647–671. [Google Scholar] [CrossRef]
- Smith, E.A.; Fuchs, E. Defining the interactions between intermediate filaments and desmosomes. J. Cell Biol. 1998, 141, 1229–1241. [Google Scholar] [CrossRef]
- Liovic, M.; D’Alessandro, M.; Tomic-Canic, M.; Bolshakov, V.N.; Coats, S.E.; Lane, E.B. Severe keratin 5 and 14 mutations induce down-regulation of junction proteins in keratinocytes. Exp. Cell Res. 2009, 315, 2995–3003. [Google Scholar] [CrossRef]
- Amar, L.S.; Shabana, A.H.; Oboeuf, M.; Martin, N.; Forest, N. Involvement of desmoplakin phosphorylation in the regulation of desmosomes by protein kinase C, in HeLa cells. Cell Adhes. Commun. 1999, 7, 125–138. [Google Scholar] [CrossRef]
- Bornslaeger, E.A.; Corcoran, C.M.; Stappenbeck, T.S.; Green, K.J. Breaking the connection: Displacement of the desmosomal plaque protein desmoplakin from cell-cell interfaces disrupts anchorage of intermediate filament bundles and alters intercellular junction assembly. J. Cell Biol. 1996, 134, 985–1001. [Google Scholar] [CrossRef]
- Lessard, J.L.; Wee, E.L.; Zimmerman, E.F. Presence of contractile proteins in mouse fetal palate prior to shelf elevation. Teratology 1974, 9, 113–125. [Google Scholar] [CrossRef] [PubMed]
- Chiquet, M.; Blumer, S.; Angelini, M.; Mitsiadis, T.A.; Katsaros, C. Mesenchymal Remodeling during Palatal Shelf Elevation Revealed by Extracellular Matrix and F-Actin Expression Patterns. Front. Physiol. 2016, 7, 392. [Google Scholar] [CrossRef] [PubMed]
- Birnbaum, S.; Reutter, H.; Mende, M.; de Assis, N.A.; Diaz-Lacava, A.; Herms, S.; Scheer, M.; Lauster, C.; Braumann, B.; Schmidt, G.; et al. Further evidence for the involvement of MYH9 in the etiology of non-syndromic cleft lip with or without cleft palate. Eur. J. Oral Sci. 2009, 117, 200–203. [Google Scholar] [CrossRef]
- Chiquet, B.T.; Hashmi, S.S.; Henry, R.; Burt, A.; Mulliken, J.B.; Stal, S.; Bray, M.; Blanton, S.H.; Hecht, J.T. Genomic screening identifies novel linkages and provides further evidence for a role of MYH9 in nonsyndromic cleft lip and palate. Eur. J. Hum. Genet. 2009, 17, 195–204. [Google Scholar] [CrossRef][Green Version]
- Moreno Uribe, L.M.; Fomina, T.; Munger, R.G.; Romitti, P.A.; Jenkins, M.M.; Gjessing, H.K.; Gjerdevik, M.; Christensen, K.; Wilcox, A.J.; Murray, J.C.; et al. A Population-Based Study of Effects of Genetic Loci on Orofacial Clefts. J. Dent. Res. 2017, 96, 1322–1329. [Google Scholar] [CrossRef]
- Liu, H.; Duncan, K.; Helverson, A.; Kumari, P.; Mumm, C.; Xiao, Y.; Carlson, J.C.; Darbellay, F.; Visel, A.; Leslie, E.; et al. Analysis of zebrafish periderm enhancers facilitates identification of a regulatory variant near human KRT8/18. eLife 2020, 9, e51325. [Google Scholar] [CrossRef]
- Lu, H.; Hesse, M.; Peters, B.; Magin, T.M. Type II keratins precede type I keratins during early embryonic development. Eur. J. Cell Biol. 2005, 84, 709–718. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, W.; Nakanishi, H.; Miyoshi, J.; Mandai, K.; Ishizaki, H.; Tanaka, M.; Togawa, A.; Takahashi, K.; Nishioka, H.; Yoshida, H.; et al. Afadin: A key molecule essential for structural organization of cell-cell junctions of polarized epithelia during embryogenesis. J. Cell Biol. 1999, 146, 1117–1132. [Google Scholar] [CrossRef]
- Lough, K.J.; Spitzer, D.C.; Bergman, A.J.; Wu, J.J.; Byrd, K.M.; Williams, S.E. Disruption of the nectin-afadin complex recapitulates features of the human cleft lip/palate syndrome CLPED1. Development 2020, 147, dev189241. [Google Scholar] [CrossRef]
- Camargo, M.; Rivera, D.; Moreno, L.; Lidral, A.C.; Harper, U.; Jones, M.; Solomon, B.D.; Roessler, E.; Vélez, J.I.; Martinez, A.F. GWAS reveals new recessive loci associated with non-syndromic facial clefting. Eur. J. Med. Genet. 2012, 55, 510–514. [Google Scholar] [CrossRef]
- El-Sibai, M.; El Hajj, J.; Al Haddad, M.; El Baba, N.; Al Saneh, M.; Daoud Khatoun, W.; Helaers, R.; Vikkula, M.; El Atat, O.; Sabbagh, J.; et al. Dysregulation of Rho GTPases in orofacial cleft patients-derived primary cells leads to impaired cell migration, a potential cause of cleft/lip palate development. Cells Dev. 2021, 165, 203656. [Google Scholar] [CrossRef] [PubMed]
- Hoebel, A.K.; Drichel, D.; van de Vorst, M.; Böhmer, A.C.; Sivalingam, S.; Ishorst, N.; Klamt, J.; Gölz, L.; Alblas, M.; Maaser, A.; et al. Candidate Genes for Nonsyndromic Cleft Palate Detected by Exome Sequencing. J. Dent. Res. 2017, 96, 1314–1321. [Google Scholar] [CrossRef] [PubMed]
- Leslie, E.J.; Mansilla, M.A.; Biggs, L.C.; Schuette, K.; Bullard, S.; Cooper, M.; Dunnwald, M.; Lidral, A.C.; Marazita, M.L.; Beaty, T.H.; et al. Expression and mutation analyses implicate ARHGAP29 as the etiologic gene for the cleft lip with or without cleft palate locus identified by genome-wide association on chromosome 1p22. Birth Defects Res. A Clin. Mol. Teratol. 2012, 94, 934–942. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Busch, T.; Eliason, S.; Anand, D.; Bullard, S.; Gowans, L.J.J.; Nidey, N.; Petrin, A.; Augustine-Akpan, E.A.; Saadi, I.; et al. Exome sequencing provides additional evidence for the involvement of ARHGAP29 in Mendelian orofacial clefting and extends the phenotypic spectrum to isolated cleft palate. Birth Defects Res. 2017, 109, 27–37. [Google Scholar] [CrossRef]
- Liu, H.; Leslie, E.J.; Carlson, J.C.; Beaty, T.H.; Marazita, M.L.; Lidral, A.C.; Cornell, R.A. Identification of common non-coding variants at 1p22 that are functional for non-syndromic orofacial clefting. Nat. Commun. 2017, 8, 14759. [Google Scholar] [CrossRef]
- Savastano, C.P.; Brito, L.A.; Faria, Á.C.; Setó-Salvia, N.; Peskett, E.; Musso, C.M.; Alvizi, L.; Ezquina, S.A.; James, C. Impact of rare variants in ARHGAP29 to the etiology of oral clefts: Role of loss-of-function vs. missense variants. Clin. Genet. 2017, 91, 683–689. [Google Scholar] [CrossRef]
- Paul, B.J.; Palmer, K.; Sharp, J.C.; Pratt, C.H.; Murray, S.A.; Dunnwald, M. ARHGAP29 Mutation Is Associated with Abnormal Oral Epithelial Adhesions. J. Dent. Res. 2017, 96, 1298–1305. [Google Scholar] [CrossRef]
- Tucci, V.; Kleefstra, T.; Hardy, A.; Heise, I.; Maggi, S.; Willemsen, M.H.; Hilton, H.; Esapa, C.; Simon, M.; Buenavista, M.T.; et al. Dominant beta-catenin mutations cause intellectual disability with recognizable syndromic features. J. Clin. Investig. 2014, 124, 1468–1482. [Google Scholar] [CrossRef]
- He, F.; Xiong, W.; Wang, Y.; Li, L.; Liu, C.; Yamagami, T.; Taketo, M.M.; Zhou, C.; Chen, Y. Epithelial Wnt/beta-catenin signaling regulates palatal shelf fusion through regulation of Tgfbeta3 expression. Dev. Biol. 2011, 350, 511–519. [Google Scholar] [CrossRef][Green Version]
- Ghoumid, J.; Stichelbout, M.; Jourdain, A.S.; Frenois, F.; Lejeune-Dumoulin, S.; Alex-Cordier, M.P.; Lebrun, M.; Guerreschi, P.; Duquennoy-Martinot, V.; Vinchon, M.; et al. Blepharocheilodontic syndrome is a CDH1 pathway-related disorder due to mutations in CDH1 and CTNND1. Genet. Med. 2017, 19, 1013–1021. [Google Scholar] [CrossRef]
- Cox, L.L.; Cox, T.C.; Moreno Uribe, L.M.; Zhu, Y.; Richter, C.T.; Nidey, N.; Standley, J.M.; Deng, M.; Blue, E.; Chong, J.X.; et al. Mutations in the Epithelial Cadherin-p120-Catenin Complex Cause Mendelian Non-Syndromic Cleft Lip with or without Cleft Palate. Am. J. Hum. Genet. 2018, 102, 1143–1157. [Google Scholar] [CrossRef]
- Alharatani, R.; Ververi, A.; Beleza-Meireles, A.; Ji, W.; Mis, E.; Patterson, Q.T.; Griffin, J.N.; Bhujel, N.; Chang, C.A.; Dixit, A.; et al. Novel truncating mutations in CTNND1 cause a dominant craniofacial and cardiac syndrome. Hum. Mol. Genet. 2020, 29, 1900–1921. [Google Scholar] [CrossRef]
- Brito, L.A.; Yamamoto, G.L.; Melo, S.; Malcher, C.; Ferreira, S.G.; Figueiredo, J.; Alvizi, L.; Kobayashi, G.S.; Naslavsky, M.S.; Alonso, N.; et al. Rare Variants in the Epithelial Cadherin Gene Underlying the Genetic Etiology of Nonsyndromic Cleft Lip with or without Cleft Palate. Hum. Mutat. 2015, 36, 1029–1033. [Google Scholar] [CrossRef]
- Bureau, A.; Parker, M.M.; Ruczinski, I.; Taub, M.A.; Marazita, M.L.; Murray, J.C.; Mangold, E.; Noethen, M.M.; Ludwig, K.U.; Hetmanski, J.B.; et al. Whole exome sequencing of distant relatives in multiplex families implicates rare variants in candidate genes for oral clefts. Genetics 2014, 197, 1039–1044. [Google Scholar] [CrossRef]
- Du, S.; Yang, Y.; Yi, P.; Luo, J.; Liu, T.; Chen, R.; Liu, C.J.; Ma, T.; Li, Y.; Wang, C.; et al. A Novel CDH1 Mutation Causing Reduced E-Cadherin Dimerization Is Associated with Nonsyndromic Cleft Lip With or Without Cleft Palate. Genet. Test. Mol. Biomark. 2019, 23, 759–765. [Google Scholar] [CrossRef]
- Vogelaar, I.P.; Figueiredo, J.; van Rooij, I.A.; Simões-Correia, J.; van der Post, R.S.; Melo, S.; Seruca, R.; Carels, C.E.; Ligtenberg, M.J.; Hoogerbrugge, N. Identification of germline mutations in the cancer predisposing gene CDH1 in patients with orofacial clefts. Hum. Mol. Genet. 2013, 22, 919–926. [Google Scholar] [CrossRef]
- Selvanathan, A.; Nixon, C.Y.; Zhu, Y.; Scietti, L.; Forneris, F.; Uribe, L.M.M.; Lidral, A.C.; Jezewski, P.A.; Mulliken, J.B.; Murray, J.C.; et al. CDH1 Mutation Distribution and Type Suggests Genetic Differences between the Etiology of Orofacial Clefting and Gastric Cancer. Genes 2020, 11, 391. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Higuera, I.; Manchado, E.; Dubus, P.; Cañamero, M.; Méndez, J.; Moreno, S.; Malumbres, M. Genomic stability and tumour suppression by the APC/C cofactor Cdh1. Nat. Cell Biol. 2008, 10, 802–811. [Google Scholar] [CrossRef]
- Shao, R.; Liu, J.; Yan, G.; Zhang, J.; Han, Y.; Guo, J.; Xu, Z.; Yuan, Z.; Liu, J.; Malumbres, M.; et al. Cdh1 regulates craniofacial development via APC-dependent ubiquitination and activation of Goosecoid. Cell Res. 2016, 26, 699–712. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tunggal, J.A.; Helfrich, I.; Schmitz, A.; Schwarz, H.; Günzel, D.; Fromm, M.; Kemler, R.; Krieg, T.; Niessen, C.M. E-cadherin is essential for in vivo epidermal barrier function by regulating tight junctions. EMBO J. 2005, 24, 1146–1156. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, K.; Hu, D.; Bustos, T.; Zlotogora, J.; Richieri-Costa, A.; Helms, J.A.; Spritz, R.A. Mutations of PVRL1, encoding a cell-cell adhesion molecule/herpesvirus receptor, in cleft lip/palate-ectodermal dysplasia. Nat. Genet. 2000, 25, 427–430. [Google Scholar] [CrossRef]
- Barron, M.J.; Brookes, S.J.; Draper, C.E.; Garrod, D.; Kirkham, J.; Shore, R.C.; Dixon, M.J. The cell adhesion molecule nectin-1 is critical for normal enamel formation in mice. Hum. Mol. Genet. 2008, 17, 3509–3520. [Google Scholar] [CrossRef][Green Version]
- Yoshida, T.; Iwata, T.; Takai, Y.; Birchmeier, W.; Yamato, M.; Okano, T. Afadin requirement for cytokine expressions in keratinocytes during chemically induced inflammation in mice. Genes Cells 2014, 19, 842–852. [Google Scholar] [CrossRef]
- Brancati, F.; Fortugno, P.; Bottillo, I.; Lopez, M.; Josselin, E.; Boudghene-Stambouli, O.; Agolini, E.; Bernardini, L.; Bellacchio, E.; Iannicelli, M.; et al. Mutations in PVRL4, encoding cell adhesion molecule nectin-4, cause ectodermal dysplasia-syndactyly syndrome. Am. J. Hum. Genet. 2010, 87, 265–273. [Google Scholar] [CrossRef] [PubMed]
- Saadi, I.; Alkuraya, F.S.; Gisselbrecht, S.S.; Goessling, W.; Cavallesco, R.; Turbe-Doan, A.; Petrin, A.L.; Harris, J.; Siddiqui, U.; Grix, A.W., Jr.; et al. Deficiency of the cytoskeletal protein SPECC1L leads to oblique facial clefting. Am. J. Hum. Genet. 2011, 89, 44–55. [Google Scholar] [CrossRef]
- Hall, E.G.; Wenger, L.W.; Wilson, N.R.; Undurty-Akella, S.S.; Standley, J.; Augustine-Akpan, E.A.; Kousa, Y.A.; Acevedo, D.S.; Goering, J.P.; Pitstick, L.; et al. SPECC1L regulates palate development downstream of IRF6. Hum. Mol. Genet. 2020, 29, 845–858. [Google Scholar] [CrossRef]
- Goering, J.P.; Wenger, L.W.; Stetsiv, M.; Moedritzer, M.; Hall, E.G.; Isai, D.G.; Jack, B.M.; Umar, Z.; Rickabaugh, M.K.; Czirok, A.; et al. In-frame deletion of SPECC1L microtubule association domain results in gain-of-function phenotypes affecting embryonic tissue movement and fusion events. Hum. Mol. Genet. 2021, 31, 18–31. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, K.; Kumari, P.; Sepulveda Rincon, L.; Gu, R.; Ji, Y.; Kumar, S.; Zhou, C.J. Wnt signaling in orofacial clefts: Crosstalk, pathogenesis and models. Dis. Model. Mech. 2019, 12, dmm037051. [Google Scholar] [CrossRef]
- Nopoulos, P.; Richman, L.; Andreasen, N.C.; Murray, J.C.; Schutte, B. Abnormal brain structure in adults with Van der Woude syndrome. Clin. Genet. 2007, 71, 511–517. [Google Scholar] [CrossRef]
- Kousa, Y.A.; Zhu, H.; Fakhouri, W.D.; Lei, Y.; Kinoshita, A.; Roushangar, R.R.; Patel, N.K.; Agopian, A.J.; Yang, W.; Leslie, E.J.; et al. The TFAP2A-IRF6-GRHL3 genetic pathway is conserved in neurulation. Hum. Mol. Genet. 2019, 28, 1726–1737. [Google Scholar] [CrossRef] [PubMed]
- Dixon, M.J.; Marazita, M.L.; Beaty, T.H.; Murray, J.C. Cleft lip and palate: Understanding genetic and environmental influences. Nat. Rev. Genet. 2011, 12, 167–178. [Google Scholar] [CrossRef]
- Campbell, H.K.; Maiers, J.L.; DeMali, K.A. Interplay between tight junctions & adherens junctions. Exp. Cell Res. 2017, 358, 39–44. [Google Scholar]
- Tessier, P. Anatomical classification facial, cranio-facial and latero-facial clefts. J. Maxillofac. Surg. 1976, 4, 69–92. [Google Scholar] [CrossRef]
- Shah, J.; Guerrera, D.; Vasileva, E.; Sluysmans, S.; Bertels, E.; Citi, S. PLEKHA7: Cytoskeletal adaptor protein at center stage in junctional organization and signaling. Int. J. Biochem. Cell. Biol. 2016, 75, 112–116. [Google Scholar] [CrossRef] [PubMed]
- Kondo, S.; Schutte, B.C.; Richardson, R.J.; Bjork, B.C.; Knight, A.S.; Watanabe, Y.; Howard, E.; de Lima, R.L.; Daack-Hirsch, S.; Sander, A.; et al. Mutations in IRF6 cause Van der Woude and popliteal pterygium syndromes. Nat. Genet. 2002, 32, 285–289. [Google Scholar] [CrossRef] [PubMed]
- Zucchero, T.M.; Cooper, M.E.; Maher, B.S.; Daack-Hirsch, S.; Nepomuceno, B.; Ribeiro, L.; Caprau, D.; Christensen, K.; Suzuki, Y.; Machida, J.; et al. Interferon regulatory factor 6 (IRF6) gene variants and the risk of isolated cleft lip or palate. N. Engl. J. Med. 2004, 351, 769–780. [Google Scholar] [CrossRef] [PubMed]
- Ingraham, C.R.; Kinoshita, A.; Kondo, S.; Yang, B.; Sajan, S.; Trout, K.J.; Malik, M.I.; Dunnwald, M.; Goudy, S.L.; Lovett, M.; et al. Abnormal skin, limb and craniofacial morphogenesis in mice deficient for interferon regulatory factor 6 (Irf6). Nat. Genet. 2006, 38, 1335–1340. [Google Scholar] [CrossRef] [PubMed]
- Antiguas, A.; DeMali, K.A.; Dunnwald, M. IRF6 Regulates the Delivery of E-Cadherin to the Plasma Membrane. J. Investig. Dermatol. 2021; in press. [Google Scholar] [CrossRef] [PubMed]
- Biggs, L.C.; Naridze, R.; DeMali, K.A.; Lusche, D.F.; Kuhl, S.; Soll, D.R.; Schutte, B.C.; Dunnwald, M. Interferon Regulatory Factor 6 regulates keratinocyte migration. J. Cell Sci. 2014, 127, 2840–2848. [Google Scholar] [CrossRef] [PubMed]
- Fukata, M.; Kaibuchi, K. Rho-family GTPases in cadherin-mediated cell-cell adhesion. Nat. Rev. Mol. Cell Biol. 2001, 2, 887–897. [Google Scholar] [CrossRef] [PubMed]
- Hall, A. Rho GTPases and the control of cell behaviour. Biochem. Soc. Trans. 2005, 33, 891–895. [Google Scholar] [CrossRef] [PubMed]
- Tagashira, T.; Fukuda, T.; Miyata, M.; Nakamura, K.; Fujita, H.; Takai, Y.; Hirata, K.I.; Rikitake, Y. Afadin Facilitates Vascular Endothelial Growth Factor-Induced Network Formation and Migration of Vascular Endothelial Cells by Inactivating Rho-Associated Kinase Through ArhGAP29. Arter. Thromb. Vasc. Biol. 2018, 38, 1159–1169. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
