Artificial-Intelligence-Based Imaging Analysis of Stem Cells: A Systematic Scoping Review
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Identifying the Research Question
- Population: stem cells;
- Concept: AI-based technique;
- Context: imaging analysis.
2.2. Searching Strategy
2.3. Eligibility Criteria
2.4. Study Selection and Data Extraction
3. Results
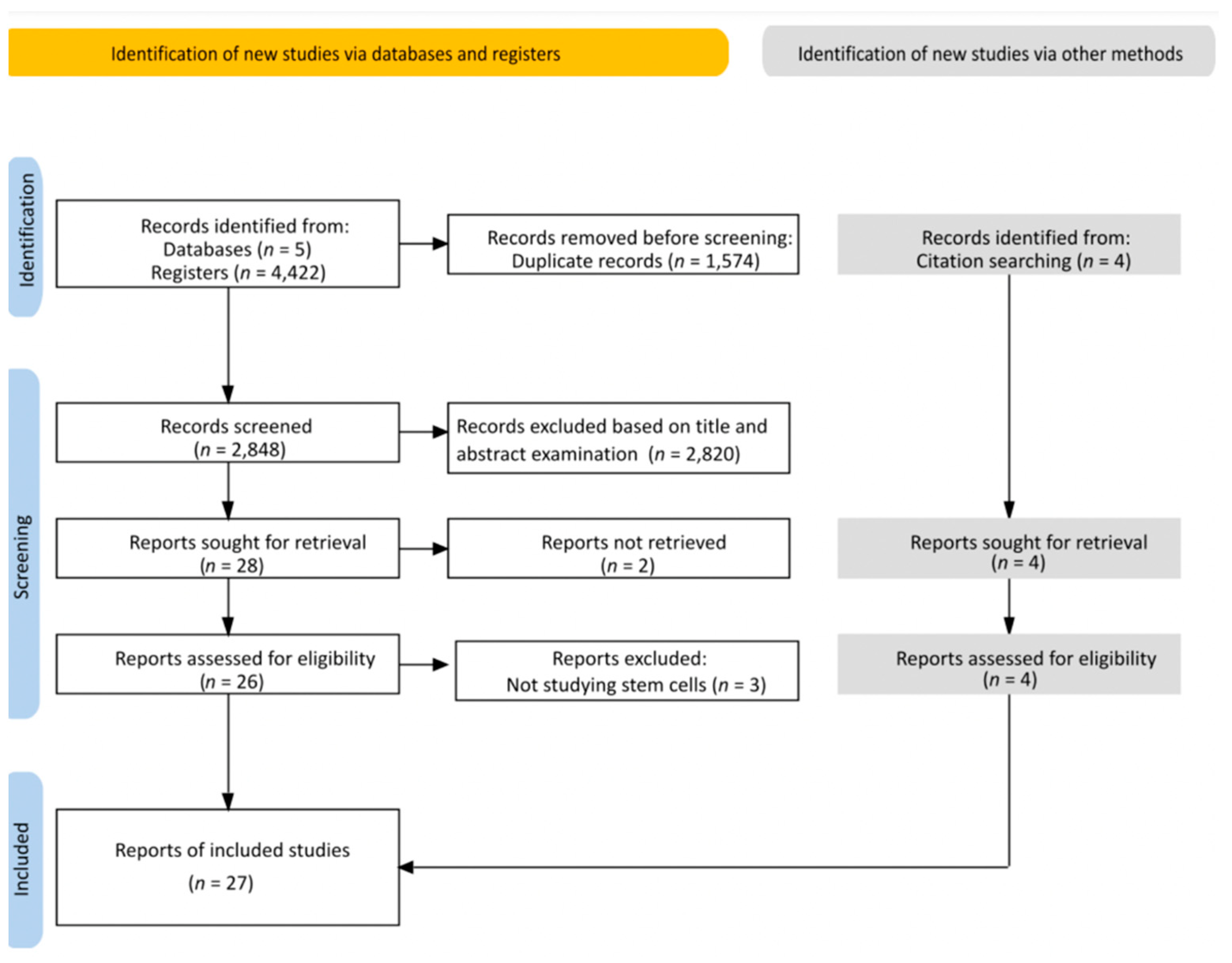
3.1. Search Result
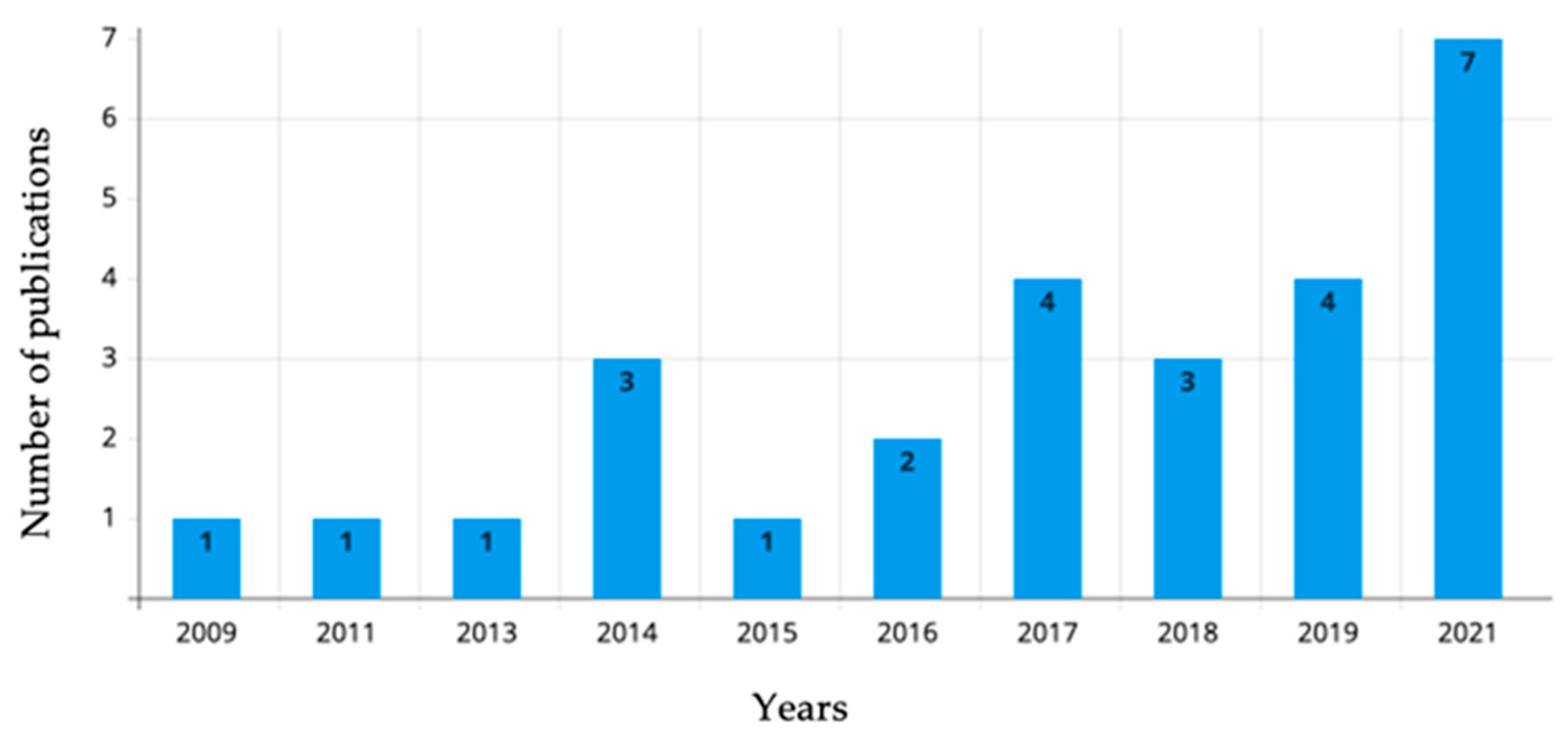
3.2. Extracted Data
4. Discussion
4.1. iPSC
4.2. ESC
4.3. Other Stem cells
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Liu, G.; David, B.T.; Trawczynski, M.; Fessler, R.G. Advances in Pluripotent Stem Cells: History, Mechanisms, Technologies, and Applications. Stem Cell Rev. Rep. 2020, 16, 3–32. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, M.; Thangaraj, S.R.; Ramasubramanian, K.; Thangaraj, P.P.; Ramasubramanian, K.V. Exploring the Current Trends of Artificial Intelligence in Stem Cell Therapy: A Systematic Review. Cureus 2021, 12, e20083. [Google Scholar] [CrossRef] [PubMed]
- Jankowski, M.; Dompe, C.; Sibiak, R.; Wąsiatycz, G.; Mozdziak, P.; Jaśkowski, J.M.; Antosik, P.; Kempisty, B.; Dyszkiewicz-Konwińska, M. In Vitro Cultures of Adipose-Derived Stem Cells: An Overview of Methods, Molecular Analyses, and Clinical Applications. Cells 2020, 9, 1783. [Google Scholar] [CrossRef] [PubMed]
- Ramakrishna, R.R.; Abd Hamid, Z.; Wan Zaki, W.; Huddin, A.B.; Mathialagan, R. Stem cell imaging through convolutional neural networks: Current issues and future directions in artificial intelligence technology. PeerJ 2020, 8, e10346. [Google Scholar] [CrossRef] [PubMed]
- Cable, J.; Fuchs, E.; Weissman, I.; Jasper, H.; Glass, D.; Rando, T.A.; Blau, H.; Debnath, S.; Oliva, A.; Park, S.; et al. Adult stem cells and regenerative medicine-a symposium report. Ann. N. Y. Acad. Sci. 2020, 1462, 27–36. [Google Scholar] [CrossRef]
- Malik, N.; Rao, M.S. A review of the methods for human iPSC derivation. Methods Mol. Biol. 2013, 997, 23–33. [Google Scholar] [CrossRef]
- Bacakova, L.; Zarubova, J.; Travnickova, M.; Musilkova, J.; Pajorova, J.; Slepicka, P.; Kasalkova, N.S.; Svorcik, V.; Kolska, Z.; Motarjemi, H.; et al. Stem cells: Their source, potency and use in regenerative therapies with focus on adipose-derived stem cells—A review. Biotechnol. Adv. 2018, 36, 1111–1126. [Google Scholar] [CrossRef] [PubMed]
- Zakrzewski, W.; Dobrzyński, M.; Szymonowicz, M.; Rybak, Z. Stem cells: Past, present, and future. Stem Cell Res. Ther. 2019, 10, 68. [Google Scholar] [CrossRef]
- Menasche, P.; Vanneaux, V.; Hagege, A.; Bel, A.; Cholley, B.; Cacciapuoti, I.; Parouchev, A.; Benhamouda, N.; Tachdjian, G.; Tosca, L.; et al. Human embryonic stem cell-derived cardiac progenitors for severe heart failure treatment: First clinical case report. Eur. Heart J. 2015, 36, 2011–2017. [Google Scholar] [CrossRef]
- Schwartz, S.D.; Regillo, C.D.; Lam, B.L.; Eliott, D.; Rosenfeld, P.J.; Gregori, N.Z.; Hubschman, J.P.; Davis, J.L.; Heilwell, G.; Spirn, M.; et al. Human embryonic stem cell-derived retinal pigment epithelium in patients with age-related macular degeneration and Stargardt’s macular dystrophy: Follow-up of two open-label phase 1/2 studies. Lancet 2015, 385, 509–516. [Google Scholar] [CrossRef]
- Dantuma, E.; Merchant, S.; Sugaya, K. Stem cells for the treatment of neurodegenerative diseases. Stem Cell Res. Ther. 2010, 1, 37. [Google Scholar] [CrossRef] [PubMed]
- Longo, U.G.; Ronga, M.; Maffulli, N. Achilles tendinopathy. Sports Med. Arthrosc. 2009, 17, 112–126. [Google Scholar] [CrossRef] [PubMed]
- Widuchowski, W.; Widuchowsk, J.; Trzaska, T. Articular cartilage defects: Study of 25,124 knee arthroscopies. Knee 2007, 14, 177–182. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, K.; Ohta, H.; Kurimoto, K.; Aramaki, S.; Saitou, M. Reconstitution of the mouse germ cell specification pathway in culture by pluripotent stem cells. Cell 2011, 146, 519–532. [Google Scholar] [CrossRef] [PubMed]
- Sadri-Ardekani, H.; Atala, A. Testicular tissue cryopreservation and spermatogonial stem cell transplantation to restore fertility: From bench to bedside. Stem Cell Res. Ther. 2014, 5, 68. [Google Scholar] [CrossRef]
- Kroon, E.; Martinson, L.A.; Kadoya, K.; Bang, A.G.; Kelly, O.G.; Eliazer, S.; Young, H.; Richardson, M.; Smart, N.G.; Cunningham, J.; et al. Pancreatic endoderm derived from human embryonic stem cells generates glucose-responsive insulin-secreting cells in vivo. Nat. Biotechnol. 2008, 26, 443–452. [Google Scholar] [CrossRef]
- Lee, G.; Papapetrou, E.P.; Kim, H.; Chambers, S.M.; Tomishima, M.J.; Fasano, C.A.; Ganat, Y.M.; Menon, J.; Shimizu, F.; Viale, A.; et al. Modelling pathogenesis and treatment of familial dysautonomia using patient-specific iPSCs. Nature 2009, 461, 402–406. [Google Scholar] [CrossRef]
- Imamura, K.; Izumi, Y.; Banno, H.; Uozumi, R.; Morita, S.; Egawa, N.; Ayaki, T.; Nagai, M.; Nishiyama, K.; Watanabe, Y.; et al. Induced pluripotent stem cell-based Drug Repurposing for Amyotrophic lateral sclerosis Medicine (iDReAM) study: Protocol for a phase I dose escalation study of bosutinib for amyotrophic lateral sclerosis patients. BMJ 2019, 9, e033131. [Google Scholar] [CrossRef]
- De Masi, C.; Spitalieri, P.; Murdocca, M.; Novelli, G.; Sangiuolo, F. Application of CRISPR/Cas9 to human-induced pluripotent stem cells: From gene editing to drug discovery. Hum. Genom. 2020, 14, 25. [Google Scholar] [CrossRef]
- Amisha; Malik, P.; Pathania, M.; Rathaur, V.K. Overview of artificial intelligence in medicine. J. Fam. Med. Prim. Care 2019, 8, 2328–2331. [Google Scholar] [CrossRef]
- Peters, M.D.; Godfrey, C.M.; Khalil, H.; McInerney, P.; Parker, D.; Soares, C.B. Guidance for conducting systematic scoping reviews. Int. J. Evid.-Based Healthc. 2015, 13, 141–146. [Google Scholar] [CrossRef] [PubMed]
- Tricco, A.C.; Lillie, E.; Zarin, W.; O’Brien, K.K.; Colquhoun, H.; Levac, D.; Moher, D.; Peters, M.; Horsley, T.; Weeks, L.; et al. PRISMA Extension for Scoping Reviews (PRISMA-ScR): Checklist and Explanation. Ann. Intern. Med. 2018, 169, 467–473. [Google Scholar] [CrossRef] [PubMed]
- Issa, J.; Abou Chaar, M.; Kempisty, B.; Mozdziak, P.; Dyszkiewicz-Konwińska, M. Artificial intelligence-based imaging analysis of stem cells: A systematic scoping review protocol. Res. Sq. 2022, preprint. [Google Scholar] [CrossRef]
- Clark, J.; Glasziou, P.; Del Mar, C.; Bannach-Brown, A.; Stehlik, P.; Scott, A.M. A full systematic review was completed in 2 weeks using automation tools: A case study. J. Clin. Epidemiol. 2020, 121, 81–90. [Google Scholar] [CrossRef] [PubMed]
- Fischbacher, B.; Hedaya, S.; Hartley, B.J.; Wang, W.; Lallos, G.; Hutson, D.; Zimmer, M.; Brammer, J.; The NYSCF Global Stem Cell Array Team; Paull, D. Modular deep learning enables automated identification of monoclonal cell lines. Nat. Mach. Intell. 2021, 3, 632–640. [Google Scholar] [CrossRef]
- Guan, B.X.; Bhanu, B.; Theagarajan, R.; Liu, H.; Talbot, P.; Weng, N. Human embryonic stem cell classification: Random network with autoencoded feature extractor. J. Biomed. Opt. 2021, 26, 052913. [Google Scholar] [CrossRef]
- Guo, J.; Wang, P.; Sozen, B.; Qiu, H.; Zhu, Y.; Zhang, X.; Ming, J.; Zernicka-Goetz, M.; Na, J. Machine learning-assisted high-content analysis of pluripotent stem cell-derived embryos in vitro. Stem Cell Rep. 2021, 16, 1331–1346. [Google Scholar] [CrossRef]
- Imamura, K.; Yada, Y.; Izumi, Y.; Morita, M.; Kawata, A.; Arisato, T.; Nagahashi, A.; Enami, T.; Tsukita, K.; Kawakami, H.; et al. Prediction Model of Amyotrophic Lateral Sclerosis by Deep Learning with Patient Induced Pluripotent Stem Cells. Ann. Neurol. 2021, 89, 1226–1233. [Google Scholar] [CrossRef]
- Joy, D.A.; Libby, A.; McDevitt, T.C. Deep neural net tracking of human pluripotent stem cells reveals intrinsic behaviors directing morphogenesis. Stem Cell Rep. 2021, 16, 1317–1330. [Google Scholar] [CrossRef]
- Mota, S.M.; Rogers, R.E.; Haskell, A.W.; McNeill, E.P.; Kaunas, R.; Gregory, C.A.; Giger, M.L.; Maitland, K.C. Automated mesenchymal stem cell segmentation and machine learning-based phenotype classification using morphometric and textural analysis. J. Med. Imaging 2021, 8, 014503. [Google Scholar] [CrossRef]
- Zhu, Y.; Huang, R.; Wu, Z.; Song, S.; Cheng, L.; Zhu, R. Deep learning-based predictive identification of neural stem cell differentiation. Nat. Commun. 2021, 12, 2614. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.H.; Abe, K.; Yokota, H.; Sudo, K.; Nakamura, Y.; Chu, S.L.; Hsu, C.Y.; Tsai, M.D. Human Induced Pluripotent Stem Cell Reprogramming Prediction in Microscopy Images using LSTM based RNN. In Proceedings of the Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Berlin, Germany, 23–27 July 2019; pp. 2416–2419. [Google Scholar] [CrossRef]
- Orita, K.; Sawada, K.; Koyama, R.; Ikegaya, Y. Deep learning-based quality control of cultured human-induced pluripotent stem cell-derived cardiomyocytes. J. Pharmacol. Sci. 2019, 140, 313–316. [Google Scholar] [CrossRef] [PubMed]
- Waisman, A.; La Greca, A.; Möbbs, A.M.; Scarafía, M.A.; Santín Velazque, N.L.; Neiman, G.; Moro, L.N.; Luzzani, C.; Sevlever, G.E.; Guberman, A.S.; et al. Deep Learning Neural Networks Highly Predict Very Early Onset of Pluripotent Stem Cell Differentiation. Stem Cell Rep. 2019, 12, 845–859. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Shao, X.; Peng, Y.; Teng, Y.; Saravanan, K.M.; Zhang, H.; Li, H.; Wei, Y. A novel machine learning based approach for iPS progenitor cell identification. PLoS Comput. Biol. 2019, 15, e1007351. [Google Scholar] [CrossRef] [PubMed]
- Kavitha, M.S.; Kurita, T.; Ahn, B.C. Critical texture pattern feature assessment for characterizing colonies of induced pluripotent stem cells through machine learning techniques. Comput. Biol. Med. 2018, 94, 55–64. [Google Scholar] [CrossRef] [PubMed]
- Kusumoto, D.; Lachmann, M.; Kunihiro, T.; Yuasa, S.; Kishino, Y.; Kimura, M.; Katsuki, T.; Itoh, S.; Seki, T.; Fukuda, K. Automated Deep Learning-Based System to Identify Endothelial Cells Derived from Induced Pluripotent Stem Cells. Stem Cell Rep. 2018, 10, 1687–1695. [Google Scholar] [CrossRef]
- Theagarajan, R.; Guan, B.X.; Bhanu, B. DeephESC: An Automated System for Generating and Classification of Human Embryonic Stem Cells. In Proceedings of the 2018 24th International Conference on Pattern Recognition (ICPR), Beijing, China, 20–24 August 2018; pp. 3826–3831. [Google Scholar] [CrossRef]
- Buggenthin, F.; Buettner, F.; Hoppe, P.S.; Endele, M.; Kroiss, M.; Strasser, M.; Schwarzfischer, M.; Loeffler, D.; Kokkaliaris, K.D.; Hilsenbeck, O.; et al. Prospective identification of hematopoietic lineage choice by deep learning. Nat. Methods 2017, 14, 403–406. [Google Scholar] [CrossRef]
- Chang, Y.H.; Abe, K.; Yokota, H.; Sudo, K.; Nakamura, Y.; Lin, C.-Y.; Tsai, M.D. Human induced pluripotent stem cell region recognition in microscopy images using Convolutional Neural Networks. In Proceedings of the Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Jeju, Korea, 11–15 July 2017; pp. 4058–4061. [Google Scholar] [CrossRef]
- Fan, K.; Zhang, S.; Zhang, Y.; Lu, J.; Holcombe, M.; Zhang, X. A Machine Learning Assisted, Label-free, Non-invasive Approach for Somatic Reprogramming in Induced Pluripotent Stem Cell Colony Formation Detection and Prediction. Sci. Rep. 2017, 7, 13496. [Google Scholar] [CrossRef]
- Li, C.; Huang, X.; Jiang, T.; Xu, N. Full-automatic computer aided system for stem cell clustering using content-based microscopic image analysis. Biocybern. Biomed. Eng. 2017, 37, 540–558. [Google Scholar] [CrossRef]
- Joutsijoki, H.; Haponen, M.; Rasku, J.; Aalto-Setälä, K.; Juhola, M. Machine Learning Approach to Automated Quality Identification of Human Induced Pluripotent Stem Cell Colony Images. Comput. Math. Methods Med. 2016, 2016, 3091039. [Google Scholar] [CrossRef]
- Wuttisarnwattana, P.; Gargesha, M.; van’t Hof, W.; Cooke, K.R.; Wilson, D.L. Automatic Stem Cell Detection in Microscopic Whole Mouse Cryo-Imaging. IEEE Trans. Med. Imaging 2016, 35, 819–829. [Google Scholar] [CrossRef] [PubMed]
- Bajcsy, P.; Simon, M.; Florczyk, S.J.; Simon, C.G., Jr.; Juba, D.; Brady, M.C. A method for the evaluation of thousands of automated 3D stem cell segmentations. J. Microsc. 2015, 260, 363–376. [Google Scholar] [CrossRef] [PubMed]
- Lou, X.; Kang, M.; Xenopoulos, P.; Muñoz-Descalzo, S.; Hadjantonakis, A.K. A rapid and efficient 2D/3D nuclear segmentation method for analysis of early mouse embryo and stem cell image data. Stem Cell Rep. 2014, 2, 382–397. [Google Scholar] [CrossRef] [PubMed]
- Maddah, M.; Loewke, K. Automated, non-invasive characterization of stem cell-derived cardiomyocytes from phase-contrast microscopy. In Medical Image Computing and Computer-Assisted Intervention: MICCAI, Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Boston, MA, USA, 14–18 September 2014; Springer: Cham, Switzerland, 2014; Volume 17, pp. 57–64. [Google Scholar] [CrossRef]
- Maddah, M.; Loewke, K. Dynamic morphology-based characterization of stem cells enabled by texture-based pattern recognition from phase-contrast images. In Proceedings of the 2014 IEEE 11th International Symposium on Biomedical Imaging (ISBI), Beijing, China, 29 April–2 May 2014; pp. 77–80. [Google Scholar] [CrossRef]
- Paduano, V.; Tagliaferri, D.; Falco, G.; Ceccarelli, M. Automated Identification and Location Analysis of Marked Stem Cells Colonies in Optical Microscopy Images. PLoS ONE 2013, 8, e80776. [Google Scholar] [CrossRef] [PubMed]
- Faustino, G.M.; Gattass, M.; de Lucenaa, C.J.P.; Camposb, P.B.; Rehenb, S.K. A Graph-mining Algorithm for Automatic Detection and Counting of Embryonic Stem Cells in Fluorescence Microscopy Images. Integr. Comput. Aided Eng. 2011, 18, 91–106. [Google Scholar] [CrossRef]
- Faustino, G.M.; Gattass, M.; Rehen, S.; de Lucena, C.J.P. Automatic embryonic stem cells detection and counting method in fluorescence microscopy images. In Proceedings of the 2009 IEEE International Symposium on Biomedical Imaging: From Nano to Macro, Boston, MA, USA, 28 June–1 July 2009; pp. 799–802. [Google Scholar] [CrossRef]

| Inclusion Criteria | Exclusion Criteria |
|---|---|
| Studies involving any type of Stem cells (iPSCs, ECSs, ACSs, PSCs) | Studies investigating different types of cells rather than stem cells |
| Studies using AI-based imaging analysis | Studies using AI technology for other purposes than imaging analysis |
| Published studies in any language | Reviews |
| No date restriction | Preprints and conference papers |
| Full-text accessible | Full-text not accessible |
| Author, Study, Location & Year of Publication | Study Aim | Cell Type | Sample Size | Algorithm | Findings |
|---|---|---|---|---|---|
| Fischbacher et al., USA, 2021. [25] | Automatic detection and identification of colony presence and clonality | Human-induced pluripotent stem cells (hiPSCs) | Approximately 30,000 images | Monoqlo RetinaNet ResNet | The algorithm was capable of analyzing the data volumes in less than an hour. |
| Guan et al., USA, 2021. [26] | Developing a deep learning classifiers for the classification of human embryonic stem cells (hESCs) on a video dataset | hESCs | 27,603 unlabeled grayscale images and 3559 labeled grayscale images | Random network (RandNet) | The proposed approach achieved a classification accuracy of 97.23 ± 0.94%. |
| Guo et al., China, 2021. [27] | Setting up a workflow to use machine-learning-assisted high-content analysis to study embryo-like structures | Mouse iPSCs, ESCs, and trophectoderm stem cells (TSCs) | N/A | Algorithms developed by the PerkinElmer HCS system | The workflow was able to establish a robust, unbiased, and automated machine learning-based protocols. |
| Imamura et al., Japan, 2021. [28] | iPSC detection using deep learning for amyotrophic lateral sclerosis prediction | iPSCs | 4500 images for training 1350 images for validation 900 images for testing | CNNs | The algorithm achived an average accuracy of 0.90 ± 0.10 for cell classification. |
| Joy et al., USA, 2021. [29] | Training a group of neural networks to localize individual cell nucleus in an hiPSC colony, and to generate longitudinal measures of cell and cellular neighborhood properties | hiPSCs | 12 time lapse movies | FCRN-A FCRN-B U-Net Residual U-Net Count-ception | The trained group of neural networks was able to identify the characteristics of multicellular organization at the single-cell local neighborhood and whole-colony scales. |
| Mota et al., USA, 2021. [30] | Proposing an objective aproach that determines the morphological phenotypes of mesenchymal stem cells (MSCs) for culture efficacy prediction | Human bone-marrow-derived MSCs (hBMSCs) | Training dataset 71 images Validation dataset 36 images | Proposed a new algorithm generated using MATLAB | The proposed method showed 88% sensitivity and 86% precision for overall cell detection. |
| Zhu et al., China, 2021. [31] | Building a CNN system that uses unlabelled brightfield single-cell images to recognize differentiated neural stem cell (NSC) features | NSCs | 119,533 images for training 29,895 images for testing | Xception ResNet VGGNet Inception-v730 | The model estimated the proportion of final cell-type differentiation in early stages of differentiation before the common laboratory techniques were able to detect it. |
| Chang et al., Taiwan, 2019. [32] | Establishing a traceable method for human iPSC formation from CD34+ cord blood cells | CD34+ cells | 144 images | CNNs | The machine learning method provided a time-series visualization and quantitative analysis of the hiPSC induction and transition process. |
| Orita et al., Japan, 2019. [33] | Training a CNN model using bright-field images | hiPSC-derived cardiomyocytes (hiPSC-CMs) | 14,000 images for training 2000 images for validation 2000 images for testing | VGG16 | The tested model showed an average of 0.897 ± 0.01 accuracy, 0.946 ± 0.005 precision, 0.843 ± 0.02 recall, and 0.890 ± 0.01 F1-score. |
| Waisman et al., Argentina, 2019. [34] | Training CNN to distinguish the pluripotent stem cells from early-differentiating cells based on cellular morphology | Mouse embryonic stem cells (mouse ESCs) | 1116 images | ResNET50 DenseNet | The tested model was able achieve distinguishment with a 99% accuracy. |
| Zhang et al., China, 2019. [35] | Proposing a machine-learning-based approach to detect iPS progenitor cells during the early stage of reprogramming and against normal mouse embryonic fibroblasts (MEFs) in the same stage | iPS progenitor cells and MEFs | N/A | XGBoost | The model predicted iPS progenitor cells with a minimum precision of 52% and a maximum precision of 75%. |
| Kavitha et al., South Korea, 2018. [36] | Evaluating several machine learning classifiers for iPSC colony characterization based on a quantitative texture extraction | iPSC and inactive MEFs | 169 phase-contrast microscopic images | Support vector machine (SVM) Random forest (RF) Multilayer perceptron (MLP) Decision tree (DT) Adaptive boosting (Adaboost) classifier models | SVM, RF, and Adaboost delivered better classification performances than DT and MLP. The proposed automated fused statistical, shape-based, and moment-based texture pattern features that are potentially more helpful to biologists for characterizing the colonies of stem cells. |
| Kusumoto et al., Japan, 2018. [37] | Testing an automated method for identifiying iPSC-derived endothelial cells based on morphology | 640 images for training 160 images for validation 600 images for testing | LeNet AlexNet | The deep learning technique was able to detect iPSC-derived endothelial cells with 90% accuracy. | |
| Theagarajan et al., USA, 2018. [38] | Proposing a system for hESC images image classification using CNN and triplet CNN in a hierarchical system which allows for their classifications into 6 categories | hESCs | 784 images | Conv Maxpool FC Layer | The proposed system classified hESC images with 85.67% accuracy using the CNN Alone and recorded a 91.38% accuracy using the CNN and Triplet CNN and 94.11% accuracy by fusing the outputs of the CNN and triplet CNNs. |
| Buggenthin et al., Germany, 2017. [39] | Testing a deep learning method that predicts the lineage choice in the differentiating primary hematopoietic progenitors | Murine hematopoietic stem and progenitor cells (HSPCs) | 2,400,000 image patches | CNN Recurrent neural network (RNN) | Without a molecular labeling, the algorithm was able to identify cells with differentially expressed lineage-specifying genes. |
| Chang et al., Taiwan, 2017. [40] | Automatic detection and localization of human iPSC regions in brightfield microscopy images | CD34+ cells | 132 images | CNN | The automatic method successfully localized and detected human iPS cell formation, ultimately producing an iPS cell culture perk. |
| Fan et al., China, 2017. [41] | Testing a label-free and quantitative automated system for iPSCs segmentation and classification | iPSCs (human and animals) | 50 images | Modified AlexNET | No significant differences were recorded between the used algorithm and the manual method for cell classification. |
| Li et al., China, 2017. [42] | Proposing a system of multi-stage frameworks using content-based microscopic image analysis (CBMIA) | hiPSCs | 81 microscopic images | Improved supervised normalized cut (ISNC) segmentation algorithm k-means clustering algorithm | Results show that the CBMIA system was able to support a high-performing clustering result, allowing for the prediction of the stem cell differentiation process. |
| Joutsijoki et al., Finland, 2016. [43] | Assessing the automated quality of iPSC colony image identification where feeder cells are included and not included | iPSCs | 173 images | Multiclass support vector machines Scaled invariant feature transformation (SIFT) | The k-NN classifier achieved accurate results with an accuracy of 62.4%. |
| Wuttisarnwattana P et al., Thailand, 2016. [44] | Describing a novel machine-learning-based approach for detecting fluorescently labeled stem cells in cryo-imaging data | Mouse multipotent adult progenitors cells (MAPCs) | 700 fluorescent images | A novel algorithm created using MATLAB | The new tested software allowed for an accurate detection and quantification of cells anywhere in the entire whole mouse volume with single-cell sensitivity. |
| Bajcsy et al., USA, 2015. [45] | Designing algorithms that can be applied to a very large number of confocal microscopy images (z-stacks) for three-dimensional (3D) segmentation | hBMSCs | More than 1000 z-stacks | A set of six newly constructed 3D segmentation algorithms | The most accurate 3D segmentation algorithm achieved an average precision of 0.82 and accuracy of 0.84 measured by the Dice similarity index. |
| Lou et Al., USA, 2014. [46] | Developing a modular interactive nuclear segmentation (MINS) as a MATLAB/ C++-based segmentation tool tailored for counting cells and fluorescent-intensity measurements | Murine extraembryonic endoderm stem and embryonic stem cells (ESCs) | N/A | Seeded geodesic image segmentation (SGIS) | The framework achieved a balance between computational complexity and runtime. |
| Maddah et al., USA, 2014. [47] | Presenting a new method that can reliably extract and quantify beat signals from cardiomyocyte cell cultures | iPSC-derived cardiomyocytes | More than 500 videos | Hierarchical clustering algorithm | The use method was able to properly characterize stem-cell-derived cardiomyocytes. |
| Maddah et al., USA, 2014. [48] | Presenting a framework for the automated analysis of phase-contrast images of stem cells to capture and quantify morphological changes during colony growth | iPSCs | Over 500 time-lapse sequences images | N/A | The proposed novel framework demonstrated the successful classification of stem cells based on texture pattern recognition. |
| Paduano et al., Italy, 2013. [49] | Developing an analysis pipeline which can automatically process images of stem cell colonies in optical microscopy in order to study markers of embryonic stem cells (ESCs) heterogeneity | Mouse ESCs | 57 images | A proposed approach in MATLAB, CLAHE (image adjustment) Orientation matching algorithm | The tested algorithm achieved proper image processing. |
| Faustino et al., Brazil, 2011. [50] | Presenting an algorithm that counts and detects ESCs in fluorescence microscopy images | Murine ESCs | 234 images | Developed their own graph-mining algorithm | The used method achieved an average F-measure above 90%. |
| Faustino et al., Brazil, 2009. [51] | Proposing an automatic method for ESC detection and counting under fluorescence microscopy images | ESCs | 92 images | The algorithm was implemented in Java language 6.0 using the development tool Eclipse 3.2 | The used method resulted in an average of 93.97% precision, recall 92.04%, and 92.87% F-measure. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Issa, J.; Abou Chaar, M.; Kempisty, B.; Gasiorowski, L.; Olszewski, R.; Mozdziak, P.; Dyszkiewicz-Konwińska, M. Artificial-Intelligence-Based Imaging Analysis of Stem Cells: A Systematic Scoping Review. Biology 2022, 11, 1412. https://doi.org/10.3390/biology11101412
Issa J, Abou Chaar M, Kempisty B, Gasiorowski L, Olszewski R, Mozdziak P, Dyszkiewicz-Konwińska M. Artificial-Intelligence-Based Imaging Analysis of Stem Cells: A Systematic Scoping Review. Biology. 2022; 11(10):1412. https://doi.org/10.3390/biology11101412
Chicago/Turabian StyleIssa, Julien, Mazen Abou Chaar, Bartosz Kempisty, Lukasz Gasiorowski, Raphael Olszewski, Paul Mozdziak, and Marta Dyszkiewicz-Konwińska. 2022. "Artificial-Intelligence-Based Imaging Analysis of Stem Cells: A Systematic Scoping Review" Biology 11, no. 10: 1412. https://doi.org/10.3390/biology11101412
APA StyleIssa, J., Abou Chaar, M., Kempisty, B., Gasiorowski, L., Olszewski, R., Mozdziak, P., & Dyszkiewicz-Konwińska, M. (2022). Artificial-Intelligence-Based Imaging Analysis of Stem Cells: A Systematic Scoping Review. Biology, 11(10), 1412. https://doi.org/10.3390/biology11101412










