Selenium Nanoparticle-Enriched and Potential Probiotic, Lactiplantibacillus plantarum S14 Strain, a Diet Supplement Beneficial for Rainbow Trout
Simple Summary
Abstract
1. Introduction
2. Materials and Method
2.1. Animals Used
2.2. Obtention of Putative LAB Isolates from the Intestinal Content of Rainbow Trout
2.3. Search for Isolates from the Intestinal Content of Rainbow Trout Having Characteristics of LAB
2.3.1. Morphological Characterization of Isolates
2.3.2. Catalase Test
2.3.3. Carbohydrate Fermentation Tests
2.4. Search for Potential Probiotic Characteristics in the Isolated LAB Strains
2.4.1. Antibacterial Activity of the LAB Strains
2.4.2. Antibiotic Susceptibility Test of the LAB Strains
2.4.3. Hemolytic Activity of the LAB Strains
2.4.4. Hydrophobicity Assays
2.4.5. Cell Viability of the LAB Strains at a Low pH or Bile Salts
2.5. Biosynthesis and Characterization of Se0Nps-Enriched Probiotic Strain (LABstrain-Se0Nps)
2.6. DNA Isolation, 16S rDNA Gene Amplification and Sequencing of LAB Strain-Se0Nps
2.7. Effects of the LAB Strain-Se0Nps Dietary Supplementation on the Innate Immune Response, the Oxidative Status, and Productive Parameters of Rainbow Trout
2.7.1. Rainbow Trout Rearing Conditions and Experimental Design
2.7.2. Preparation of Diets
2.7.3. Rainbow Trout Sampling
2.7.4. Evaluation of ROS in White Blood Cells and Lysozyme Activity in Plasma
2.7.5. Activity of the Antioxidant Enzyme Glutathione Peroxidase (Gpx) in Plasma, Liver and Dorsal Muscle
2.7.6. Effect of Diets on Trout Growth Performance and Survival Rate
2.8. Statistics
3. Results
3.1. Isolation of LAB
3.2. Evaluation of the Attributes of a Probiotic in the LAB Strains
3.2.1. Antibacterial Activity of the LAB Strains
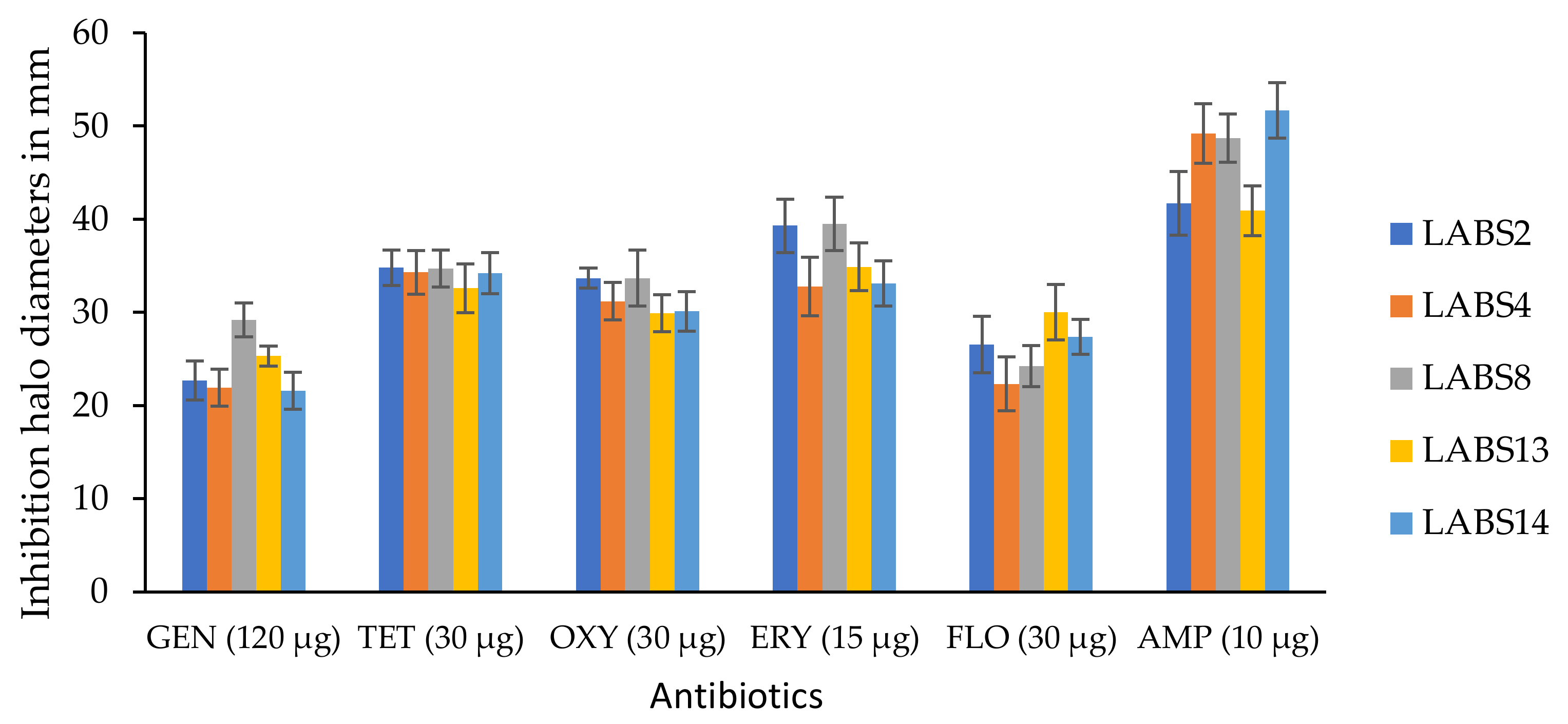
3.2.2. Antibiotic Susceptibility of the LAB Strains
3.2.3. Hemolytic Activity
3.2.4. Hydrophobicity Assays
3.2.5. Cell Viability of the LAB Strains at Low pH
3.2.6. Cell Viability of the LAB Strains in the Presence of Bile Salts
3.3. Screening for Biosynthesis of Se0Nps by LAB Strains (LABstrain-Se0Nps)
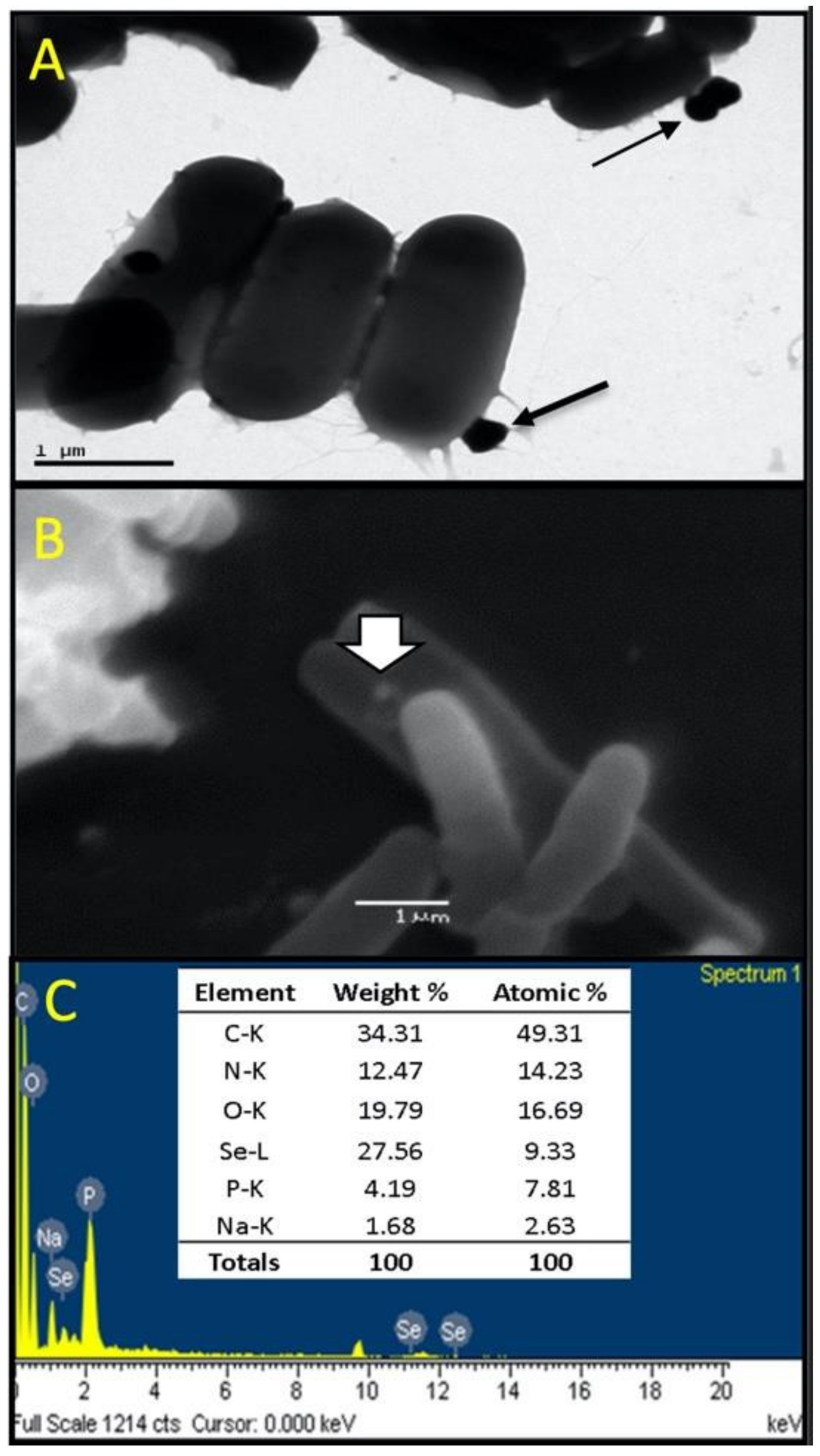
3.4. Characterization of Se Nanoparticles Produced by LABS14
3.5. Molecular Identification of LABS14 Strain by 16S rDNA Sequence Analysis
3.6. Effect of the Dietary Administration of Enriched-(LABS14-Se0Nps) as a Nutritional Supplement in Rainbow Trout (In Vivo Model)
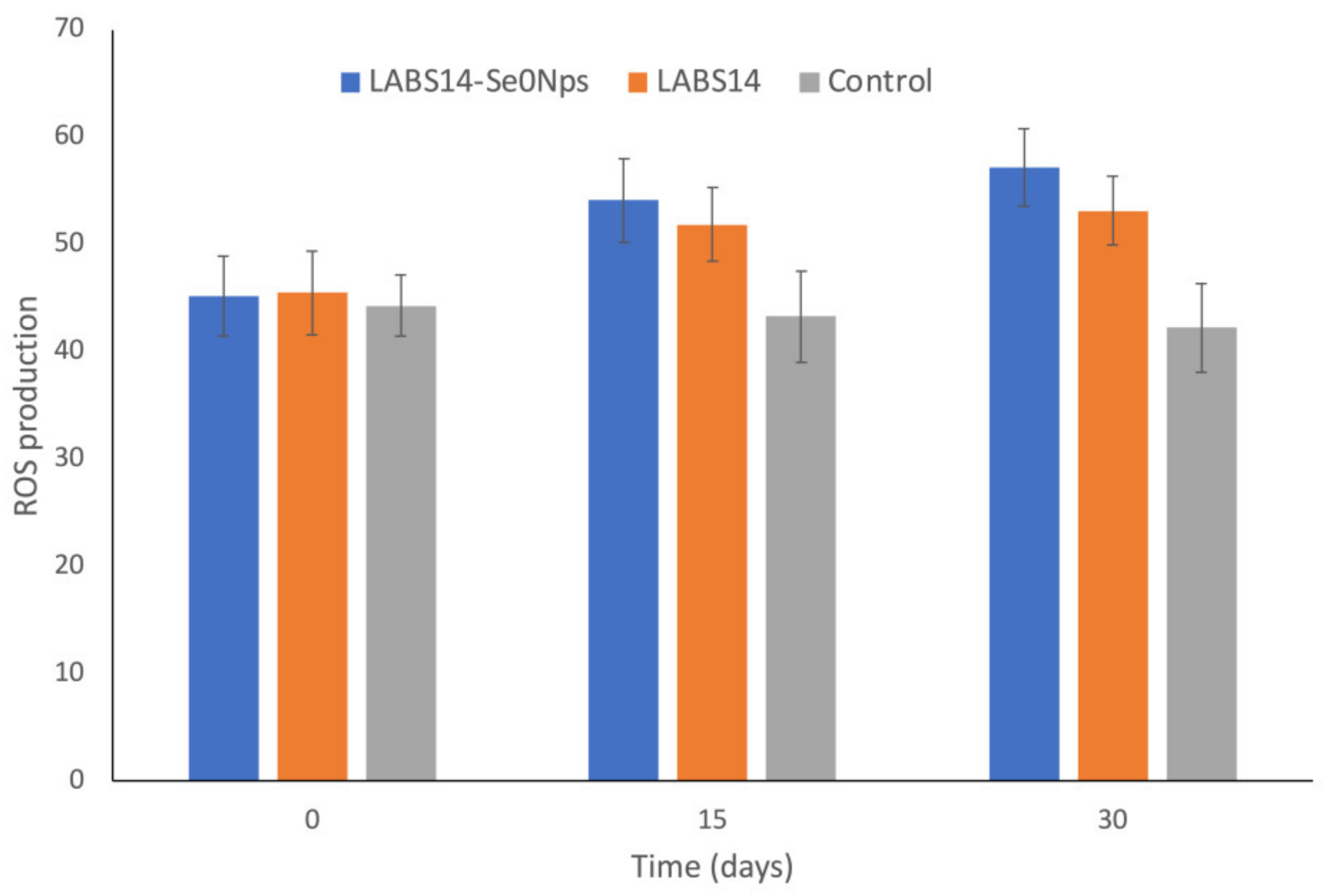
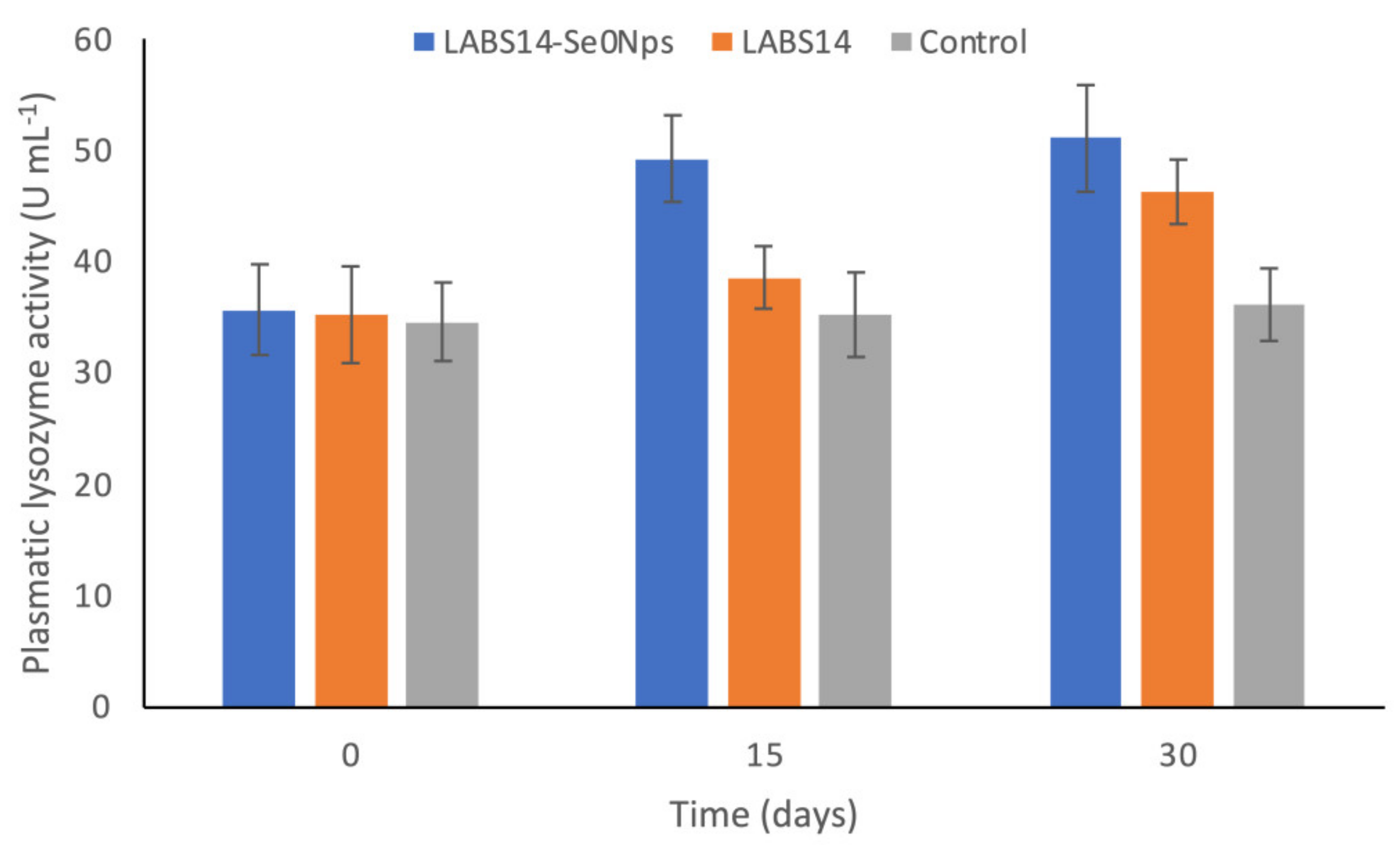
3.6.1. ROS in White Blood Cells and Lysozyme Activity in Plasma
3.6.2. Activity of the Antioxidant Enzyme Gpx
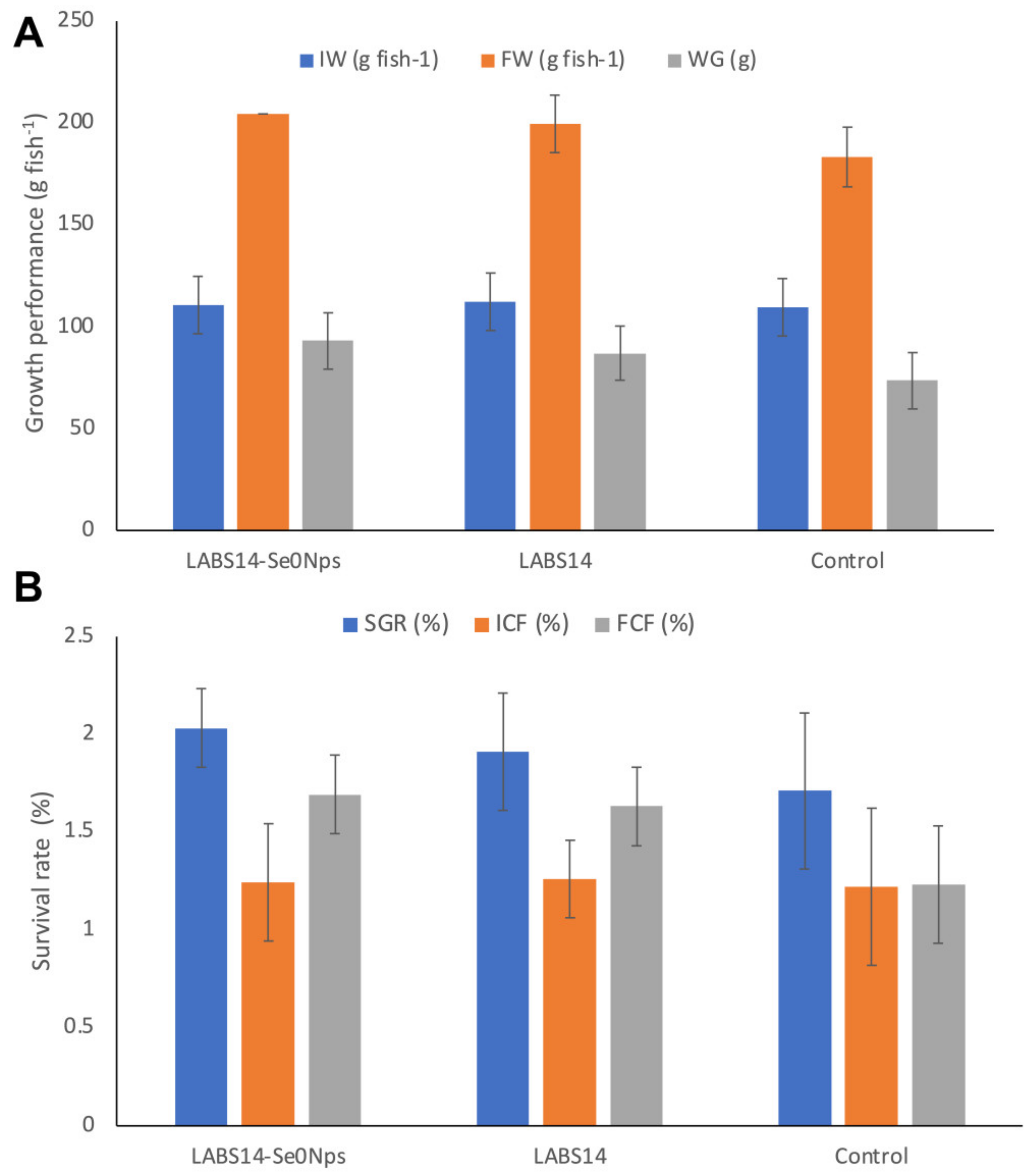
3.6.3. Growth Performance and Survival
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Iversen, A.; Asche, F.; Hermansen, Ø.; Nilstøyl, R. Production cost and competitiveness in major salmon farming countries 2003–2018. Aquaculture 2020, 522, 735089. [Google Scholar] [CrossRef]
- Valenzuela, C.A.; Ponce, C.; Zuloaga, R.; González, P.; Avendaño-Herrera, R.; Valdés, J.A.; Molina, A. Effects of crowding on the three main proteolytic mechanisms of skeletal muscle in rainbow trout (Oncorhynchus mykiss). BMC Vet. Res. 2020, 16, 294. [Google Scholar] [CrossRef] [PubMed]
- Harper, C.; Wolf, J.C. Morphologic Effects of the Stress Response in Fish. ILAR J. 2009, 50, 387–396. [Google Scholar] [CrossRef] [PubMed]
- Subpesca. Informe Ambiental de la Acuicultura; Subsecretarıa de Pesca y Acuicultura, Ministerio de Economía, Gobierno de Chile: Santiago, Chile, 2017; 91p.
- Urbina, M.A. Temporal variation on environmental variables and pollution indicators in marine sediments under sea Salmon farming cages in protected and exposed zones in the Chilean inland Southern Sea. Sci. Total Environ. 2016, 573, 841–853. [Google Scholar] [CrossRef]
- Cabello, F.C.; Godfrey, H.P. Salmon aquaculture, Piscirickettsia salmonis virulence, and One Health: Dealing with harmful synergies between heavy antimicrobial use and piscine and human health. Aquaculture 2019, 507, 451–456. [Google Scholar] [CrossRef]
- Talwar, C.; Nagar, S.; Lal, R.; Negi, R.K. Fish gut microbiome: Current approaches and future perspectives. Indian J. Microbiol. 2018, 58, 397–414. [Google Scholar] [CrossRef]
- Navarrete, P.; Mardones, P.; Opazo, R.; Espejo, R.; Romero, J. Oxytetracycline Treatment Reduces Bacterial Diversity of Intestinal Microbiota of Atlantic Salmon. J. Aquat. Anim. Health 2008, 20, 177–183. [Google Scholar] [CrossRef]
- Donati, V.L.; Madsen, L.; Middelboe, M.; Strube, M.L.; Dalsgaard, I. The Gut Microbiota of Healthy and Flavobacterium psychrophilum-Infected Rainbow Trout Fry Is Shaped by Antibiotics and Phage Therapies. Front. Microbiol. 2022, 13, 771296. [Google Scholar] [CrossRef]
- Valdés, N.; González, A.; García, V.; Tello, M. Analysis of the Microbiome of Rainbow Trout (Oncorhynchus mykiss) Exposed to the Pathogen Flavobacterium psychrophilum 10094. Microbiol. Resour. Announc. 2020, 9, e01562-19. [Google Scholar] [CrossRef]
- Wang, N.; Jiang, M.; Zhang, P.; Shu, H.; Li, Y.; Guo, Z.; Li, Y. Amelioration of Cd-induced bioaccumulation, oxidative stress and intestinal microbiota by Bacillus cereus in Carassius auratus gibelio. Chemosphere 2020, 245, 125613. [Google Scholar] [CrossRef]
- Wang, N.; Yuan, Z.; Wang, K.; Gao, D.; Liu, Z.; Liles, M.R. Consumption of florfenicol-medicated feed alters the composition of the channel catfish intestinal microbiota including enriching the relative abundance of opportunistic pathogens. Aquaculture 2019, 501, 111–118. [Google Scholar] [CrossRef]
- Sernapesca. Informe Sobre Uso de Antimicrobianos en la Salmonicultura Nacional 2019. Valparaíso: Servicio Nacional de Pesca y Acuicultura 2020. Available online: http://www.sernapesca.cl (accessed on 7 October 2022).
- FAO/WHO. Probiotics in Food. Health and Nutritional Properties and Guidelines for Evaluation; FAO Food and Nutrition Paper 85; FAO: Rome, Italy, 2006; ISSN 0254-4725. [Google Scholar]
- Sharifuzzaman, S.M.; Austin, B. Probiotics for disease control in aquaculture. In Diagnosis and Control of Diseases of Fish and Shellfish; Austin, B., Newaj-Fyzul, A., Eds.; Wiley: Hoboken, NJ, USA, 2017; Chapter 8; pp. 189–222. [Google Scholar]
- Das, S.; Mondal, K.; Haque, S. A review on application of probiotic, prebiotic and synbiotic for sustainable development of aquaculture. J. Entomol. Zool. Stud. 2017, 5, 422–429. [Google Scholar]
- Carvalho, E.D.; David, G.S.; Silva, R.J. Health and Environment in Aquaculture; IntechOpen: London, UK, 2012; 430p, Available online: https://www.intechopen.com/books/2052 (accessed on 7 October 2022).
- Merrifield, D.; Bradley, G.; Harper, G.; Baker, R.; Munn, C.; Davies, S. Assessment of the effects of vegetative and lyophilized Pediococcus acidilactici on growth, feed utilization, intestinal colonization and health parameters of rainbow trout (Oncorhynchus mykiss Walbaum). Aquac. Nutr. 2011, 17, 73–79. [Google Scholar] [CrossRef]
- Lamari, F.; Castex, M.; Larcher, T.; Ledevin, M.; Mazurais, D.; Bakhrouf, A.; Gatesoupe, F.-J. Comparison of the effects of the dietary addition of two lactic acid bacteria on the development and conformation of sea bass larvae, Dicentrarchus labrax, and the influence on associated microbiota. Aquaculture 2013, 376, 137–145. [Google Scholar] [CrossRef]
- Shahid, M.; Hussain, B.; Riaz, D.; Khurshid, M.; Ismail, M.; Tariq, M. Identification and partial characterization of potential probiotic lactic acid bacteria in freshwater Labeo rohita and Cirrhinus mrigala. Aquac. Res. 2016, 48, 1688–1698. [Google Scholar] [CrossRef]
- Quinto, E.J.; Jiménez, P.; Caro, I.; Tejero, J.; Mateo, J.; Girbés, T. Probiotic Lactic Acid Bacteria: A Review. Food Nutr. Sci. 2014, 5, 1765–1775. [Google Scholar] [CrossRef]
- Olofsson, T.C.; Butler, È.; Markowicz, P.; Lindholm, C.; Larsson, L.; Vásquez, A. Lactic acid bacterial symbionts in honeybees—An unknown key to honey’s antimicrobial and therapeutic activities. Int. Wound J. 2016, 13, 668–679. [Google Scholar] [CrossRef]
- Llewellyn, M.S.; McGinnity, P.; Dionne, M.; Letourneau, J.; Thonier, F.; Carvalho, G.R.; Creer, S.; Derome, N. The biogeography of the Atlantic salmon (Salmo salar) gut microbiome. ISME J. 2016, 10, 1280–1284. [Google Scholar] [CrossRef]
- Fidanza, M.; Panigrahi, P.; Kollmann, T.R. Lactiplantibacillus plantarum–Nomad and Ideal Probiotic. Front. Microbiol. 2021, 12, 712236. [Google Scholar] [CrossRef]
- Gildberg, A.; Johansen, A.; Bøgwald, J. Growth and survival of Atlantic salmon (Salmo salar) fry given diets supplemented with fish protein hydrolysate and lactic acid bacteria during a challenge trial with Aeromonas salmonicida. Aquaculture 1995, 138, 23–34. [Google Scholar] [CrossRef]
- Pérez-Sánchez, T.; Balcázar, J.; García, Y.; Halaihel, N.; Vendrell, D.; De Blas, I.; Merrifield, D.; Ruiz-Zarzuela, I. Identification and characterization of lactic acid bacteria isolated from rainbow trout, Oncorhynchus mykiss (Walbaum), with inhibitory activity against Lactococcus garvieae. J. Fish Dis. 2011, 34, 499–507. [Google Scholar] [CrossRef] [PubMed]
- Soltani, M.; Pakzad, K.; Taheri-Mirghaed, A.; Mirzargar, S.; Hosseini, S.P.; Yosefi, P.; Soleymani, N. Dietary Application of the Probiotic Lactobacillus plantarum 426951 Enhances Immune Status and Growth of Rainbow Trout (Oncorhynchus mykiss) Vaccinated Against Yersinia ruckeri. Probiotics Antimicrob. Proteins 2019, 11, 207–219. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Suzuki, N.; Ogra, Y. Effect of gut microflora on nutritional availability of selenium. Food Chem. 2020, 319, 126537. [Google Scholar] [CrossRef] [PubMed]
- Naderi, M.; Keyvanshokooh, S.; Salati, A.P.; Ghaedi, A. Combined or individual effects of dietary vitamin E and selenium nanoparticles on humoral immune status and serum parameters of rainbow trout (Oncorhynchus mykiss) under high stocking density. Aquaculture 2017, 474, 40–47. [Google Scholar] [CrossRef]
- Baeverfjord, G.; Prabhu, P.A.; Fjelldal, P.G.; Albrektsen, S.; Hatlen, B.; Denstadli, V.; Ytteborg, E.; Takle, H.; Lock, E.-J.; Berntssen, M.H.G.; et al. Mineral nutrition and bone health in salmonids. Rev. Aquac. 2018, 11, 740–765. [Google Scholar] [CrossRef]
- Rathore, S.S.; Murthy, H.S.; Mamun, M.A.-A.; Nasren, S.; Rakesh, K.; Kumar, B.T.N.; Abhiman, P.B.; Khandagale, A.S. Nanoselenium supplementation to ameliorate nutrition physiology, immune response, antioxidant system and disease resistance against Aeromonas hydrophila in monosex Nile tilapia (Oreochromis niloticus). Biol. Trace Elem. Res. 2021, 199, 3073–3088. [Google Scholar] [CrossRef]
- Ibrahim, M.S.; El-gendy, G.M.; Ahmed, A.I.; Elharoun, E.R.; Hassaan, M.S. Nanoselenium versus bulk selenium as a dietary supplement: Effects on growth, feed efficiency, intestinal histology, haemato-biochemical and oxidative stress biomarkers in Nile tilapia (Oreochromis niloticus Linnaeus, 1758) fingerlings. Aquac. Res. 2021, 52, 5642–5655. [Google Scholar] [CrossRef]
- Karamzadeh, M.; Yahyavi, M.; Salarzadeh, A.; Nokhbe Zare, D. The effects of different concentrations of selenium and zinc nanoparticles on growth performance, survival and chemical composition of whiteleg shrimp (Litopenaeus vannamei). Iran. Sci. Fish. J. 2021, 29, 43–51. [Google Scholar]
- Deilamy Pour, H.; Mousavi, S.M.; Zakeri, M.; Keyvanshokooh, S.; Kochanian, P. Synergistic effects of selenium and magnesium nanoparticles on growth, digestive enzymes, some serum biochemical parameters and immunity of Asian sea bass (Lates calcarifer). Biol. Trace Elem. Res. 2021, 199, 3102–3111. [Google Scholar] [CrossRef]
- Sarkar, B.; Bhattacharjee, S.; Daware, A.; Tribedi, P.; Krishnani, K.K.; Minhas, P.S. Selenium nanoparticles for stress-resilient fish and livestock. Nanoscale Res. Lett. 2015, 10, 371. [Google Scholar] [CrossRef]
- Mechlaoui, M.; Dominguez, D.; Robaina, L.; Geraert, P.-A.; Kaushik, S.; Saleh, R.; Briens, M.; Montero, D.; Izquierdo, M. Effects of different dietary selenium sources on growth performance, liver and muscle composition, antioxidant status, stress response and expression of related genes in gilthead seabream (Sparus aurata). Aquaculture 2019, 507, 251–259. [Google Scholar] [CrossRef]
- Arshad, A. Bacterial Synthesis and Applications of Nanoparticles. Nano Sci. Nano Technol. 2017, 11, 119. [Google Scholar]
- Dawood, M.A.O.; Zommara, M.; Eweedah, N.M.; Helal, A.I. The evaluation of growth performance, blood health, oxidative status and immune-related gene expression in Nile tilapia (Oreochromis niloticus) fed dietary nano selenium spheres produced by lactic acid bacteria. Aquaculture 2020, 515, 734571. [Google Scholar] [CrossRef]
- Yang, J.; Yang, H. Recent development in Se-enriched yeast, lactic acid bacteria and bifidobacterial. Crit. Rev. Food Sci. Nutr. 2021, 1–15. [Google Scholar] [CrossRef]
- Moreno-Martin, G.; Pescuma, M.; Pérez-Corona, T.; Mozzi, F.; Madrid, Y. Determination of size and mass-and number-based concentration of biogenic SeNPs synthesized by lactic acid bacteria by using a multimethod approach. Anal. Chim. Acta 2017, 992, 34–41. [Google Scholar] [CrossRef]
- Pescuma, M.; Gomez-Gomez, B.; Perez-Corona, T.; Font, G.; Madrid, Y.; Mozzi, F. Food prospects of selenium enriched-Lactobacillus acidophilus CRL 636 and Lactobacillus reuteri CRL 1101. J. Funct. Foods 2017, 35, 466–473. [Google Scholar] [CrossRef]
- Salama, H.H.; El-Sayed, N.; Abd-Rabou, N.S.; Hassan, Z.M.R. Production and use of eco-friendly selenium nanoparticles in the fortification of yoghurt. J. Food Process. Preserv. 2021, 45, e15510. [Google Scholar] [CrossRef]
- Shang, X.; Xu, W.; Zhao, Z.; Luo, L.; Zhang, Q.; Li, M.; Sun, Q.; Geng, L. Effects of exposure to cadmium (Cd) and selenium-enriched Lactobacillus plantarum in Luciobarbus capito: Bioaccumulation, antioxidant responses and intestinal microflora. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2022, 257, 109352. [Google Scholar] [CrossRef]
- Khattab, A.E.-N.; Darwish, A.M.; Othman, S.I.; Allam, A.A.; Alqhtani, H.A. Anti-inflammatory and Immunomodulatory Potency of Selenium-Enriched Probiotic Mutants in Mice with Induced Ulcerative Colitis. Biol. Trace Elem. Res. 2022, 1–15. [Google Scholar] [CrossRef]
- Office International des Epizooties (OIE). Welfare of farmed fish during transport. In OIE—Aquatic Animal Health Code; Monique Eloit: Paris, France, 2015; Chapter 7.2; Available online: https://www.woah.org (accessed on 7 October 2022).
- American Veterinary Medical Association (AVMA). Guidelines for the Euthanasia of Animals: 2020 Edition; Steven Leary: Schaumburg, IL, USA, 2020; ISBN 978-1-882691-09-8. Available online: https://www.avma.org/sites/default/files/2020-02/Guidelines-on-Euthanasia-2020.pdf (accessed on 7 October 2022).
- Huys, G.; D’Haene, K.; Swings, J. Influence of the culture medium on antibiotic susceptibility testing of food-associated lactic acid bacteria with the agar overlay disc diffusion method. Lett. Appl. Microbiol. 2002, 34, 402–406. [Google Scholar] [CrossRef]
- Saha, U.S.; Misra, R.; Tiwari, D.; Prasad, K.N. A cost-effective anaerobic culture method & its comparison with a standard method. Indian J. Med. Res. 2016, 144, 611–613. [Google Scholar] [CrossRef] [PubMed]
- Temmerman, R.; Huys, G.; Swings, J. Identification of lactic acid bacteria: Culture-dependent and culture-independent methods. Trends Food Sci. Technol. 2004, 15, 348–359. [Google Scholar] [CrossRef]
- Khalid, K. An overview of lactic acid bacteria. Int. J. Biosci. 2011, 1, 1–13. [Google Scholar]
- Procop, G.W.; Church, D.L.; Hall, G.S.; Janda, W.M.; Koneman, E.W.; Schreckenberger, P.C.; Woods, G.L. Koneman’s Color Atlas and Textbook of Diagnostic Microbiology, 7th ed.; Wolters Kluwer: Alphen aan den Rijn, The Netherlands, 2017; ISBN 9781451116595. [Google Scholar]
- Salvetti, E.; Torriani, S.; Felis, G.E. The Genus Lactobacillus: A Taxonomic Update. Probiotics Antimicrob. Proteins 2012, 4, 217–226. [Google Scholar] [CrossRef]
- Klayraung, S.; Okonogi, S. Antibacterial and Antioxidant activities of acid and bile resistant strains of Lactobacillus fermentum isolated from miang. Braz. J. Microbiol. 2009, 40, 757–766. [Google Scholar] [CrossRef] [PubMed]
- Erkus, O. Isolation, Phenotypic and Genotypic Characterization of Yoghurt Starter Bacteria. Master’s Thesis, Izmir Institute of Technology, Izmir, Turkey, 2007. [Google Scholar]
- Rondón, A.J.; Samaniego, L.M.; Bocourt, R.; Rodríguez, S.; Milián, G.; Ranilla, M.J.; Laurencio, M.; Pérez, M. Aislamiento, identificación y caracterización parcial de las propiedades probióticas de cepas de Lactobacillus sp. procedentes del tracto gastrointestinal de pollos de ceba. Cienc. Tecnol. Aliment. 2008, 6, 56–63. [Google Scholar] [CrossRef][Green Version]
- Schillinger, U.; Lücke, F. Antibacterial activity of Lactobacillus sake isolated from meat. Appl. Environ. Microbiol. 1989, 55, 1901–1906. [Google Scholar] [CrossRef]
- Geria, M.; Dambrosio, A.; Normanno, G.; Lorusso, V.; Caridi, A. Antagonistic activity of dairy lactobacilli against gram-foodborne pathogens. Acta Sci. Technol. 2014, 36, 1–6. [Google Scholar] [CrossRef]
- Alonso, S.; Castro, M.C.; Berdasco, M.; García de la Banda, I.; Moreno-Ventas, X.; Hernández de Rojas, A. Isolation and Partial Characterization of Lactic Acid Bacteria from the Gut Microbiota of Marine Fishes for Potential Application as Probiotics in Aquaculture. Probiotics Antimicrob. Proteins 2019, 11, 569–579. [Google Scholar] [CrossRef]
- M100; Performance Standard for Antimicrobial Susceptibility Testing. 32nd ed. Clinical and Laboratory Standards Institute: Malvern, PA, USA, 2022.
- Tripathi, K.D. Antimicrobial drugs, Section 12. In Essentials of Medical Pharmacology, 8th ed.; Tripathi, M., Ed.; Jaypee Brothers Medical Publishers (P) Ltd.: New Deli, India, 2019; pp. 739–906. [Google Scholar]
- Rodrigues, L.; Fortes, L.L.; Durmaz, E.; Goh, Y.J.; Sanozky-Dawes, R.B.; Klaenhammer, T.R. Characterization of Lactobacillus gasseri isolates from a breast-fed infant. Gut Microbes 2012, 3, 15–24. [Google Scholar] [CrossRef]
- Xu, H.; Jeong, H.S.; Lee, H.Y.; Ahn, J. Assessment of cell surface properties and adhesion potential of selected probiotic strains. Lett. Appl. Microbiol. 2009, 49, 434–442. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Ortiz, A.C.; Luna-González, A.; Campa-Córdova, A.I.; Escamilla-Montes, R.; Flores-Miranda, M.; Mazón-Suástegui, J.M. Isolation and characterization of potential probiotic bacteria from pustulose ark (Anadara tuberculosa) suitable for shrimp farming. Lat. Am. J. Aquat. Res. 2015, 43, 123–136. [Google Scholar] [CrossRef]
- Kaushik, J.; Kumar, A.; Duary, R.K.; Mohanty, A.K.; Grover, S.; Batish, V.K. Functional and Probiotic Attributes of an Indigenous Isolate of Lactobacillus plantarum. PLoS ONE 2009, 4, e8099. [Google Scholar] [CrossRef] [PubMed]
- Mortezaei, F.; Royan, M.; Allaf Noveirian, H.; Babakhani1, A.; Alaie Kordghashlaghi, H.; Balcazar, J.L. In vitro assessment of potential probiotic characteristics of indigenous Lactococcus lactis and Weissella oryzae isolates from rainbow trout (Oncorhynchus mykiss Walbaum). J. Appl. Microbiol. 2020, 129, 1004–1019. [Google Scholar] [CrossRef] [PubMed]
- Daza, C.; Campos, V.; Rojas, C.; Rodríguez-Llamazares, S.; Smith, C.; Mondaca, M. Reduction of selenite to elemental Selenium by Pantoea agglomerans. Gayana 2016, 80, 67–74. [Google Scholar] [CrossRef]
- Oremland, R.S.; Herbel, M.J.; Switzer-Blum, J.; Langley, S.; Beveridge, T.J.; Ajayan, P.M.; Sutto, T.; Ellis, A.V.; Curran, S. Structural and spectral features of selenium nanospheres produced by Se-respiring bacteria. Appl. Environ. Microbiol. 2004, 70, 52–60. [Google Scholar] [CrossRef]
- Dhanjal, S.; Cameotra, S.S. Aerobic biogenesis of selenium nanospheres by Bacillus cereus isolated from coalmine soil. Microb. Cell Factories 2010, 9, 52. [Google Scholar] [CrossRef]
- Torres, S.K.; Campos, V.L.; León, C.G.; Rodríguez-Llamazares, S.M.; Rojas, S.M.; González, M.; Smith, C.T.; Mondaca, M.A. Biosynthesis of selenium nanoparticles by Pantoea agglomerans and their antioxidant activity. J. Nanoparticle Res. 2012, 14, 1236. [Google Scholar] [CrossRef]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian Classifier for Rapid Assignment of rRNA Sequences into the New Bacterial Taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5277. [Google Scholar] [CrossRef]
- Campos, V.L.; León, C.; Mondaca, M.A.; Yáñez, J.; Zaror, C. Arsenic mobilization by epilithic bacterial communities associated with volcanic rocks from Camarones River Atacama Desert Northern Chile. Arch. Environ. Contam. Toxicol. 2011, 61, 185–192. [Google Scholar] [CrossRef]
- Valenzuela, A.; Campos, V.; Yañez, F.; Alveal, K.; Gutiérrez, P.; Rivas, M.; Contreras, N.; Klempau, A.; Fernandez, I.; Oyarzun, C. Application of artificial photoperiod in fish: A factor that increases susceptibility to infectious diseases? Fish Physiol. Biochem. 2012, 38, 943–950. [Google Scholar] [CrossRef] [PubMed]
- Vera, B. Bio-Obtención de Nanopartículas de Selenio y su Potencial Aplicación Como Suplemento Alimentario Inmunoestimulante en Trucha Arcoíris (Oncorhynchus mykiss). Bachelor’s Thesis, University of Concepcion, Concepción, Chile, 2016. [Google Scholar]
- Brown, L.A. Anaesthesia and restraint. In Fish Medicine, W.B.; Stoskopf, M.K., Ed.; Saunders Company, Harcourt Brace Jovanovich Inc.: Philadelphia, PA, USA, 1993; pp. 79–90. [Google Scholar]
- Coyle, S.D.; Durborow, R.M.; Tidwell, J.H. Anesthetics in aquaculture. South. Reg. Aquac. Cent. SRAC 2004, 3900, 1–6. [Google Scholar]
- Hu, Y.; Maisey, K.; Subramani, A.A.; Liu, F.; Flores-Kossack, C.; Imarai, M.; Secombes, C.J.; Wang, T. Characterisation of rainbow trout peripheral blood leucocytes prepared by hypotonic lysis of erythrocytes, and analysis of their phagocytic activity, proliferation and response to PAMPs and proinflammatory cytokines. Dev. Comp. Immunol. 2018, 88, 104–113. [Google Scholar] [CrossRef] [PubMed]
- Anderson, D.; Siwicki, A. Measuring the effects of contaminants on fish by haematological and serological methods. In Modulators of Fish Immune Responses; Stolen, J., Anderson, D., Zelikoff, S., Twerdok, L., Kaattari, S., Eds.; SOS Publications: Fair Haven, NJ, USA, 1993; pp. 95–118. [Google Scholar]
- Takemura, A.; Takano, K. Lysozyme in the ovary of tilapia (Oreochromis mossambicus): Its purification and some biological properties. Fish Physiol. Biochem. 1995, 14, 415–421. [Google Scholar] [CrossRef]
- Lawrence, R.; Burk, R. Glutathione peroxidase activity in selenium-deficient rat live. Biochem. Biophys. Res. Commun. 1976, 71, 952–958. [Google Scholar] [CrossRef]
- Fontagne’-Dicharry, S.; Godin, S.; Liu, H.; Prabhu, P.A.J.; Bouyssière, B.; Bueno, M.; Tacon, P.; Médale, F.; Kaushik, S. Influence of the forms and levels of dietary selenium on antioxidant status and oxidative stress-related parameters in rainbow trout (Oncorhynchus mykiss) fry. Br. J. Nutr. 2015, 113, 1876–1887. [Google Scholar] [CrossRef]
- Lowry, O.H.; Rosebrough, N.J.; Farr, A.L.; Randall, R.J. Protein Measurement with the Folin Phenol Reagent. J. Biol. Chem. 1951, 193, 265–275. [Google Scholar] [CrossRef]
- Naderi, M.; Keyvanshokooh, S.; Salati, A.P.; Ghaedi, A. Proteomic analysis of liver tissue from rainbow trout (Oncorhynchus mykiss) under high rearing density after administration of dietary vitamin E and selenium nanoparticles. Comp. Biochem. Physiol. Part D Genom. Proteom. 2017, 22, 10–19. [Google Scholar] [CrossRef]
- Lugert, V.; Thaller, G.; Tetens, J.; Schulz, C.; Krieter, J. A review on fish growth calculation: Multiple functions in fish production and their specific application. Rev. Aquac. 2014, 8, 30–42. [Google Scholar] [CrossRef]
- Denev, S.; Staykov, Y.; Moutafchieva, R.; Beev, G. Microbial ecology of the gastrointestinal tract of fish and the potential application of probiotics and prebiotics in finfish aquaculture. Int. Aquat. Res. 2009, 1, 1–29. [Google Scholar] [CrossRef]
- Hai, N.V. The use of probiotics in aquaculture. J. Appl. Microbiol. 2015, 19, 917–935. [Google Scholar] [CrossRef] [PubMed]
- Dawood, M.A.O.; Koshio, S.; Abdel-Daim, M.M.; Van Doan, H. Probiotic application for sustainable aquaculture. Rev. Aquac. 2019, 11, 907–924. [Google Scholar] [CrossRef]
- Valipour, A.; Nadaei, S.; Noori, A.; Khanipour, A.A.; Hoseinifar, S.H. Dietary Lactobacillus plantarum affected on some immune parameters, air-exposure stress response, intestinal microbiota, digestive enzyme activity and performance of narrow clawed crayfish (Astacus leptodactylus, Eschscholtz). Aquaculture 2019, 504, 121–130. [Google Scholar] [CrossRef]
- Zhai, Q.; Wang, H.; Tian, F.; Zhao, J.; Zhang, H.; Chen, W. Dietary Lactobacillus plantarum supplementation decreases tissue lead accumulation and alleviates lead toxicity in Nile tilapia (Oreochromis niloticus). Aquac. Res. 2017, 48, 5094–5103. [Google Scholar] [CrossRef]
- Silarudee, S.; Tongpim, S.; Charoensri, N.; Doolgindachbaporn, S. Effect of a Probiotic Lactobacillus plantarum CR1T5 Dietary Supplements on Non-specific Immunity in Black Eared Catfish (Pangasius larnaudii). J. Pure Appl. Microbiol. 2019, 13, 289–296. [Google Scholar] [CrossRef]
- Shakibaie, M.; Mohammadi-Khorsand, T.; Adeli-Sardou, M.; Jafari, M.; Amirpour-Rostami, S.; Ameri, A.; Forootanfar, H. Probiotic and antioxidant properties of selenium-enriched Lactobacillus brevis LSe isolated from an Iranian traditional dairy product. J. Trace Elem. Med. Biol 2017, 40, 1–9. [Google Scholar] [CrossRef]
- Kang, S.; Li, R.; Jin, H.; You, H.J.; Ji, G.E. Effects of Selenium- and Zinc-Enriched Lactobacillus plantarum SeZi on Antioxidant Capacities and Gut Microbiome in an ICR Mouse Model. Antioxidants 2020, 9, 1028. [Google Scholar] [CrossRef]
- Shu, G.; Mei, S.; Chen, L.; Zhang, B.; Guo, M.; Cui, X.; Chen, H. Screening, identification, and application of selenium-enriched Lactobacillus in goat milk powder and tablet. J. Food Process. Preserv. 2020, 44, e14470. [Google Scholar] [CrossRef]
- Fečkaninová, A.; Koščová, J.; Mudroňová, D.; Schusterová, P.; Cingeľová, I.; Maruščáková, C.; Popelka, P. Characterization of two novel lactic acid bacteria isolated from the intestine of rainbow trout (Oncorhynchus mykiss, Walbaum) in Slovakia. Aquaculture 2019, 506, 294–301. [Google Scholar] [CrossRef]
- Savadogo, A.; Ouattara, C.; Bassole, I.; Traore, A. Antimicrobial Activities of Lactic Acid Bacteria Strains Isolated from Burkina Faso Fermented Milk. Pak. J. Nutr. 2004, 3, 174–179. [Google Scholar]
- Tebyanian, H.; Bakhtiari, A.; Karami, A.; Kariminik, A. Antimicrobial Activity of some Lactobacillus Species against Intestinal Pathogenic Bacteria. Int. Lett. Nat. Sci. 2017, 65, 10–15. [Google Scholar] [CrossRef]
- Saulnier, D.M.; Spinler, J.K.; Gibson, G.R.; Versalovic, J. Mechanisms of probiosis and prebiosis: Considerations for enhanced functional foods. Curr. Opin. Biotechnol. 2009, 20, 135–141. [Google Scholar] [CrossRef] [PubMed]
- Shokryazdan, P.; Sieo, C.; Kalavathy, R.; Liang, J.; Alitheen, N.; Jahromi, M.; Ho, Y. Probiotic Potential of Lactobacillus Strains with Antimicrobial Activity against Some Human Pathogenic Strains. BioMed Res. Int. 2014, 2014, 927268. [Google Scholar] [CrossRef]
- European Food Safety Authority (EFSA). Panel on Additives and Products or Substances used in Animal Feed (FEEDAP). Guidance on the assessment of bacterial susceptibility to antimicrobials of human and veterinary importance. EFSA J. 2012, 10, 2740. [Google Scholar]
- Bujnakova, D.; Strakova, E. Safety, probiotic and technological properties of Lactobacilli isolated from unpasteurised ovine and caprine cheeses. Ann. Microbiol. 2017, 67, 813–826. [Google Scholar] [CrossRef]
- Rowe, G.E.; Welch, R.A. Assays of hemolytic toxins. Methods Enzym. 1994, 235, 657–667. [Google Scholar]
- Ramachandran, G. Gram-positive and Gram-negative bacterial toxins in sepsis: A brief review. Virulence 2013, 5, 213–218. [Google Scholar] [CrossRef]
- Yasmin, I.; Saeed, M.; Khan, W.A.; Khaliq, A.; Chughtai, M.F.J.; Iqbal, R.; Tehseen, S.; Naz, S.; Liaqat, A.; Mehmood, T.; et al. In Vitro Probiotic Potential and Safety Evaluation (Hemolytic, Cytotoxic Activity) of Bifidobacterium Strains Isolated from Raw Camel Milk. Microorganisms 2020, 8, 354. [Google Scholar] [CrossRef]
- Sharma, K.; Sharma, N.; Sharma, R. Identification and evaluation of in vitro probiotic attributes of novel and potential strains of lactic acid bacteria isolated from traditional dairy products of North-West Himalayas. J. Clin. Microbiol. Biochem. Technol. 2016, 2, 18–25. [Google Scholar] [CrossRef]
- Tokatlı, M.; Gülgör, G.; Bağder Elmaci, S.; Arslankoz İşleyen, N.; Özçelik, F. In vitro properties of potential probiotic indigenous lactic acid bacteria originating from traditional pickles. BioMed Res. Int. 2015, 2015, 315819. [Google Scholar] [CrossRef]
- Sica, M.G.; Brugnoni, L.I.; Marucci, P.L.; Cubitto, M.A. Characterization of probiotic properties of lactic acid bacteria isolated from an estuarine environment for application in rainbow trout (Oncorhynchus mykiss, Walbaum) farming. Antonie Van Leeuwenhoek 2012, 101, 869–879. [Google Scholar] [CrossRef] [PubMed]
- Bravo, J.P.; Hernandez, A.J.; Morales, G.; Dantagnan, P.; Márquez, L. Digestive coordination of the gastric function in Atlantic salmon Salmo salar juveniles. Lat. Am. J. Aquat. Res. 2018, 46, 1083–1090. [Google Scholar] [CrossRef]
- Lückstädt, C. The use of acidifiers in fish nutrition. CAB Rev. Perspect. Agric. Vet. 2008, 3, 1–8. [Google Scholar] [CrossRef]
- Fečkaninová, A.; Koščová, J.; Mudroňová, D.; Popelka, P.; Toropilová, J. The use of probiotic bacteria against Aeromonas infections in salmonid aquaculture. Aquaculture 2017, 469, 1–8. [Google Scholar] [CrossRef]
- Iorizzo, M.; Albanese, G.; Letizia, F.; Testa, B.; Tremonte, P.; Vergalito, F.; Lombardi, S.J.; Succi, M.; Coppola, R.; Sorrentino, E. Probiotic Potentiality from Versatile Lactiplantibacillus plantarum strains as resource to enhance freshwater fish health. Microorganisms 2022, 10, 463. [Google Scholar] [CrossRef]
- Fontana, L.; Brito, M.B.; Diaz, J.P.; Quezada, S.M.; Gil, A. Sources, isolation, characterization and evaluation of probiotics. Br. J. Nutr. 2013, 109, 35–50. [Google Scholar] [CrossRef]
- Begley, M.; Hill, C.; Gahan, C.G. Bile salt hydrolase activity in probiotics. Appl. Environ. Microbiol. 2006, 72, 1729–1738. [Google Scholar] [CrossRef]
- Martínez, F.G.; Moreno-Martin, G.; Pescuma, M.; Madrid-Albarrán, Y.; Mozzi, F. Biotransformation of Selenium by Lactic Acid Bacteria: Formation of Seleno-Nanoparticles and Seleno-Amino. Front. Bioeng. Biotechnol. 2020, 8, 506. [Google Scholar] [CrossRef]
- Ogasawara, Y.; Lacourciere, G.M.; Ishii, K.; Stadtman, T.C. Characterization of potential selenium-binding proteins in the selenophosphate synthetase system. Proc. Natl. Acad. Sci. USA 2005, 102, 1012–1016. [Google Scholar] [CrossRef]
- Ravanal, J. Determinación de Proteínas que Participan en el Control del Tamaño de Nanopartículas de Selenio Elemental Producidas por Pantoea Agglomerans. Bachelor’s Thesis, University of Concepcion, Concepción, Chile, 2015. [Google Scholar]
- Xia, S.K.; Chen, L.; Liang, J.Q. Enriched selenium and its effects on growth and biochemical composition in Lactobacillus bulgaricus. J. Agric. Food Chem. 2007, 55, 2413–2417. [Google Scholar] [CrossRef]
- Eszenyi, P.; Sztrik, A.; Babka, B.; Prokisch, J. Elemental, Nano-sized (100–500 nm) selenium production by probiotic lactic acid bacteria. Int. J. Biosci. Biochem. Bioinform. 2011, 1, 148–152. [Google Scholar] [CrossRef]
- Zhang, W.; Chen, Z.; Liu, H.; Zhang, L.; Gao, P.; Li, D. Biosynthesis and structural characteristics of selenium nanoparticles by Pseudomonas alcaliphila. Colloids Surf. B Biointerfaces 2011, 88, 196–201. [Google Scholar] [CrossRef] [PubMed]
- Hosnedlova, B.; Kepinska, M.; Skalickova, S.; Fernandez, C.; Ruttkay-Nedecky, B.; Peng, Q.; Baron, M.; Melcova, M.; Opatrilova, R.; Zidkova, J.; et al. Nano-selenium and its nanomedicine applications: A critical review. Int. J. Nanomed. 2018, 13, 2107–2128. [Google Scholar] [CrossRef]
- Daming, R.; Yingyu, W.; Zilai, W.; Jun, C.; Hekui, L.; Jingye, Z. Complete DNA sequence and analysis of two cryptic plasmids isolated from Lactobacillus plantarum. Plasmid 2003, 50, 70–73. [Google Scholar] [CrossRef]
- Enferadi, M.H.N.; Mohammadizadeh, F.; Soltani, M.; Bahri, A.H.; Sheikhzadeh, N. Effects of Lactobacillus plantarum on Growth Performance, Proteolytic Enzymes Activity and Intestine Morphology in Rainbow Trout (Oncorhynchus mykiss). Turk. J. Fish. Aquat. Sci. 2018, 18, 351–356. [Google Scholar] [CrossRef]
- Medina, M.; Sotil, G.; Flores, V.; Fernández, C.; Sandoval, N. In vitro assessment of some probiotic properties and inhibitory activity against Yersinia ruckeri of bacteria isolated from rainbow trout Oncorhynchus mykiss (Walbaum). Aquac. Rep. 2020, 18, 100447. [Google Scholar] [CrossRef]
- Uribe-Querol, E.; Rosales, C. Phagocytosis: Our Current Understanding of a Universal Biological Process. Front. Immunol. 2020, 11, 1066. [Google Scholar] [CrossRef] [PubMed]
- Dalmo, R.A.; Ingebrigtsen, K.; Bøgwald, J. Non-specific defence mechanisms in fish, with particular reference to the reticuloendothelial system (RES). J. Fish Dis. 1997, 20, 241–273. [Google Scholar] [CrossRef]
- Rodríguez, A.; Esteban, M.A.; Meseguer, J. Phagocytosis and peroxidase release by seabream (Sparus aurata L.) leucocytes in response to yeast cells. Anat. Rec. A Discov. Mol. Cell. Evol. Biol. 2003, 272A, 415–423. [Google Scholar] [CrossRef]
- Soltani, M.; Abdy, E.; Alishahi, M.; Taheri Mirghaed, A.; Hosseini-Shekarabi, P. Growth performance, immune-physiological variables and disease resistance of common carp (Cyprinus carpio) orally subjected to different concentrations of Lactobacillus plantarum. Aquacult. Int. 2017, 25, 1913–1933. [Google Scholar] [CrossRef]
- Chen, L.; Pan, D.-D.; Zhou, J.; Jiang, Y.-Z. Protective effect of selenium-enriched lactobacillus on CCl4-induced liver injury in mice and its possible mechanisms. World J. Gastroenterol. 2005, 11, 5795–5800. [Google Scholar] [CrossRef] [PubMed]
- Saurabh, S.; Sahoo, P.K. Lysozyme: An important defence molecule of fish innate immune system. Aquac. Res. 2008, 39, 223–239. [Google Scholar] [CrossRef]
- Fast, M.D.; Simsa, D.E.; Burka, J.F.; Mustafa, A.; Ross, N.W. Skin morphology and humoral non-specific defence parameters of mucus and plasma in rainbow trout, coho and Atlantic salmon. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2002, 132, 645–657. [Google Scholar] [CrossRef]
- Kane, A.M.; Soltani, M.; Ebrahimzahe-Mousavi, H.; Pakzad, K. Influence of probiotic, Lactobacillus plantarum on serum biochemical and immune parameters in vaccinated rainbow trout (Oncorhynchus mykiss) against streptococcosis/lactococosis. Int. J. Aquat. Biol. 2016, 4, 285–294. [Google Scholar] [CrossRef]
- Hang, B.T.; Balami, S.; Phuong, N.T. Effect of Lactobacillus plantarum on growth performance, immune responses, and disease resistance of striped catfish (Pangasianodon hypophthalmus). AACL Bioflux 2022, 15, 174–187. [Google Scholar]
- Son, V.M.; Chang, C.-C.; Wu, M.-C.; Guu, Y.-K.; Chiu, C.-H.; Cheng, W. Dietary administration of the probiotic, Lactobacillus plantarum, enhanced the growth, innate immune responses, and disease resistance of the grouper Epinephelus coioides. Fish Shellfish Immunol. 2009, 26, 691–698. [Google Scholar] [CrossRef]
- Yanez-Lemus, F.; Moraga, R.; Mercado, L.; Jara-Gutierrez, C.; Smith, C.; Aguayo, P.; Sanchez-Alonzo, K.; García-Cancino, A.; Valenzuela, A.; Campos, L. Selenium nanoparticles biosynthesized by Pantoea agglomerans and their effects on cellular and physiological parameters in the rainbow trout Oncorhynchus mykiss. Biology 2022, 11, 463. [Google Scholar] [CrossRef]
- Le, K.T.; Fotedar, R. Bioavailability of selenium from different dietary sources in yellowtail kingfish (Seriola lalandi). Aquaculture 2014, 420, 57–62. [Google Scholar] [CrossRef]
- Dawood, M.A.O.; Zommara, M.; Eweedah, N.M.; Helal, A.I.; Aboel-Darag, M.A. The potential role of nano-selenium and vitamin C on the performances of Nile tilapia (Oreochromis niloticus). Environ. Sci. Pollut. Res. 2020, 27, 9843–9852. [Google Scholar] [CrossRef]
- Shang, X.; Wang, B.; Sun, Q.; Zhang, Y.; Lu, Y.; Liu, S.; Li, Y. Selenium-enriched Bacillus subtilis reduces the effects of mercury-induced on inflammation and intestinal microbes in carp (Cyprinus carpio var. specularis). Fish Physiol. Biochem. 2022, 48, 215–226. [Google Scholar] [CrossRef]
- Lushchak, V.I.; Bagnyukova, T.V. Effects of different environmental oxygen levels on free radical processes in fish. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2006, 144, 283–289. [Google Scholar] [CrossRef] [PubMed]
- Magnoni, L.; Novais, S.C.; Silva, C.O.; Lemos, M.F.; Ozorio, R.; Geurden, I.; Leguen, I.; Prunet, P.; Eding, E.; Schrama, J. The impact of nutritional and environmental stressors on the immune response, oxidative stress and energy use of rainbow trout (Oncorhynchus mykiss). In Proceedings of the International Meeting on Marine Research 2016, Peniche, Portugal, 14–15 July 2016. [Google Scholar] [CrossRef]
- Mengistu, B.M.; Bitsue, H.K.; Huang, K. The Effects of Selenium-Enriched Probiotics on Growth Performance, Oocysts Shedding, Intestinal Cecal Lesion Scores, Antioxidant Capacity, and mRNA Gene Expression in Chickens Infected with Eimeria tenella. Biol. Trace Elem. Res. 2021, 199, 278–291. [Google Scholar] [CrossRef] [PubMed]
- Tollerz Bratteby, U. Factors Explaining Variation in the Fecundity of Female Baltic Salmon (Salmo salar)–The Influence of Length, Body Condition and Growth Rate at Sea. Master’s Thesis, Swedish University of Agricultural Sciences, Öregrund, Sweden, 2019. [Google Scholar]
- Melo-Bolívar, J.; Ruiz, R.; Hume, M.; Villamil, L.; Alzate, A.; Cañas, B.; Pérez-Munguia, S.; Hernandez-Mendoza, H.; Pérez-Conde, C.; Gutiérrez, A.M.; et al. Evaluation of the Inorganic Selenium Biotransformation in Selenium-Enriched Yogurt by HPLC-ICP-MS. J. Agric. Food Chem. 2007, 55, 9776–9783. [Google Scholar] [CrossRef]
- Van Zyl, W.; Deane, S.; Dicks, L. Molecular insights into probiotic mechanisms of action employed against intestinal pathogenic bacteria. Gut Microbes 2020, 12, e1831339. [Google Scholar] [CrossRef]
- Ringø, E. Probiotics in shellfish aquaculture. Aquac. Fish. 2020, 5, 1–27. [Google Scholar] [CrossRef]
- Van Doan, H.; Hoseinifar, S.H.; Ringø, E.; Esteban, M.A.; Dadar, M.; Dawood, M.A.O.; Faggio, C. Host-Associated Probiotics: A Key Factor in Sustainable Aquaculture. Rev. Fish. Sci. Aquac. 2019, 28, 14–16. [Google Scholar] [CrossRef]
- Jatobá, A.; Pereira, M.O.; Vieira, L.M.; Bitencourt, M.; Rodrigues, E.; Fachini, F.A.; Moraes, A.V. Action time and feed frequency of Lactobacillus plantarum for Nile tilapia. Arq. Bras. Med. Vet. Zootec. 2018, 70, 327–332. [Google Scholar] [CrossRef]
- Parthasarathy, R.; Ravi, D. Probiotic bacteria as growth promoter and biocontrol agent against Aeromonas hydrophila in Catla catla (Hamilton, 1822). Indian J. Fish. 2011, 58, 87–93. [Google Scholar]
- Giri, S.S.; Sukumaran, V.; Oviya, M. Potential probiotic Lactobacillus plantarum VSG3 improves the growth, immunity, and disease resistance of tropical freshwater fish, Labeo rohita. Fish Shellfish Immunol. 2013, 34, 660–666. [Google Scholar] [CrossRef]
- El-Kader, M.F.A.; El-Bab, A.F.F.; Abd-Elghany, M.F.; Abdel-Warith, A.-W.A.; Younis, E.M.; Dawood, M.A.O. Selenium Nanoparticles Act Potentially on the Growth Performance, Hemato-Biochemical Indices, Antioxidative, and Immune-Related Genes of European Seabass (Dicentrarchus labrax). Biol. Trace Elem. Res. 2021, 199, 3126–3134. [Google Scholar] [CrossRef]
- Zhou, X.; Wang, Y.; Gu, Q.; Li, W. Effects of different dietary selenium sources (selenium nanoparticle and selenomethionine) on growth performance, muscle composition and glutathione peroxidase enzyme activity of crucian carp (Carassius auratus gibelio). Aquaculture 2009, 291, 78–81. [Google Scholar] [CrossRef]
- Yang, L.; Wang, J.; Huang, K.; Liu, Q.; Liu, G.; Xu, X.; Zhang, H.; Zhu, M. Selenium-enriched Bacillus subtilis yb-114246 improved growth and immunity of broiler chickens through modified ileal bacterial composition. Sci. Rep. 2021, 11, 21690. [Google Scholar] [CrossRef] [PubMed]

| Antibiotic (µg) | Interpretive Categories and Inhibition Diameter Breakpoints, Nearest Whole mm | Antibiotic Class | Mechanism of Action | ||
|---|---|---|---|---|---|
| S | I | R | |||
| GEN (10) | ≥15 | 14–13 | ≤12 | Aminoglycosides | Inhibitor of protein synthesis |
| TET (30) | ≥15 | 14–12 | ≤11 | Tetracycline | Inhibitor of protein synthesis |
| OXY (30) | ≥15 | 14–12 | ≤11 | Tetracycline | Inhibitor of protein synthesis |
| ERY (15) | ≥22 | 21–16 | ≤15 | Macrolides | Inhibitor of protein synthesis |
| FLO (30) | ≥19 | 18–15 | ≤14 | Amphenicols | Inhibitor of protein synthesis |
| AMP (10) | ≥17 | 16–14 | ≤13 | Β-Lactams | Inhibitor of the cell wall synthesis |
| Compound | % | Compound | % | Compound | % |
|---|---|---|---|---|---|
| Crude protein | 39–43% | Lipids | 10–16% | Ash | 9–12% |
| Moisture | 7–13% | Calcium | 1–2% | Fiber | 3–4% |
| Phosphate | 1–1.4% |
| Isolate | Gram | Morphology | Catalase |
|---|---|---|---|
| S1 | positive | coccoid | positive |
| S2 | positive | rod-shaped | negative |
| S3 | positive | coccoid | positive |
| S4 | positive | rod-shaped | negative |
| S5 | positive | coccoid | positive |
| S6 | positive | coccoid | positive |
| S7 | negative | rod-shaped | N/A |
| S8 | positive | rod-shaped | negative |
| S9 | negative | rod-shaped | N/A |
| S10 | positive | coccoid | positive |
| S11 | positive | coccoid | positive |
| S12 | positive | rod-shaped | negative |
| S13 | positive | rod-shaped | negative |
| S14 | positive | rod-shaped | negative |
| S15 | positive | rod-shaped | positive |
| S16 | positive | rod-shaped | negative |
| Isolate | Carbohydrate | |||||
|---|---|---|---|---|---|---|
| Glucose | Fructose | Galactose | Ribose | Xylose | Arabinose | |
| S2 | + | + | − | − | − | − |
| S4 | +g | +g | − | − | − | − |
| S8 | +g | +g | − | + | + | + |
| S12 | +g | +g | − | + | + | + |
| S13 | + | + | − | − | − | − |
| S14 | + | +g | − | + | + | + |
| LAB Strains | Inhibition Halo (in mm) of Reference Bacterial Strains | |||
|---|---|---|---|---|
| S. aureus ATCC 25923 | B. subtilis ATCC 6633 | E. coli ATCC 25922 | P. aeruginosa ATCC 10145 | |
| LABS2 | 16.7 ± 0.9 a | 18.9 ± 0.4 a | 18.6 ± 1.0 a | 13.9± 0.5 a |
| LABS4 | 30.8 ± 0.7 b | 25.3 ± 0.7 b | 16.2 ± 0.3 b | 31.6± 0.8 b |
| LABS8 | 20.8 ± 0.4 c | 18.3 ± 0.4 a | 8.9 ± 0.4 c | 12.6 ± 0.4 a |
| LABS12 | - | 12.8 ± 0.7 c | - | - |
| LABS13 | 20.5 ± 0.8 c | 16.5 ± 0.7 d | 11.5 ± 0.4 d | 12.4 ± 1.1 a |
| LABS14 | 26.3 ± 0.4 d | 20.9 ± 1.1 e | 12.9 ± 0.3 de | 18.9 ± 0.3 c |
| LAB Strain | Type of Hemolysis | ||
|---|---|---|---|
| 24 h | 48 h | 72 h | |
| LABS2 | Alpha | Alpha | Alpha |
| LABS4 | Gamma | Gamma | Gamma |
| LABS8 | Alpha | Alpha | Alpha |
| LABS13 | Beta | Beta | Beta |
| LABS14 | Gamma | Gamma | Gamma |
| LAB Strain | Acid Resistance | Bile Salts Resistance | ||
|---|---|---|---|---|
| pH 3 (Viability %) | Control | Bile Salt (Viability %) | Control | |
| LABS4 | * 8 × 108 ± 0.06 (57.1) | 14 × 108 ± 0.12 | * 9 × 108 ± 0.07 (69.2) | 13 × 108 ± 0.18 |
| LABS14 | * 12 × 108 ± 0.10 (74.8) | 16 × 108 ± 0.09 | * 12 × 108 ± 0.14 (82.3) | 15 × 108 ± 0.03 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yanez-Lemus, F.; Moraga, R.; Smith, C.T.; Aguayo, P.; Sánchez-Alonzo, K.; García-Cancino, A.; Valenzuela, A.; Campos, V.L. Selenium Nanoparticle-Enriched and Potential Probiotic, Lactiplantibacillus plantarum S14 Strain, a Diet Supplement Beneficial for Rainbow Trout. Biology 2022, 11, 1523. https://doi.org/10.3390/biology11101523
Yanez-Lemus F, Moraga R, Smith CT, Aguayo P, Sánchez-Alonzo K, García-Cancino A, Valenzuela A, Campos VL. Selenium Nanoparticle-Enriched and Potential Probiotic, Lactiplantibacillus plantarum S14 Strain, a Diet Supplement Beneficial for Rainbow Trout. Biology. 2022; 11(10):1523. https://doi.org/10.3390/biology11101523
Chicago/Turabian StyleYanez-Lemus, Francisco, Rubén Moraga, Carlos T. Smith, Paulina Aguayo, Kimberly Sánchez-Alonzo, Apolinaria García-Cancino, Ariel Valenzuela, and Víctor L. Campos. 2022. "Selenium Nanoparticle-Enriched and Potential Probiotic, Lactiplantibacillus plantarum S14 Strain, a Diet Supplement Beneficial for Rainbow Trout" Biology 11, no. 10: 1523. https://doi.org/10.3390/biology11101523
APA StyleYanez-Lemus, F., Moraga, R., Smith, C. T., Aguayo, P., Sánchez-Alonzo, K., García-Cancino, A., Valenzuela, A., & Campos, V. L. (2022). Selenium Nanoparticle-Enriched and Potential Probiotic, Lactiplantibacillus plantarum S14 Strain, a Diet Supplement Beneficial for Rainbow Trout. Biology, 11(10), 1523. https://doi.org/10.3390/biology11101523







