Insights into Bacterial Communities and Diversity of Mangrove Forest Soils along the Upper Gulf of Thailand in Response to Environmental Factors
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Determination of Soil and Water Physicochemical Parameters
2.3. Soil DNA Extraction and Illumina NGS
2.4. Data Processing and Bioinformatics Analyses
2.5. Statistical Analyses
3. Results
3.1. Soil and Surface Water Physicochemical Parameters
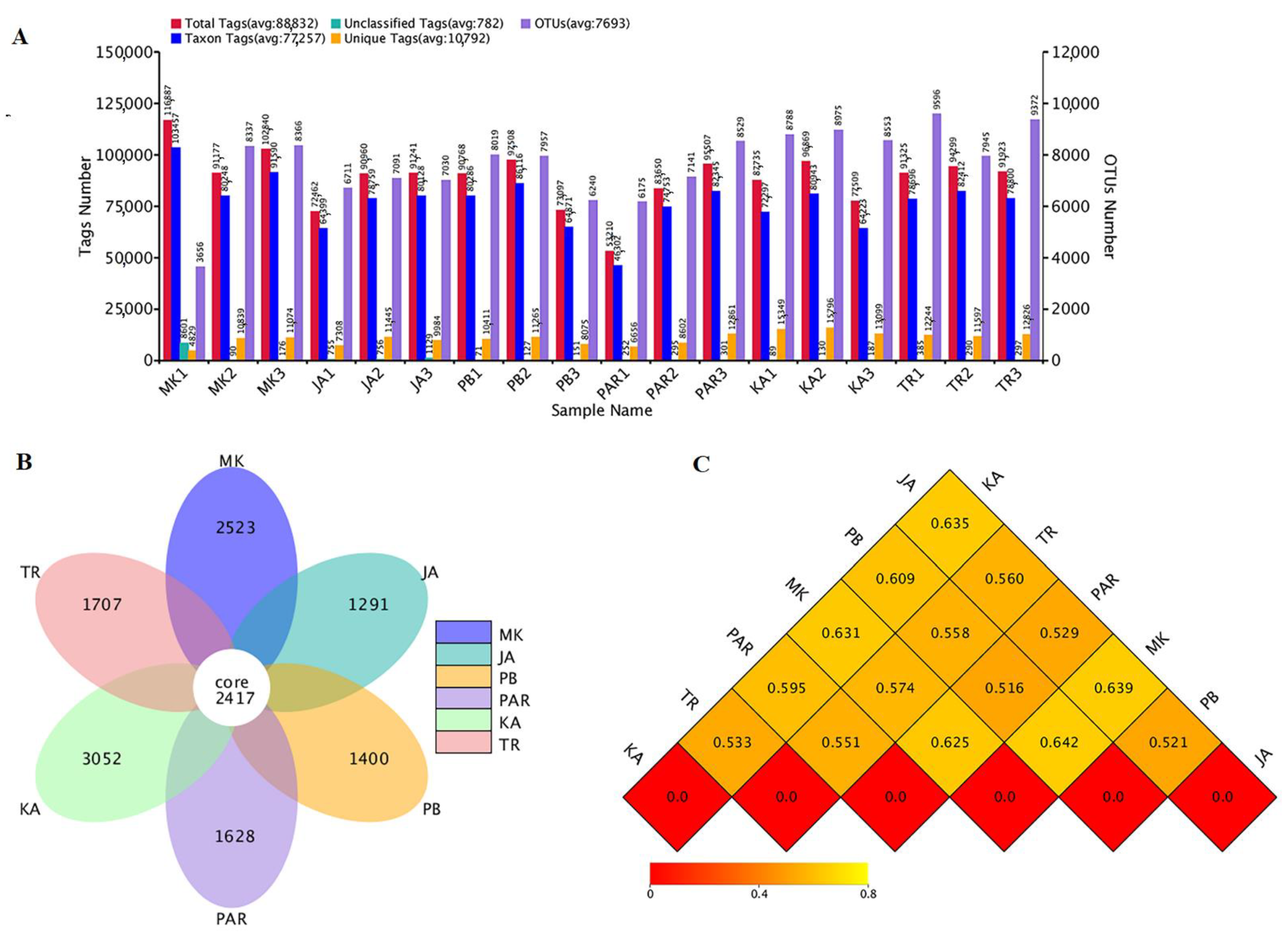
3.2. Sequence Analyses and Bacterial Diversity Indices
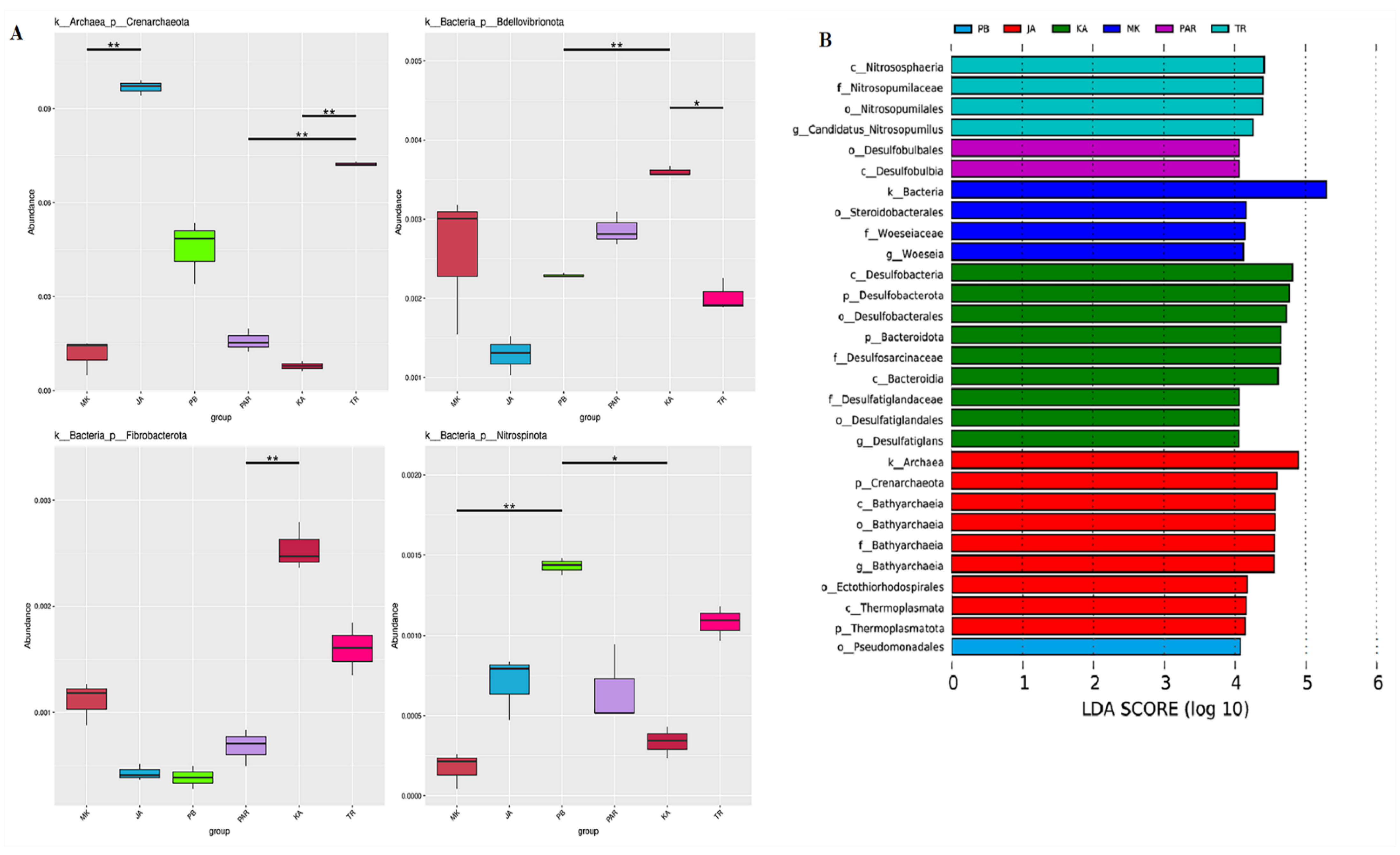
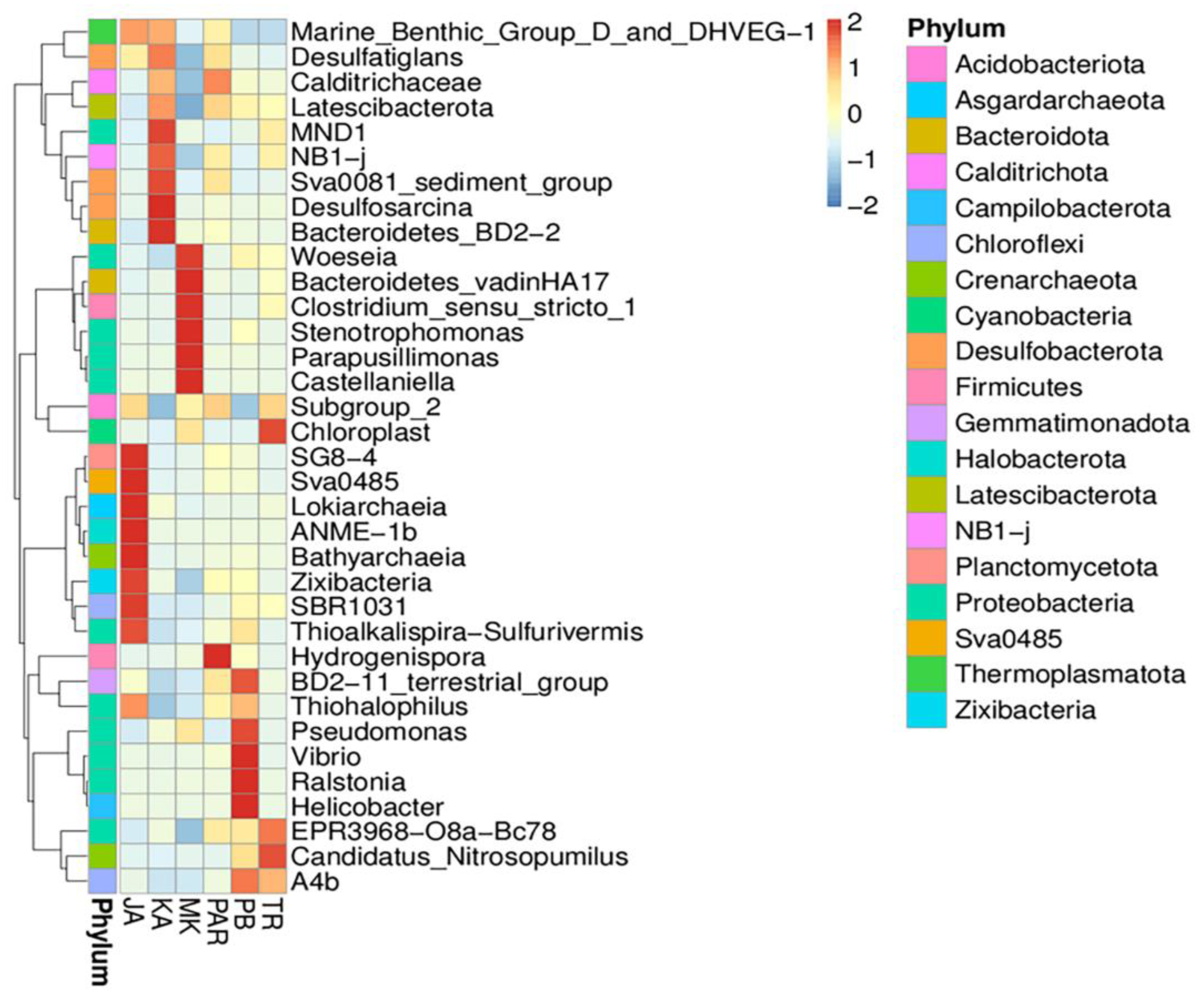
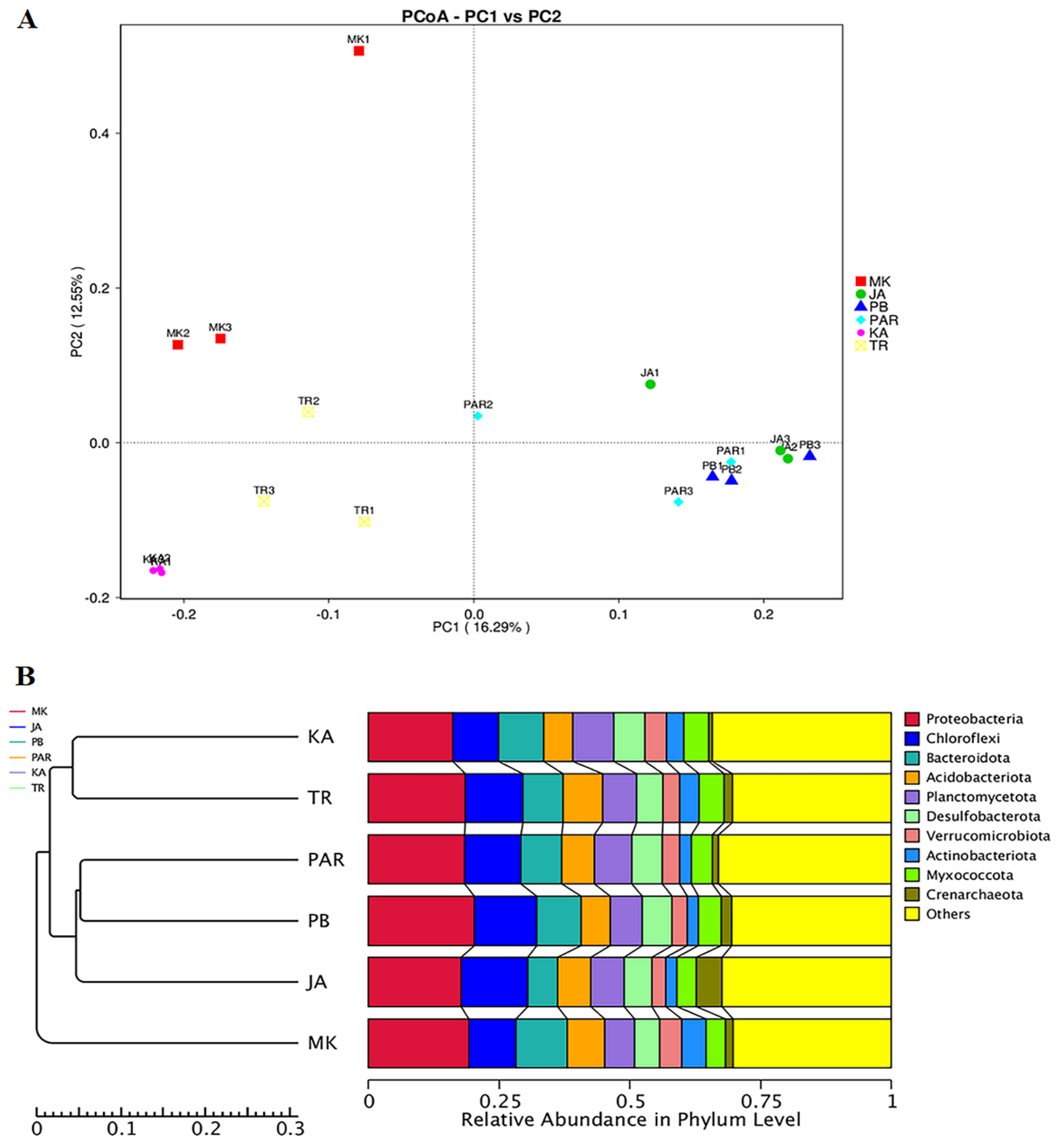
3.3. Illumina NGS and Bacterial Community Structure
3.4. Effect of Environmental Factors on the Distribution of Bacterial Community Structure
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Liang, J.B.; Chen, Y.Q.; Lan, C.Y.; Tam, N.F.Y.; Zan, Q.J.; Huang, L.N. Recovery of novel bacterial diversity from mangrove sediment. Mar. Biol. 2007, 150, 739–747. [Google Scholar] [CrossRef]
- Thatoi, H.; Behera, B.C.; Mishra, R.R.; Dutta, S.K. Biodiversity and biotechnological potential of microorganisms from mangrove ecosystems: A review. Ann. Microbiol. 2013, 63, 1–19. [Google Scholar] [CrossRef]
- Hu, B.; Liao, J.; Zhang, Q.; Ding, S.; He, M.; Qiao, Y.; Zhang, Z.; Shang, C.; Chen, S. Diversity and vertical distribution of sedimentary bacterial communities and its association with metal bioavailability in three distinct mangrove reserves of South China. Water 2022, 14, 971. [Google Scholar] [CrossRef]
- Pupin, B.; Nahas, E. Microbial populations and activities of mangrove, restinga and Atlantic forest soils from Cardoso Island, Brazil. J. Appl. Microbiol. 2014, 116, 851–864. [Google Scholar] [CrossRef] [PubMed]
- Clark, M.W.; McConchie, D.; Lewis, D.W.; Saenger, P. Redox stratification and heavy metal partitioning in Avicennia-dominated mangrove sediments: A geochemical model. Chem. Geol. 1998, 149, 147–171. [Google Scholar] [CrossRef]
- Alongi, D.M. The dynamics of benthic nutrient pools and fluxes in tropical mangrove forests. J. Mar. Res. 1996, 54, 123–148. [Google Scholar] [CrossRef]
- Liu, M.; Huang, H.; Bao, S.; Tong, Y. Microbial community structure of soils in Bamenwan mangrove wetland. Sci. Rep. 2019, 9, 8406. [Google Scholar] [CrossRef]
- Pramanik, A.; Sengupta, S.; Bhattacharyya, M. Microbial diversity and community analysis of the Sundarbans mangrove, a world heritage site. In Microbial Diversity in the Genomic Era, 1st ed.; Das, S., Dash, H.R., Eds.; Academic Press: Cambridge, MA, USA, 2018; pp. 65–76. [Google Scholar]
- Holguin, G.; Vazquez, P.; Bashan, Y. The role of sediment microorganisms in the productivity, conservation, and rehabilitation of mangrove ecosystems: An overview. Biol. Fertil. Soils 2001, 33, 265–278. [Google Scholar] [CrossRef]
- Hou, L.J.; Liu, M.; Xu, S.Y.; Ou, D.N.; Yu, J.; Cheng, S.B.; Lin, X.; Yang, Y. The effects of semi-lunar spring and neap tidal change on nitrification, denitrification, and N2O vertical distribution in the intertidal sediments of the Yangtze estuary, China. Estuar. Coast. Shelf Sci. 2007, 73, 607–616. [Google Scholar] [CrossRef]
- Hossain, M.Z.; Aziz, C.B.; Saha, M.L. Relationships between soil physico-chemical properties and total viable bacterial counts in Sunderban mangrove forests, Bangladesh. Dhaka Univ. J. Biol. Sci. 2012, 21, 169–175. [Google Scholar] [CrossRef]
- Chancham, C. Wind resource assessment in the northern gulf of Thailand using atmospheric modeling and climatic database. J. Renew. Energy. Smart Grid Technol. 2016, 11, 9–15. [Google Scholar]
- Nimnoi, P.; Pongsilp, N. Marine bacterial communities in the upper gulf of Thailand assessed by Illumina next-generation sequencing platform. BMC Microbiol. 2020, 20, 19. [Google Scholar] [CrossRef] [PubMed]
- Department of Marine and Coastal Resources, Thailand. The Oceanography of the Gulf of Thailand. Available online: https://km.dmcr.go.th/en/c_51/d_1132 (accessed on 1 October 2022).
- Alongi, D.M.; Wattayakorn, G.; Pfitzner, J.; Tirendi, F.; Zagorskis, I.; Brunskill, G.J.; Davidson, A.; Clough, B.F. Organic carbon accumulation and metabolic pathways in sediments of mangrove forests in southern Thailand. Mar. Geol. 2001, 179, 85–103. [Google Scholar] [CrossRef]
- Pumijumnong, N. Mangrove forests in Thailand. In Mangrove Ecosystems of Asia, 1st ed.; Faridah-Hanum, I., Latiff, A., Hakeem, K., Ozturk, M., Eds.; Springer: New York, NY, USA, 2014; pp. 61–79. [Google Scholar]
- Buajan, S.; Pumijumnong, N. Distribution of heavy metals in mangrove sediment at the Tha Chin estuary, Samut Sakhon province, Thailand. J. Environ. Res. 2010, 32, 61–77. [Google Scholar]
- Buajan, S.; Pumijumnong, N. Seasonal cambial activity of some mangrove trees in Inner Gulf of Thailand in dependence on climate. Songklanakarin J. Sci. Technol. 2012, 34, 337–344. [Google Scholar]
- Vu, M.T.; Geraldi, A.; Do, H.D.K.; Luqman, A.; Nguyen, H.D.; Fauzia, F.N.; Amalludin, F.I.; Sadila, A.Y.; Wijaya, N.H.; Santoso, H.; et al. Soil mineral composition and salinity are the main factors regulating the bacterial community associated with the roots of coastal sand dune halophytes. Biology 2022, 11, 695. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, A.; Saha, R.; Bhadury, P. Metagenomic insights into surface water microbial communities of a South Asian mangrove ecosystem. PeerJ 2022, 10, e13169. [Google Scholar] [CrossRef]
- Li, T.; Liu, T.; Zheng, C.; Kang, C.; Yang, Z.; Yao, X.; Song, F.; Zhang, R.; Wang, X.; Xu, N.; et al. Changes in soil bacterial community structure as a result of incorporation of Brassica plants compared with continuous planting eggplant and chemical disinfection in greenhouses. PLoS ONE 2017, 12, e0173923. [Google Scholar] [CrossRef]
- Jackson, M.L. Soil Chemical Analysis, 1st ed.; Parallel Press: Madison, WI, USA, 1967; pp. 510–594. [Google Scholar]
- Bouyoucos, G.J. Hydrometer method improved for making particle size analyses of soils. Agron. J. 1962, 54, 464–465. [Google Scholar] [CrossRef]
- Walkley, A.; Black, I.A. An examination of the Degtjareff method for determining soil organic matter, and a proposed modification of the chromic acid titration method. Soil Sci. 1934, 37, 29–38. [Google Scholar] [CrossRef]
- Bray, R.H.; Kurtz, L.T. Determination of total, organic, and available forms of phosphorus in soils. Soil Sci. 1945, 59, 39–46. [Google Scholar] [CrossRef]
- American Public Health Association (APHA). Standard Methods for the Examination of Water and Wastewater; American Public Health Association; American Water Works Association and Water Environment Federation: Washington, DC, USA, 2012. [Google Scholar]
- Rasheed, M.A.; Rao, P.L.S.; Radha, B.A.; Lakshmi, M.; Dayal, A.M. Assessment of groundwater quality using ICP-MS in the eastern region of Adilabad district, Andhra Pradesh, India. J. Environ. Prot. Ecol. 2011, 12, 425–433. [Google Scholar]
- Abdul, R.M.; Mutnuri, L.; Dattatreya, P.J.; Mohan, D.A. Assessment of drinking water quality using ICP-MS and microbiological methods in the Bholakpur area, Hyderabad, India. Environ. Monit. Assess. 2012, 184, 1581–1592. [Google Scholar] [CrossRef]
- Apprill, A.; McNally, S.; Parsons, R.; Weber, L. Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquat. Microb. Ecol. 2015, 75, 129–137. [Google Scholar] [CrossRef]
- Parada, A.E.; Needham, D.M.; Fuhrman, J.A. Every base matters: Assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ. Microbiol. 2016, 18, 1403–1414. [Google Scholar] [CrossRef]
- Magoč, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Haas, B.J.; Gevers, D.; Earl, A.M.; Feldgarden, M.; Ward, D.V.; Giannoukos, G.; Ciulla, D.; Tabbaa, D.; Highlander, S.K.; Sodergren, E.; et al. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 2011, 21, 494–504. [Google Scholar] [CrossRef]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mother: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2013; Available online: http://www.R-project.org/ (accessed on 1 March 2022).
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis, 2nd ed.; Hadley Wickham: Houston, TX, USA, 2009; pp. 11–32. [Google Scholar]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef]
- Hammer, Ø.; Harper, D.A.T.; Ryan, P.D. PAST: Paleontological statistics software package for education and data analysis. Palaeontol. Electron. 2001, 4, 1–9. [Google Scholar]
- Alongi, D.M.; Christoffersen, P.; Tirendi, F. The influence of forest type on microbial-nutrient relationships in tropical mangrove sediments. J. Exp. Mar. Biol. Ecol. 1993, 171, 201–223. [Google Scholar] [CrossRef]
- Kathiresan, K.; Selvam, M.M. Evaluation of beneficial bacteria from mangrove soil. Bot. Mar. 2006, 49, 86–88. [Google Scholar] [CrossRef]
- Mishra, R.R.; Swain, M.R.; Dangar, T.K.; Thatoi, H. Diversity and seasonal fluctuation of predominant microbial communities in Bhitarkanika, a tropical mangrove ecosystem in India. Rev. Biol. Trop. 2012, 60, 909–924. [Google Scholar]
- Zhou, Z.; Meng, H.; Liu, Y.; Gu, J.D.; Li, M. Stratified bacterial and archaeal community in mangrove and intertidal wetland mudflats revealed by high throughput 16S rRNA gene sequencing. Front. Microbiol. 2017, 8, 2148. [Google Scholar] [CrossRef]
- Essien, J.P.; Antai, S.P.; Benson, N.U. Microbial population dynamics as a function of sediment salinity gradients in the Qua lboe Estuary Mangrove swamp (Nigeria). Res. J. Microbiol. 2006, 1, 255–265. [Google Scholar]
- Gonzalez-Acosta, B.; Bashan, Y.; Hernandez- Saavedra, N.Y.; Ascencio, F.; De la Cruz-Aguero, G. Seasonal seawater temperature as the major determinant for populations of culturable bacteria in the sediments of an intact mangrove in an arid region. FEMS Microbiol. Ecol. 2006, 55, 311–321. [Google Scholar] [CrossRef]
- Alzubaidy, H.; Essack, M.; Malas, T.B.; Bokhari, A.; Motwalli, O.; Kamanu, F.K.; Jamhor, S.A.; Mokhtar, N.A.; Antunes, A.; Simões, M.F.; et al. Rhizosphere microbiome metagenomics of gray mangroves (Avicennia marina) in the Red Sea. Gene 2016, 576, 626–636. [Google Scholar] [CrossRef]
- Zhang, Y.; Dong, J.; Yang, B.; Ling, J.; Wang, Y.; Zhang, S. Bacterial community structure of mangrove sediments in relation to environmental variables accessed by 16S rRNA gene-denaturing gradient gel electrophoresis fingerprinting. Sci. Mar. 2009, 73, 487–498. [Google Scholar] [CrossRef]
- Rocha, L.L.; Colares, G.B.; Nogueira, V.L.R.; Paes, F.A.; Melo, V.M.M. Distinct habitats select particular bacterial communities in mangrove sediments. Int. J. Microbiol. 2016, 2016, 3435809. [Google Scholar] [CrossRef]
- Lertprasert, N. Accumulation and Distribution of Heavy Metals in Water, Sediment, Ipomoea Aquatica Forsk., and Rhizophora Apiculata Blume, in the Phi Lok Canal System, Samut Songkhram Province. Available online: https://graduate.mahidol.ac.th/engine/current-students/detail/abstract_view.php?id=4637127&fac=24&prg=2401M&gp=6 (accessed on 25 March 2022).
- Caregnato, F.F.; Koller, C.E.; MacFarlane, G.R.; Moreira, J. The glutathione antioxidant system as a biomarker suite for the assessment of heavy metal exposure and effect in the grey mangrove, Avicennia marina (Forsk.) Vierh. Mar. Pollut. Bull. 2008, 56, 1119–1127. [Google Scholar] [CrossRef]
- MacFarlane, G.R.; Burchett, M.D. Photosynthetic pigments and peroxidase activity as indicators of heavy metal stress in the grey mangrove, Avicennia marina (Forsk.) Vierh. Mar. Pollut. Bull. 2001, 42, 233–240. [Google Scholar] [CrossRef]
- Kinuthia, G.K.; Ngure, V.; Beti, D.; Lugalia, R.; Wangila, A.; Kamau, L. Levels of heavy metals in wastewater and soil sample from open drainage channels in Nairobi, Kenya: Community health implication. Sci. Rep. 2020, 10, 8434. [Google Scholar] [CrossRef]
- United States of Environmental Protection Agency (U.S. EPA). Effluent Limitations Guideline and Standards (ELG) Database. Available online: https://owapps.epa.gov/elg/results (accessed on 1 October 2022).
- World Health Organization (WHO). Regional Office for the Eastern Mediterranean. A Compendium of Standards for Wastewater Reuse in the Eastern Mediterranean Region. Available online: https://apps.who.int/iris/handle/10665/116515 (accessed on 1 October 2022).
- Rahman, M.A.; Hashem, M.A.; Rana, M.S.; Islam, M.R. Manganese in potable water of nine districts, Bangladesh: Human health risk. Environ. Sci. Pollut. Res. 2021, 28, 45663–45675. [Google Scholar] [CrossRef]
- Rojas, A.; Holguin, G.; Glick, B.R.; Bashan, Y. Synergism between Phyllobacterium sp. (N2-fixer) and Bacillus licheniformis (P-solubilizer), both from a semiarid mangrove rhizosphere. FEMS Microbiol. Ecol. 2001, 35, 181–187. [Google Scholar] [CrossRef]
- Custodio, M.; Espinoza, C.; Peñaloza, R.; Peralta-Ortiz, T.; Sánchez-Suárez, H.; Ordinola-Zapata, A.; Vieyra-Peña, E. Microbial diversity in intensively farmed lake sediment contaminated by heavy metals and identification of microbial taxa bioindicators of environmental quality. Sci. Rep. 2022, 12, 80. [Google Scholar] [CrossRef]
- Bisessar, S. Effect of heavy metals on microorganisms in soils near a secondary lead smelter. Water Air Soil Pollut. 1982, 17, 305–308. [Google Scholar] [CrossRef]
- Aoyama, M.; Nagumo, T. Effects of heavy metal accumulation in apple orchard soils on microbial biomass and microbial activities. Soil Sci. Plant Nutr. 1997, 43, 601–612. [Google Scholar] [CrossRef][Green Version]
- Andreote, F.D.; Jiménez, D.J.; Chaves, D.; Dias, A.C.F.; Luvizotto, D.M.; Dini-Andreote, F.; Fasanella, C.C.; Lopez, M.V.; Baena, S.; Taketani, R.G.; et al. The microbiome of Brazilian mangrove sediments as revealed by metagenomics. PLoS ONE 2012, 7, e38600. [Google Scholar] [CrossRef]
- Fiebig, A.; Herrou, J.; Willett, J.; Crosson, S. General stress signaling in the Alphaproteobacteria. Ann. Rev. Genet. 2015, 49, 603–625. [Google Scholar] [CrossRef]
- Zilius, M.; Bonaglia, S.; Broman, E.; Chiozzini, V.G.; Samuiloviene, A.; Nascimento, F.J.A.; Cardini, U.; Bartoli, M. N2 fixation dominates nitrogen cycling in a mangrove fiddler crab holobiont. Sci. Rep. 2020, 10, 13966. [Google Scholar] [CrossRef]
- Zhao, M.; Xue, K.; Wang, F.; Liu, S.; Bai, S.; Sun, B.; Zhou, J.; Yang, Y. Microbial mediation of biogeochemical cycles revealed by simulation of global changes with soil transplant and cropping. ISME J. 2014, 8, 2045–2055. [Google Scholar] [CrossRef]
- Pongsilp, N.; Nimnoi, P. Inoculation of Ensifer fredii strain LP2/20 immobilized in agar results in growth promotion and alteration of bacterial community structure of Chinese kale planted soil. Sci. Rep. 2020, 10, 15857. [Google Scholar] [CrossRef]
- Pongsilp, N.; Nimnoi, P. Paenibacillus sp. strain OL15 immobilized in agar as a potential bioremediator for waste lubricating oil-contaminated soils and insights into soil bacterial communities affected by inoculations of the strain and environmental factors. Biology 2022, 11, 727. [Google Scholar] [CrossRef]
- Behera, B.C.; Mishra, R.R.; Dutta, S.K.; Thatoi, H.N. Sulphur oxidising bacteria in mangrove ecosystem: A review. Afr. J. Biotechnol. 2014, 13, 2897–2907. [Google Scholar]
- Tourna, M.; Stieglmeier, M.; Spang, A.; Könneke, M.; Schintlmeister, A.; Urich, T.; Engel, M.; Schloter, M.; Wagner, M.; Richter, A.; et al. Nitrososphaera viennensis, an ammonia oxidizing archaeon from soil. Proc. Natl. Acad. Sci. 2011, 108, 8420–8425. [Google Scholar] [CrossRef]
- Burdorf, L.D.W.; Tramper, A.; Seitaj, D.; Meire, L.; Hidalgo-Martinez, S.; Zetsche, E.M.; Boschker, H.T.S.; Meysman, F.J.R. Long-distance electron transport occurs globally in marine sediments. Biogeosciences 2017, 14, 683–701. [Google Scholar] [CrossRef]
- Pfeffer, C.; Larsen, S.; Song, J.; Dong, M.; Besenbacher, F.; Meyer, R.L.; Kjeldsen, K.U.K.; Schreiber, L.; Gorby, Y.A.; El-Naggar, M.Y.; et al. Filamentous bacteria transport electrons over centimetre distances. Nature 2012, 491, 218–221. [Google Scholar] [CrossRef]
- Reguera, G. Bacterial power cords. Nature 2012, 491, 201–202. [Google Scholar] [CrossRef]
- Holmer, M.; Storkholm, P. Sulphate reduction and sulphur cycling in lake sediments: A review. Freshw. Biol. 2001, 46, 431–451. [Google Scholar] [CrossRef]
- Mendes, R.; Garbeva, P.; Raaijmakers, J.M. The rhizosphere microbiome: Significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol. Rev. 2013, 37, 634–663. [Google Scholar] [CrossRef]
- Jehlička, J.; Osterrothová, K.; Oren, A.; Edwards, H.G.M. Raman spectrometric discrimination of flexirubin pigments from two genera of Bacteroidetes. FEMS Microbiol. Lett. 2013, 348, 97–102. [Google Scholar] [CrossRef]
- Romano, R.G.; Bendia, A.G.; Moreira, J.C.F.; Franco, D.C.; Signori, C.N.; Yu, T.; Wang, F.; Jovane, L.; Pellizari, V.H. Bathyarchaeia occurrence in rich methane sediments from a Brazilian ría. Estuar. Coast. Shelf Sci. 2021, 263, 107631. [Google Scholar] [CrossRef]
- Loh, H.Q.; Hervé, V.; Brune, A. Metabolic potential for reductive acetogenesis and a movel energy-converting [NiFe] hydrogenase in Bathyarchaeia from termite guts-A genome-centric analysis. Front. Microbiol. 2021, 11, 635786. [Google Scholar] [CrossRef]
- Hu, W.; Pan, J.; Wang, B.; Guo, J.; Li, M.; Xu, M. Metagenomic insights into the metabolism and evolution of a new Thermoplasmata order (Candidatus gimiplasmatales). Environ. Microbiol. 2020, 23, 3695–3709. [Google Scholar] [CrossRef]
- Taylor, H.B.; Kurtz, H.D., Jr. Microbial community structure shows differing levels of temporal stability in intertidal beach sands of the grand strand region of South Carolina. PLoS ONE 2020, 15, e0229387. [Google Scholar] [CrossRef]
- Bergo, N.M.; Bendia, A.G.; Ferreira, J.C.N.; Murton, B.J.; Bandini, F.P.; Pellizari, V.H. Microbial diversity of deep-sea ferromanganese crust field in the Rio Grande Rise, Southwestern Atlantic Ocean. Microb. Ecol. 2021, 82, 344–355. [Google Scholar] [CrossRef]
- Cavalcanti, J.A.D.; Santos, R.V. Ferromanganese crust: Is a type of cenozoic black stromatolite in seabed? The case of the Rio Grande Rise, South Atlantic Ocean. Int. J. Paleobiol. Paleontol. 2022, 5, 000129. [Google Scholar]
- Meng, S.; Peng, T.; Liu, X.; Wang, H.; Huang, T.; Gu, J.D.; Hu, Z. Ecological role of bacteria involved in the biogeochemical cycles of mangroves based on functional genes detected through GeoChip 5.0. mSphere. 2022, 7, e0093621. [Google Scholar] [CrossRef] [PubMed]
- Jing, H.; Xiao, X.; Zhang, Y.; Li, Z.; Jian, H.; Luo, Y.; Han, Z. Composition and ecological roles of the core microbiome along the Abyssal-Hadal transition zone sediments of the Mariana Trench. Microbiol. Spectr. 2022, 10, e01988-21. [Google Scholar] [CrossRef] [PubMed]
- Vuillemin, A.; Coskun, Ö.K.; Orsi, W.D. Microbial activities and selection from surface ocean to subseafloor on the Namibian Continental Shelf. Appl. Environ. Microbiol. 2022, 88, e0021622. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, T.T.H.; Pereira, C.F.; Melo, W.G.P.; Menegatti, C.; Andrade, P.H.M.; Groppo, M.; Lacava, P.T.; Currie, C.R.; Pupo, M.T. Insights into the ecological role of Pseudomonas spp. in an ant-plant symbiosis. Front. Microbiol. 2021, 12, 621274. [Google Scholar] [CrossRef]
- Li, Y.; Jing, H.; Xia, X.; Cheung, S.; Suzuki, K.; Liu, H. Metagenomic insights into the microbial community and nutrient cycling in the Western Subarctic Pacific Ocean. Front. Microbiol. 2018, 9, 623. [Google Scholar] [CrossRef]
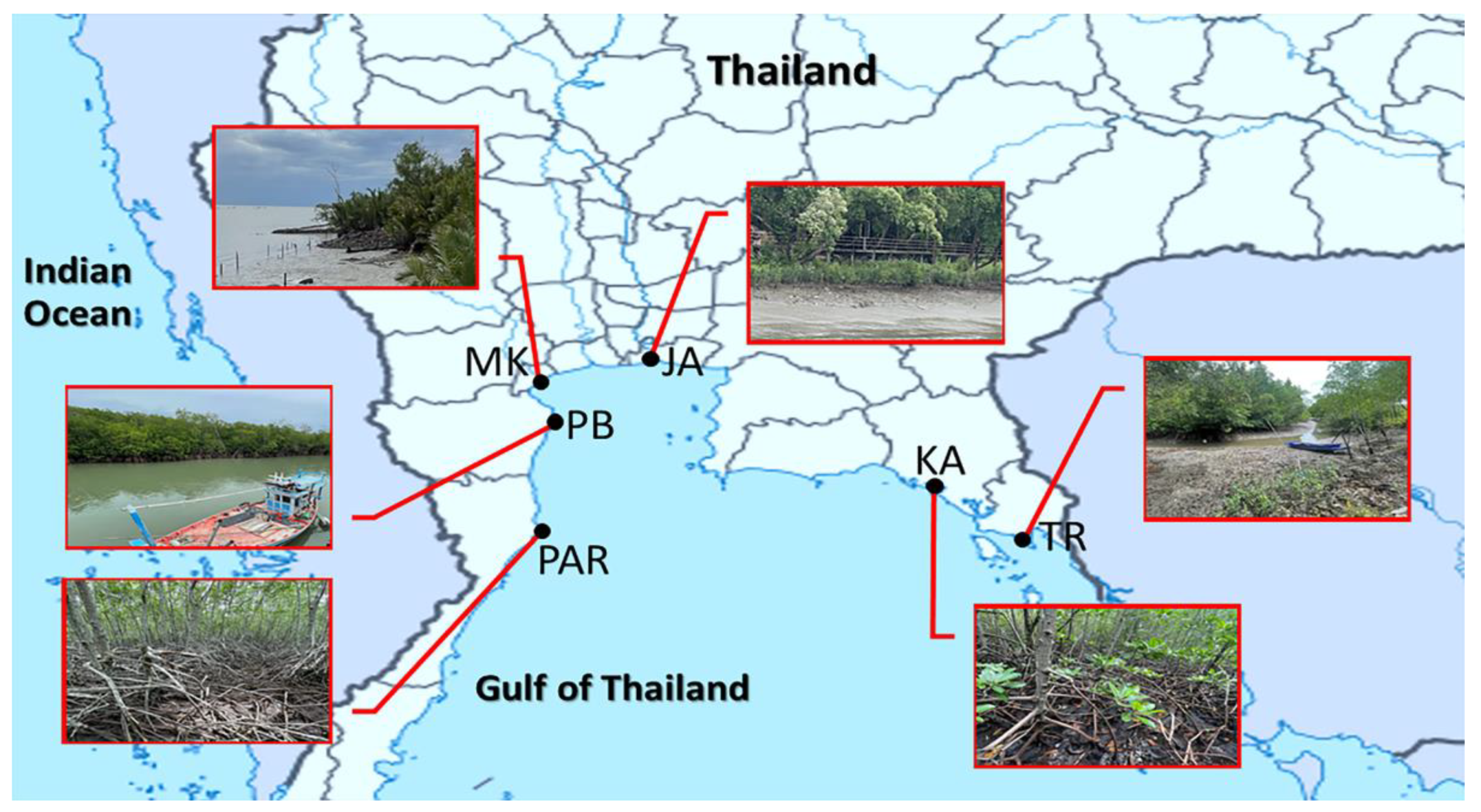
| Site Code | Place | District, Province | Sampling Date | Latitude | Longitude |
|---|---|---|---|---|---|
| TR | Black Sand Beach | Laemngop, Trat | 21 August 2021 | 12.1723 N | 102.4076 E |
| KA | Kungkrabaen Bay | Thamai, Chanthaburi | 21 August 2021 | 12.5719 N | 101.9011 E |
| JA | Chaopraya Estuary | Phrasamutchedi, Samutprakan | 7 August 2021 | 13.3702 N | 99.9919 E |
| MK | Mae Klong Estuary | Mueang, Samutsongkhram | 7 August 2021 | 13.5997 N | 100.5866 E |
| PB | Laemphakbia Promontory | Laemphakbia, Phetchaburi | 14 August 2021 | 13.0413 N | 100.0902 E |
| PAR | Pranburi forest park | Pranburi, Prachuapkhirikhan | 14 August 2021 | 12.4236 N | 99.9813 E |
| Parameter * | Soil Samples from Site | |||||
|---|---|---|---|---|---|---|
| TR | KA | JA | MK | PB | PAR | |
| pH | 6.28 ± 0.01 c ** | 5.52 ± 0.02 b | 6.91 ± 0.01 d | 7.12 ± 0.00 e | 7.40 ± 0.01 f | 4.98 ± 0.00 a |
| EC (dS/m) | 19.20 ± 0.00 b | 39.71 ± 0.01 e | 12.33 ± 0.01 a | 27.21 ± 0.01 c | 31.60 ± 0.00 d | 44.51 ± 0.01 f |
| OM (g/kg) | 0.57 ± 0.01 e | 1.85 ± 0.19 f | 0.35 ± 0.00 c | 0.32 ± 0.01 b | 0.19 ± 0.00 a | 0.52 ± 0.00 d |
| N (mg/kg) | 20.00 ± 0.00 e | 45.00 ± 0.01 f | 16.00 ± 0.00 d | 14.00 ± 0.00 c | 9.000 ± 0.00 a | 12.60 ± 0.01 b |
| P (mg/kg) | 20.98 ± 0.05 a | 90.98 ± 0.14 f | 74.43 ± 0.39 e | 32.82 ± 0.17 b | 72.62 ± 0.21 d | 34.77 ± 0.27 c |
| K (mg/kg) | 793.44 ± 5.84 d | 1022.06 ± 16.17 e | 729.60 ± 14.61 c | 732.96 ± 7.90 c | 641.78 ± 19.18 b | 523.70 ± 11.00 a |
| Ca (mg/kg) | 1686.20 ± 96.88 b | 2880.05 ± 12.73 c | 1838.63 ± 16.32 b | 6715.81 ± 37.57 e | 6020.12 ± 125.42 d | 972.03 ± 0.51 a |
| Mg (mg/kg) | 1550.79 ± 7.52 b | 3614.57 ± 29.35 d | 1737.39 ± 116.96 c | 1438.67 ± 24.34 ab | 1297.53 ± 35.96 a | 1440.86 ± 32.79 ab |
| Cd (mg/kg) | ND *** | ND | ND | ND | ND | ND |
| Cr (mg/kg) | ND | ND | ND | ND | ND | ND |
| Ag (mg/kg) | ND | ND | ND | ND | ND | ND |
| Se (mg/kg) | ND | ND | ND | ND | ND | ND |
| Cu (mg/kg) | 4.40 ± 0.07 c | 9.05 ± 0.33 d | 11.53 ± 0.17 e | 2.97 ± 0.08 b | 1.00 ± 0.01 a | 3.95 ± 0.06 c |
| Al (mg/kg) | ND | ND | ND | ND | ND | ND |
| Ba (mg/kg) | ND | ND | ND | ND | ND | ND |
| Hg (mg/kg) | ND | ND | ND | ND | ND | ND |
| Mn (mg/kg) | 101.96 ± 2.60 c | 5.54 ± 0.17 a | 191.14 ± 2.21 e | 157.81 ± 6.31 d | 105.03 ± 3.65 c | 21.75 ± 0.55 b |
| As (mg/kg) | ND | ND | ND | ND | ND | ND |
| Zn (mg/kg) | 1.92 ± 0.13 b | 5.94 ± 0.33 d | 21.43 ± 0.31 e | 3.76 ± 0.19 c | 0.92 ± 0.02 a | 5.68 ± 0.09 d |
| Pb (mg/kg) | ND | ND | ND | ND | ND | ND |
| Fe (mg/kg) | 251.92 ± 9.32 c | 273.26 ± 5.37 d | 256.25 ± 7.57 cd | 119.91 ± 1.57 b | 58.01 ± 0.05 a | 383.34 ± 11.43 e |
| Sand (%) | 28.92 | 10.14 | 24.18 | 32.04 | 31.74 | 58.09 |
| Silt (%) | 59.74 | 56.89 | 60.39 | 46.58 | 58.18 | 31.24 |
| Clay (%) | 11.33 | 32.97 | 15.44 | 21.38 | 10.08 | 10.67 |
| Soil texture | Silt Loam | Silty Clay Loam | Silt Loam | Loam | Silt Loam | Sandy Loam |
| Parameter * | Surface Sample from Site | |||||
|---|---|---|---|---|---|---|
| TR | KA | JA | MK | PB | PAR | |
| pH | 7.660 ± 0.017 c ** | 7.536 ± 0.005 a | 7.880 ± 0.020 e | 8.010 ± 0.010 f | 7.603 ± 0.015 b | 7.800 ± 0.017 d |
| NaCl (%) | 0.000 | 1.000 ± 0.000 a | 0.000 | 0.000 | 3.000 ± 0.000 c | 2.000 ± 0.000 b |
| Cd (mg/L) | ND *** | ND | ND | ND | ND | ND |
| Cr (mg/L) | ND | <LOQ *** | ND | <LOQ | ND | ND |
| Ag (mg/L) | ND | 0.026 ± 0.001 b | ND | ND | 0.011 ± 0.000 a | ND |
| Se (mg/L) | ND | ND | ND | ND | ND | ND |
| Cu (mg/L) | 0.171 ± 0.006 | ND | ND | ND | ND | ND |
| Al (mg/L) | 0.013 ± 0.000 a | 0.028 ± 0.001 c | <LOQ | <LOQ | 0.019 ± 0.000 b | 0.013 ± 0.000 a |
| Ba (mg/L) | ND | 0.014 ± 0.000 a | 0.027 ± 0.000 c | 0.039 ± 0.001 d | 0.019 ± 0.000 b | 0.056 ± 0.001 e |
| Hg (mg/L) | ND | ND | ND | ND | ND | ND |
| Mn (mg/L) | 0.122 ± 0.007 c | 0.032 ± 0.001 b | 1.296 ± 0.02 d | 0.010 ± 0.000 a | <LOQ | <LOQ |
| As (mg/L) | <LOQ | <LOQ | <LOQ | 0.023 ± 0.000 | <LOQ | <LOQ |
| Zn (mg/L) | 0.011 ± 0.000 a | <LOQ | <LOQ | 0.015 ± 0.000 b | <LOQ | <LOQ |
| Pb (mg/L) | <LOQ | <LOQ | 0.012 ± 0.001 a | <LOQ | 0.016 ± 0.000 b | 0.018 ± 0.001 c |
| Fe (mg/L) | ND | ND | ND | ND | ND | <LOQ |
| Soil Samples Form Site | Number of Observed Species * | Diversity Indices * | Richness Indices * | ||
|---|---|---|---|---|---|
| Shanon–Weaver | Simpson | Chao1 | ACE | ||
| TR | 7563.66 ± 657.49 a ** | 11.20 ± 0.25 a | 0.99 ± 0.00 a | 10,203.01 ± 907.83 a | 10,573.65 ± 928.85 a |
| KA | 7496.66 ± 110.56 a | 11.27 ± 0.05 a | 0.99 ± 0.00 a | 9619.66 ± 951.11 a | 10,072.83 ± 784.60 a |
| JA | 5881.33 ± 126.47 a | 10.63 ± 0.12 a | 0.99 ± 0.00 a | 7541.69 ± 871.15 a | 7872.85 ± 728.95 a |
| MK | 5814.00 ± 2415.35 a | 10.04 ± 1.75 a | 0.98 ± 0.02 a | 7697.23 ± 3638.31 a | 8051.35 ± 3787.22 a |
| PB | 6260.00 ± 811.15 a | 10.67 ± 0.39 a | 0.99 ± 0.01 a | 8118.93 ± 1757.09 a | 8521.46 ± 1736.08 a |
| PAR | 6295.66 ± 931.50 a | 11.05 ± 0.19 a | 0.99 ± 0.00 a | 7748.19 ± 2109.44 a | 7892.24 ± 2140.54 a |
| Correlations | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Factors | Correlation | Observed Species | Bacterial Richness | Bacterial Community | Gamma-Proteobacteria | Desulfobacteria | Bacteroidia | Anaerolineae | Alpha-Proteobacteria | Bathyarchaeia | Acidobacteriae | Nitrososphaeria | Clostridia | Thermoplasmata |
| Soil pH | rs | −0.327 | −0.146 | −0.508 * | 0.569 * | −0.708 ** | −0.160 | 0.522 * | 0.058 | 0.177 | −0.226 | 0.329 | 0.187 | −0.406 |
| Sig. | 0.185 | 0.564 | 0.031 | 0.014 | 0.001 | 0.526 | 0.026 | 0.820 | 0.483 | 0.367 | 0.183 | 0.458 | 0.095 | |
| Soil EC | rs | 0.102 | 0.001 | 0.322 | −0.048 | 0.540 * | 0.551 * | −0.415 | −0.139 | −0.392 | −0.062 | −0.396 | 0.141 | −0.007 |
| Sig. | 0.686 | 0.997 | 0.193 | 0.851 | 0.021 | 0.018 | 0.087 | 0.583 | 0.108 | 0.807 | 0.104 | 0.578 | 0.977 | |
| Soil OM | rs | 0.602 ** | 0.408 | 0.649 ** | −0.558 * | 0.647 ** | 0.199 | −0.534 * | −0.096 | −0.406 | 0.049 | −0.207 | −0.414 | 0.342 |
| Sig. | 0.008 | 0.093 | 0.004 | 0.016 | 0.004 | 0.428 | 0.023 | 0.705 | 0.095 | 0.848 | 0.409 | 0.088 | 0.165 | |
| Soil N | rs | 0.556 * | 0.398 | 0.515 * | −0.598 ** | 0.387 | 0.141 | −0.403 | −0.243 | −0.347 | −0.025 | −0.185 | −0.394 | 0.408 |
| Sig. | 0.017 | 0.102 | 0.029 | 0.009 | 0.113 | 0.577 | 0.097 | 0.330 | 0.158 | 0.921 | 0.462 | 0.106 | 0.093 | |
| Soil P | rs | −0.066 | −0.143 | −0.064 | −0.320 | 0.539 * | 0.156 | −0.110 | −0.598 ** | 0.091 | −0.527 * | −0.463 | −0.279 | 0.680 ** |
| Sig. | 0.794 | 0.572 | 0.801 | 0.195 | 0.021 | 0.536 | 0.665 | 0.009 | 0.720 | 0.025 | 0.053 | 0.262 | 0.002 | |
| Soil K | rs | 0.519 * | 0.426 | 0.381 | −0.278 | 0.156 | 0.245 | −0.315 | −0.191 | −0.478 * | −0.226 | −0.082 | −0.358 | 0.154 |
| Sig. | 0.027 | 0.078 | 0.119 | 0.265 | 0.537 | 0.328 | 0.203 | 0.448 | 0.045 | 0.367 | 0.748 | 0.145 | 0.542 | |
| Soil Ca | rs | −0.146 | −0.049 | −0.259 | 0.317 | −0.492 * | 0.404 | 0.139 | −0.282 | −0.325 | −0.207 | −0.146 | 0.387 | −0.232 |
| Sig. | 0.565 | 0.848 | 0.299 | 0.200 | 0.038 | 0.097 | 0.581 | 0.257 | 0.188 | 0.409 | 0.565 | 0.113 | 0.354 | |
| Soil Mg | rs | 0.404 | 0.247 | 0.383 | −0.666 ** | 0.465 | 0.065 | −0.321 | −0.441 | −0.143 | −0.086 | −0.302 | −0.397 | 0.668 ** |
| Sig. | 0.097 | 0.324 | 0.117 | 0.003 | 0.052 | 0.798 | 0.194 | 0.067 | 0.570 | 0.735 | 0.223 | 0.103 | 0.002 | |
| Soil Fe | rs | 0.195 | 0.005 | 0.356 | −0.496 * | 0.697 ** | 0.129 | −0.513 * | −0.092 | −0.152 | 0.127 | −0.404 | −0.234 | 0.513 * |
| Sig. | 0.438 | 0.984 | 0.147 | 0.036 | 0.001 | 0.610 | 0.030 | 0.717 | 0.548 | 0.616 | 0.097 | 0.349 | 0.030 | |
| Soil Mn | rs | −0.511 * | −0.360 | −0.659 ** | 0.261 | −0.717 ** | −0.463 | 0.529 * | 0.073 | 0.480 * | 0.119 | 0.232 | 0.207 | −0.030 |
| Sig. | 0.030 | 0.142 | 0.003 | 0.295 | 0.001 | 0.053 | 0.024 | 0.773 | 0.044 | 0.639 | 0.354 | 0.409 | 0.906 | |
| Soil Cu | rs | 0.199 | 0.071 | 0.168 | −0.655 ** | 0.393 | −0.293 | −0.270 | −0.216 | 0.115 | 0.038 | −0.245 | −0.456 | 0.730 ** |
| Sig. | 0.428 | 0.779 | 0.504 | 0.003 | 0.107 | 0.237 | 0.279 | 0.390 | 0.650 | 0.880 | 0.327 | 0.057 | 0.001 | |
| Soil Zn | rs | −0.137 | −0.269 | −0.074 | −0.516 * | 0.410 | −0.107 | −0.284 | −0.404 | 0.189 | −0.032 | −0.574 * | −0.210 | 0.914 ** |
| Sig. | 0.586 | 0.281 | 0.769 | 0.028 | 0.091 | 0.671 | 0.253 | 0.096 | 0.452 | 0.900 | 0.013 | 0.403 | 0.000 | |
| Water pH | rs | −0.467 | −0.366 | −0.467 | 0.114 | −0.551 * | −0.233 | 0.215 | 0.061 | 0.302 | 0.411 | −0.118 | 0.416 | 0.107 |
| Sig. | 0.051 | 0.135 | 0.051 | 0.653 | 0.018 | 0.353 | 0.392 | 0.810 | 0.224 | 0.090 | 0.641 | 0.086 | 0.671 | |
| Water NaCl | rs | −0.143 | −0.157 | −0.023 | 0.263 | 0.370 | 0.230 | 0.017 | −0.023 | 0.003 | −0.363 | −0.023 | 0.003 | −0.183 |
| Sig. | 0.571 | 0.535 | 0.927 | 0.291 | 0.131 | 0.359 | 0.948 | 0.927 | 0.990 | 0.139 | 0.927 | 0.990 | 0.467 | |
| Water Al | rs | 0.360 | 0.253 | 0.438 | −0.054 | 0.652 ** | 0.418 | −0.270 | −0.168 | −0.327 | −0.450 | −0.095 | −0.285 | −0.039 |
| Sig. | 0.143 | 0.310 | 0.069 | 0.833 | 0.003 | 0.085 | 0.278 | 0.504 | 0.185 | 0.061 | 0.709 | 0.252 | 0.878 | |
| Water Mn | rs | 0.221 | 0.181 | 0.090 | −0.477 * | −0.020 | −0.389 | 0.038 | −0.119 | 0.196 | 0.072 | 0.069 | −0.371 | 0.442 |
| Sig. | 0.378 | 0.472 | 0.721 | 0.045 | 0.937 | 0.110 | 0.881 | 0.638 | 0.436 | 0.778 | 0.784 | 0.129 | 0.066 | |
| Water As | rs | −0.129 | −0.043 | −0.158 | 0.244 | −0.589 * | 0.330 | −0.043 | −0.101 | −0.330 | 0.273 | −0.244 | 0.560 * | −0.187 |
| Sig. | 0.609 | 0.865 | 0.531 | 0.329 | 0.010 | 0.180 | 0.865 | 0.691 | 0.180 | 0.273 | 0.329 | 0.016 | 0.458 | |
| Water Zn | rs | 0.189 | 0.267 | 0.108 | 0.193 | −0.634 ** | 0.122 | 0.007 | 0.234 | −0.345 | 0.441 | 0.197 | 0.349 | −0.475 * |
| Sig. | 0.452 | 0.284 | 0.671 | 0.443 | 0.005 | 0.628 | 0.977 | 0.351 | 0.161 | 0.067 | 0.434 | 0.156 | 0.046 | |
| Water Pb | rs | −0.561 * | −0.526 * | −0.419 | 0.221 | 0.140 | −0.288 | 0.263 | 0.104 | 0.537 * | −0.081 | 0.048 | 0.072 | 0.027 |
| Sig. | 0.015 | 0.025 | 0.083 | 0.379 | 0.580 | 0.246 | 0.292 | 0.681 | 0.022 | 0.749 | 0.851 | 0.776 | 0.917 | |
| Bacterial Class | Bacterial Number in Soil Sample from Site * | |||||
|---|---|---|---|---|---|---|
| TR | KA | JA | MK | PB | PAR | |
| Gamma-Proteobacteria | 19.34 ± 0.70 a ** | 16.52 ± 2.53 a | 17.68 ± 5.31 a | 28.93 ± 14.55 a | 29.54 ± 3.46 a | 19.66 ± 1.90 a |
| Desulfobacteria | 3.16 ± 1.17 a | 15.66 ± 1.30 c | 3.73 ± 0.65 a | 1.87 ± 0.68 a | 3.51 ± 0.08 a | 8.09 ± 0.34 b |
| Bacteroidia | 5.29 ± 2.07 ab | 10.78 ± 0.52 b | 2.91 ± 1.49 a | 8.64 ± 3.35 b | 6.10 ± 1.45 ab | 6.24 ± 2.22 ab |
| Anaerolineae | 7.33 ± 1.44 a | 4.24 ± 0.76 a | 8.51 ± 2.01 a | 6.35 ± 3.16 a | 8.99 ± 0.94 a | 5.85 ± 1.24 a |
| Alpha-Proteobacteria | 8.74 ± 1.41 a | 5.79 ± 0.10 a | 6.27 ± 0.50 a | 6.93 ± 2.18 a | 7.53 ± 1.40 a | 7.32 ± 1.49 a |
| Bathyarchaeia | 1.04 ± 0.37 ab | 0.49 ± 0.13 a | 9.10 ± 0.24 c | 0.72 ± 0.33 ab | 1.59 ± 0.71 b | 1.14 ± 0.16 ab |
| Acidobacteriae | 3.93 ± 2.39 a | 0.61 ± 0.12 a | 3.17 ± 4.48 a | 3.15 ± 0.15 a | 0.90 ± 0.78 a | 4.20 ± 3.26 a |
| Nitrososphaeria | 6.18 ± 0.41 c | 0.28 ± 0.04 a | 0.56 ± 0.60 a | 0.42 ± 0.23 a | 2.93 ± 0.31 b | 0.44 ± 0.21 a |
| Clostridia | 0.91 ± 1.45 ab | 0.15 ± 0.03 a | 0.43 ± 0.57 a | 3.95 ± 2.61 b | 0.39 ± 0.19 a | 0.74 ± 0.57 ab |
| Thermoplasmata | 0.26 ± 0.28 a | 2.12 ± 0.65 b | 3.39 ± 0.87 c | 0.58 ± 0.88 a | 0.16 ± 0.06 a | 1.29 ± 0.19 ab |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nimnoi, P.; Pongsilp, N. Insights into Bacterial Communities and Diversity of Mangrove Forest Soils along the Upper Gulf of Thailand in Response to Environmental Factors. Biology 2022, 11, 1787. https://doi.org/10.3390/biology11121787
Nimnoi P, Pongsilp N. Insights into Bacterial Communities and Diversity of Mangrove Forest Soils along the Upper Gulf of Thailand in Response to Environmental Factors. Biology. 2022; 11(12):1787. https://doi.org/10.3390/biology11121787
Chicago/Turabian StyleNimnoi, Pongrawee, and Neelawan Pongsilp. 2022. "Insights into Bacterial Communities and Diversity of Mangrove Forest Soils along the Upper Gulf of Thailand in Response to Environmental Factors" Biology 11, no. 12: 1787. https://doi.org/10.3390/biology11121787
APA StyleNimnoi, P., & Pongsilp, N. (2022). Insights into Bacterial Communities and Diversity of Mangrove Forest Soils along the Upper Gulf of Thailand in Response to Environmental Factors. Biology, 11(12), 1787. https://doi.org/10.3390/biology11121787






