Environment-Dependent Heterosis and Transgressive Gene Expression in Reciprocal Hybrids between the Channel Catfish Ictalurus punctatus and the Blue Catfish Ictalurus furcatus
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Fish Maintenance and RNA-Seq Sample Collection
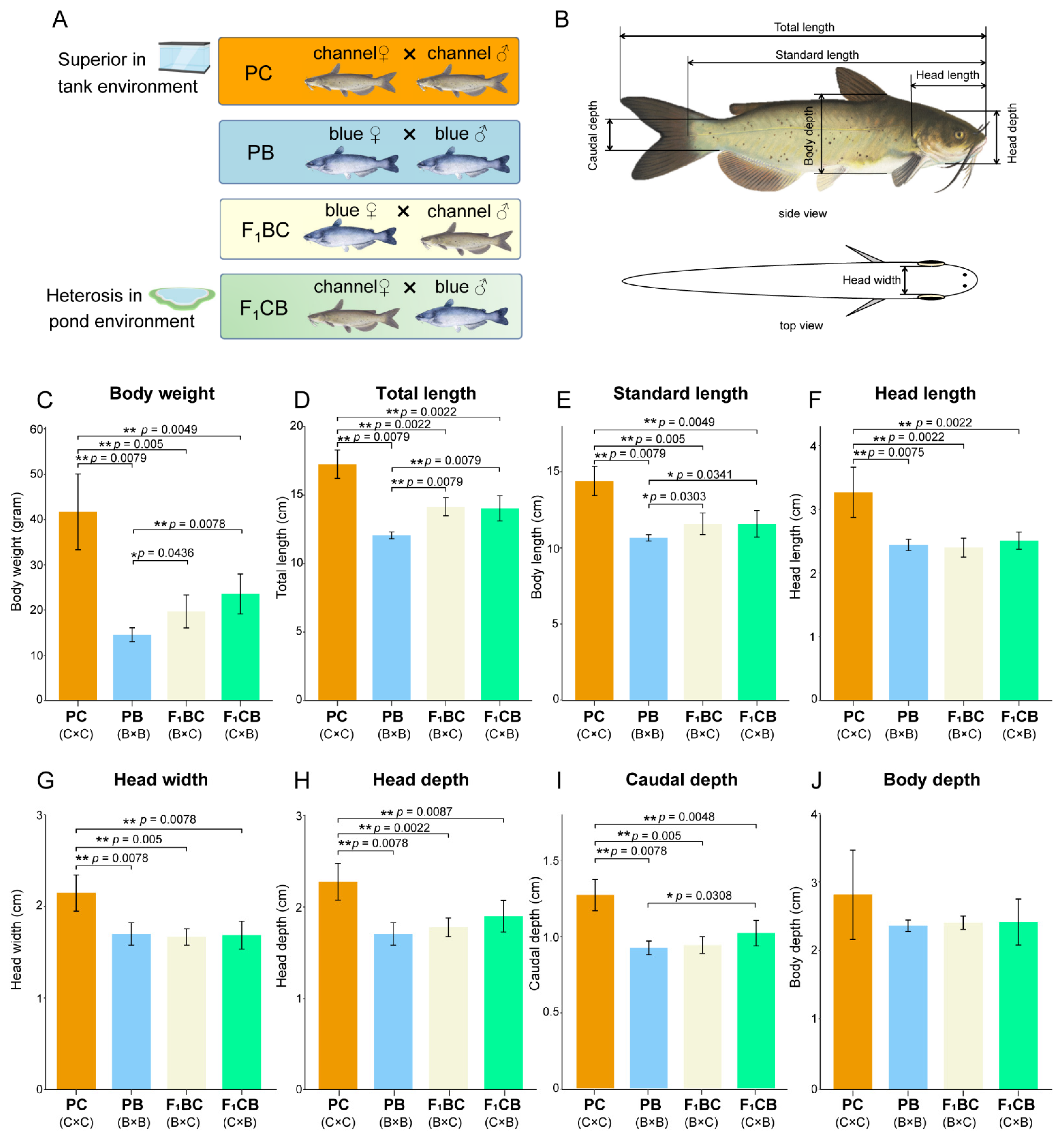
2.2. Morphometric Measurements
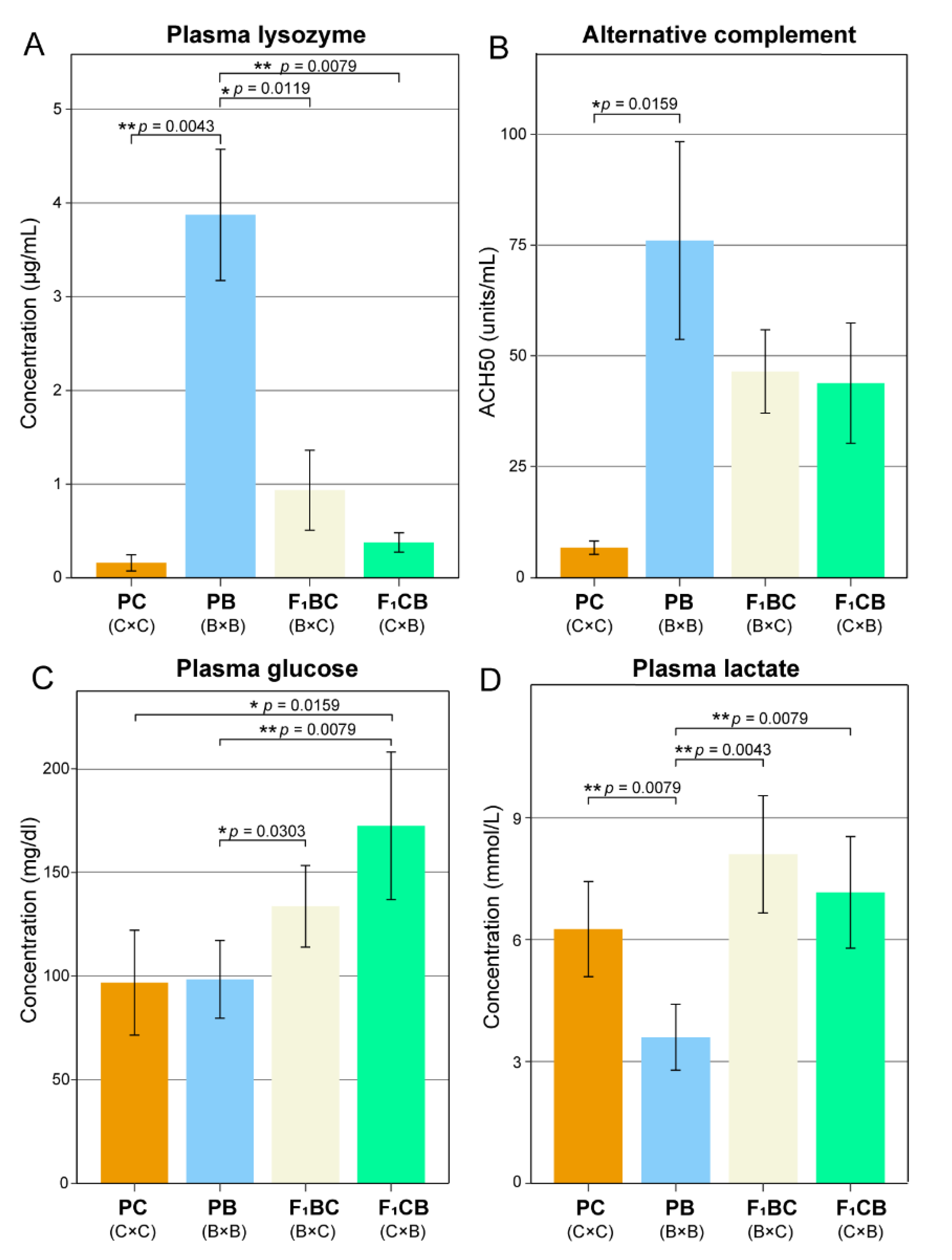
2.3. Biochemical and Immunological Assays
2.4. Total RNA Extraction, RNA-Seq Library Preparation, and Sequencing
2.5. RNA-Seq Data Analysis and Identification of Differentially Expressed Genes among Channel Catfish, Blue Catfish, and Their Reciprocal F1 Hybrids
2.6. Gene Ontology and Functional Enrichment Analysis for DEGs among PC, PB, and Reciprocal Hybrids F1BC and F1CB
2.7. Identification and Functional Pathway Analysis of Transgressive Genes in Reciprocal F1 Hybrids
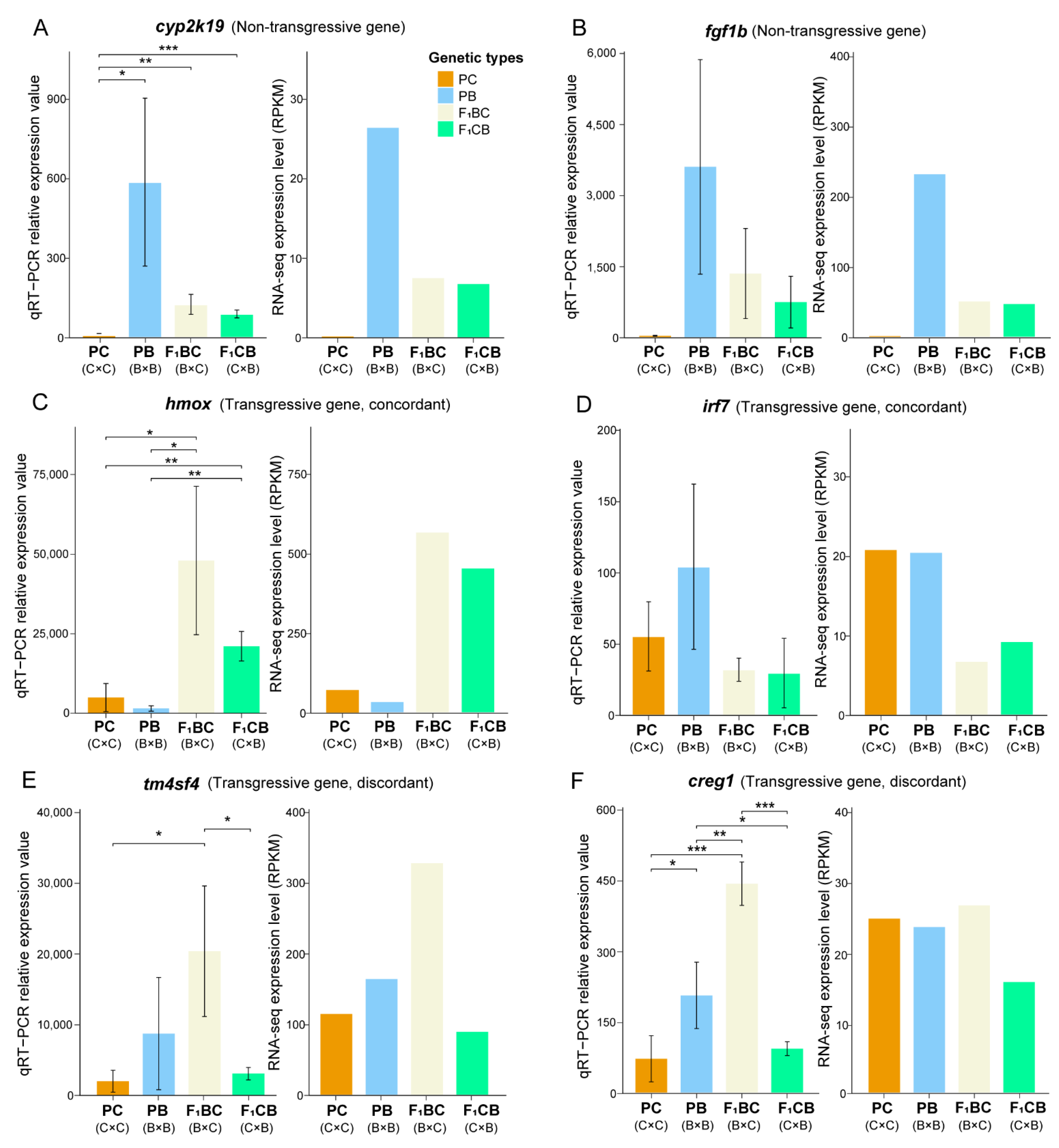
2.8. Quantitative Reverse Transcription PCR Validation of DEGs and Transgressive Genes
2.9. Statistical Analysis
3. Results
3.1. Environment-Dependent Heterobeltiosis—Channel Catfish Is Superior in Aquarium Culture
3.2. Low Level of Innate Immunity and Complement Activities in Channel Catfish Raised in the Aquarium Environment
3.3. Transgressive Effects in Metabolism—F1 Hybrids Have Significantly Higher Blood Glucose Lactate Levels Than both Channel and Blue Catfish Parents
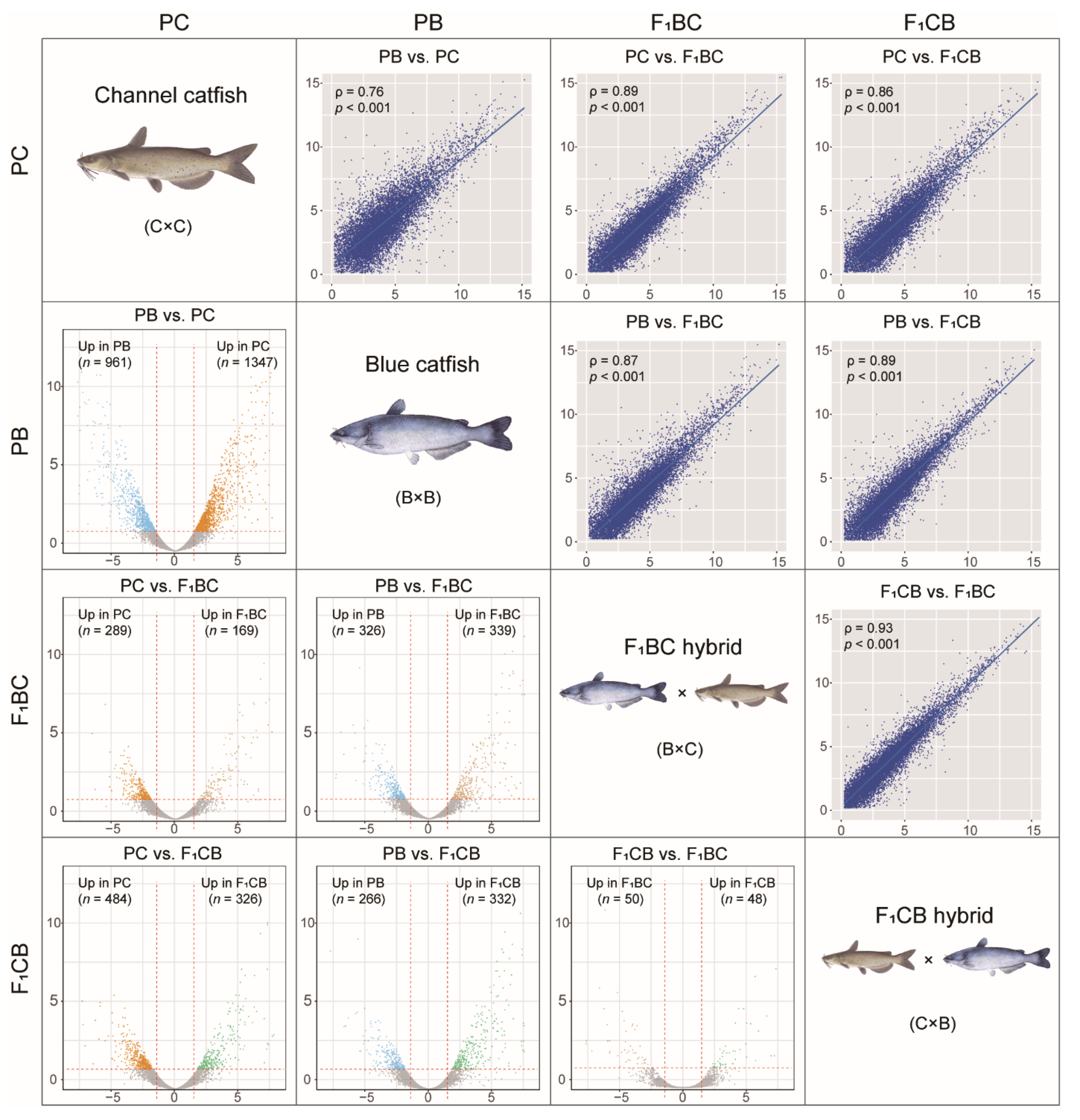
3.4. Transcriptome Analysis in the Channel and Blue Catfish Parents and the Reciprocal F1 Hybrids Revealed >2000 Differentially Expressed Genes
3.5. Fatty Acid Metabolism and Transport Genes Were Significantly Upregulated in Channel Catfish Compared to Blue Catfish and Hybrids
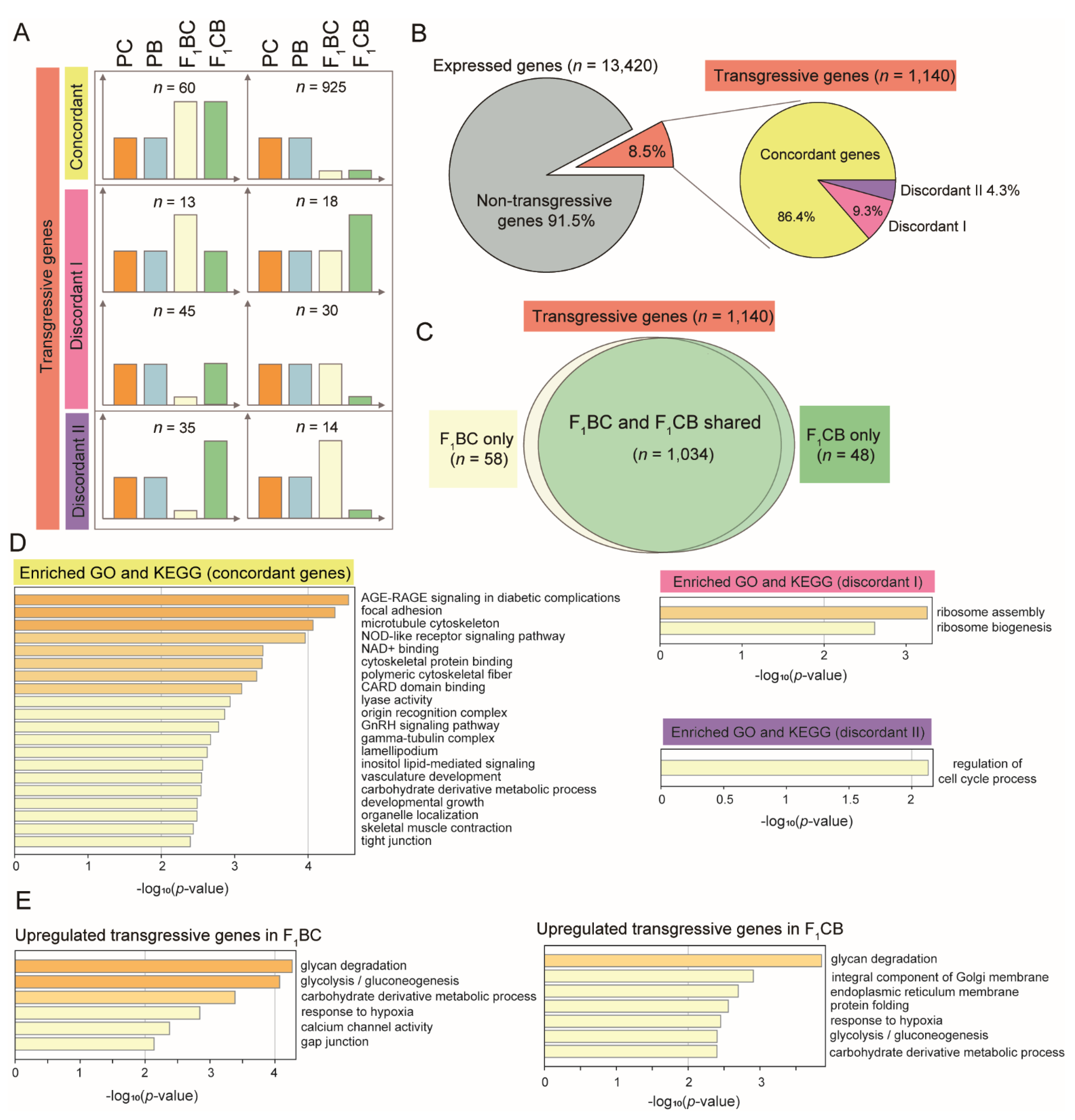
3.6. One Thousand Genes Displayed Transgressive Expression Patterns in the Liver of F1 Hybrid Catfish
3.7. Concordant Transgressive Genes in Hybrid Catfish Were Enriched for Cytoskeleton Functions, Stress, and Immune-Related Pathways
3.8. Overrepresentation of Glycan Degradation Function among Upregulated Transgressive Genes Provided a Potential Mechanism for the Blood Glucose Elevation in F1 Hybrids
4. Discussion
4.1. The Phenomenon of Environment-Dependent Heterosis in Hybrid Catfish
4.2. The Biological Robustness and Prevalence of Transgressive Genes in Channel-Blue Catfish Hybrid System
4.3. The Superiority in Tank Growth May Be Associated with Low Immune Activity and Stress Levels in Channel Catfish
4.4. Fatty Acid and Lipid Metabolism Are Enriched in Channel Catfish
4.5. High Blood Glucose in Hybrids Is Likely to Be due to Glycan Degradation in the Liver
4.6. Toward a Better Understanding of the Environment-Dependent Heterobeltiosis in Hybrid Catfish
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Darwin, C. The Works of Charles Darwin, Volume 25: The Effects of Cross and Self Fertilization in the Vegetable Kingdom; NYU Press: New York, NY, USA, 2010. [Google Scholar]
- Shull, G.H. What is “ heterosis”? Genetics 1948, 33, 439. [Google Scholar] [CrossRef]
- Batte, M.; Nyine, M.; Uwimana, B.; Swennen, R.; Akech, V.; Brown, A.; Hovmalm, H.P.; Geleta, M.; Ortiz, R. Significant progressive heterobeltiosis in banana crossbreeding. BMC Plant Biol. 2020, 20, 489. [Google Scholar] [CrossRef] [PubMed]
- Dunham, R.A.; Smitherman, R.O. Crossbreeding channel catfish for improvement of body weight in earthen ponds. Growth 1983, 47, 97–103. [Google Scholar] [PubMed]
- Dunham, R.A. Aquaculture and Fisheries Biotechnology: Genetic Approaches, 2nd ed.; CABI: Wallingford, UK, 2011; pp. 1–495. [Google Scholar]
- Chen, Z.J. Molecular mechanisms of polyploidy and hybrid vigor. Trends Plant Sci. 2010, 15, 57–71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Troyer, A.F. Development of hybrid corn and the seed corn industry. In Handbook of Maize; Springer: Berlin/Heidelberg, Germany, 2009; pp. 87–114. [Google Scholar]
- Williams, J.; Aguilar, I.; Rekaya, R.; Bertrand, J. Estimation of breed and heterosis effects for growth and carcass traits in cattle using published crossbreeding studies. J. Anim. Sci. 2010, 88, 460–466. [Google Scholar] [CrossRef] [PubMed]
- Buckley, F.; Lopez-Villalobos, N.; Heins, B. Crossbreeding: Implications for dairy cow fertility and survival. Animal 2014, 8, 122–133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferreira, V.; Rosa, G.; Berger, Y.; Thomas, D. Survival in crossbred lambs: Breed and heterosis effects. J. Anim. Sci. 2015, 93, 912–919. [Google Scholar] [CrossRef] [PubMed]
- Lalev, M.; Mincheva, N.; Oblakova, M.; Hristakieva, P.; Ivanova, I. Estimation of heterosis, direct and maternal additive effects from crossbreeding experiment involving two White Plymouth Rock lines of chickens. Biotechnol. Anim. Husb. 2014, 30, 103–114. [Google Scholar] [CrossRef]
- Dagilis, A.J.; Kirkpatrick, M.; Bolnick, D.I. The evolution of hybrid fitness during speciation. PLoS Genet. 2019, 15, e1008125. [Google Scholar] [CrossRef] [Green Version]
- Carroll, S.B. Evo-devo and an expanding evolutionary synthesis: A genetic theory of morphological evolution. Cell 2008, 134, 25–36. [Google Scholar] [CrossRef] [Green Version]
- Hodgins-Davis, A.; Rice, D.P.; Townsend, J.P. Gene expression evolves under a house-of-cards model of stabilizing selection. Mol. Biol. Evol. 2015, 32, 2130–2140. [Google Scholar] [CrossRef] [Green Version]
- Mack, K.L.; Nachman, M.W. Gene regulation and speciation. Trends Genet. 2017, 33, 68–80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haerty, W.; Singh, R.S. Gene regulation divergence is a major contributor to the evolution of Dobzhansky–Muller incompatibilities between species of Drosophila. Mol. Biol. Evol. 2006, 23, 1707–1714. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Banho, C.A.; Merel, V.; Oliveira, T.Y.K.; Carareto, C.M.A.; Vieira, C. Comparative transcriptomics between Drosophila mojavensis and D. arizonae reveals transgressive gene expression and underexpression of spermatogenesis-related genes in hybrid testes. Sci. Rep. 2021, 11, 9844. [Google Scholar] [CrossRef]
- Crow, J.F. Dominance and overdominance. Genet. Exploit. Heterosis Crops 1999, 49–58. [Google Scholar] [CrossRef]
- Malone, J.H.; Michalak, P. Gene expression analysis of the ovary of hybrid females of Xenopus laevis and X. muelleri. BMC Evol. Biol. 2008, 8, 82. [Google Scholar] [CrossRef] [Green Version]
- Bolnick, D.I.; Turelli, M.; Lopez-Fernández, H.; Wainwright, P.C.; Near, T.J. Accelerated mitochondrial evolution and “Darwin’s corollary”: Asymmetric viability of reciprocal F1 hybrids in Centrarchid fishes. Genetics 2008, 178, 1037–1048. [Google Scholar] [CrossRef] [Green Version]
- Davidson, J.H.; Balakrishnan, C.N. Gene regulatory evolution during speciation in a songbird. G3 Genes Genomes Genet. 2016, 6, 1357–1364. [Google Scholar] [CrossRef] [Green Version]
- Turelli, M.; Moyle, L.C. Asymmetric postmating isolation: Darwin’s corollary to Haldane’s rule. Genetics 2007, 176, 1059–1088. [Google Scholar] [CrossRef] [Green Version]
- Brideau, N.J.; Flores, H.A.; Wang, J.; Maheshwari, S.; Wang, X.; Barbash, D.A. Two Dobzhansky-Muller genes interact to cause hybrid lethality in Drosophila. Science 2006, 314, 1292–1295. [Google Scholar] [CrossRef] [Green Version]
- Lippman, Z.B.; Zamir, D. Heterosis: Revisiting the magic. Trends Genet. 2007, 23, 60–66. [Google Scholar] [CrossRef]
- Dobzhansky, T. Studies on hybrid sterility. II. Localization of sterility factors in Drosophila pseudoobscura hybrids. Genetics 1936, 21, 113. [Google Scholar] [CrossRef]
- Wei, X.; Zhang, J. The optimal mating distance resulting from heterosis and genetic incompatibility. Sci. Adv. 2018, 4, eaau5518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mendel, G. Experiments in Plant Hybridisation; Harvard University Press: Cambridge, MA, USA, 1965. [Google Scholar]
- Larièpe, A.; Mangin, B.; Jasson, S.; Combes, V.; Dumas, F.; Jamin, P.; Lariagon, C.; Jolivot, D.; Madur, D.; Fievet, J. The genetic basis of heterosis: Multiparental quantitative trait loci mapping reveals contrasted levels of apparent overdominance among traits of agronomical interest in maize (Zea mays L.). Genetics 2012, 190, 795–811. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, L.; Lu, K.; Chen, Z.; Mu, T.; Hu, Z.; Li, X. Dominance, overdominance and epistasis condition the heterosis in two heterotic rice hybrids. Genetics 2008, 180, 1725–1742. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, G.; Zhan, W.; Chen, H.; Xing, Y. Dominance and epistasis are the main contributors to heterosis for plant height in rice. Plant Sci. 2014, 215, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Swanson-Wagner, R.A.; Jia, Y.; DeCook, R.; Borsuk, L.A.; Nettleton, D.; Schnable, P.S. All possible modes of gene action are observed in a global comparison of gene expression in a maize F1 hybrid and its inbred parents. Proc. Natl. Acad. Sci. USA 2006, 103, 6805–6810. [Google Scholar] [CrossRef] [Green Version]
- Crow, J.F. Mid-century controversies in population genetics. Annu. Rev. Genet. 2008, 42, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Bartley, D.M.; Rana, K.; Immink, A.J. The use of inter-specific hybrids in aquaculture and fisheries. Rev. Fish Biol. Fish. 2000, 10, 325–337. [Google Scholar] [CrossRef]
- Hulata, G. A Review of Genetic-Improvement of the Common Carp (Cyprinus carpio L.) and Other Cyprinids by Crossbreeding, Hybridization and Selection. Aquaculture 1995, 129, 143–155. [Google Scholar] [CrossRef]
- Steffens, W.; Jahnichen, H.; Fredrich, F. Possibilities of Sturgeon Culture in Central-Europe. Aquaculture 1990, 89, 101–122. [Google Scholar] [CrossRef]
- Amiri, B.M.; Maebayashi, M.; Hara, A.; Adachi, S.; Yamauchi, K. Ovarian development and serum sex steroid and vitellogenin profiles in the female cultured sturgeon hybrid, the bester. J. Fish Biol. 1996, 48, 1164–1178. [Google Scholar] [CrossRef]
- Henderson-Arzapalo, A.; Colura, R.L.; Maciorowski, A.F. A Comparison of Black Drum, Red Drum, and their Hybrid in Saltwater Pond Culture. J. World Aquac. Soc. 1994, 25, 289–296. [Google Scholar] [CrossRef]
- Tripp-Valdez, M.A.; Cicala, F.; Galindo-Sanchez, C.E.; Chacon-Ponce, K.D.; Lopez-Landavery, E.; Diaz, F.; Re-Araujo, D.; Lafarga-De la Cruz, F. Growth Performance and Transcriptomic Response of Warm-Acclimated Hybrid Abalone Haliotis rufescens (female symbol) × H. corrugata (male symbol). Mar. Biotechnol. 2021, 23, 62–76. [Google Scholar] [CrossRef] [PubMed]
- Harrell, R.M.; Webster, D.W. An overview of Morone culture. Dev. Aquac. Fish. Sci. 1997, 30, 1–10. [Google Scholar]
- Dunham, R.A.; Elaswad, A. Catfish biology and farming. Annu. Rev. Anim. Biosci. 2018, 6, 305–325. [Google Scholar] [CrossRef]
- Dunham, R.A.; Umali, G.M.; Beam, R.; Kristanto, A.H.; Trask, M. Comparison of production traits of NWAC103 channel catfish, NWAC103 channel catfish× blue catfish hybrids, Kansas Select 21 channel catfish, and blue catfish grown at commercial densities and exposed to natural bacterial epizootics. N. Am. J. Aquac. 2008, 70, 98–106. [Google Scholar] [CrossRef]
- Brown, T.W.; Chappell, J.A.; Boyd, C.E. A commercial-scale, in-pond raceway system for Ictalurid catfish production. Aquac. Eng. 2011, 44, 72–79. [Google Scholar] [CrossRef]
- Bosworth, B.G. Effects of winter feeding on growth, body composition, and processing traits of co-cultured Blue Catfish, Channel Catfish, and Channel Catfish × Blue Catfish hybrids. N. Am. J. Aquac. 2012, 74, 553–559. [Google Scholar] [CrossRef]
- Dunham, R.A.; Smitherman, R.O.; Webber, C. Relative tolerance of channel x blue hybrid and channel catfish to low oxygen concentrations. Progress. Fish-Cult. 1983, 45, 55–57. [Google Scholar] [CrossRef]
- Arias, C.R.; Cai, W.; Peatman, E.; Bullard, S.A. Catfish hybrid Ictalurus punctatus× I. furcatus exhibits higher resistance to columnaris disease than the parental species. Dis. Aquat. Org. 2012, 100, 77–81. [Google Scholar]
- Dunham, R.; Masser, M. Production of Hybrid Catfish; Southern Regional Aquaculture Center: Stoneville, MI, USA, 2012; Volume 436. [Google Scholar]
- Bott, L.B.; Roy, L.A.; Hanson, T.R.; Chappell, J.; Whitis, G.N. Research verification of production practices using intensive aeration at a hybrid catfish operation. N. Am. J. Aquac. 2015, 77, 460–470. [Google Scholar] [CrossRef]
- Kumar, G.; Engle, C.R.; Hanson, T.R.; Tucker, C.S.; Brown, T.W.; Bott, L.B.; Roy, L.A.; Boyd, C.E.; Recsetar, M.S.; Park, J.; et al. Economics of alternative catfish production technologies. J. World Aquac. Soc. 2018, 49, 1039–1057. [Google Scholar] [CrossRef]
- Torrans, L.; Ott, B. Effect of grading fingerling hybrid catfish (♀ channel catfish × ♂ blue catfish) on growth, production, feed conversion, and food fish size distribution. N. Am. J. Aquac. 2018, 80, 187–192. [Google Scholar] [CrossRef]
- Domínguez, A.; Albornoz, J. Environment-dependent heterosis in Drosophila melanogaster. Génétique Sélection Évolution 1987, 19, 37–48. [Google Scholar] [CrossRef] [PubMed]
- Sankaran, K.; Gurnani, S. On the variation in the catalytic activity of lysozyme in fishes. Indian J. Biochem. Biophys. 1972, 9, 162–165. [Google Scholar] [PubMed]
- Welker, T.L.; Lim, C.; Klesius, P.; Liu, K. Evaluation of Distiller’s Dried Grains with Solubles from Different Grain Sources as Dietary Protein for Hybrid Tilapia, Oreochromis niloticus (♀) × Oreochromis aureus (♂). J. World Aquac. Soc. 2014, 45, 625–637. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 5 January 2022).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.; Liu, S.; Yao, J.; Bao, L.; Zhang, J.; Li, Y.; Jiang, C.; Sun, L.; Wang, R.; Zhang, Y. The channel catfish genome sequence provides insights into the evolution of scale formation in teleosts. Nat. Commun. 2016, 7, 11757. [Google Scholar] [CrossRef]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013, 14, R36. [Google Scholar] [CrossRef] [Green Version]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef] [Green Version]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [Green Version]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate—A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B-Stat. Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef]
- Saurabh, S.; Sahoo, P.K. Lysozyme: An important defence molecule of fish innate immune system. Aquac. Res. 2008, 39, 223–239. [Google Scholar] [CrossRef]
- Holland, M.C.; Lambris, J.D. The complement system in teleosts. Fish Shellfish Immunol. 2002, 12, 399–420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Falconer, D. Selection in different environments: Effects on environmental sensitivity (reaction norm) and on mean performance. Genet. Res. 1990, 56, 57–70. [Google Scholar] [CrossRef] [Green Version]
- Bryant, J.; López-Villalobos, N.; Pryce, J.; Holmes, C.; Johnson, D.; Garrick, D. Effect of environment on the expression of breed and heterosis effects for production traits. J. Dairy Sci. 2007, 90, 1548–1553. [Google Scholar] [CrossRef] [Green Version]
- Guy, J.A.; Jerry, D.R.; Rowland, S.J. Heterosis in fingerlings from a diallel cross between two wild strains of silver perch (Bidyanus bidyanus). Aquac. Res. 2009, 40, 1291–1300. [Google Scholar] [CrossRef]
- Dunham, R.A.; Brummett, R.E.; Ella, M.O.; Smitherman, R.O. Genotype-environment interactions for growth of blue, channel and hybrid catfish in ponds and cages at varying densities. Aquaculture 1990, 85, 143–151. [Google Scholar] [CrossRef]
- Bomblies, K.; Weigel, D. Hybrid necrosis: Autoimmunity as a potential gene-flow barrier in plant species. Nat. Rev. Genet. 2007, 8, 382–393. [Google Scholar] [CrossRef] [PubMed]
- Vaid, N.; Laitinen, R.A.E. Diverse paths to hybrid incompatibility in Arabidopsis. Plant J. 2019, 97, 199–213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McClintock, B. The significance of responses of the genome to challenge. Science 1984, 226, 792–801. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.; Peatman, E.; Abernathy, J.; Waldbieser, G.; Lindquist, E.; Richardson, P.; Lucas, S.; Wang, M.; Li, P.; Thimmapuram, J. Assembly of 500,000 inter-specific catfish expressed sequence tags and large scale gene-associated marker development for whole genome association studies. Genome Biol. 2010, 11, R8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dunham, R.A.; Argue, B.J. Reproduction among Channel Catfish, Blue Catfish, and Their F1 and F2 Hybrids. Trans. Am. Fish. Soc. 2000, 129, 222–231. [Google Scholar] [CrossRef]
- Sahoo, P. Innate/natural resistance and non-lymphoid defence mechanisms in fish. In Fish and Shellfish Immunology, An Introduction; Swain, P., Sahoo, P.K., Ayyappan, S., Eds.; Narendra Publishing House: Dehli, India, 2006; pp. 37–46. [Google Scholar]
- Sunyer, J.O.; Tort, L. Natural hemolytic and bactericidal activities of sea bream Sparus aurata serum are effected by the alternative complement pathway. Vet. Immunol. Immunopathol. 1995, 45, 333–345. [Google Scholar] [CrossRef]
- Small, B.C.; Bilodeau, A.L. Effects of cortisol and stress on channel catfish (Ictalurus punctatus) pathogen susceptibility and lysozyme activity following exposure to Edwardsiella ictaluri. Gen. Comp. Endocrinol. 2005, 142, 256–262. [Google Scholar] [CrossRef]
- Nayak, S.; Portugal, I.; Zilberg, D. Analyzing complement activity in the serum and body homogenates of different fish species, using rabbit and sheep red blood cells. Vet. Immunol. Immunopathol. 2018, 199, 39–42. [Google Scholar] [CrossRef]
- Wolters, W.R.; Johnson, M.R. Enteric Septicemia Resistance in Blue Catfish and Three Channel Catfish Strains. J. Aquat. Anim. Health 1994, 6, 329–334. [Google Scholar] [CrossRef]
- Wolters, W.R.; Wise, D.J.; Klesius, P.H. Survival and Antibody Response of Channel Catfish, Blue Catfish, and Channel Catfish Female × Blue Catfish Male Hybrids after Exposure to Edwardsiella ictaluri. J. Aquat. Anim. Health 1996, 8, 249–254. [Google Scholar] [CrossRef]
- Li, C.; Beck, B.; Su, B.; Terhune, J.; Peatman, E. Early mucosal responses in blue catfish (Ictalurus furcatus) skin to Aeromonas hydrophila infection. Fish Shellfish Immunol. 2013, 34, 920–928. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Yuan, Z.; Tan, S.; Jin, Y.; Yang, Y.; Shi, H.; Wang, W.; Niu, D.; Gao, L.; Jiang, W.; et al. A Review of Molecular Responses of Catfish to Bacterial Diseases and Abiotic Stresses. Front. Physiol. 2018, 9, 1113. [Google Scholar] [CrossRef] [Green Version]
- Zhang, D.; Xu, D.-H.; Shoemaker, C. Experimental induction of motile Aeromonas septicemia in channel catfish (Ictalurus punctatus) by waterborne challenge with virulent Aeromonas hydrophila. Aquac. Rep. 2016, 3, 18–23. [Google Scholar] [CrossRef] [Green Version]
- Zhang, D.; Moreira, G.S.; Shoemaker, C.; Newton, J.C.; Xu, D.H. Detection and quantification of virulent Aeromonas hydrophila in channel catfish tissues following waterborne challenge. FEMS Microbiol. Lett. 2016, 363, fnw080. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.-L.; Zhang, C.-M.; Fan, Q.-X.; Shen, Z.-G. Comparative Study of Growth, Feed Utilization, and Immunity of Hybrid Catfish and All-Male Yellow Catfish. N. Am. J. Aquac. 2020, 82, 71–74. [Google Scholar] [CrossRef]
- Soengas, J.; Fuentes, J.; Otero, J.; Andrés, M.; Aldegunde, M. Seasonal changes in carbohydrate metabolism in the rainbow trout (Oncorhynchus mykiss) and their relationship to changes in gill (Na+-K+)-ATPase activity. Aquaculture 1992, 108, 369–380. [Google Scholar] [CrossRef]
- Conde-Sieira, M.; Aguilar, A.J.; López-Patiño, M.A.; Míguez, J.M.; Soengas, J.L. Stress alters food intake and glucosensing response in hypothalamus, hindbrain, liver, and Brockmann bodies of rainbow trout. Physiol. Behav. 2010, 101, 483–493. [Google Scholar] [CrossRef]
- Polakof, S.; Panserat, S.; Soengas, J.L.; Moon, T.W. Glucose metabolism in fish: A review. J. Comp. Physiol. B 2012, 182, 1015–1045. [Google Scholar] [CrossRef]
- Schroeter, J.C.; Fenn, C.M.; Small, B.C. Elucidating the roles of gut neuropeptides on channel catfish feed intake, glycemia, and hypothalamic NPY and POMC expression. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2015, 188, 168–174. [Google Scholar] [CrossRef]
- Small, B.C. Effect of isoeugenol sedation on plasma cortisol, glucose, and lactate dynamics in channel catfish Ictalurus punctatus exposed to three stressors. Aquaculture 2004, 238, 469–481. [Google Scholar] [CrossRef]
- Caillouet, C.W., Jr. Hyperactivity, blood lactic acid and mortality in channel catfish. Iowa Agric. Home Econ. Exp. Stn. Res. Bull. 1967, 35, 1. [Google Scholar]
- Offermanns, S. Hydroxy-Carboxylic Acid Receptor Actions in Metabolism. Trends Endocrinol. Metab. 2017, 28, 227–236. [Google Scholar] [CrossRef] [PubMed]
- Jia, X.; Zhai, T. Integrated analysis of multiple microarray studies to identify novel gene signatures in non-alcoholic fatty liver disease. Front. Endocrinol. 2019, 10, 599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, T.-Q.; Ren, N.; Jin, L.; Cheng, K.; Kash, S.; Chen, R.; Wright, S.D.; Taggart, A.K.; Waters, M.G. Role of GPR81 in lactate-mediated reduction of adipose lipolysis. Biochem. Biophys. Res. Commun. 2008, 377, 987–991. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Wu, J.; Zhu, J.; Kuei, C.; Yu, J.; Shelton, J.; Sutton, S.W.; Li, X.; Yun, S.J.; Mirzadegan, T. Lactate inhibits lipolysis in fat cells through activation of an orphan G-protein-coupled receptor, GPR81. J. Biol. Chem. 2009, 284, 2811–2822. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moon, T.W. Glucose intolerance in teleost fish: Fact or fiction? Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2001, 129, 243–249. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, H.; Bruce, T.J.; Su, B.; Li, S.; Dunham, R.A.; Wang, X. Environment-Dependent Heterosis and Transgressive Gene Expression in Reciprocal Hybrids between the Channel Catfish Ictalurus punctatus and the Blue Catfish Ictalurus furcatus. Biology 2022, 11, 117. https://doi.org/10.3390/biology11010117
Wang H, Bruce TJ, Su B, Li S, Dunham RA, Wang X. Environment-Dependent Heterosis and Transgressive Gene Expression in Reciprocal Hybrids between the Channel Catfish Ictalurus punctatus and the Blue Catfish Ictalurus furcatus. Biology. 2022; 11(1):117. https://doi.org/10.3390/biology11010117
Chicago/Turabian StyleWang, Haolong, Timothy J. Bruce, Baofeng Su, Shangjia Li, Rex A. Dunham, and Xu Wang. 2022. "Environment-Dependent Heterosis and Transgressive Gene Expression in Reciprocal Hybrids between the Channel Catfish Ictalurus punctatus and the Blue Catfish Ictalurus furcatus" Biology 11, no. 1: 117. https://doi.org/10.3390/biology11010117
APA StyleWang, H., Bruce, T. J., Su, B., Li, S., Dunham, R. A., & Wang, X. (2022). Environment-Dependent Heterosis and Transgressive Gene Expression in Reciprocal Hybrids between the Channel Catfish Ictalurus punctatus and the Blue Catfish Ictalurus furcatus. Biology, 11(1), 117. https://doi.org/10.3390/biology11010117








