Simple Summary
Heat shock proteins (Hsps) are ubiquitous and conserved in almost all living organisms and are involved in a wide spectrum of cellular responses against diverse environmental stresses. However, our knowledge about the coordinated Hsp co-chaperon interaction is still limited, especially in aquatic animals facing dynamic water environments. In this study, we provided the systematic analysis of 95 Hsp genes (LmHsps) in spotted sea bass (Lateolabrax maculatus), an important aquaculture species in China, under salinity change and alkalinity stress through in silico analysis. The coordinated expression of LmHsps in response to salinity change and alkalinity stress in the gills was determined. Our results confirmed the diverse regulated expression of Hsps in L. maculatus, and that the responses to alkalinity stress may have arisen through the adaptive recruitment of LmHsp40-70-90 co-chaperons. Our results provide vital insights into the function and adaptation of aquatic animal Hsps in response to salinity-alkalinity stress.
Abstract
The heat shock protein (Hsp) superfamily has received accumulated attention because it is ubiquitous and conserved in almost all living organisms and is involved in a wide spectrum of cellular responses against diverse environmental stresses. However, our knowledge about the Hsp co-chaperon network is still limited in non-model organisms. In this study, we provided the systematic analysis of 95 Hsp genes (LmHsps) in the genome of spotted sea bass (Lateolabrax maculatus), an important aquaculture species in China that can widely adapt to diverse salinities from fresh to sea water, and moderately adapt to high alkaline water. Through in silico analysis using transcriptome and genome database, we determined the expression profiles of LmHsps in response to salinity change and alkalinity stress in L. maculatus gills. The results revealed that LmHsps were sensitive in response to alkalinity stress, and the LmHsp40-70-90 members were more actively regulated than other LmHsps and may also be coordinately interacted as co-chaperons. This was in accordance with the fact that members of LmHsp40, LmHsp70, and LmHsp90 evolved more rapidly in L. maculatus than other teleost lineages with positively selected sites detected in their functional domains. Our results revealed the diverse and cooperated regulation of LmHsps under alkaline stress, which may have arisen through the functional divergence and adaptive recruitment of the Hsp40-70-90 co-chaperons and will provide vital insights for the development of L. maculatus cultivation in alkaline water.
1. Introduction
Heat shock proteins (Hsps) are highly conserved stress proteins existing broadly from prokaryotes to mammals with chaperone activity [1,2], which play essential roles in maintaining the stability of cell structure and mitigating the effect of protein misfolding and aggregation in almost all aspects of protein metabolism [3]. The Hsp superfamily could be classified into Hsp10 (Hspe, 10 kDa), small Hsp (sHsp/Hspb, 20–30 kDa), Hsp40 (Dnaj, 40 kDa), Hsp60 (Hspd, 60 kDa), Hsp70 (Hspa, 70 kDa), Hsp90 (Hspc, 83~90 kDa), and Hsp100/110 (Hsph, 100–110 kDa) according to their relative molecular mass, structure homology, and function [4]. For example, sHsps can bind a large range of non-native substrate proteins to form a sHsp substrate complex prior to irreversible aggregation, which becomes the target of ATP-dependent Hsp70–Hsp100 disaggregation activity [5]. Hsp40s, also known as J-domain proteins (Dnajs), have a key role in the process of transferring substrates to their Hsp70 partners by stimulating the ATP hydrolysis of Hsp70 [6]. Hsp10/60s are co-chaperones mainly implicated in mitochondrial protein import and macromolecular assembly, which function together to facilitate the correct folding of imported proteins [7]. Hsp70s and Hsp90s are both ATP-dependent molecular chaperones, which perform numerous functions in a wide variety of cellular processes, including the protection of proteins from stresses [8]. In addition, the Hsp chaperons also cooperate with each other to constitute a dynamic and functionally versatile network for diverse cellular functions [9]. For instance, Hsp70 and Hsp40 could interact with a substrate protein and co-chaperones to facilitate the transfer of the substrate to Hsp90 [9,10], while Hsp90 and Hsp70 could also directly collaborate with each other [10]. In contrast, sHsps function independently of ATP so as to prevent the formation of large insoluble protein aggregates [11].
With the increasing impacts of climate change and anthropogenic stresses, the function of the heat-shock system affecting organisms’ evolutionary fitness in diverse environments is a growing and important topic [12]. A large number of studies have focused on Hsp families in animals living in a highly dynamic aquatic environment. For instance, changes in abiotic factors (temperature, salinity, pH, dissolved oxygen, heavy metals, or other pollutants) [13,14,15] as well as biotic pathogen and toxic algae infection [16,17,18,19,20] could cause stresses in aquatic animals. Accordingly, Hsps are expressed significantly and act as chaperones to help cells maintain normal physiological activities related to growth, reproduction, and physiological homeostasis. In particular, salinity changes in aquatic environments can increase the lysozyme activity, mucosal production, and immune defense in teleosts, and high alkalinity is harmful to most fishes, leading to health damage and even degradation of biomolecules [21]. Recently, although numerous studies have strongly implicated a critical role of Hsps in salinity-alkalinity adaptation [22,23,24], most of them are only focused on responses for a limited number of Hsps rather than the full complement of genes that comprise the Hsp co-chaperone network in salinity-alkalinity adaptation. Therefore, with the saline-alkaline water aquaculture becoming a promising way to accommodate the growing need for the aquaculture industry, it is important to investigate the physiological change and molecular response in fishes adapting to saline-alkaline waters, which could also help to understand their adaptation in the aquatic ecosystem.
Spotted sea bass (Lateolabrax maculatus) is an economically important fish with a high nutritional value that is widely distributed in the coastal and estuarine areas of China, Japan, and the Korean peninsula [25]. L. maculatus has an excellent ability to adapt to a broad variety of salinity environments ranging from fresh to sea water [26]; therefore, its aquaculture is viable in both freshwater ponds and sea water cages in China. Additionally, L. maculatus has been proved to be able to survive in high alkaline water (carbonate alkalinity = 10 mmol/L) for a long period of time [27], and the aquaculture of L. maculatus in alkaline water has been under development recently. To explore the function of Hsp superfamily in L. maculatus (LmHsp) under salinity change and alkalinity stress, in this study, 95 LmHsp genes were identified in L. maculatus genome, and their phylogeny, conserved structure, expression profile, possible co-chaperon network under stress, as well as molecular evolution pattern were investigated. Our results provide comprehensive insights for further understanding the environmental adaptation of Hsp co-chaperons in L. maculatus and will help to establish the alkaline water aquaculture of L. maculatus.
2. Materials and Methods
2.1. Genome-Wide Identification and Annotation of Hsp Families in L. maculatus
To identify LmHsp genes, the coding sequences (cds) and amino acid (aa) sequences of L. maculatus genome (PRJNA408177) were blast searched using available Hsp cds and aa sequences from nine fish species, including zebrafish (Danio rerio), channel catfish (Ictalurus punetaus), stickleback (Gasterosteus aculeatus), medaka (Oryzias latipes), tilapia (Oreochromis niloticus), turbot (Scophthalmus maximus), fugu (Takifugu rubripes), Asian sea bass (Lates calcarifer), spotted gar (Lepisosteus oculatus), and four tetrapod species, including human (Homo sapiens), mouse (Mus musculus), chicken (Gallus gallus), and Xenopus (Xenopus tropicalis) from Ensemble (http://asia.ensembl.org accessed on 9 November 2021) and NCBI (http://www.ncbi.nlm.nih.gov accessed on 9 November 2021). These sequences were used as query to perform BLASTN and BLASTP (e-value = 1 × 10−5) against the sequence resources of L. maculatus. The conserved domains of candidate Hsp sequences were predicted using SMART [28] and Pfam [29], and the sequences without a corresponding complete Hsp domain were excluded from further analysis. When a gene could not be accurately distinguished, it was named after the zebrafish orthologs with letter “L” (meaning “like”).
For the identified LmHsp aa sequences, the Multiple Em for Motif Elicitation (MEME, https://meme-suite.org/meme accessed on 18 November 2021) program was applied to evaluate their conserved motifs, with the parameters of any number of repetitions, optimum width of motifs from 6 to 200 with 4 motifs for sHsp, 5 motifs for Hsp40, 3 motifs for Hsp10/60, 13 motifs for Hsp70, 6 motifs for Hsp90, and 3 motifs for Hsp100. The Gene Structure Display Sever (GSDS 2.0, http://gsds.gao-lab.org accessed on 20 November 2021) was used to visualize the exon-intron of LmHsp genes, and the diagrammatic sketches were illustrated using TBtools v1.09 [30].
2.2. Phylogenetic Analysis of Hsp Families
All Hsp aa sequences from L. maculatus and other selected vertebrates mentioned before were used to construct phylogenetic trees. Multiple protein sequence alignments were conducted with Muscle [31]. According to the Bayesian information criterion (BIC), the best models selected for phylogeny construction were JTT + F + G4 model for sHsp, VT + G4 model for Hsp40, LG + I + G4 model for Hsp60, JTT + G4 model for Hsp70, LG + G4 model for Hsp90, and JTT + I for Hsp100. The Maximum Likelihood (ML) phylogenetic trees of each Hsp family were generated using IQ-Tree [32], respectively, with a bootstrap of 1000 replicates. Furthermore, iTOL [33] was used to visualize the phylogenetic trees.
2.3. Expression of LmHsp Genes through Transcriptome Analysis
To investigate the expression profiles of LmHsp genes in L. maculatus tissues, transcriptome data of seven adult tissues were obtained from NCBI (brain-SRR7528887, stomach-SRR7528884, spleen-SRR7528888, liver-SRR7528886, gill-SRR7528883, testis-SRR7528885, and ovary-SRR2937376). Through the Hisat and StringTie pipeline [34], the Fragments Per Kilobase of exon per Million mapped reads (FPKM) of each LmHsp gene were obtained. To understand the expression dynamics of LmHsps under stresses, the RNA-seq data was also obtained from NCBI as gill tissues under salinity change (PRJNA515986) and alkalinity stress (PRJNA611641), respectively. The former data represented L. maculatus specimens (158.23 ± 18.77 g) originated from sea water (13.5–14.5 °C, pH 7.8–8.15, salinity 30 ppt) with salinity acclimation in fresh water (0 ppt, control group), brackish water (15 ppt), and sea water (30 ppt) for 30 days, respectively [35]. The latter data represented L. maculatus specimens (body weight: 140.32 ± 2.56 g) acclimated in fresh water (pH 7.8–8.15) for 30 days (control group) and then challenged with alkalinity stress (carbonate alkalinity = 18 mmol/L with NaHCO3 and Na2CO3 added to fresh water) at 0 h, 12 h, 24 h, and 72 h [27]. All the experimental groups contained three individuals for biological replicate, which represented a good correlation among groups from principal component analysis (PCA) (Figure S1). These RNA-seq data were processed with the Hisat and StringTie pipeline and the log2 (fold change) (log2FC) for LmHsp genes between the test and control group was calculated by edgeR [36]. TBtools was employed to draw heatmaps with log2 (FPKM + 1) and log2FC values, respectively. In addition, the trend analysis of gene expression was conducted to cluster LmHsp genes with similar expression patterns along the test time points under alkalinity stress via online toolkit OmicShare (https://www.omicshare.com/ accessed on 17 Feburary 2022).
2.4. Protein-Protein Interaction and Co-Expression Analysis
Protein-protein interaction (PPI) networks among Hsp families were analyzed via the online STRING 11.5 tool (https://string-db.org/ accessed on 10 October 2021) for human and zebrafish, with the parameter minimum required interaction score of 0.7. For L. maculatus Hsps in response to alkalinity stress, the expression correlation between each LmHsp gene was evaluated using pairwise Pearson’s correlation coefficient (PCC) with their FPKM and the correlation coefficient value of 0.7. The networks were visualized using Cytoscape [37].
2.5. Molecular Evolution Analysis
The cds of each Hsp family was aligned by Muscle, and MEGA7 [38] was used to generate ML trees to determine their phylogeny. The EasyCodeML v1.21 [39] was used to perform the molecular evolution analysis, with the site model (SM), branch model (BM), and branch-site model (BSM) tests in the PAML package [40]. Firstly, the ratio of non-synonymous to synonymous substitution (dN/dS, ω) and the likelihood ratio tests (LRTs) were employed to evaluate the selective pressures in each LmHsp family. Six SMs were used to test the positive selection in each codon: M0 assumes to have the same ω for all codons, M3 assumes the ω of all codons showing a simple discrete division trend; M1a assumes only conservative sites (0 < ω < 1) and neutral sites (ω = 1) for all codons, while M2a is considered to increase the existence of positive sites (ω > 1) for all codons on the basis of M1a; the ω of all codons in M7 are assumed to belong to the matrix (0, 1) with a beta distribution, while M8 adds another type of ω (ω > 1) on the basis of M7. LRTs were used to judge whether the paired models (M0 vs. M3; M1a vs. M2a; M7 vs. M8) are significantly different [40], and to estimate whether there are positive selected sites (PSSs) (M2a vs. M1a and M8 vs. M7) under the premise of significant p value (p < 0.05).
Furthermore, BM were conducted to test the selective pressure of each LmHsp family. Phylogeny was constructed with all LmHsp cds, and each Hsp family was marked as the foreground branch (ω1), respectively, to explore the evolution rate compared with the remaining LmHsp families (background branch, ω0). BSM were conducted to test the selective pressure of single LmHsp gene (foreground branch, ω1) with 10 fish species (background branch, ω0), including spotted sea bass (L. maculatus), zebrafish (D. rerio), channel catfish (I. punetaus), stickleback (G. aculeatus), medaka (O. latipes), tilapia (O. niloticus), turbot (S. maximus), fugu (T. rubripes), Asian sea bass (L. calcarifer), and spotted gar (L. oculatus). With positive selection suggested (p < 0.05), the PSSs were further verified with high BEB posterior probabilities (>0.95). The PSSs were then retrieved from the aa sequences within functional Hsp domains from the Pfam database. 3D Structure prediction analysis was performed via Phyre2 online tool (https://www.sbg.bio.ic.ac.uk/phyre2/ accessed on 29 December 2021) [41] and modified by PyMol software (https://pymol.org/2/ accessed on 30 December 2021).
3. Results and Discussion
3.1. Genomic Landscape, Functional Domain, and Phylogeny of Hsp Superfamily in L. maculatus
Through genome-wide screen, 95 Hsp genes (LmHsps) were identified in the L. maculatus genome, including 12 sHsps (LmHspb), 50 Hsp40s (LmDnaj), 9 Hsp10/60s (LmHspe, LmHspd, Lmcct), 17 Hsp70s (LmHspa), 5 Hsp90s (LmHspc), and 2 Hsp100s (LmXlp) (Figure 1 and Table S1). The general features of LmHsps were listed in Table S2, with their length ranging from 56 to 2220 amino acids (aa), and exon numbers between 1 and 53. The copy number of each LmHsp family was generally conserved among the selected teleost and tetrapod species (Figure 1 and Table S1), some of which (sHsp, Hsp40 and Hsp90) were more in teleosts than that in the tetrapod lineages, most likely due to the teleost specific genome duplication (TSGD) event in the teleost lineages [42].
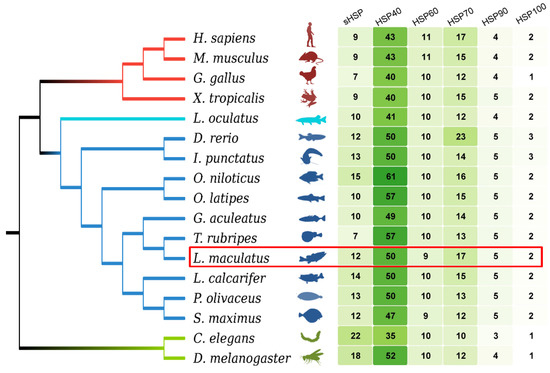
Figure 1.
Genome-wide identification of Hsp families in 10 teleost species and other metazoan species, with L. maculatus in the red frame. Branches with different colors represent different metazoan groups. The gene numbers are illustrated with the heatmap.
Specifically, 12 sHsp genes were identified in the L. maculatus genome, which were clustered into 8 sub-families, but absent in other 4 sub-families as Hspb1, Hspb2, Hspb3, and Hspb9 (Figure S2A). The Hsp30s was not identified in the tetrapods, but was only present in teleosts with independent duplication. In addition, as the ubiquitous family of chaperones, the structure of sHsps can be defined into three domains [43]: a conserved α-crystallin domain (ACD) with N-terminal region (NTR) and C-terminal region (CTR) on either side [44]. Most LmsHsps comprised three conserved motifs (motif 1/2/4, Figure 2A) except Hsp30L_10022466, while Cryab, Hspb11, Hsp30L_10002822, and Hsp30L_10002821 contained one extra motif 3, suggesting strong structure conservation among LmsHsp genes (Figure 2A).

Figure 2.
Conserved domain architecture of LmHsp families, as (A) LmsHsps, (B) LmHsp40s, (C) LmHsp10/60s, (D) LmHsp70s, (E) LmHsp90s, and (F) LmHsp100s, showing along their phylogenetic relationships. The horizontal grey bars represent amino acid sequences without predicted functional domains, whereas the colored boxes represent the regions with successfully predicted motifs. The orange boxes represent the corresponding HSP domain of each family, the green boxes represent the ATPase domains, and the other boxes represent specifically annotated domains. The domains that appear only in some family members are marked with dashed frames.
A total of 50 Hsp40 genes (Dnajs) were identified in the L. maculatus genome, which were clustered into 42 sub-families (Figure S2B). Most LmHsp40s comprised 3 conserved motifs (motifs 1/2/3, Figure 2B) as the major DnaJ domain, while 10 LmHsp40s had extra motifs 4 or 5 in their sequences. Moreover, one Hsp10 (Hspe1) and eight Hsp60s (Hspd1 and 7 Ccts) were identified, with Cct8 missing in the L. maculatus genome. The phylogeny supported the evolutionary relationship of LmHspd1 and LmHspe1, respectively, clustered with orthologous genes of other teleost species (Figure 2C and Figure S2C). There were three conserved motifs identified among LmHsp60 members as the Cpn60_TCP1 domain, while none were found in LmHsp10 (Figure 2C).
Seventeen LmHsp70 genes were identified in the L. maculatus genome, which contained more duplicated copies, mainly from the Hspa1, Hspa8, and Hspa12 sub-families (Table S1 and Figure 2D). Almost all the Hsp70 sub-families were found between human and L. maculatus except for Hspa2, Hspa6, Hspa7, and Hsph1, which exist in human [45] but not in fish species apart from spotted gar, zebrafish, and stickleback (Figure S2D). Thirteen conserved motifs were identified in LmHsp70s, and genes with closer phylogenetic relationship had similar conserved motifs. For example, motifs 9 and 11 were only annotated in Hspa12 sub-family (Figure 2D and Figure S2D), which directly confirmed the fact that this sub-family was distantly related to other Hsp70 genes in vertebrates [45]. Another noteworthy gene was Hsp4a1, which only contained one specific motif 13, and was significantly different from other LmHsp70s, such as Hspa4a2. The vertebrate Hsp70s were mainly scattered into nine clads, among which eight sub-families were easily distinguished, except for the sub-families Hspa1-2-6-7-8 and Hsc70 (Heat shock cognate 71 kDa), all clustering together (Figure S2D). This was in accordance with the fact that Hspa1-2-6-7-8 and Hsc70 shared a common domain called HSP1_2_6_8Nucleoid Binding Domain [46] with very similar conserved motifs (Figure S2D).
In addition, a total of five Hsp90s and two Hsp100s were collected from the L. maculatus genome, which was similar to that in other teleosts. The vertebrate Hsp90 genes can be classified into four sub-families, supporting the four clads (Hsp90aa1, Hsp90ab1, Hsp90b1 and Trap1) in their phylogeny (Figure S2E). Teleost Hsp90aa1 genes were clustered into two sub-clads, Hsp90aa1.1 and Hsp90aa1.2, further supporting that they were originated from the TSGD in the teleost lineage [42] (Figure S2E). Moreover, all the LmHsp90 members contain multi-introns, and their detected six motifs are conserved with each other (Figure 2E). In total, all this information revealed the generally conserved sequence and function in teleost Hsp families.
3.2. Diverse Expression of LmHsp Genes among L. maculatus Normal Tissues
Gene expression profiling facilitates the understanding of the function and evolution of Hsp families. Here, we conducted transcriptome analysis with RNA-seq data from seven L. maculatus adult tissues (brain, stomach, liver, spleen, gill, ovary, and testis), which illustrated diverse expression profiles of the 95 LmHsp genes across tissues. In particular, LmHsp10/60s were mostly highly expressed among tissues, while LmsHsps were lowly expressed (Figure 3). Interestingly, Hspa8a and Hsp90ab1 were the most abundantly expressed across all tissues, suggesting their essential role in maintaining the normal physiological homeostasis in diverse tissues, whereas Dnajb9, Dnajc5b, and Hspb7L were not expressed in any tissues (Figure 3). Moreover, some Hsp genes showed biased expression in certain tissues. For example, Cryab was only expressed in brain, Hspb11 was highly expressed in gill, and both spleen and liver had higher Hspa1.1 and Hspa1.2 expression than other tissues (Figure 3), which may indicate the tissue specific function of these Hsp members as chaperons. In addition, almost all sHsp genes (except Hspb8) were weakly expressed or even not expressed in ovary, while Cryaa, Hspb6, Hspb7, and Hsp30L were highly expressed in testis (Figure 3).
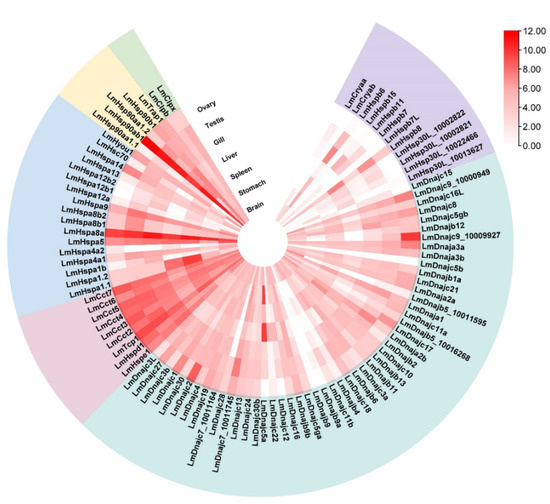
Figure 3.
Expression profile of LmHsp genes among adult tissues of L. maculatus. The log2 (FPKM + 1) values are represented as 0–12 according to the colored scale bar, and the heatmap is ranked with each LmHsp families.
3.3. Regulated Expression of LmHsp Genes in Response to Salinity Change and Alkalinity Stress
To investigate the functional response of LmHsps to environmental changes, the regulated expression of 95 LmHsp genes was characterized under salinity or alkalinity stress in gills where the ion secretion and uptake happen. As chaperons, Hsps may help to maintain the homeostasis by interacting with the osmotic stress denatured proteins in gill cells. Firstly, L. maculatus could be well adapted to wide salinity ranges, therefore, with freshwater acclimation, the expression of LmHsp genes may be weakly regulated (Figure 4A and Table S3). For example, only Hspa12b1 and three Hsp40s (Dnajc3L, Dnajc3a, and Dnajc5b) were significantly influenced in the brackish water group, and only Dnajc5ga was up-regulated in the sea water group (Figure 4A), which suggested that the gill tissues were already adapted to the stimulation caused by freshwater acclimation. Moreover, even though not significantly regulated, the expression of almost all sHsp members (except Hspb7) were reduced, and the expression of most Hsp70s were increased, whereas most Hsp90 members (Hsp90ab1, Hsp90b1, and Trap1) did not respond to the stimulation (Figure 4A), suggesting possible functional diversification among the LmHsp families.
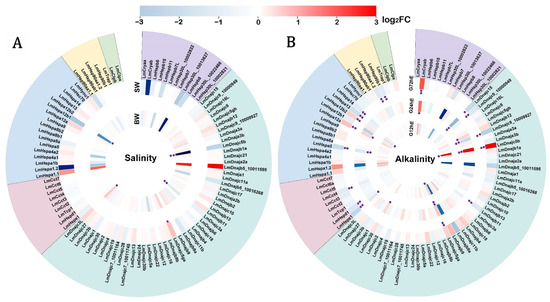
Figure 4.
Regulated expression of LmHsp genes in gills of L. maculatus in response to (A) salinity change and (B) alkalinity stress. The heatmap was based on the log2FC values and ranked with each LmHsp family. BW and SW represent brackish and sew water groups compared to the freshwater group, while G12hE, G24hE, and G72hE indicate the alkalinity experiment duration for 12 h, 24 h, and 72 h, compared to the blank control 0 h, respectively. * indicates |log2FC| > 0.7 with p < 0.05, ** indicates |log2FC| > 0.7 with both p value and false discovery rate (FDR) < 0.05.
The alkalinity challenge revealed that LmHsps in gills were sensitive to alkalinity stress within a short period of time (Figure 4B and Table S3), which was most likely in accordance with the good adaptation of L. maculatus to diverse salinities rather than alkalinity. For example, under alkalinity stress, the number of significantly regulated LmHsp genes (|log2FC| > 0.7 and p < 0.05) steadily increased along the test time points (Figure 4B). In particular, only 3 LmHsps (Hspa12b2, Dnajc5ga, and Dnajc5b) were up-regulated at 12 h exposure, and 10 LmHsps (Cryab, Hspe1, Dnajb12, Dnajc5b, Dnajc5ga, Dnajc9_10009927, Dnajc15, Dnajc30, Hspa5, and Hspa12a) were significantly influenced at 24 h, while at 72 h, 17 LmHsp genes (Cryab, Hspe1, Hspd1, Cct4, Dnajb11, Dnajc5b, Dnajc5ga, Dnajc9_10000949, Dnajc9_10009927, Dnajc15, Hspa5, Hspa12a, Hspa12b2, Hspa13, Hspa14, Hsp90aa1.2, and Hsp90b1) were significantly regulated (Figure 4B). Of these regulated LmHsps, 12 were down-regulated (Hspe1, Hspd1, Cct4, Dnajb11, Dnajc9_10000949, Dnajc9_10009927, Dnajc15, Hspa5, Hspa13, Hspa14, Hsp90aa1.2, and Hsp90b1) with log2FC ranging from −0.72 to −2.59, whereas 7 were up-regulated (Cryab, Dnajb12, Dnajc5b, Dnajc5ga, Dnajc30, Hspa12a, and Hspa12b2) with log2FC ranging from 0.80 to 12.04 (Table S3), which was mostly in the LmHsp40, LmHsp70, and LmHsp90 families but less in LmsHsp and LmHsp10/60 families. Hsp70 and Hsp90 are the two highly conserved ATP-dependent molecular chaperones that fold and remodel proteins in almost every cellular process with co-chaperon Dnajs [8,9]. The regulation of LmHsp40s (Dnajs), LmHsp70s, and LmHsp90s indicated that they could play more essential roles in response to alkalinity stress than other LmHsps. Under alkalinity stress, the ionic balance disruption occurred due to the hyperosmotic environment with protein damage [47], which can increase the levels of some LmHsp40s and LmHsp70s (Figure 4B). However, there were more down-regulated LmHsps rather than up-regulated ones, possibly due to the certain degree of adaptation to alkalinity stress in L. maculatus, which could survive in alkaline water for more than two weeks to two months [27]. Several studies also reported down-regulation of Hsps in marine organisms under diverse stimuli, such as scallops Chlamys farreri and Patinopecten yessoensis Hsp70B2 genes in response to toxic dinoflagellates [16,17], ascidian Ciona savignyi Hsp20/60/70/90 genes under both low and high salinity stresses [48], and European seabass Dicentrarchus labrax Hsp70 in low salinity conditions [47]. Further investigation is warranted with a possible delayed response in LmHsp expression level as well as the regulatory heat shock factor (Hsf) of these LmHsps. In addition, the expression of Hspa8a and Hsp90ab1 did not exhibit differential regulation (Figure 4B), although they showed high abundance in both normal and challenged gill tissues (Figure 3 and Table S3), indicating that these genes did not participate in the response to alkalinity stress but may be needed to maintain the physiological homeostasis.
3.4. Coordinated Regulation of LmHsp Co-Chaperones under Alkalinity Stress
When the Hsps repair misfolded proteins in a simplified model, Hsp70 initially facilitates the re-folding of hydrophobic residue stretches into their native state together with Hsp40s (Dnajs), whereas Hsp90 catalyzes a step later in protein folding and activation [8,11]. With the possible co-chaperon among Hsp40s, Hsp70s, and Hsp90s, to further excavate their associated regulation, the expression trend and Pearson’s correlation coefficient (PCC) of LmHsp40-70-90 genes under alkalinity stress were further investigated. As a result, 24 LmHsp40-70-90 genes were enriched into 4 expression profiles (Figure 5A and Table S4), among which 10 LmHsps were enriched in profile 0 (declined expression) and 9 LmHsps were enriched in profile 4 (raised expression) (Figure 5B). In profile 0, the expression of two LmHsp70s (LmHspa5 and LmHspa13), two LmHsp90s (LmHsp90aa1.2 and LmHsp90b1), and six LmHsp40s (LmDnajc15, LmDnajc9_10000949, LmDnajc9_10009927, LmDnajb5_10016268, LmDnajb11, and LmDnajb9a) were continuously down-regulated (Figure 5B), while in profile 4, the expression of three LmHsp70s (LmHspa1.1, LmHspa12a, and LmHspa12b2) and six LmHsp40s (LmDnajb12, LmDnajc5b, LmDnajb4, LmDnajc5ga, LmDnajc22, and LmDnajc30) were continuously up-regulated (Figure 5B). These transcriptional profiles may be suggestive of coordinated regulation of the Hsp40-70-90 genes and could thus comprise the possible component of alkalinity adaptation in L. maculatus, which warrants further verification.
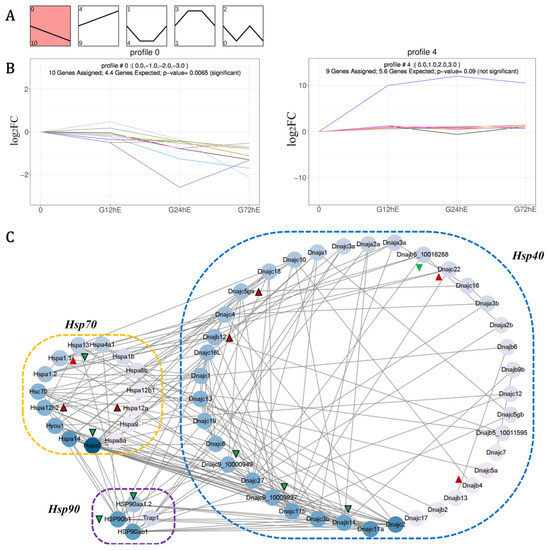
Figure 5.
The expression trend and coordinated regulation of LmHsp40-70-90 co-chaperons in response to alkalinity stress. (A) Expression patterns enriched in five profiles with the number of LmHsps indicated; (B) Expression trends of LmHsp genes enriched in the down-regulated profile 0 and up-regulated profile 4; (C) Protein-protein interaction (PPI) according to the PCC of LmHsp expression under alkalinity stress. Green and red triangles represent Hsps from profiles 0 and 4, respectively, and triangles with black frame are the significantly regulated LmHsps in response to alkalinity stress.
In addition, according to PCC > 0.7, 15 LmHsp40-70-90 members from profile 0 (8 Hsps) and profile 4 (7 Hsps) may be coordinately regulated with each other (Figure 5C and Table S5). For example, the up-regulated LmHspa12b2 was positively correlated with LmDnajb12 and negatively correlated with LmDnajc9, while the down-regulated LmHsp90b1 and LmHsp90aa1.2 were positively correlated with LmDnajc9 or LmDnajb11 (Figure 5C and Table S5), which may indicate LmDnajc9 as the key co-chaperon of LmHspa12b2, LmHsp90b1, and LmHsp90aa1.2. Moreover, the down-regulated LmHspa5 also co-expressed with LmDnajb11, LmDnajc9, and LmHsp90b1, while negatively related to LmHspa12b2 and LmDnajc5ga. These protein-protein interaction correlations among Hsp40-70-90 members could also be observed in model organisms such as zebrafish and human (Figure S3), indicating functional conservation of these Hsp co-chaperons. Moreover, the expression trends and PCC relationship also revealed that the down-regulated co-expression of LmHsp40-70-90 members may reflect their corporation, whereas the up-regulation of LmHsp40s and atypical LmHspa12s may supplement the loss caused by down-regulation (Figure 5C).
3.5. Evolutionary Dynamics of LmHsp Families in L. maculatus
To gain insights of whether the regulated LmHsps may function under adaptive evolution in dynamic aquatic environments, the molecular evolution of LmHsp families was evaluated through a series of model tests, including site model (SM), branch model (BM), and branch-site model (BSM) via PAML. Firstly, the ω values (dN/dS) of each LmHsp family were obtained via the SM tests, which revealed that the evolutionary rate (ω) of the LmsHsp family was generally higher than that of other LmHsp families (Figure 6A and Table S6). For example, the LmsHsp family exhibited rapid evolution with ω > 1, while the LmHsp70 and LmHsp90 families were more conserved during evolution (ω < 0.1) (Figure 6A). In addition, with BM tests for the 95 LmHsp genes labeling each family as the foreground branch (ω1), respectively, it confirmed that in L. maculatus, sHsps represented faster molecular evolution rates than other Hsp families (p < 0.01), whereas LmHsp70s had the opposite evolutionary pattern with high sequence conservation (Figure 6B and Table S6). This was in line with the results obtained in the SM tests (Figure 6A). Moreover, LmHsp90s represented rapid evolution in the BM test but not in the SM test. These results suggested that although most Hsp genes were deemed to be highly conserved among teleost species, the molecular evolutionary rates of sHsps and Hsp90s were generally faster in L. maculatus.
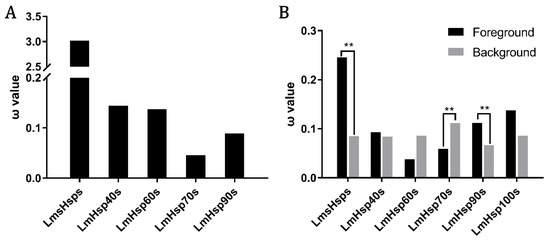
Figure 6.
Molecular evolution of LmHsp genes in L. maculatus. (A) ω values of each LmHsp family from site model tests; (B) Branch-model tests of each LmHsp family from L. maculatus, black and gray columns indicate ω values of each LmHsp family (ω1, foreground branch) and other LmHsp families (ω0, background branch), respectively. ** indicates p < 0.01.
Accordingly, the BSM tests of LmsHsps, LmHsp70s and LmHsp90s, which represented significant altered evolutionary rates in L. maculatus (Figure 6B), were further conducted against Hsps of other teleost lineages, and the putative positively selected sites (PSSs) were detected in 10 genes (p < 0.01, BEB probability > 0.95), including Hspa4a1, Hspa4a2, Hspa8a, Hspa9, Hspa12b1, Hspa12b2, Hsc70, Hsp90aa1.2, Hsp90ab1, and Hsp90b1 (Table S7). The genes with PSS detected were mainly found in seven Hsp70 and three Hsp90 members but not in sHsps (Table S7), among which Hspa12b2, Hsp90aa1.2, and Hsp90b1 were also significantly regulated in response to alkalinity stress (Figure 5C). Therefore, in an adverse aquatic environment, the sHsp family may be under stronger evolutionary pressure in both L. maculatus and other teleosts (Figure 6), whereas in L. maculatus, some members of Hsp70 and Hsp90 families may evolve more rapidly than other teleosts (Table S7), possibly demonstrating the good adaptation of L. maculatus to the environmental changes, such as water temperature, salinity, and alkalinity.
In addition, with BSM tests of the 15 putative correlated Hsp40-70-90 co-chaperons in response to alkalinity stress (Table 1 and Figure 5C), the PSSs in the four significantly regulated Hsps, Hsp90aa1.2, Hsp90b1, Hspa12b2, and Dnajc9, were detected in their conserved functional domains (Figure 7). Hsp90 chaperones normally exist as a dimer, and each part contains three domains: the N-terminal domain (NTD) with ATP hydrolytic activity, the middle domain (MD) to interact with Hsp70, and the C-terminal domain (CTD) to participate in identification and interaction of clients [49]. The eukaryotic Hsp90 proteins also contain a C-terminal extension of the MEEVD motif to help them bind to co-chaperones [8,49]. The fast-evolved PSSs of Hsp90aa1.2 and Hsp90b1 were mainly present in the MD and CTD (Figure 7A,B,E), which could most likely affect their interaction efficiency with Hsp70 proteins. Moreover, gene duplication is one of the most essential ways to develop functional divergence through environmental adaptation [42]. Both Hsp90 and Hsp70 function in an ATP-dependent scenario and require the involvement of a protein-named activator of Hsp90 ATPase homolog (Hsp90aa1), which interacts with Hsp90 and Hsp70 to stimulate ATPase activity [8,11]. Two putative Hsp90aa1 genes were identified in L. maculatus due to TSGD, while only one (LmHsp90aa1.2) was significantly induced in response to alkalinity (Figure 4B). Therefore, there can be functional diversification between the duplicated Hsp90aa1 genes in L. maculatus, with Hsp90aa1.2 both evolving significantly fast and regulated under alkalinity stress, but not in Hsp90aa1.1 (Figure 4 and Table 1).

Table 1.
Branch-site model tests for the 15 coordinated regulated LmHsp40-70-90 co-chaperons (Figure 5C) between L. maculatus and other teleost species.
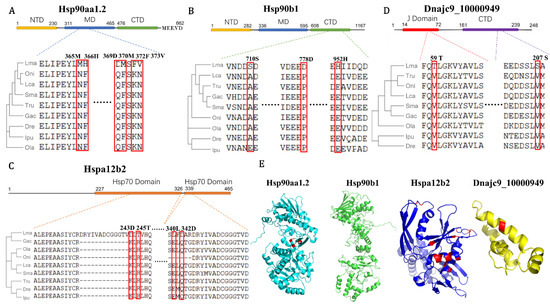
Figure 7.
Examples of positively selected sites (PSSs) in (A) Hsp90aa1.2, (B) Hsp90b1, (C) Hspa12b2, and (D) Dnajc9 functional domains from branch-site model tests. The partial aa sequence alignments of the selected vertebrates with their phylogeny are presented. Red frames indicate PSSs detected with their location at the bar above. Omitted multiple sequence alignment between the two segments are represented by dots. NTD, N-terminal domain; MD, middle domain; and CTD, C-terminal domain. (E) Schematic diagram of 3D structure of the above-mentioned LmHsp members with PSSs labeled in red. Only 710 S was predicted in the Hsp90b1 3D structure.
Moreover, Hsp70s are regarded as the most well-known member of the Hsp superfamily, which can act as co-chaperones of Hsp40s and Hsp90s involved in the folding and remodeling of various proteins [9]. Generally, the typical Hsp70 members consist of two regions, a conserved nucleotide-binding domain (NBD) with an ATP binding site, and a substrate-binding domain (SBD), which can mediate substrate or co-chaperone binding [50]. Those lacking NBD or SBD, such as Hspa4 and Hspa12 (Figure 2D), are considered atypical members of the Hsp70 family [45]. Here, only the atypical Hspa12 genes in L. maculatus were up-regulated under alkalinity stress, while other typical Hsp70s (LmHspa5, LmHspa13, and LmHspa14) were mostly down-regulated together with their co-chaperon Hsp90s, indicating possible functional diversification. There are two Hsp70 domains detected in LmHspa12b2, and the PSSs mainly exist in its C-terminal domain (Figure 7C,E), indicating its possible binding specificity alternation. Moreover, PSSs were also detected in the J domain and CTD of LmDnajc9, which is essential in stimulating Hsp70 ATPase activity (Figure 7D,E) and also as the putative key co-chaperon with LmHspa12b2 and LmHsp90aa1.2 (Figure 5C). Therefore, these fast evolved loci in functional domains may contribute to the Hsps functional co-evolution in L. maculatus (Figure 7E), and it may also suggest their vital correlated roles in response to diverse environmental stresses in L. maculatus through adaptive evolution. Further investigation is warranted among these Hsp40-70-90 co-chaperons to better understand their function and adaptive evolution in L. maculatus.
4. Conclusions
Hsps contribute to mediate stress responses by refolding damaged proteins to prevent their aggregation. This study provided a systematic genomic and transcriptomic survey of 95 Hsp genes in spotted sea bass (L. maculatus), which has good adaptability in saline-alkaline waters. Diverse expression regulation of LmHsps was observed in responses to alkalinity stress, which was majorly responded in coordinated LmHsp40-70-90 families through the adaptive recruitment of these co-chaperons. These findings are useful for understanding the diverse functions of Hsp co-chaperon network and adaptive evolution in teleost species. Further investigation on the functions of teleost Hsp network will provide a more detailed explanation about their adaptation mechanisms against different environmental challenges and warrant a better feasibility regarding the cultivation of L. maculatus in alkaline water.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/biology11030353/s1, Table S1: Detailed copy number of Hsp family genes in selected vertebrate genomes; Table S2: Summarized information of LmHsp genes; Table S3: Differential expression of LmHsps under salinity change and alkalinity stress by edgeR analysis; Table S4: Trend analysis for the LmHsp40-70-90 gene expression under alkalinity stress; Table S5: Protein-protein interaction (PPI) network for the LmHsp40-70-90 gene expression under alkalinity stress; Table S6: Molecular evolution analysis of the LmHsp genes among teleost lineages via PAML; Table S7: Branch-site model tests of the LmHsp genes with other teleost species via PAML; Figure S1: Principal component analysis (PCA) with FPKM values of all transcriptome samples under salinity change and alkalinity stress; Figure S2: Structure, motif and phylogeny of LmHsp families, showing phylogenetic relationship along with their conserved motifs. (A) LmsHsps, (B) LmHsp40s, (C) LmHsp10/60s, (D) LmHsp70s, (E) LmHsp90s, and (F) LmHsp100s. Abbreviations: Hsa: H. sapiens; Mmu: M. musculus; Gga: G. Gallus; Xtr: X. tropicalis; Loc: L. oculatus; Dre: D. rerio; Ipu: I. punctatus; Ola: O. latipes; Gac: G. aculeatus; Sma: S. maximus; Tru: T. rubripes; Oni: O. niloticus; Lca: L. calcarifer; Lma: L. maculatus; Figure S3: Protein-protein interaction (PPI) of Hsps from zebrafish and human.
Author Contributions
J.C. and Q.Z. designed the study, X.L., S.L., Y.W. and W.L. analyzed the data. J.C. and X.L. conducted the manuscript writing. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the China Agriculture Research System (CARS-47-G06) and the National Marine Genetic Resource Center, China.
Institutional Review Board Statement
This study was conducted in accordance with the Institutional Animal Care and Use Committee of Ocean University of China (IACUC-OUC), and it does not contain any studies with human participants.
Informed Consent Statement
Not applicable.
Data Availability Statement
The transcriptome datasets used in this study can be found in the NCBI Sequence Read Archive (SRA) BioProject PRJNA611641 and PRJNA515986, as well as L. maculatus tissue transcriptomes, including brain-SRR7528887, stomach-SRR7528884, spleen-SRR7528888, liver-SRR7528886, gill-SRR7528883, testis-SRR7528885 and ovary-SRR2937376.
Conflicts of Interest
The authors declare that they have no conflict of interest.
References
- Ritossa, F.M. A new puffing pattern induced by heat shock and DNP in Drosophila. Experientia 1962, 18, 515–523. [Google Scholar] [CrossRef]
- Whitley, D.; Goldberg, S.P.; Jordan, W.D. Heat shock proteins: A review of the molecular chaperones. J. Vasc. Surg. 1999, 29, 748–751. [Google Scholar] [CrossRef] [Green Version]
- Basu, N.; Todgham, A.E.; Ackerman, P.A.; Bibeau, M.R.; Nakano, K.; Schulte, P.M.; Iwama, G.K. Heat shock protein genes and their functional significance in fish. Gene 2002, 295, 173–183. [Google Scholar] [CrossRef]
- Feder, M.E.; Hofmann, G.E. Heat-shock proteins, molecular chaperones, and the stress response: Evolutionary and ecological physiology. Annu. Rev. Physiol. 1999, 61, 243–282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dabbaghizadeh, A.; Tanguay, R.M. Structural and functional properties of proteins interacting with small heat shock proteins. Cell Stress Chaperones 2020, 25, 629–637. [Google Scholar] [CrossRef]
- Faust, O.; Abayev-Avraham, M.; Wentink, A.S.; Maurer, M.; Nillegoda, N.B.; London, N.; Bukau, B.; Rosenzweig, R. HSP40 proteins use class-specific regulation to drive HSP70 functional diversity. Nature 2020, 587, 489–494. [Google Scholar] [CrossRef] [PubMed]
- Bie, A.S.; Cömert, C.; Körner, R.; Corydon, T.J.; Palmfeldt, J.; Hipp, M.S.; Hartl, F.U.; Bross, P. An inventory of interactors of the human HSP60/HSP10 chaperonin in the mitochondrial matrix space. Cell Stress Chaperones 2020, 25, 407–416. [Google Scholar] [CrossRef]
- Genest, O.; Wickner, S.; Doyle, S.M. Hsp90 and Hsp70 chaperones: Collaborators in protein remodeling. J. Biol. Chem. 2019, 294, 2109–2120. [Google Scholar] [CrossRef] [Green Version]
- Rosenzweig, R.; Nillegoda, N.B.; Mayer, M.P.; Bukau, B. The Hsp70 chaperone network. Nat. Rev. Mol. Cell Biol. 2019, 20, 665–680. [Google Scholar] [CrossRef]
- Mayer, M.P.; Le Breton, L. Hsp90: Breaking the symmetry. Mol. Cell 2015, 58, 8–20. [Google Scholar] [CrossRef] [Green Version]
- Jiang, F.; Chang, G.; Li, Z.; Abouzaid, M.; Du, X.; Hull, J.; Ma, W.; Lin, Y. The HSP/co-chaperone network in environmental cold adaptation of Chilo suppressalis. Int. J. Biol. Macromol. 2021, 187, 780–788. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Feder, M.E.; Kang, L. Evolution of heat-shock protein expression underlying adaptive responses to environmental stress. Mol. Ecol. 2018, 27, 3040–3054. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, B.M.; Rhee, J.S.; Jeong, C.B.; Seo, J.S.; Park, G.S.; Lee, Y.M.; Lee, J.S. Heavy metals induce oxidative stress and trigger oxidative stress-mediated heat shock protein (hsp) modulation in the intertidal copepod Tigriopus japonicus. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2014, 166, 65–74. [Google Scholar] [CrossRef] [PubMed]
- Cantinha, R.D.S.; Borrely, S.I.; Oguiura, N.; Pereira, C.A.d.B.; Rigolon, M.M.; Nakano, E. HSP70 expression in Biomphalaria glabrata snails exposed to cadmium. Ecotoxicol. Environ. Saf. 2017, 140, 18–23. [Google Scholar] [CrossRef]
- Ulaje, S.A.; Lluch-Cota, S.E.; Sicard, M.T.; Ascencio, F.; Cruz-Hernández, P.; Racotta, I.S.; Rojo-Arreola, L. Litopenaeus vannamei oxygen consumption and HSP gene expression at cyclic conditions of hyperthermia and hypoxia. J. Therm. Biol. 2020, 92, 102666. [Google Scholar] [CrossRef]
- Cheng, J.; Xun, X.; Kong, Y.; Wang, S.; Yang, Z.; Li, Y.; Kong, D.; Wang, S.; Zhang, L.; Hu, X.; et al. Hsp70 gene expansions in the scallop Patinopecten yessoensis and their expression regulation after exposure to the toxic dinoflagellate Alexandrium catenella. Fish Shellfish Immunol. 2016, 58, 266–273. [Google Scholar] [CrossRef]
- Hu, B.; Li, M.; Yu, X.; Xun, X.; Lu, W.; Li, X.; Li, Y.; Lou, J.; Wang, S.; Zhang, L.; et al. Diverse expression regulation of Hsp70 genes in scallops after exposure to toxic Alexandrium dinoflagellates. Chemosphere 2019, 234, 62–69. [Google Scholar] [CrossRef]
- Lei, Q.N.; Wu, Y.Y.; Liang, H.Y.; Wang, Z.X.; Zheng, Z.; Deng, Y.W. Molecular cloning and expression analysis of heat shock protein 20 (HSP20) from the pearl oyster Pinctada martensii. Genet. Mol. Res. 2016, 15, 10. [Google Scholar] [CrossRef]
- Xie, Y.; Song, L.; Weng, Z.; Liu, S.; Liu, Z. Hsp90, Hsp60 and sHsp families of heat shock protein genes in channel catfish and their expression after bacterial infections. Fish Shellfish Immunol. 2015, 44, 642–651. [Google Scholar] [CrossRef]
- Song, L.; Zhang, J.; Li, C.; Yao, J.; Jiang, C.; Li, Y.; Liu, S.; Liu, Z. Genome-wide identification of hsp40 genes in channel catfish and their regulated expression after bacterial infection. PLoS ONE 2014, 9, e115752. [Google Scholar] [CrossRef] [Green Version]
- Song, L.; Zhao, Y.; Song, Y.; Zhao, L.; Ma, C.; Zhao, J. Effects of saline-alkaline water on growth performance, nutritional processing, and immunity in Nile tilapia (Oreochromis niloticus). Aquaculture 2021, 544, 737036. [Google Scholar] [CrossRef]
- Islam, M.J.; Kunzmann, A.; Slater, M.J. Extreme winter cold-induced osmoregulatory, metabolic, and physiological responses in European seabass (Dicentrarchus labrax) acclimatized at different salinities. Sci. Total Environ. 2021, 771, 145202. [Google Scholar] [CrossRef]
- Islam, M.J.; Slater, M.J.; Kunzmann, A. What metabolic, osmotic and molecular stress responses tell us about extreme ambient heatwave impacts in fish at low salinities: The case of European seabass, Dicentracus labrax. Sci. Total Environ. 2020, 749, 141458. [Google Scholar] [CrossRef]
- Zhang, M.; Li, L.; Liu, Y.; Gao, X. Effects of sudden drop in salinity on osmotic pressure regulation and antioxidant defense mechanism of Scapharca subcrenata. Front. Physiol. 2020, 11, 884. [Google Scholar]
- Yokogawa, K.; Seki, S. Morphological and genetic differences between Japanese and Chinese sea bass of the genus Lateolabrax. Jpn. J. Ichthyol. 1995, 41, 437–445. [Google Scholar]
- Tian, Y.; Wen, H.; Qi, X.; Zhang, X.; Li, Y. Identification of mapk gene family in Lateolabrax maculatus and their expression profiles in response to hypoxia and salinity challenges. Gene 2019, 684, 20–29. [Google Scholar] [CrossRef]
- Wang, L.Y.; Tian, Y.; Wen, H.S.; Yu, P.; Liu, Y.; Qi, X.; Gao, Z.C.; Zhang, K.Q.; Li, Y. Slc4 gene family in spotted sea bass. (Lateolabrax maculatus): Structure, evolution, and expression profiling in response to alkalinity stress and salinity changes. Genes 2020, 11, 1271. [Google Scholar] [CrossRef]
- Schultz, J.; Copley, R.R.; Doerks, T.; Ponting, C.P.; Bork, P. SMART: A web-based tool for the study of genetically mobile domains. Nucleic Acids Res. 2000, 28, 231–234. [Google Scholar] [CrossRef]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [Green Version]
- Trifinopoulos, J.; Nguyen, L.T.; von Haeseler, A.; Minh, B.Q. W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016, 44, W232–W235. [Google Scholar]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016, 44, W242–W245. [Google Scholar] [CrossRef]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef]
- Tian, Y.; Wen, H.; Qi, X.; Zhang, X.; Liu, S.; Li, B.; Sun, Y.; Li, J.; He, F.; Yang, W.; et al. Characterization of full-length transcriptome sequences and splice variants of Lateolabrax maculatus by single-molecule long-read sequencing and their involvement in salinity regulation. Front. Genet. 2019, 10, 1126. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. EdgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [Green Version]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Gao, F.; Chen, C.; Arab, D.A.; Du, Z.; He, Y.; Ho, S. EasyCodeML: A visual tool for analysis of selection using CodeML. Ecol. Evol. 2019, 9, 3891–3898. [Google Scholar] [CrossRef] [Green Version]
- Yang, Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007, 24, 1586–1591. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J.E. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Glasauer, S.M.; Neuhauss, S.C. Whole-genome duplication in teleost fishes and its evolutionary consequences. Mol. Genet. Genom. 2014, 289, 1045–1060. [Google Scholar] [CrossRef] [Green Version]
- Kriehuber, T.; Rattei, T.; Weinmaier, T.; Bepperling, A.; Haslbeck, M.; Buchner, J. Independent evolution of the core domain and its flanking sequences in small heat shock proteins. FASEB J. 2010, 24, 3633–3642. [Google Scholar] [CrossRef]
- Haslbeck, M.; Weinkauf, S.; Buchner, J. Small heat shock proteins: Simplicity meets complexity. J. Biol. Chem. 2019, 294, 2121–2132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brocchieri, L.; Macario, E.C.d.; Macario, A.J. Hsp70 genes in the human genome: Conservation and differentiation patterns predict a wide array of overlapping and specialized functions. BMC Evol. Biol. 2008, 8, 19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, L.; Li, C.; Xie, Y.; Liu, S.; Zhang, J.; Yao, J.; Jiang, C.; Li, Y.; Liu, Z. Genome-wide identification of Hsp70 genes in channel catfish and their regulated expression after bacterial infection. Fish Shellfish Immunol. 2016, 49, 154–162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kokou, F.; Con, P.; Barki, A.; Nitzan, T.; Slosman, T.; Mizrahi, I.; Cnaani, A. Short- and long-term low-salinity acclimation effects on the branchial and intestinal gene expression in the European seabass (Dicentrarchus labrax). Comp. Biochem. Physiol. A 2019, 231, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Li, S.; Gao, Y.; Zhan, A. Genome-wide identification, characterization and expression analyses of heat shock protein-related genes in a highly invasive ascidian Ciona savignyi. Front. Physiol. 2018, 9, 1043. [Google Scholar] [CrossRef] [Green Version]
- Hoter, A.; El-Sabban, M.E.; Naim, H.Y. The HSP90 Family: Structure, regulation, function, and implications in health and disease. Int. J. Mol. Sci. 2018, 19, 2560. [Google Scholar] [CrossRef] [Green Version]
- Murphy, M.E. The HSP70 family and cancer. Carcinogenesis 2013, 34, 1181–1188. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).