Characterization of miRNAs in Embryonic, Larval, and Adult Lumpfish Provides a Reference miRNAome for Cyclopterus lumpus
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Fish Holding
2.2. Ethics Statement
2.3. Sample Collection
2.4. RNA Extraction
2.5. High-Throughput Sequencing (HTS)
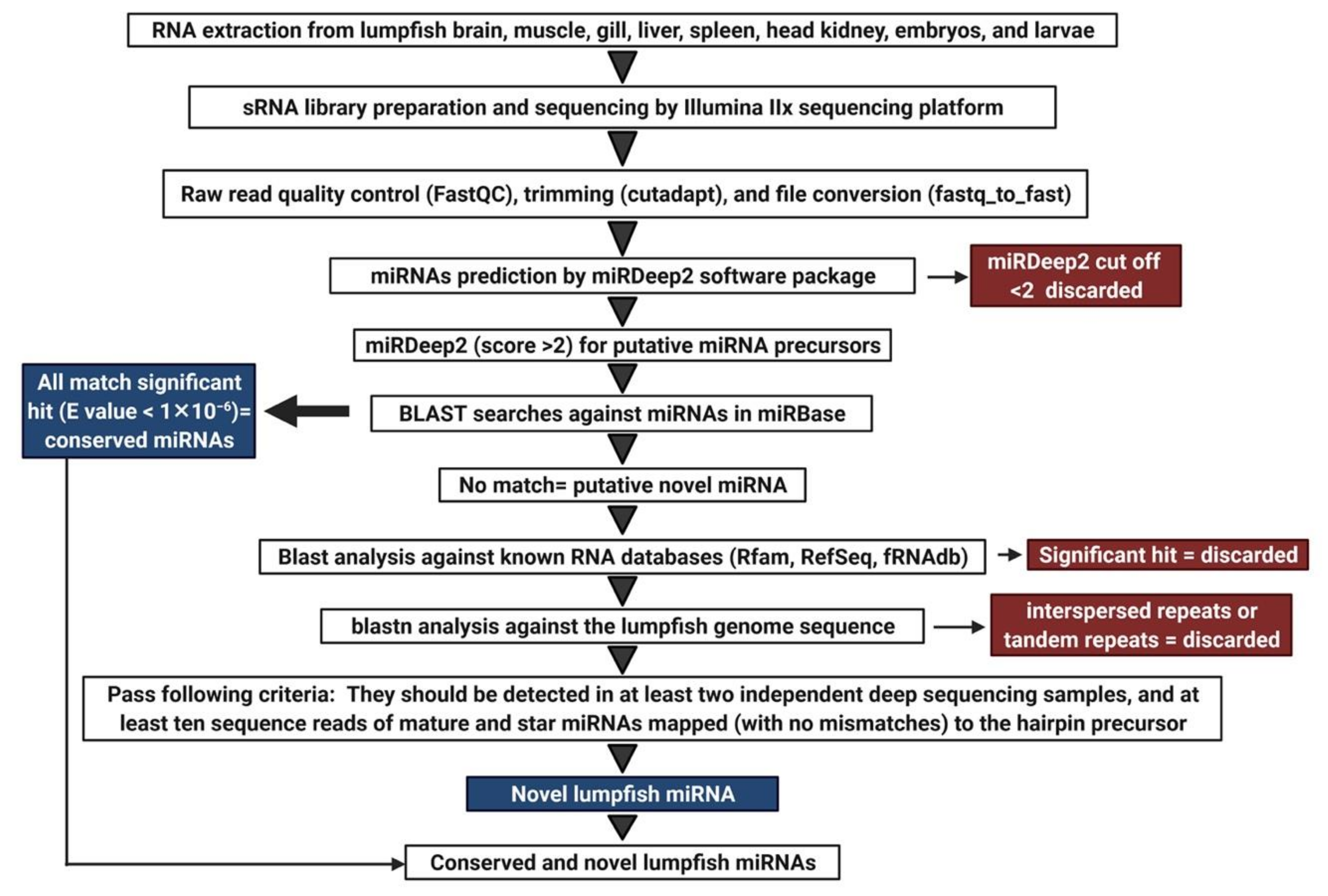
2.6. Analysis of Sequencing Data
2.7. Disclosing Putative Differentially Expressed and Organ and Developmental Stage Enriched miRNAs
2.8. RT-qPCR
3. Results
3.1. Total RNA Extraction, Library Preparation, and Small RNA Sequencing
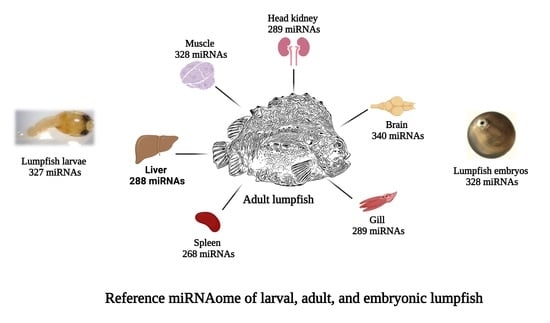
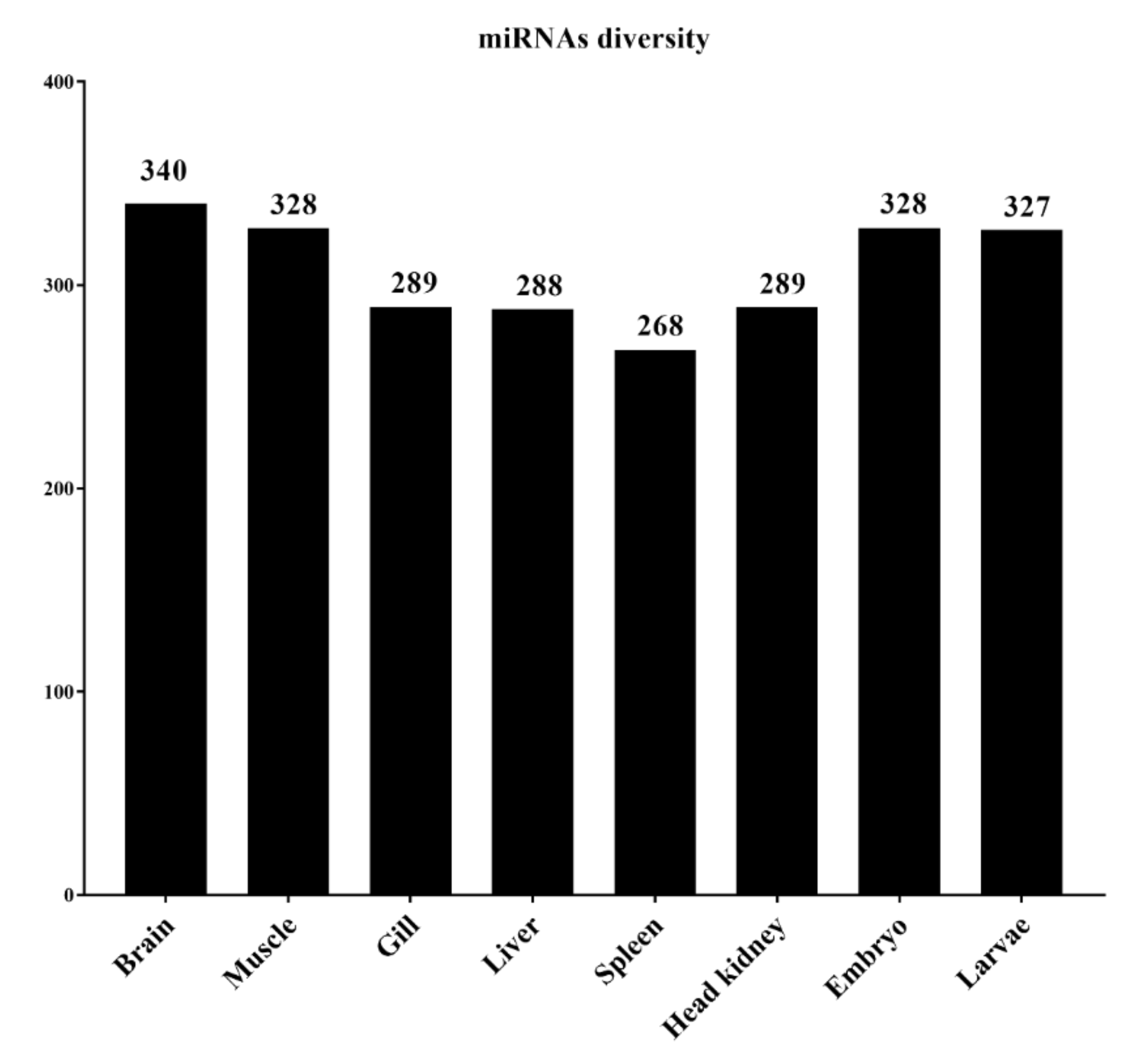
3.2. Characterization of Lumpfish miRNA
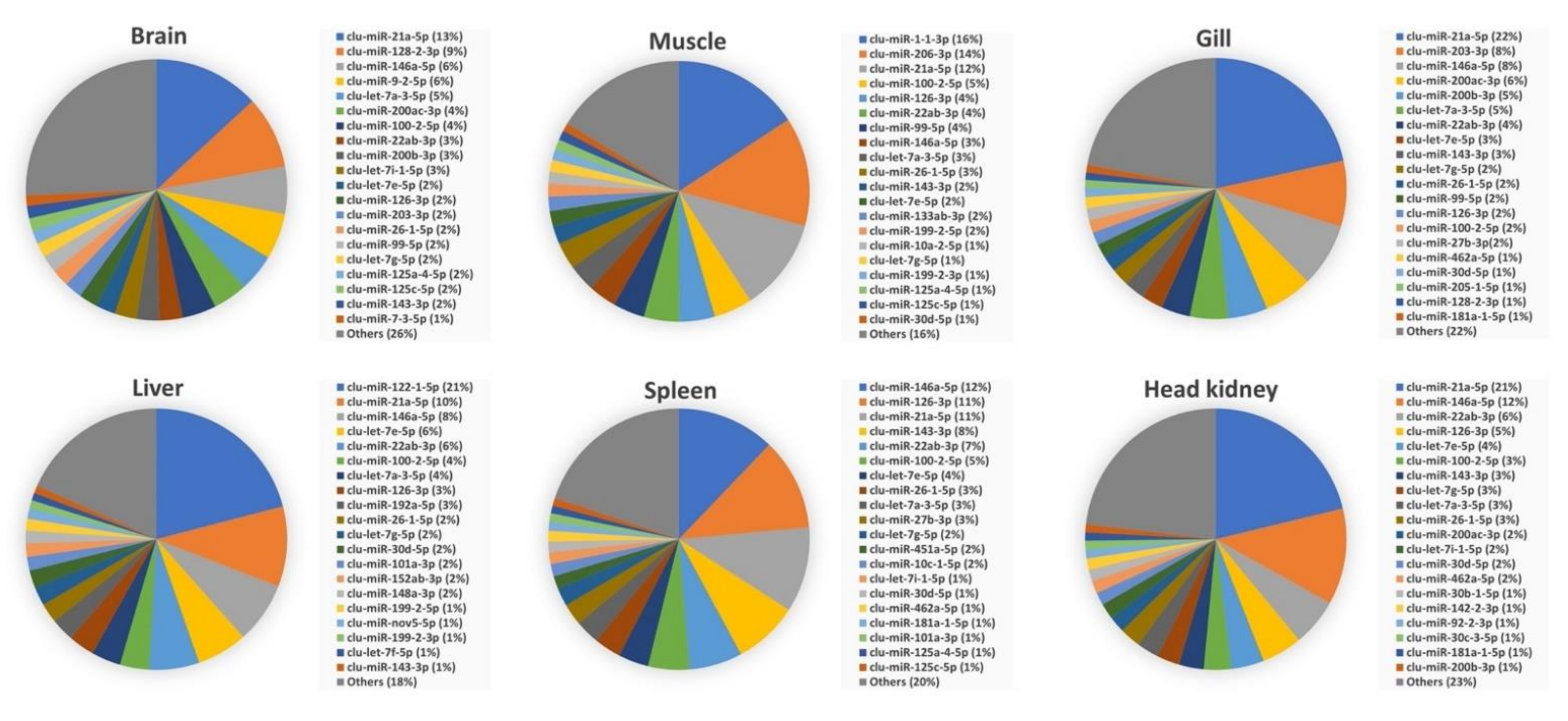
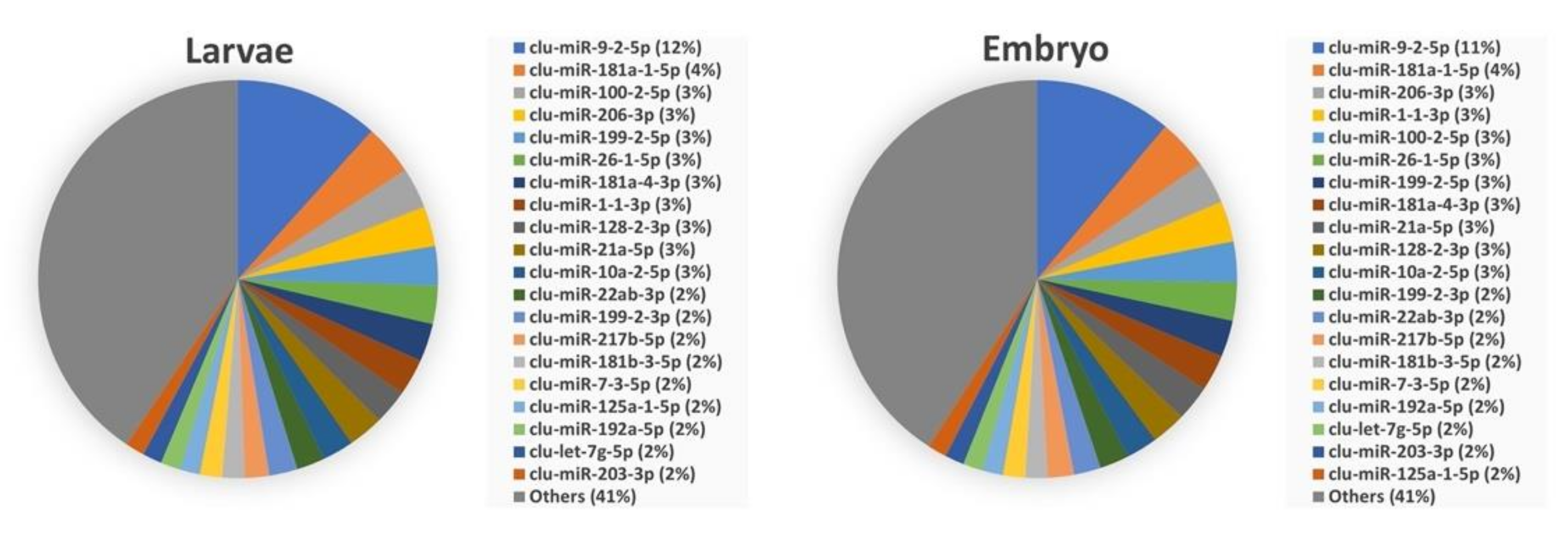
3.3. Abundance of miRNAs within Organs and Developmental Stages
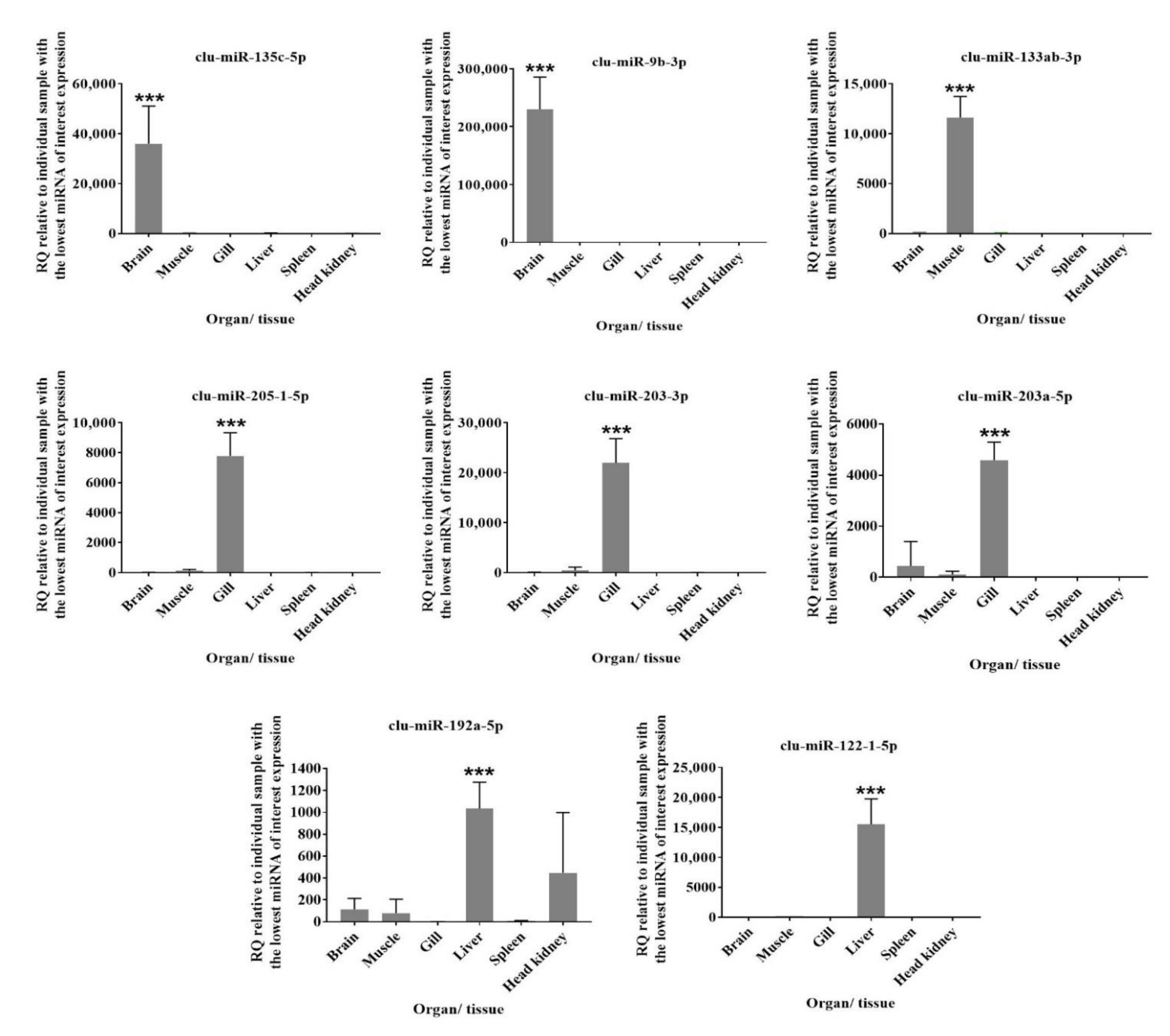
3.4. Comparison of Mature miRNA Expression between Organs and Early Developmental Stages
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bhaskaran, M.; Mohan, M. MicroRNAs: History, biogenesis, and their evolving role in animal development and disease. Vet. Pathol. 2014, 51, 759–774. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Michlewski, G.; Caceres, J.F. Post-transcriptional control of miRNA biogenesis. RNA 2019, 25, 1–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krol, J.; Loedige, I.; Filipowicz, W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010, 11, 597–610. [Google Scholar] [CrossRef]
- Saliminejad, K.; Khorram Khorshid, H.R.; Soleymani Fard, S.; Ghaffari, S.H. An overview of microRNAs: Biology, functions, therapeutics, and analysis methods. J. Cell. Physiol. 2019, 234, 5451–5465. [Google Scholar] [CrossRef]
- Bronevetsky, Y.; Ansel, K.M. Regulation of miRNA biogenesis and turnover in the immune system. Immunol. Rev. 2013, 253, 304–316. [Google Scholar] [CrossRef] [Green Version]
- Matsuyama, H.; Suzuki, H.I. Systems and Synthetic microRNA Biology: From Biogenesis to Disease Pathogenesis. Int. J. Mol. Sci. 2019, 21, 132. [Google Scholar] [CrossRef] [Green Version]
- De Sousa, M.C.; Gjorgjieva, M.; Dolicka, D.; Sobolewski, C.; Foti, M. Deciphering miRNAs’ Action through miRNA Editing. Int. J. Mol. Sci. 2019, 20, 6249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef]
- Hammond, S.M. An overview of microRNAs. Adv. Drug Deliv. Rev. 2015, 87, 3–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ravi, V.; Venkatesh, B. Rapidly evolving fish genomes and teleost diversity. Curr. Opin. Genet. Dev. 2008, 18, 544–550. [Google Scholar] [CrossRef]
- Bizuayehu, T.T.; Babiak, I. MicroRNA in Teleost Fish. Genome Biol. Evol. 2014, 6, 1911–1937. [Google Scholar] [CrossRef] [Green Version]
- Xue, X.; Woldemariam, N.T.; Caballero-Solares, A.; Umasuthan, N.; Fast, M.D.; Taylor, R.G.; Rise, M.L.; Andreassen, R. Dietary Immunostimulant CpG Modulates MicroRNA Biomarkers Associated with Immune Responses in Atlantic Salmon (Salmo salar). Cells 2019, 8, 1592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, N.C.; Christian, S.L.; Woldemariam, N.T.; Clow, K.A.; Rise, M.L.; Andreassen, R. Characterization of miRNAs in Cultured Atlantic Salmon Head Kidney Monocyte-Like and Macrophage-Like Cells. Int. J. Mol. Sci. 2020, 21, 3989. [Google Scholar] [CrossRef] [PubMed]
- Woldemariam, N.T.; Agafonov, O.; Sindre, H.; Høyheim, B.; Houston, R.D.; Robledo, D.; Bron, J.E.; Andreassen, R. miRNAs Predicted to Regulate Host Anti-viral Gene Pathways in IPNV-Challenged Atlantic Salmon Fry Are Affected by Viral Load, and Associated with the Major IPN Resistance QTL Genotypes in Late Infection. Front. Immunol. 2020, 11, 2113. [Google Scholar] [CrossRef] [PubMed]
- Andreassen, R.; Woldemariam, N.T.; Egeland, I.O.; Agafonov, O.; Sindre, H.; Hoyheim, B. Identification of differentially expressed Atlantic salmon miRNAs responding to salmonid alphavirus (SAV) infection. BMC Genom. 2017, 18, 349. [Google Scholar] [CrossRef]
- Andreassen, R.; Høyheim, B. miRNAs associated with immune response in teleost fish. Dev. Comp. Immunol. 2017, 75, 77–85. [Google Scholar] [CrossRef] [PubMed]
- Eslamloo, K.; Inkpen, S.M.; Rise, M.L.; Andreassen, R. Discovery of microRNAs associated with the antiviral immune response of Atlantic cod macrophages. Mol. Immunol. 2018, 93, 152–161. [Google Scholar] [CrossRef] [Green Version]
- Andreassen, R.; Worren, M.M.; Høyheim, B. Discovery and characterization of miRNA genes in atlantic salmon (Salmo salar) by use of a deep sequencing approach. BMC Genom. 2013, 14, 482. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Woldemariam, N.T.; Agafonov, O.; Høyheim, B.; Houston, R.D.; Taggart, J.B.; Andreassen, R. Expanding the miRNA Repertoire in Atlantic Salmon; Discovery of IsomiRs and miRNAs Highly Expressed in Different Tissues and Developmental Stages. Cells 2019, 8, 42. [Google Scholar] [CrossRef] [Green Version]
- Andreassen, R.; Rangnes, F.; Sivertsen, M.; Chiang, M.; Tran, M.; Worren, M.M. Discovery of miRNAs and Their Corresponding miRNA Genes in Atlantic Cod (Gadus morhua): Use of Stable miRNAs as Reference Genes Reveals Subgroups of miRNAs That Are Highly Expressed in Particular Organs. PLoS ONE 2016, 11, e0153324. [Google Scholar] [CrossRef]
- Ma, H.; Hostuttler, M.; Wei, H.; Rexroad, C.E., 3rd; Yao, J. Characterization of the rainbow trout egg microRNA transcriptome. PLoS ONE 2012, 7, e39649. [Google Scholar]
- Bizuayehu, T.T.; Fernandes, J.M.O.; Johansen, S.D.; Babiak, I. Characterization of Novel Precursor miRNAs Using Next Generation Sequencing and Prediction of miRNA Targets in Atlantic Halibut. PLoS ONE 2013, 8, e61378. [Google Scholar] [CrossRef] [Green Version]
- Xu, Z.; Chen, J.; Li, X.; Ge, J.; Pan, J.; Xu, X. Identification and Characterization of MicroRNAs in Channel Catfish (Ictalurus punctatus) by Using Solexa Sequencing Technology. PLoS ONE 2013, 8, e54174. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Shi, Z.; Wu, M.; Zhang, J.; Jia, L.; Chen, X. Identification and Differential Expression of MicroRNAs during Metamorphosis of the Japanese Flounder (Paralichthys olivaceus). PLoS ONE 2011, 6, e22957. [Google Scholar] [CrossRef]
- Xia, J.H.; He, X.P.; Bai, Z.Y.; Yue, G.H. Identification and Characterization of 63 MicroRNAs in the Asian Seabass Lates calcarifer. PLoS ONE 2011, 6, e17537. [Google Scholar] [CrossRef] [PubMed]
- Robledo, D.; Martin, A.P.; Álvarez-Dios, J.A.; Bouza, C.; Pardo, B.G.; Martínez, P. First characterization and validation of turbot microRNAs. Aquaculture 2017, 472, 76–83. [Google Scholar] [CrossRef]
- Chen, P.Y.; Manninga, H.; Slanchev, K.; Chien, M.; Russo, J.J.; Ju, J.; Sheridan, R.; John, B.; Marks, D.S.; Gaidatzis, D.; et al. The developmental miRNA profiles of zebrafish as determined by small RNA cloning. Genes Dev. 2005, 19, 1288–1293. [Google Scholar] [CrossRef] [Green Version]
- Desvignes, T.; Batzel, P.; Sydes, J.; Eames, B.F.; Postlethwait, J.H. miRNA analysis with Prost! reveals evolutionary conservation of organ-enriched expression and post-transcriptional modifications in three-spined stickleback and zebrafish. Sci. Rep. 2019, 9, 3913. [Google Scholar] [CrossRef] [Green Version]
- DFO. Aquaculture in Atlantic Canada—Atlantic Salmon. 2021. Available online: https://www.canada.ca/en/atlantic-canada-opportunities/services/factsheetsandbrochures35.html (accessed on 21 January 2021).
- DFO. Evaluation of Bacterial Kidney Disease (BKD) Impacts on the Canadian Salmon Aquaculture Industry Final Report; Fisheries and Oceans Canada (DFO): Ottawa, ON, Canada, 2010; Available online: https://www.cahs-bc.ca/wp-content/uploads/2019/03/BKDWhitePaper_FinalApr2010_WithAppendices.pdf (accessed on 21 January 2021).
- Akazawa, N.; Alvial, A.; Blanc, P.P.; Burgos, J.; Chamberlain, G.; Forster, J.; Hoang, T.; Ibarra, R.; Khoa, L.; Kibenge, F.; et al. Reducing Disease Risk in Aquaculture—World Bank Report Number 88257-GLB; World Bank: Washington, DC, USA, 2014. [Google Scholar]
- Brooker, A.J.; Papadopoulou, A.; Gutierrez, C.; Rey, S.; Davie, A.; Migaud, H. Sustainable production and use of cleaner fish for the biological control of sea lice: Recent advances and current challenges. Vet. Rec. 2018, 183, 383. [Google Scholar] [CrossRef] [Green Version]
- Lam, C.T.; Rosanowski, S.M.; Walker, M.; St-Hilaire, S. Sea lice exposure to non-lethal levels of emamectin benzoate after treatments: A potential risk factor for drug resistance. Sci. Rep. 2020, 10, 932–938. [Google Scholar] [CrossRef] [Green Version]
- Karbowski, C.; Finstad, B.; Karbowski, N.; Hedger, R. Sea lice in Iceland: Assessing the status and current implications for aquaculture and wild salmonids. Aquac. Environ. Interact. 2019, 11, 149–160. [Google Scholar] [CrossRef]
- Aaen, S.M.; Helgesen, K.O.; Bakke, M.J.; Kaur, K.; Horsberg, T.E. Drug resistance in sea lice: A threat to salmonid aquaculture. Trends Parasitol. 2015, 31, 72–81. [Google Scholar] [CrossRef] [PubMed]
- Abolofia, J.; Asche, F.; Wilen, J.E. The Cost of Lice: Quantifying the Impacts of Parasitic Sea Lice on Farmed Salmon. Mar. Resour. Econ. 2017, 32, 329–349. [Google Scholar] [CrossRef]
- Overton, K.; Dempster, T.; Oppedal, F.; Kristiansen, T.S.; Gismervik, K.; Stien, L.H. Salmon lice treatments and salmon mortality in Norwegian aquaculture: A review. Rev. Aquac. 2019, 11, 1398–1417. [Google Scholar] [CrossRef] [Green Version]
- Barker, S.E.; Bricknell, I.R.; Covello, J.; Purcell, S.; Fast, M.D.; Wolters, W.; Bouchard, D.A. Sea lice, Lepeophtheirus salmonis (Krøyer 1837), infected Atlantic salmon (Salmo salar L.) are more susceptible to infectious salmon anemia virus. PLoS ONE 2019, 14, e0209178. [Google Scholar] [CrossRef] [Green Version]
- Torrissen, O.; Jones, S.; Asche, F.; Guttormsen, A.; Skilbrei, O.T.; Nilsen, F.; Horsberg, T.E.; Jackson, D. Salmon lice—Impact on wild salmonids and salmon aquaculture. J. Fish Dis. 2013, 36, 171–194. [Google Scholar] [CrossRef] [Green Version]
- Costello, M.J. The global economic cost of sea lice to the salmonid farming industry. J. Fish Dis. 2009, 32, 115–118. [Google Scholar] [CrossRef]
- Barrett, L.T.; Overton, K.; Stien, L.H.; Oppedal, F.; Dempster, T. Effect of cleaner fish on sea lice in Norwegian salmon aquaculture: A national scale data analysis. Int. J. Parasitol. 2020, 50, 787–796. [Google Scholar] [CrossRef]
- Imsland, A.K.D.; Hanssen, A.; Nytrø, A.V.; Reynolds, P.; Jonassen, T.M.; Hangstad, T.A.; Elvegård, T.A.; Urskog, T.C.; Mikalsen, B. It works! Lumpfish can significantly lower sea lice infestation in large-scale salmon farming. Biol. Open 2018, 7, bio036301. [Google Scholar] [CrossRef] [Green Version]
- Vasquez, I.; Cao, T.; Chakraborty, S.; Gnanagobal, H.; O’Brien, N.; Monk, J.; Boyce, D.; Westcott, J.D.; Santander, J. Comparative Genomics Analysis of Vibrio anguillarum Isolated from Lumpfish (Cyclopterus lumpus) in Newfoundland Reveal Novel Chromosomal Organizations. Microorganisms 2020, 8, 1666. [Google Scholar] [CrossRef]
- Chakraborty, S.; Cao, T.; Hossain, A.; Gnanagobal, H.; Vasquez, I.; Boyce, D.; Santander, J. Vibrogen-2 vaccine trial in lumpfish (Cyclopterus lumpus) against Vibrio anguillarum. J. Fish Dis. 2019, 42, 1057–1064. [Google Scholar] [CrossRef]
- Dang, M.; Cao, T.; Vasquez, I.; Hossain, A.; Gnanagobal, H.; Kumar, S.; Hall, J.; Monk, J.; Boyce, D.; Westcott, J.; et al. Oral Immunization of Larvae and Juvenile of Lumpfish (Cyclopterus lumpus) against Vibrio anguillarum Does Not Influence Systemic Immunity. Vaccines 2021, 9, 819. [Google Scholar] [CrossRef]
- Boyce, D.; Ang, K.P.; Prickett, R. Cunner and lumpfish as cleaner fish species in Canada. In Cleaner Fish Biology and Aquaculture Applications; Treasurer, J.W., Ed.; 5M Publishing: Sheffield, UK, 2018. [Google Scholar]
- Shwe, A.; Østbye, T.-K.K.; Krasnov, A.; Ramberg, S.; Andreassen, R. Characterization of Differentially Expressed miRNAs and Their Predicted Target Transcripts during Smoltification and Adaptation to Seawater in Head Kidney of Atlantic Salmon. Genes 2020, 11, 1059. [Google Scholar] [CrossRef]
- Knutsen, T.M.; Kirubakaran, G.T.; Mommens, M.; Moen, T. Lumpfish (Cyclopterus lumpus) Draft Genome Assembly; Figshare: Iasi, Romania, 2018. [Google Scholar]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Friedlander, M.R.; Chen, W.; Adamidi, C.; Maaskola, J.; Einspanier, R.; Knespel, S.; Rajewsky, N. Discovering microRNAs from deep sequencing data using miRDeep. Nat. Biotechnol. 2008, 26, 407–415. [Google Scholar] [CrossRef]
- Friedländer, M.R.; Mackowiak, S.D.; Li, N.; Chen, W.; Rajewsky, N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012, 40, 37–52. [Google Scholar] [CrossRef]
- Kozomara, A.; Griffiths-Jones, S. miRBase: Annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014, 42, D68–D73. [Google Scholar] [CrossRef] [Green Version]
- Griffiths-Jones, S.; Grocock, R.J.; Van Dongen, S.; Bateman, A.; Enright, A.J. miRBase: MicroRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006, 34 (Supplement S1), D140–D144. [Google Scholar] [CrossRef] [PubMed]
- Ambros, V.; Bartel, B.; Bartel, D.P.; Burge, C.B.; Carrington, J.C.; Chen, X.; Dreyfuss, G.; Eddy, S.R.; Griffiths-Jones, S.; Marshall, M.; et al. A uniform system for microRNA annotation. RNA 2003, 9, 277–279. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johansen, I.; Andreassen, R. Validation of miRNA genes suitable as reference genes in qPCR analyses of miRNA gene expression in Atlantic salmon (Salmo salar). BMC Res. Notes 2014, 7, 945. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruijter, J.M.; Ramakers, C.; Hoogaars, W.M.H.; Karlen, Y.; Bakker, O.; van den Hoff, M.J.B.; Moorman, A.F.M. Amplification efficiency: Linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res. 2009, 37, e45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- miRBase: The MicroRNA Database. Available online: http://www.mirbase.org (accessed on 15 November 2012).
- Dantzer, R.; Wollman, E.E. Relationships between the brain and the immune system. J. Société Biol. 2003, 197, 81–88. [Google Scholar] [CrossRef]
- Dantzer, R. Neuroimmune Interactions: From the Brain to the Immune System and Vice Versa. Physiol. Rev. 2018, 98, 477–504. [Google Scholar] [CrossRef]
- Brennan, G.; Henshall, D.C. MicroRNAs as regulators of brain function and targets for treatment of epilepsy. Nat. Rev. Neurol. 2020, 16, 506–519. [Google Scholar] [CrossRef]
- Tessmar-Raible, K.; Raible, F.; Christodoulou, F.; Guy, K.; Rembold, M.; Hausen, H.; Arendt, D. Conserved Sensory-Neurosecretory Cell Types in Annelid and Fish Forebrain: Insights into Hypothalamus Evolution. Cell 2007, 129, 1389–1400. [Google Scholar] [CrossRef] [Green Version]
- Giraldez, A.J.; Cinalli, R.M.; Glasner, M.E.; Enright, A.; Thomson, J.M.; Baskerville, S.; Hammond, S.M.; Bartel, D.P.; Schier, A.F. MicroRNAs Regulate Brain Morphogenesis in Zebrafish. Science 2005, 308, 833–838. [Google Scholar] [CrossRef] [Green Version]
- Radhakrishnan, B.; Anand, A.A.P. Role of miRNA-9 in Brain Development. J. Exp. Neurosci. 2016, 10, JEN.S32843. [Google Scholar] [CrossRef] [Green Version]
- Zhao, J.; Zhou, Y.; Guo, M.; Yue, D.; Chen, C.; Liang, G.; Xu, L. MicroRNA-7: Expression and function in brain physiological and pathological processes. Cell Biosci. 2020, 10, 1–12. [Google Scholar] [CrossRef]
- Vaz, C.; Wee, C.W.; Lee, G.P.S.; Ingham, P.W.; Tanavde, V.; Mathavan, S. Deep sequencing of small RNA facilitates tissue and sex associated microRNA discovery in zebrafish. BMC Genom. 2015, 16, 950. [Google Scholar] [CrossRef] [Green Version]
- Tan, C.L.; Plotkin, J.L.; Venø, M.T.; von Schimmelmann, M.; Feinberg, P.; Mann, S.; Handler, A.; Kjems, J.; Surmeier, D.J.; O’Carroll, D.; et al. MicroRNA-128 Governs Neuronal Excitability and Motor Behavior in Mice. Science 2013, 342, 1254–1258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsuyama, J.; Bunt, J.; Richards, L.J.; Iwanari, H.; Mochizuki, Y.; Hamakubo, T.; Shimazaki, T.; Okano, H. MicroRNA-153 Regulates the Acquisition of Gliogenic Competence by Neural Stem Cells. Stem Cell Rep. 2015, 5, 365–377. [Google Scholar] [CrossRef] [Green Version]
- Wanet, A.; Tacheny, A.; Arnould, T.; Renard, P. miR-212/132 expression and functions: Within and beyond the neuronal compartment. Nucleic Acids Res 2012, 40, 4742–4753. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Howe, J.; Li, E.S.; Streeter, S.E.; Rahme, G.; Chipumuro, E.; Russo, G.B.; Litzky, J.F.; Hills, L.B.; Rodgers, K.; Skelton, P.; et al. MiR-338-3p regulates neuronal maturation and suppresses glioblastoma proliferation. PLoS ONE 2017, 12, e0177661. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valenzuela, C.A.; Zuloaga, R.; Poblete-Morales, M.; Vera-Tobar, T.; Mercado, L.; Avendaño-Herrera, R.; Valdés, J.A.; Molina, A. Fish skeletal muscle tissue is an important focus of immune reactions during pathogen infection. Dev. Comp. Immunol. 2017, 73, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Brzeszczyńska, J.; Brzeszczyński, F.; Hamilton, D.F.; McGregor, R.; Simpson, A.H.R.W. Role of microRNA in muscle regeneration and diseases related to muscle dysfunction in atrophy, cachexia, osteoporosis, and osteoarthritis. Bone Jt. Res. 2020, 9, 798–807. [Google Scholar] [CrossRef]
- Bhattacharya, M.; Sharma, A.R.; Sharma, G.; Patra, B.C.; Nam, J.-S.; Chakraborty, C.; Lee, S.-S. The crucial role and regulations of miRNAs in zebrafish development. Protoplasma 2017, 254, 17–31. [Google Scholar] [CrossRef]
- Yu, H.; Lu, Y.; Li, Z.; Wang, Q. microRNA-133: Expression, Function and Therapeutic Potential in Muscle Diseases and Cancer. Curr. Drug Targets 2014, 15, 817–828. [Google Scholar] [CrossRef]
- Zhang, R.; Li, R.; Lin, Y. Identification and characterization of microRNAs in the muscle of Schizothorax prenanti. Fish Physiol. Biochem. 2017, 121, 207–1064. [Google Scholar] [CrossRef]
- Horak, M.; Novak, J.; Bienertova-Vasku, J. Muscle-specific microRNAs in skeletal muscle development. Dev. Biol. 2016, 410, 1–13. [Google Scholar] [CrossRef]
- Yu, Y.; Wang, Q.; Huang, Z.; Ding, L.; Xu, Z. Immunoglobulins, Mucosal Immunity and Vaccination in Teleost Fish. Front. Immunol. 2020, 11, 567941. [Google Scholar] [CrossRef]
- Zhang, Q.-L.; Dong, Z.-X.; Luo, Z.-W.; Jiao, Y.-J.; Guo, J.; Deng, X.-Y.; Wang, F.; Chen, J.-Y.; Lin, L.-B. MicroRNA profile of immune response in gills of zebrafish (Danio rerio) upon Staphylococcus aureus infection. Fish Shellfish. Immunol. 2019, 87, 307–314. [Google Scholar] [CrossRef] [PubMed]
- Juanchich, A.; Bardou, P.; Rué, O.; Gabillard, J.-C.; Gaspin, C.; Bobe, J.; Guiguen, Y. Characterization of an extensive rainbow trout miRNA transcriptome by next generation sequencing. BMC Genom. 2016, 17, 164. [Google Scholar] [CrossRef]
- Su, H.; Fan, J.; Ma, D.; Zhu, H. Identification and Characterization of Osmoregulation Related MicroRNAs in Gills of Hybrid Tilapia Under Three Types of Osmotic Stress. Front. Genet. 2021, 12, 361. [Google Scholar] [CrossRef]
- Ng, H.M.; Ho, J.C.H.; Nong, W.; Hui, J.H.L.; Lai, K.P.; Wong, C.K.C. Genome-wide analysis of MicroRNA-messenger RNA interactome in ex-vivo gill filaments, Anguilla japonica. BMC Genom. 2020, 21, 208. [Google Scholar] [CrossRef]
- Sales, C.F.; Silva, R.F.; Amaral, M.G.C.; Domingos, F.F.T.; Ribeiro, R.I.M.A.; Thomé, R.G.; Santos, H.B. Comparative histology in the liver and spleen of three species of freshwater teleost. Neotrop. Ichthyol. 2017, 15. [Google Scholar] [CrossRef] [Green Version]
- Dai, Z.; Yang, T.; Song, G. The roles of miRNAs in liver diseases. Non-Coding RNA Investig. 2019, 3, 25. [Google Scholar] [CrossRef]
- Østbye, T.K.; Woldemariam, N.T.; Lundberg, C.E.; Berge, G.M.; Ruyter, B.; Andreassen, R. Modulation of hepatic miRNA expression in Atlantic salmon (Salmo salar) by family background and dietary fatty acid composition. J. Fish Biol. 2021, 98, 1172–1185. [Google Scholar] [CrossRef] [PubMed]
- Lagos-Quintana, M.; Rauhut, R.; Yalcin, A.; Meyer, J.; Lendeckel, W.; Tuschl, T. Identification of Tissue-Specific MicroRNAs from Mouse. Curr. Biol. 2002, 12, 735–739. [Google Scholar] [CrossRef] [Green Version]
- Trattner, S.; Vestergren, A.S. Tissue distribution of selected microRNA in Atlantic salmon. Eur. J. Lipid Sci. Technol. 2013, 115, 1348–1356. [Google Scholar] [CrossRef]
- Liu, X.-L.; Cao, H.-X.; Wang, B.-C.; Xin, F.-Z.; Zhang, R.-N.; Zhou, D.; Yang, R.-X.; Zhao, Z.-H.; Pan, Q.; Fan, J.-G. miR-192-5p regulates lipid synthesis in non-alcoholic fatty liver disease through SCD-1. World J. Gastroenterol. 2017, 23, 8140–8151. [Google Scholar] [CrossRef]
- Su, H.; Yang, J.-R.; Xu, T.; Huang, J.; Xu, L.; Yuan, Y.; Zhuang, S.-M. MicroRNA-101, Down-regulated in Hepatocellular Carcinoma, Promotes Apoptosis and Suppresses Tumorigenicity. Cancer Res. 2009, 69, 1135–1142. [Google Scholar] [CrossRef] [Green Version]
- Bronte, V.; Pittet, M.J. The Spleen in Local and Systemic Regulation of Immunity. Immunity 2013, 39, 806–818. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaattari, S.L.; Irwin, M.J. Salmonid spleen and anterior kidney harbor populations of lymphocytes with different B cell repertoires. Dev. Comp. Immunol. 1985, 9, 433–444. [Google Scholar] [CrossRef]
- Press, C.; Evensen, Ø. The morphology of the immune system in teleost fishes. Fish Shellfish Immunol. 1999, 9, 309–318. [Google Scholar] [CrossRef] [Green Version]
- Li, G.; Zhao, Y.; Wen, L.; Liu, Z.; Yan, F.; Gao, C. Identification and Characterization of MicroRNAs in the Spleen of Common Carp Immune Organ. J. Cell. Biochem. 2014, 115, 1768–1778. [Google Scholar] [CrossRef]
- Cao, Y.; Wang, D.; Li, S.; Zhao, J.; Xu, L.; Liu, H.; Lu, T.; Mou, Z. A transcriptome analysis focusing on splenic immune-related mciroRNAs of rainbow trout upon Aeromonas salmonicida subsp. salmonicida infection. Fish Shellfish. Immunol. 2019, 91, 350–357. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Wang, Y.-Y.; Fu, H.-C.; Huang, H.-Z. MicroRNA expression and analysis of immune-related putative target genes in ISKNV-infected spleen of mandarin fish (Siniperca chuatsi). Aquaculture 2021, 547, 737450. [Google Scholar] [CrossRef]
- Grimes, J.A.; Prasad, N.; Levy, S.; Cattley, R.; Lindley, S.; Boothe, H.W.; Henderson, R.A.; Smith, B.F. A comparison of microRNA expression profiles from splenic hemangiosarcoma, splenic nodular hyperplasia, and normal spleens of dogs. BMC Vet. Res. 2016, 12, 272. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Z.J.; Zhang, Y.P.; Li, Y.; Zheng, H.W.; Zheng, Y.S.; Liu, C.J. Distinct expression pattern of miRNAs in Marek’s disease virus infected-chicken splenic tumors and non-tumorous spleen tissues. Res. Vet. Sci. 2014, 97, 156–161. [Google Scholar] [CrossRef] [PubMed]
- He, J.-J.; Ma, J.; Wang, J.-L.; Xu, M.-J.; Zhu, X.-Q. Analysis of miRNA expression profiling in mouse spleen affected by acute Toxoplasma gondii infection. Infect. Genet. Evol. 2016, 37, 137–142. [Google Scholar] [CrossRef] [PubMed]
- Arribas, A.J.; Gomez-Abad, C.; Sánchez-Beato, M.; Martinez, N.; DiLisio, L.; Casado, F.; Cruz, M.A.; Algara, P.; Piris, M.A.; Mollejo, M. Splenic marginal zone lymphoma: Comprehensive analysis of gene expression and miRNA profiling. Mod. Pathol. 2013, 26, 889–901. [Google Scholar] [CrossRef]
- Huang, L.; Ma, J.; Sun, Y.; Lv, Y.; Lin, W.; Liu, M.; Tu, C.; Zhou, P.; Gu, W.; Su, S.; et al. Altered splenic miRNA expression profile in H1N1 swine influenza. Arch. Virol. 2015, 160, 979–985. [Google Scholar] [CrossRef] [PubMed]
- Pase, L.; Layton, J.E.; Kloosterman, W.P.; Carradice, D.; Waterhouse, P.M.; Lieschke, G.J. miR-451 regulates zebrafish erythroid maturation in vivo via its target gata2. Blood 2009, 113, 1794–1804. [Google Scholar] [CrossRef] [Green Version]
- Giraldez, A.J.; Mishima, Y.; Rihel, J.; Grocock, R.J.; Van Dongen, S.; Inoue, K.; Enright, A.J.; Schier, A.F. Zebrafish MiR-430 Promotes Deadenylation and Clearance of Maternal mRNAs. Science 2006, 312, 75–79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takacs, C.M.; Giraldez, A.J. miR-430 regulates oriented cell division during neural tube development in zebrafish. Dev. Biol. 2016, 409, 442–450. [Google Scholar] [CrossRef] [Green Version]
| miRNAs | Primer Sequences (5′ to 3′) |
|---|---|
| clu-miR-25-3p | CATTGCACTTGTCTCGGTCTGA |
| clu-miR-17-1-5p | CAAAGTGCTTACAGTGCAGGTA |
| clu-miR-122-1-5p | TGGAGTGTGACAATGGTGTTTG |
| clu-miR-133ab-3p | TTTGGTCCCCTTCAACCAGCTGT |
| clu-miR-205-1-5p | TCCTTCATTCCACCGGAGTCTG |
| clu-miR-135c-5p | TATGGCTTTTTATTCCTATGTG |
| clu-miR-203-3p | GTGAAATGTTTAGGACCACTTG |
| clu-miR-203a-5p | AGTGGTTCTCAACAGTTCAACA |
| clu-miR-192a-5p | ATGACCTATGAATTGACAGCCA |
| clu-miR-9b-3p | TAAAGCTAGAGAACCGAATGTA |
| Organ 1 | miRNAs 2 | Log2FC 3 |
|---|---|---|
| Brain | clu-miR-31-3p | 6.14 |
| Brain | clu-miR-153c-3p | 5.96 |
| Brain | clu-miR-153a-3p | 5.33 |
| Brain | clu-miR-1788-5p | 4.91 |
| Brain | clu-miR-212b-1-5p | 4.10 |
| Brain | clu-miR-212b-1-3p | 3.14 |
| Brain | clu-miR-128-2-3p | 3.49 |
| Brain | clu-miR-338-1-3p | 3.40 |
| Brain | clu-miR-132-1-5p | 3.08 |
| Muscle | clu-miR-133b-3p | 6.08 |
| Muscle | clu-miR-133ab-3p | 5.45 |
| Muscle | clu-miR-1-1-3p | 5.23 |
| Muscle | clu-miR-1-3-5p | 3.56 |
| Gill | clu-miR-31-5p | 6.91 |
| Gill | clu-miR-1788-3p | 6.21 |
| Gill | clu-miR-203-3p | 5.13 |
| Gill | clu-miR-203a-5p | 4.61 |
| Gill | clu-miR-375-1-3p | 4.82 |
| Gill | clu-miR-205-1-3p | 4.16 |
| Gill | clu-miR-200b-3p | 3.8 |
| Gill | clu-miR-200b-5p | 3.36 |
| Liver | clu-miR-122-1-5p | 8.23 |
| Liver | clu-miR-122-1-3p | 7.65 |
| Liver | clu-miR-nov3-3p | 6.78 |
| Liver | clu-miR-nov3-5p | 4.68 |
| Liver | clu-miR-nov1-5p | 5.58 |
| Liver | clu-miR-101b-3p | 4.83 |
| Liver | clu-miR-101b-5p | 4.41 |
| Liver | clu-miR-722-3p | 4.71 |
| Liver | clu-miR-722-5p | 4.38 |
| Liver | clu-miR-92b-3p | 4.04 |
| Liver | clu-miR-92b-5p | 3.91 |
| Liver | clu-miR-192a-5p | 3.75 |
| Liver | clu-miR-94a-5p | 3.43 |
| Liver | clu-miR-152ab-3p | 3.37 |
| Liver | clu-miR-nov5-5p | 3.36 |
| Spleen | clu-miR-2187b-5p | 5.10 |
| Spleen | clu-miR-2187b-3p | 3.47 |
| Spleen | clu-miR-460-5p | 3.27 |
| Embryos/Larvae 1 | miRNAs 2 | Log2FC 3 |
|---|---|---|
| Embryos | clu-miR-430b-5-5p | 5.61 |
| Embryos | clu-miR-430b-4-3p | 4.51 |
| Embryos | clu-miR-430b-1-3p | 4.37 |
| Embryos | clu-miR-190b-5p | 5.45 |
| Embryos | clu-miR-726-5p | 4.91 |
| Embryos | clu-miR-184ab-2-3p | 4.77 |
| Embryos | clu-miR-184ab-3p | 4.77 |
| Embryos | clu-miR-301b-5p | 4.73 |
| Embryos | clu-miR-301b-1-5p | 4.40 |
| Embryos | clu-miR-124-1-5p | 4.40 |
| Embryos | clu-miR-217b-5p | 4.23 |
| Embryos | clu-miR-217a-5p | 4.13 |
| Embryos | clu-miR-216a-1-5p | 4.20 |
| Larvae | clu-miR-124-1-5p | 4.41 |
| Larvae | clu-miR-130-1-5p | 3.15 |
| Larvae | clu-miR-130-6-5p | 3.62 |
| Larvae | clu-miR-183-5p | 4.09 |
| Larvae | clu-miR-184ab-2-3p | 4.71 |
| Larvae | clu-miR-184ab-3p | 4.71 |
| Larvae | clu-miR-190b-5p | 5.44 |
| Larvae | clu-miR-194b-3p | 3.46 |
| Larvae | clu-miR-196a-1-5p | 3.95 |
| Larvae | clu-miR-216a-1-5p | 4.00 |
| Larvae | clu-miR-217a-5p | 4.11 |
| Larvae | clu-miR-217b-5p | 4.11 |
| Larvae | clu-miR-301b-1-5p | 4.38 |
| Larvae | clu-miR-301b-3p | 3.44 |
| Larvae | clu-miR-301b-5p | 4.70 |
| Larvae | clu-miR-430a-12-3p | 3.97 |
| Larvae | clu-miR-430a-3-3p | 3.97 |
| Larvae | clu-miR-430b-1-3p | 4.28 |
| Larvae | clu-miR-430b-4-3p | 4.40 |
| Larvae | clu-miR-430b-5-5p | 5.41 |
| Larvae | clu-miR-459-3p | 4.01 |
| Larvae | clu-miR-726-5p | 4.64 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chakraborty, S.; Woldemariam, N.T.; Visnovska, T.; Rise, M.L.; Boyce, D.; Santander, J.; Andreassen, R. Characterization of miRNAs in Embryonic, Larval, and Adult Lumpfish Provides a Reference miRNAome for Cyclopterus lumpus. Biology 2022, 11, 130. https://doi.org/10.3390/biology11010130
Chakraborty S, Woldemariam NT, Visnovska T, Rise ML, Boyce D, Santander J, Andreassen R. Characterization of miRNAs in Embryonic, Larval, and Adult Lumpfish Provides a Reference miRNAome for Cyclopterus lumpus. Biology. 2022; 11(1):130. https://doi.org/10.3390/biology11010130
Chicago/Turabian StyleChakraborty, Setu, Nardos T. Woldemariam, Tina Visnovska, Matthew L. Rise, Danny Boyce, Javier Santander, and Rune Andreassen. 2022. "Characterization of miRNAs in Embryonic, Larval, and Adult Lumpfish Provides a Reference miRNAome for Cyclopterus lumpus" Biology 11, no. 1: 130. https://doi.org/10.3390/biology11010130
APA StyleChakraborty, S., Woldemariam, N. T., Visnovska, T., Rise, M. L., Boyce, D., Santander, J., & Andreassen, R. (2022). Characterization of miRNAs in Embryonic, Larval, and Adult Lumpfish Provides a Reference miRNAome for Cyclopterus lumpus. Biology, 11(1), 130. https://doi.org/10.3390/biology11010130