Simple Summary
The selection of varieties more resilient to disease and climate change requires generating new genetic diversity for breeding. The main mechanism for reshuffling genetic information is through the recombination of chromosomes during meiosis. We showed in oilseed rape (Brassica napus, AACC, 2n = 4x = 38), which is a natural hybrid formed from a cross between turnip (B. rapa, AA, 2n = 2x = 20) and cabbage (B. oleracea, CC, 2n = 2x = 18), that there is significantly more crossovers occurring along the entire A chromosomes in allotriploid AAC (crossbetween B. napus and B. rapa) than in diploid AA or allotetraploid AACC hybrids. We demonstrated that these allotriploid AAC hybrids are highly efficient to introduce new variability within oilseed rape varieties, notably by enabling the introduction of small genomic regions carrying genes controlling agronomically interesting traits.
Abstract
Meiotic recombination is the main tool used by breeders to generate biodiversity, allowing genetic reshuffling at each generation. It enables the accumulation of favorable alleles while purging deleterious mutations. However, this mechanism is highly regulated with the formation of one to rarely more than three crossovers, which are not randomly distributed. In this study, we showed that it is possible to modify these controls in oilseed rape (Brassica napus, AACC, 2n = 4x = 38) and that it is linked to AAC allotriploidy and not to polyploidy per se. To that purpose, we compared the frequency and the distribution of crossovers along A chromosomes from hybrids carrying exactly the same A nucleotide sequence, but presenting three different ploidy levels: AA, AAC and AACC. Genetic maps established with 202 SNPs anchored on reference genomes revealed that the crossover rate is 3.6-fold higher in the AAC allotriploid hybrids compared to AA and AACC hybrids. Using a higher SNP density, we demonstrated that smaller and numerous introgressions of B. rapa were present in AAC hybrids compared to AACC allotetraploid hybrids, with 7.6 Mb vs. 16.9 Mb on average and 21 B. rapa regions per plant vs. nine regions, respectively. Therefore, this boost of recombination is highly efficient to reduce the size of QTL carried in cold regions of the oilseed rape genome, as exemplified here for a QTL conferring blackleg resistance.
1. Introduction
Meiotic recombination shuffles parental alleles to produce new allelic combinations in the progenies, hence producing new genetic diversity at each generation. This biological mechanism is a key evolutionary process that is commonly used in plant breeding to accumulate favorable alleles and purge deleterious mutations [1,2,3]. This phenomenon occurs during meiosis, a specialized eukaryotic cell division that gives rise to haploid gametes via a single round of DNA replication followed by two rounds of chromosome segregation [4]. The balanced segregation of homologs during the first meiotic division is ensured by meiotic recombination through crossovers that temporarily connect homologous chromosomes until metaphase I. For successful chromosome segregation and hence the production of viable gametes, such joined homolog pairs require at least one crossover. Besides this crucial role, crossovers result in reciprocal exchanges between homologous non-sister chromatids, generating new allelic combinations in gametes.
However, crossover formation is under tight genetic control [4]. Typically, eukaryotes experience one obligatory crossover per chromosome pair and per meiosis, but rarely more than three, despite a large number of generated DNA double strand breaks (DSBs). For instance, out of the 150 to 250 DSBs generated per meiosis, only ~10 result in crossovers in Arabidopsis, the others giving rise to non-crossovers (NCOs) [5,6]. In the last decade, several proteins were highlighted to promote the repair of DSBs into NCOs in A. thaliana (e.g., FANCM, RECQ4, FIGL1, HCR1), thereby limiting the overall number of crossovers generated per meiosis [7,8,9,10]. Furthermore, crossovers are unevenly distributed along chromosomes [4], with a gradient from the telomere to the centromere, as exemplified in bread wheat [11], maize [12] or potato [13]. Fine characterization of crossover distributions has pointed out links with genomic and epigenetic features, revealing that crossovers preferentially occur close to gene promoters and terminators in euchromatic regions that are hypomethylated and enriched in H3K4me3 histone marks [14,15,16,17]. On the contrary, they are greatly reduced or even prevented in regions that are heavily methylated and enriched in H3K9me2, such as pericentromeres [18,19,20], or regions exhibiting sequence variations (e.g., insertions, deletions, inversions, translocation, etc.) [21]. Consequently, most crossovers occur in a minor proportion of the chromosome, as exemplified in A. thaliana, for which 80% of crossovers formed are observed within less than 26% of the genome [14].
Despite the strong regulation of meiotic recombination, extensive variations are observed for crossover number and distribution both between and within species [22,23]. Factors responsible for these modifications have the potential to profoundly influence selective responses and adaptation while accelerating plant breeding programs [24,25,26,27]. In a non-exhaustive list of examples, environmental conditions (e.g., abiotic stress, temperature) (for a review, see [28]), sex of meiosis [29,30], genetic background [31,32,33] and ploidy level (for a review, see [34]) have been linked to variations in crossover level and/or pattern. The most striking in terms of crossover reshuffling and promising in terms of breeding applications is the last factor listed, with the particular case of Brassica AAC allotriploids (2n = 3x = 29). Indeed, in these viable and fertile hybrids, resulting from the cross between the rapeseed B. napus (AACC, 2n = 4x = 38) and its B. rapa diploid progenitor (AA, 2n = 2x = 20), crossover frequency obtains an unprecedent boost on the A07 chromosome when compared to the results obtained from diploid or allotetraploid hybrids [35]. The A chromosomes show an average recombination frequency that is 3.4-fold higher compared to diploid AA hybrids, carrying identical A genomes, when AAC plants are used as female parents [36]. Moreover, the authors pointed out that this crossover boost is strikingly associated with dramatic changes in the shape of recombination landscapes. Indeed, AAC allotriploids frequently exhibit crossovers in genomic regions that are normally totally deprived of any recombination event in AA diploids, such as the pericentromeres. The molecular mechanisms responsible for this unique recombination landscape observed in Brassica AAC allotriploids are currently being investigated but are yet to be deciphered. Nevertheless, it is feasible to recover an AACC genomic structure from AAC plants [37], enabling us to broaden the oilseed rape genetic diversity that has been severely eroded in recent decades due to a high selective pressure applied for yield and seed quality traits [38]. Interspecific crosses between B. napus and its B. rapa diploid progenitor, combined with reshaped homologous recombination, would facilitate small introgressions of targeted valuable loci deriving from B. rapa while preserving the agronomic value of B. napus cultivars. Recently, simulations conducted with AA and AAC Brassica genetic maps tend to confirm this putative benefit [39,40]. The authors demonstrated the genetic gain resulting from the use of allotriploids in long-term selection programs, at least in non/low-recombining pericentromeric regions. So far, these analyses have been limited to the comparison between AA and AAC hybrids as no comparable genetic maps have been generated to date for allotetraploid AACC hybrids with the same A genetic background as in AAC hybrids. This prevents addressing the role played by polyploidy per se in this recombination pattern, as well as the true potential of using allotriploids compared to allotetraploids in oilseed rape breeding programs. Moreover, the potential of using AAC allotriploids (compared to allotetraploids) in reducing the size of a quantitative trait loci (QTL) identified in B. napus remains to be assessed.
In the present study, we analyzed how this unique recombination pattern observed in Brassica AAC allotriploid hybrids can be used for oilseed rape breeding. Firstly, we tested the impact of polyploidy per se vs. AAC allotriploidy by comparing the recombination profile in AA, AAC and AACC hybrids carrying an identical A genotype. We confirmed that the modification of meiosis control only occurs in AAC allotriploid hybrids. Secondly, compared to a previous study [36], we densified the genetic maps from the AAC or AACC hybrids (10 times more SNPs) and finely compared the size and distribution of B. rapa introgressions observed along each A chromosome, depending on the hybrid ploidy level. Finally, we highlighted the interest of using allotriploids in breeding programs, by taking the example of a large resistance QTL against Leptosphaeria maculans [41] present in a cold recombining pericentromeric region.
2. Materials and Methods
2.1. Plant Material
We compared crossover rate between homologous A chromosomes of Brassica hybrids presenting three different ploidy levels but sharing a genetically identical A genome sequence (Figure 1). To that purpose, we used the plant material already described by Pelé et al. [36]: (1) the diploid AnAr (2n = 2x = 20) F1 hybrid, which was obtained by crossing the diploid AnAn (2n = 2x = 20) genotype extracted from the French variety B. napus cv. Darmor AnAnCnCn (2n = 4x = 38) [42] with the pure inbred line B. rapa ssp. pekinensis cv. Chiifu-401-42 (male, ArAr, 2n = 2x = 20), was backcrossed with the Korean variety B. napus cv. Yudal and gave rise to 329 plants; (2) the allotriploid hybrid AnArCn (2n = 3x = 29), which was obtained by crossing the allotetraploid B. napus cv. Darmor AnAnCnCn (2n = 4x = 38) and the pure inbred line B. rapa ssp. pekinensis cv. Chiifu (male, ArAr, 2n = 2x = 20), was then backcrossed with the Korean variety B. napus cv. Yudal and gave rise to 109 plants. This plant material was complemented by backcrossing the AnArCn hybrid (2n = 3x = 29) to B. napus cv. Darmor (131 plants selected among 234). To produce the allotetraploid hybrid AnArCnCo (2n = 4x = 38), we first crossed the pure inbred line B. rapa cv. Chiifu (ArAr, 2n = 2x = 20) with the doubled haploid B. oleracea cv. HDEM (CoCo, 2n = 2x = 18) and then performed embryo rescue on the obtained amphihaploid (ArCo, 2n = 19) as described in [43]. This hybrid spontaneously doubled its genomes and gave rise to the resynthesized allotetraploid ArArCoCo (2n = 4x = 38), hereafter referred as ChEM. This latter plant was crossed with B. napus cv. Darmor as female, and the AnArCnCo F1 hybrid gave rise to 213 plants after backcrossing with B. napus cv. Darmor as male. All parental accessions were provided by the Biological Resource Center BrACySol (Ploudaniel, France).
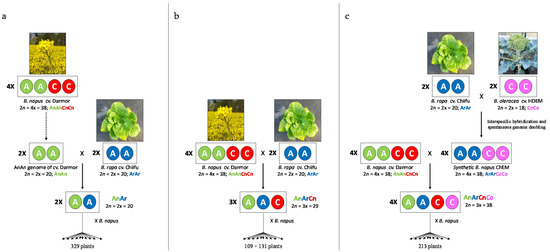
Figure 1.
Production of (a) AnAr diploid, (b) AnArCn allotriploid and (c) AnArCnCo allotetraploid hybrids sharing the same A genotype and their derived progenies.
2.2. Flow Cytometry and Cytogenetic Studies
Chromosome numbers were assessed in leaves by flow cytometry as described by [37]. For the establishment of meiotic behavior, samples of young floral buds were fixed in Carnoy’s solution (alcohol:chloroform:acetic acid, 6:3:1) for 24 h at room temperature and stored until use in 50% ethanol at 4 °C. Anthers were then squashed and stained with 1% aceto-carmine. Chromosome pairing was assessed per plant from 20 pollen mother cells (PMCs) at metaphase I. For analyzing pairing between A and C chromosomes at metaphase I, BAC FISH was performed as described in [37] using the B. oleracea BoB014O06 BAC [44] as “genomic in situ hybridization-like” (GISH-like) to specifically distinguish all C chromosomes.
2.3. DNA Extraction and SNP Genotyping
Genomic DNA was extracted from lyophilized young leaves with the sbeadex maxi plant kit (LGC Genomics, Teddington Middlesex, UK) on the oKtopure robot at the GENTYANE platform (INRAE, Clermont-Ferrand, France). Genotyping data were obtained using the 202 SNP markers already defined [36], and revealed by BiomarkTM HD system (Fluidigm technology) and KASParTM chemistry (GENTYANE platform INRAE, Clermont-Ferrand, France), as well as from the Brassica 15K Illumina Infinium SNP array (SGS-TraitGenetics GmbH, Gatersleben, Germany). The context sequences of each SNP marker were physically localized on the reference genome B. rapa Chiifu v1.5 [45] and B. napus cv. Darmor-bzh v10 [46] using BLASTn (ver. 2.9.0, min. e-value 1 × 10−20) [47] and by keeping the best blast hit obtained for a given subgenome (minimum percentage of alignment and identity: 80%). SNPs that were polymorphic between the parental genotypes (i.e., AA in B. rapa and BB in B. napus, or vice versa) were selected for either the establishment of genetic maps or assessment of introgressed regions from the diploid parent into the B. napus genome.
2.4. Genetic Maps
The first genetic maps were established separately for the AnAr, the AnArCn and the AnArCnCo populations using the CarthaGene software (v. 1.2.3, [48]). Establishment of linkage groups and SNP ordering were examined using a logarithm of odds score (LOD) threshold of 4.0 and a maximum recombination frequency of 0.4, as described in [36]. Potential double crossover supported by only one genetic marker and with a physical distance between these two events below 500 kb was corrected as missing data, as described in [21]. After these few corrections, the final genetic maps were created using the Kosambi function to evaluate the genetic distances in centimorgans (cM) between linked SNP markers [49].
2.5. Characterization of the B. rapa Introgressions
Introgressions were determined by two methods, if either one SNP or at least two consecutive SNP markers were detected as heterozygous in each plant from the backcross progeny. To estimate the length of the introgresssed regions detected by only one SNP and to prevent the underestimation of the smallest introgressions, we considered the position of the consecutive previous and following SNPs. The positions of the first and last heterozygous SNPs were used to infer the size of introgressed regions defined by at least two consecutive SNPs, preventing the overestimation of introgressions. Each SNP can be present in introgressions of different sizes in different plants of a progeny and we calculated per population for each SNP position the average size of the introgressions carrying this marker.
2.6. Localization of a QTL of Interest
A quantitative trait locus (QTL) involved in blackleg (Leptosphaeria maculans) resistance [41] and present on the B. napus cv. Darmor A01 chromosome was selected for its pericentromeric localization. The context sequences of the SNP markers (derived from Brassica 60 K Illumina infinium array) flanking this QTL were retrieved and physically localized on the B. napus reference genome Darmor-bzh v10 [46] using BLASTn [47].
2.7. Inferring the Position of Centromeric and Pericentromeric Regions
The centromeric regions were defined by blasting several centromeric-specific repeats (CentBr1, CentBr2, TR238, TR805, PCRBr and CRB; [50,51]) against B. napus cv. Darmor-bzh v10 [46] and by examining the plot density of the BLASTn results (e-value less than 1 × 10−20). The pericentromeric borders for each chromosome were inferred by using the mean gene density along the chromosomes (as presented in [52]) and defined as the regions surrounding the centromere with a gene density below the chromosome average.
2.8. Statistical Analyses
The heterogeneity of crossover rates among progenies was assessed for every interval between consecutive SNP markers using a 2-by-2 chi-squared analysis considering a significance threshold of 5%. Additionally, the heterogeneity of crossover rates among progenies was evaluated at chromosome and genome scales using 2-by-2 chi-squared tests. For these tests, a conservative Bonferroni-corrected threshold of 5% was applied, using the number of intervals between adjacent SNP markers per A chromosome or for the A genome (as described in [36]).
The relationships between the relative size of introgressions normalized per A chromosome (%) vs. their relative distance from the centromeres (%) were studied by regression analyses, using the Spearman rank correlation, for linear (y = ax + b) and order 2 polynomial (y = ax² + bx + c) regressions (as described in [36]).
3. Results
3.1. Impact of Brassica Hybrid Genomic Structure (Diploid, Allotriploid or Allotetraploid) on Homologous Recombination Frequency and Distribution
To accurately compare the homologous recombination profile between hybrids of different ploidy levels in Brassica, we generated AnAr, AnArCn and AnArCnCo F1 hybrids that present the same A genome nucleotidic sequence originating from B. napus cv. Darmor (AnAnCnCn) and B. rapa cv. Chiifu (ArAr) (Figure 1). The diploid AnAr and the allotriploid AnArCn F1 hybrids have been described by [36]. Here, we also studied the meiotic behavior of different allotriploid AnArCn hybrids (Table S1) and observed that they generally showed the expected meiotic configuration with 9 C univalents and 10 A bivalents as revealed by GISH-like analyses (Figure 2a). To produce the backcrossed progeny, we selected an AAC hybrid showing more than 80% of pollen mother cells with the expected configurations. In the backcross progeny of this hybrid, we were able to retrieve two plants with an AACC genomic structure over the 234 plants generated. Moreover, to create the allotetraploid AnArCnCo hybrid, we first produced the resynthesized B. napus ChEM by crossing B. rapa cv. Chiifu with B. oleracea cv. HDEM, followed by genome doubling of the resulting AC amphihaploid hybrid. Meiotic stability of the resynthesized allotetraploid was thereafter studied, revealing a relatively regular meiotic behavior with 70% of pollen mother cells in metaphase I exhibiting 19 bivalents (Table S1). These results were in agreement with GISH-like analyses, which revealed a regular pairing (Figure 2b) with rare A and C chromosome pairing (Figure 2c). After crossing the resynthesized B. napus ChEM to B. napus cv. Darmor, we selected the most stable F1 hybrid (Table S1, Figure 2d) and produced 213 progeny plants via a backcross to B. napus cv. Darmor. This progeny was genotyped with the same 202 SNPs used in [36], allowing a reliable comparison of the genetic maps derived from AnAr, AnArCn and AnArCnCo hybrids.
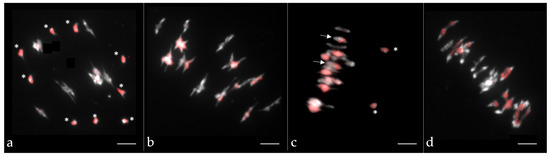
Figure 2.
Chromosome pairing at metaphase I in pollen mother cells of (a) the allotriploid AnArCn, (b,c) the synthetic B. napus ChEM ArArCoCo-S0 and (d) the allotetraploid AnArCnCo hybrid. C chromosomes are labeled in red via the use of the Bob014O06 BAC clone (GISH-like). Univalents are indicated by a * and A–C bivalents by an arrow. Chromosomes were counterstained with DAPI. Scale bars represent 5 μm.
In total, cumulated genetic maps of the 10 A chromosomes of AnAr, AnArCn and AnArCnCo hybrids showed 830.2, 2902.3 and 790.8 cM, respectively. Pairwise comparison indicated a similar level of recombination at the whole A genome scale between the AnAr and AnArCnCo hybrids (Bonferroni-corrected chi-square test, p = 0.18). However, significant variations were detected for 3x vs. 2x and 3x vs. 4x. In both cases, A genome-wide crossover rate was on average 3.6-fold higher in the allotriploid hybrids (Bonferroni-corrected chi-square test, p < 2.2 × 10−16). After anchoring the polymorphic SNPs on the B. rapa cv. Chiifu v1.5 Ar genome, crossover distribution was analyzed by assessing heterogeneity of crossover rates among progenies for every interval between adjacent SNP markers. Similar recombination landscapes were thus identified between the AnAr and AnArCnCo hybrids as they evidenced significant differences in crossover rate in only two intervals out of 192 (1%), spanning 3.19 and 1.53 Mb on chromosomes A02 and A06, respectively (Bonferroni-corrected chi-square test, p < 0.05). However, comparison of AnArCn with AnAr and AnArCnCo hybrids revealed significant differences in crossover rate in more than 60% (133 and 118 over 192, respectively) of the intervals (p < 0.05) (Figure 3). Importantly, from the 20 intervals surrounding the centromeric regions (1 Mb from each centromeric border) of the 10 A chromosomes, we noticed that the recombination frequencies were always higher in the allotriploids compared to the diploid and allotetraploid hybrids. Significant differences were detected for 18 and 10 intervals in the AnArCn vs. AnAr and AnArCn vs. AnArCnCo comparisons, respectively (p < 0.05). Therefore, we concluded that recombination landscapes along A chromosomes of the allotriploid hybrid differed in a similar way, in regard to pericentromeric regions, to those of the diploid and allotetraploid hybrids.
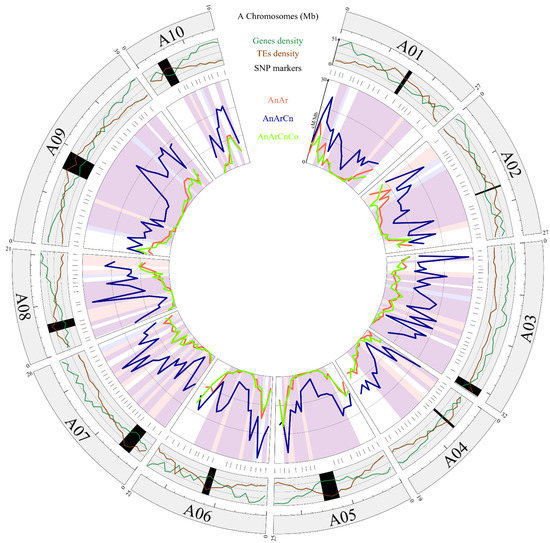
Figure 3.
Homologous recombination landscape in diploid, allotriploid and allotetraploid hybrids. The first circle represents the 10 chromosomes of B. rapa cv. Chiifu v1.5. Within the second circle, the gene and transposable element (TE) densities along each A chromosome are mentioned, with black rectangles indicating the putative centromere positions [53]. In the inner circle, the recombination rates (cM/Mb) of AnAr (2x), AnArCn (3x) and AnArCnCo (4x) are shown as red, blue and green lines, respectively. Significant differences within an interval are highlighted in peach, light blue and purple for 2x vs. 3x, 4x vs. 3x, or 4x and 2x vs. 3x, respectively. Above the recombination landscape, the physical position of each polymorph SNP used for these comparisons is indicated.
3.2. Impact of Recombination on the Size and Distribution of Introgression within Oilseed Rape
To obtain a finer analysis of the crossover distribution in the allotriploid and allotetraploid hybrids, we then analyzed their progenies using the 15K SNP Illumina infinium array. A total of 2340 polymorphic SNP markers were physically anchored on the B. napus cv. Darmor-bzh v10 A genome, increasing the number of SNPs used for the comparison by more than ten times. On average, a polymorphic marker was observed every 150 kb along each A chromosome. Using this larger number of markers, we obtained genetic maps of 3045.4 and 827.7 cM for AnArCn and AnArCnCo hybrids, respectively. We thus improved the accuracy of our comparison and confirmed a higher recombination frequency in AnArCn (3.7-fold) compared to AnArCnCo hybrids (p < 2.2 × 10−16).
Given this extraordinary reshuffling of meiotic recombination occurring in allotriploids, we analyzed the size and distribution of Ar introgressions within the An oilseed rape genome according to the genomic structure of the F1 hybrids. The overall introgressed B. rapa genetic diversity introduced per generation is similar in allotriploid and allotetraploid hybrids (50% and 48%). However, as expected, with a larger number of crossovers occurring per chromosome pair and per meiosis, we observed that the mean introgression sizes were significantly smaller when arising from the meiosis of allotriploid compared to allotetraploid hybrids, with an average of 7.6 Mb vs. 16.9 Mb, respectively (Mann–Whitney–Wilcoxon test, p = 6.4 × 10−104) (Figure 4 and Figure 5a). As a second effect of crossover boost, significantly more introgressions from B. rapa were detected in backcross progeny of allotriploid hybrids with on average 21 heterozygous regions (on the 10 A chromosomes) per plant vs. nine regions in the backcross progeny of the allotetraploid hybrid (Mann–Whitney–Wilcoxon test, p = 2.3 × 10−53) (Figure 5b). Interestingly, the size of introgressions varied along the chromosome arms. The largest introgressions tended to colocalize around pericentromeric regions while the smallest were more frequently observed on chromosome extremities (Figure 4). In an A genome-wide approach, using the relative size of introgressions normalized per A chromosome (%) and their relative distance from the centromeres (%), the regression analyses revealed a positive linear relationship within AnArCo (R² = 0.66) and a positive binomial relationship within AnArCnCo (R² = 0.68) hybrids (Figure 5c; Spearman rank correlation, p < 2.2 × 10−16). For both AnArCn and AnArCnCo hybrids, these results demonstrate that the size of introgressions increases toward the centromeres, which is in agreement with the crossover distribution along the chromosome arms: the higher rates of recombination result in smaller sizes of introgressions. However, the significant binomial regression unraveled in the AnArCnCo hybrid translates to a less continuous gradient of introgression size from the telomere to the centromere. Strikingly, 70% of the chromosome arms carried introgressions, representing about 40% of the chromosome size in this hybrid. Analyses conducted at the scale of individual A chromosomes revealed similar results (Figure S1, Table S2). The only exception is the A03 chromosome for which no significant gradient for the size of introgressions was observed in both AnArCn and AnArCnCo hybrids. This might be due to the presence of the nucleolus organizer region on the short arm of A03 [54]. For this reason, this chromosome was not considered when performing the A genome-wide approach.
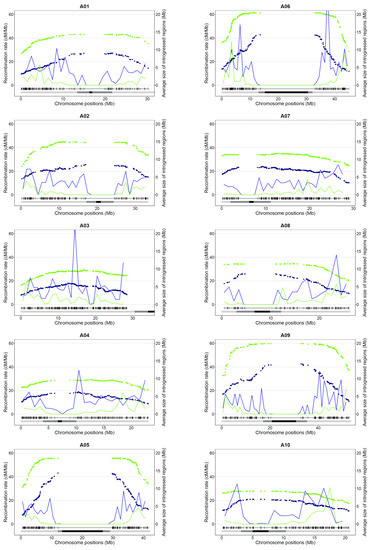
Figure 4.
Recombination and introgression patterns along the ten A chromosomes of the allotriploid (AnArCn) and allotetraploid (AnArCnCo) hybrids. For each A chromosome (A01 to A10), the distribution of the recombination (in cM/Mb, indicated by lines) and average size of introgressions (in Mb, represented by dots) at each marker position in the progeny of allotriploid (blue) and allotetraploid hybrid (green) are presented for all A chromosomes. Below each graph, the physical positions of polymorph SNPs along each B. napus cv. Darmor-bzh v10 chromosome are indicated. The position of pericentromeres and centromeres (inferred in this study) are illustrated as light gray and black boxes, respectively.
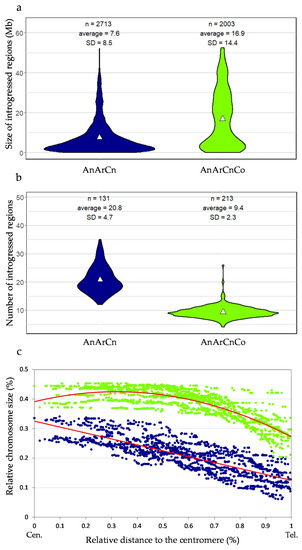
Figure 5.
Characterization of introgressions in the AnArCn and AnArCnCo populations. (a) Size of introgressions, (b) number of introgressions. White triangle indicates mean. (c) Correlation between the relative introgression size and their relative position from the centromere (Cen.) to the telomere (Tel.) analyzed in the progeny of the AnArCn hybrid (blue) and AnArCnCo hybrid (green). Regression lines are indicated in red.
3.3. Interest in Using the Modified Recombination Pattern Oberved in AAC Allotriploid Hybrid to Reduce the Size of a QTL Present in a B. napus Pericentromeric Region
To highlight the potential of using the modified recombination landscape observed in Brassica AAC allotriploids for breeding purposes, we chose as an example a QTL responsible for blackleg resistance that is present in the A01 pericentromeric region [41], at 11.8–26.7 Mb on the B. napus cv. Darmor-bzh v10 reference genome. The comparison of the recombination profiles from backcrossed progenies of allotriploid and allotetraploid hybrids revealed that significantly more crossovers were formed during the meiosis of the AnArCn hybrid compared to the AnArCnCo hybrid in this particular region, with 131 vs. 24 crossovers (corresponding to 58 and 16 different haplotypes), respectively (chi-square test, p = 1.75 × 10−32). At least one recombination event was detected in 70.23% vs. 11.27% of the AnArCn and AnArCnCo progenies, respectively, highlighting the highest diversity of gametes produced by the AnArCn hybrid compared to the AnArCnCo hybrid. It allowed us to break down the QTL into five regions of 0.02 to 10.7 Mb in the AnArCnCo population, and into 13 regions of 0.004 to only 4.96 Mb in the AnArCn population (Figure 6). It is important to note that the 4.96 Mb region that appeared to be deprived of crossovers in the AnArCn hybrid corresponds to a region lacking polymorphic markers, preventing the detection of the putative presence of crossovers in this region. Altogether, these results support the high potential of AAC allotriploids to reduce QTL confidence intervals.
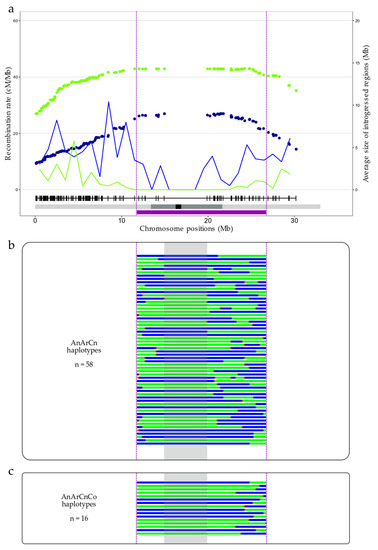
Figure 6.
Crossover localization in a pericentromeric region carrying a QTL conferring resistance to Leptosphaeria maculans. (a) Recombination rate (in cM/Mb, as lines) and the average size of introgression (in Mb, as dots) along the A01 chromosome, for the AnArCn (green) and AnArCnCo (blue) hybrids. The resistance QTL is represented by a purple box with its borders delimited by purple dashed lines. The different possible haplotypes found in the backcross progeny of (b) the AnArCn (131 plants) or (c) the AnArCnCo hybrids (213 plants) are displayed, with the homozygous B. napus cv. Darmor regions in green and the introgressed regions of B. rapa cv. Chiifu in blue. A 4.96 Mb region surrounding the centromere and deprived of markers is highlighted in gray. Red stars symbolize the crossover positions.
4. Discussion
The production of Brassica AAC allotriploid hybrids, derived from the direct cross between B. napus and its diploid B. rapa progenitor, offers a unique opportunity to introgress into B. napus small regions of interest derived from B. rapa but also to highly shuffle B. napus diversity and decrease the size of interesting B. napus QTL. Indeed, this allotriploid structure allows the modification of recombination control, with an increased crossover frequency (×3.7) compared to allotetraploids, and, more interestingly, with the formation of crossovers all along the A chromosomes even in usually cold regions such as pericentromeres. These results are in agreement with previous data [36] comparing diploid and allotriploid hybrids. However, our results differ from those obtained by Leflon et al. [35], who predicted a two-fold increase in the recombination frequency in allotetraploid compared to diploid hybrids, using 18 markers of the A07 chromosome without physical anchoring on the A07 chromosome. These different results may be partly explained by the difference in marker density between the two studies, as well as by the use of different genetic backgrounds. Indeed, Pelé et al. [36] observed a slight difference (×1.4) in the recombination rates depending on the origin of the AAC allotriploid hybrid. Nevertheless, both studies agreed with the fact that AAC allotriploidy and not allopolyploidy per se can deeply modify the homologous recombination frequency and distribution.
4.1. Comparison of Breeding Strategies
Two breeding strategies can be used to introduce into oilseed rape genetic diversity or interesting traits derived from one of its diploid progenitors, B. rapa: either direct crosses between B. napus and B. rapa or a bridge with a resynthesized oilseed rape. For the latter, it has been clearly demonstrated that homoeologous pairing occurs during the first meiosis of synthetics [43,55,56]. Such homoeologous rearrangements between A and C chromosomes are at the origin of genomic instability with univalent and multivalent formation, resulting in reduced fertility [57,58,59,60]. Despite this genomic instability, it has been shown in several instances that homoeologous exchanges were used (sometimes inadvertently) and useful for B. napus breeding (for a review, see [61]). Indeed, recent sequencing data revealed that the absence of glucosinolate in seeds, a highly selected trait in oilseed rape, is partially due to a non-reciprocal translocation between A and C genomes [62]. Similarly, other translocations between A and C genomes were shown to impact several important agronomic traits, such as flowering precocity [63], disease resistance [64] or even seed quality [65]. The major difficulty in the case of exchanges between A and C genomes is to then restore the meiotic stability by decreasing the size of the introgressed region inserted in one genome to the other one. Major traits present in B. napus progenitors were not only introduced from resynthesized tetraploids via homoeologous recombination but also via homologous recombination. This was notably the case when introducing a clubroot resistance locus present in a B. rapa accession into B. napus, leading to the B. napus variety Mendel, which is resistant to this disease [66,67]. However, results from the present study demonstrate that homologous recombination remains limited using resynthesized B. napus, making it difficult to restrict the introgression to the locus of interest. Large introgression sizes arising from this strategy, as we evidenced here, might bring undesirable loci that could reduce the agronomical value of B. napus elite varieties.
On the contrary, we showed that direct crosses between B. napus and B. rapa significantly improve the genetic shuffling of diversity all along the A genome. This strategy has also been largely used to generate new oilseed rape lines by backcrossing and/or selfing AAC hybrids [68,69], producing new lines with interesting heterosis. For example, the Chinese elite variety and reference genome Zs11 was obtained through this strategy [70]. The frequent use of B. rapa in Asian B. napus breeding programs may partially explain why Asian oilseed rape varieties appear as a specific group of oilseed rape varieties and why it is still possible to detect B. rapa introgression in this material [71]. Similarly, direct crosses between B. napus and B. rapa have been performed to introduce in B. napus major resistance genes to clubroot [72] or blackleg [73].
4.2. Optimization of Breeding Strategies
We showed that the occurrence of crossovers all along the A chromosomes, and especially in the normally cold pericentromeric regions, offers opportunities to introduce a small region of interest from B. rapa but also to reduce the B. napus QTL size. Tourrette et al. [40] found, using the data we previously obtained in Brassica allotriploids [36], that it is possible to decrease by a factor 10 the linkage drag in the cold region in BC3S1 after foreground selection for three backcrosses and background selection at the last selfing generation. This strategy seems, however, less efficient if the region of interest is located in a hot recombining region. Using an example of a large QTL for blackleg resistance [41] present in pericentromeric region of the A01 chromosome, we showed that the recombination rate in AAC hybrids may significantly reduce the QTL size. This result suggests that the simulation [40] can be transposed to oilseed rape breeding and can be highly efficient. The AAC hybrids offer the possibility to limit the introduction from B. rapa to the region of interest as well as to favor the cloning of QTL carried by B. napus in a cold recombinant region by breaking down the linkage disequilibrium. Thus, it will be of particular interest to more frequently apply this strategy in order to rapidly and more efficiently identify the genes underlying the different QTL identified for disease resistance or other important traits for B. napus (for a review, see [74]). Taking advantage of the high quality of recent B. napus genome assemblies [46,75], it becomes possible to define new markers, especially in pericentromeric regions, allowing a better assessment of recombination in these cold regions.
4.3. Development of the Strategy on Other Models
Whether the reshaping of homologous recombination observed in AAC allotriploids similarly occurs in CCA allotriploids, resulting from the cross between B. napus and its other diploid progenitor B. oleracea, remains to be deciphered (currently in progress). Even if these later CCA hybrids are more difficult to generate [76], improved recombination between homologous C chromosomes would also strongly benefit B. napus breeding programs. In fact, different studies revealed that it could be useful to introduce into B. napus new diversity from B. oleracea [77], such as clubroot resistance traits [78], and conversely from B. napus to B. oleracea [79]. It will also be interesting to determine if this modified homologous recombination landscape is also present in the other possible allotriploids from the U triangle [80], e.g., either AAB (deriving from B. juncea × B. rapa) or CCB (B. carinata × B. oleracea) hybrids, and thus possibly be useful for Brassica breeding.
5. Conclusions
Meiotic recombination is a key process that generates new genetic diversity and enables the combination of favorable alleles. However, it is strictly regulated, both in frequency and distribution. In this study, we were able to demonstrate that an AAC allotriploid hybrid presents a higher recombination rate and modified distribution compared to AA or AACC hybrids (with the latter hybrids presenting a similar recombination landscape). The boost of recombination observed in AAC hybrids allows the introduction of more and smaller genomic regions from B. rapa to B. napus compared to AACC hybrids, with a decreasing gradient of the introgression size from the centromere to the telomere in both hybrids. For breeders, the introgression of smaller genomic regions highlights the interest in using AAC allotriploid hybrids to further break the linkage disequilibrium. Additionally, the unique recombination landscape observed in AAC allotriploid hybrids will facilitate the identification of candidate genes underlying QTL of agronomical interests, most particularly in the normally cold pericentromeric regions.
Supplementary Materials
The following supplementary materials are available online at https://www.mdpi.com/article/10.3390/biology10080771/s1, Figure S1: Correlations between the relative size of introgressions (%) and their relative position to the centromere (%) for the ten A chromosomes, Table S1: Meiotic behavior of ArArCoCo-S0 synthetic and AnArCnCo, AnArCn F1 hybrids, Table S2: Correlation metrics between the relative size of introgressions (%) and their relative position to the centromere (%).
Author Contributions
Conceptualization: F.B., A.P., M.R.-G. and A.-M.C.; plant creation and care: G.T. and M.G.; genotyping data analyses: C.T., F.B., A.P., M.L.-T., R.D. and C.F.; bioinformatic analyses: L.M. and J.M.; molecular cytogenetic analyses: G.T., V.H., O.C., F.B., F.E. and A.-M.C.; writing: F.B., A.P., M.R.-G. and A.-M.C.; funding acquisition: M.R.-G. and A.-M.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the ANR-JCJC project Stirrer (ANR-19-CE20-0013, awarded to M.R.-G.). This research was also partly funded by grants from the INRAE “Biology and Plant Breeding” department (“Recipe” and “Recoptic” projects) awarded to A.-M.C. and M.R.-G. F.B.’s Ph.D. scholarship is funded by the Brittany State and the INRAE “Biology and Plant Breeding” department.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Genotyping data of the 202 SNP used for the AA and AAC hybrids were retrieved from Pelé et al. (2017) (available online: https://journals.plos.org/plosgenetics/article?id=10.1371/journal.pgen.1006794#sec024, accessed on February 2021).
Acknowledgments
We thank the Molecular Cytogenetic platform (Rennes, France, https://www6.rennes.inrae.fr/igepp/L-IGEPP/Plateformes/Cytogenetique-moleculaire (accessed on 11 July 2021)) for cytogenetic analyses, UMR INRA 1095 “GENTYANE platform” (Clermont-Ferrand, France, http://gentyane.clermont.inra.fr/ (accessed on 11 July 2021)) for the DNA extraction of segregated populations and KASPar genotyping, and the platform TraitGenetics (Gatersleben, Germany, http://www.traitgenetics.com/ (accessed on 11 July 2021)) for genotyping using the Brassica 15K Illumina infinium SNP array. We acknowledge the BrACySol BRC (INRA Ploudaniel, France, https://www6.rennes.inrae.fr/igepp/L-IGEPP/Plateformes/BrACySol (accessed on 11 July 2021)) that provided the seeds of the parental lines. We would also like to thank all the technical staff of the greenhouse for management of the plant material (especially L. Charlon, P. Rolland, J.-P. Constantin, J.-M. Lucas and F. Letertre).
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Felsenstein, J. The Evolutionary Advantage of Recombination. Genetics 1974, 78, 737–756. [Google Scholar] [CrossRef] [PubMed]
- Barton, N.H.; Charlesworth, B. Why Sex and Recombination? Science 1998, 281, 1986–1990. [Google Scholar] [CrossRef] [PubMed]
- Wijnker, E.; de Jong, H. Managing Meiotic Recombination in Plant Breeding. Trends Plant Sci. 2008, 13, 640–646. [Google Scholar] [CrossRef] [PubMed]
- Mercier, R.; Mézard, C.; Jenczewski, E.; Macaisne, N.; Grelon, M. The Molecular Biology of Meiosis in Plants. Annu. Rev. Plant Biol. 2015, 66. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Moran, E.; Santos, J.-L.; Jones, G.H.; Franklin, F.C.H. ASY1 Mediates AtDMC1-Dependent Interhomolog Recombination during Meiosis in Arabidopsis. Genes Dev. 2007, 21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferdous, M.; Higgins, J.D.; Osman, K.; Lambing, C.; Roitinger, E.; Mechtler, K.; Armstrong, S.J.; Perry, R.; Pradillo, M.; Cuñado, N.; et al. Inter-Homolog Crossing-Over and Synapsis in Arabidopsis Meiosis Are Dependent on the Chromosome Axis Protein AtASY3. PLoS Genet. 2012, 8. [Google Scholar] [CrossRef] [Green Version]
- Crismani, W.; Girard, C.; Froger, N.; Pradillo, M.; Santos, J.L.; Chelysheva, L.; Copenhaver, G.P.; Horlow, C.; Mercier, R. FANCM Limits Meiotic Crossovers. Science 2012, 336, 1588–1590. [Google Scholar] [CrossRef] [Green Version]
- Girard, C.; Chelysheva, L.; Choinard, S.; Froger, N.; Macaisne, N.; Lehmemdi, A.; Mazel, J.; Crismani, W.; Mercier, R. AAA-ATPase FIDGETIN-LIKE 1 and Helicase FANCM Antagonize Meiotic Crossovers by Distinct Mechanisms. PLoS Genet. 2015, 11, e1005369. [Google Scholar] [CrossRef] [Green Version]
- Séguéla-Arnaud, M.; Crismani, W.; Larchevêque, C.; Mazel, J.; Froger, N.; Choinard, S.; Lemhemdi, A.; Macaisne, N.; Van Leene, J.; Gevaert, K.; et al. Multiple Mechanisms Limit Meiotic Crossovers: TOP3α and Two BLM Homologs Antagonize Crossovers in Parallel to FANCM. Proc. Natl. Acad. Sci. USA 2015, 112, 4713–4718. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nageswaran, D.C.; Kim, J.; Lambing, C.; Kim, J.; Park, J.; Kim, E.-J.; Cho, H.S.; Kim, H.; Byun, D.; Park, Y.M.; et al. High Crossover Rate1 Encodes Protein Phosphatase X1 and Restricts Meiotic Crossovers in Arabidopsis. Nat. Plants 2021, 7, 452–467. [Google Scholar] [CrossRef]
- Saintenac, C.; Falque, M.; Martin, O.C.; Paux, E.; Feuillet, C.; Sourdille, P. Detailed Recombination Studies along Chromosome 3B Provide New Insights on Crossover Distribution in Wheat (Triticum Aestivum L.). Genetics 2009, 181. [Google Scholar] [CrossRef] [Green Version]
- Anderson, L.K.; Doyle, G.G.; Brigham, B.; Carter, J.; Hooker, K.D.; Lai, A.; Rice, M.; Stack, S.M. High-Resolution Crossover Maps for Each Bivalent of Zea Mays Using Recombination Nodules. Genetics 2003, 165, 849–865. [Google Scholar] [CrossRef] [PubMed]
- Felcher, K.J.; Coombs, J.J.; Massa, A.N.; Hansey, C.N.; Hamilton, J.P.; Veilleux, R.E.; Buell, C.R.; Douches, D.S. Integration of Two Diploid Potato Linkage Maps with the Potato Genome Sequence. PLoS ONE 2012, 7, e36347. [Google Scholar] [CrossRef] [Green Version]
- Choi, K.; Zhao, X.; Kelly, K.A.; Venn, O.; Higgins, J.D.; Yelina, N.E.; Hardcastle, T.J.; Ziolkowski, P.A.; Copenhaver, G.P.; Franklin, F.C.H.; et al. Arabidopsis Meiotic Crossover Hotspots Overlap with H2A.Z Nucleosomes at Gene Promoters. Nat. Genet. 2013, 45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, K.; Henderson, I.R. Meiotic Recombination Hotspots—A Comparative View. Plant J. 2015, 83, 52–61. [Google Scholar] [CrossRef]
- Choi, K.; Zhao, X.; Tock, A.J.; Lambing, C.; Underwood, C.J.; Hardcastle, T.J.; Serra, H.; Kim, J.; Cho, H.S.; Kim, J.; et al. Nucleosomes and DNA Methylation Shape Meiotic DSB Frequency in Arabidopsis Thaliana Transposons and Gene Regulatory Regions. Genome Res. 2018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marand, A.P.; Jansky, S.H.; Zhao, H.; Leisner, C.P.; Zhu, X.; Zeng, Z.; Crisovan, E.; Newton, L.; Hamernik, A.J.; Veilleux, R.E.; et al. Meiotic Crossovers Are Associated with Open Chromatin and Enriched with Stowaway Transposons in Potato. Genome Biol. 2017, 18, 203. [Google Scholar] [CrossRef] [Green Version]
- Colome-Tatche, M.; Cortijo, S.; Wardenaar, R.; Morgado, L.; Lahouze, B.; Sarazin, A.; Etcheverry, M.; Martin, A.; Feng, S.; Duvernois-Berthet, E.; et al. Features of the Arabidopsis Recombination Landscape Resulting from the Combined Loss of Sequence Variation and DNA Methylation. Proc. Natl. Acad. Sci. USA 2012, 109, 16240–16245. [Google Scholar] [CrossRef] [Green Version]
- Yelina, N.E.; Lambing, C.; Hardcastle, T.J.; Zhao, X.; Santos, B.; Henderson, I.R. DNA Methylation Epigenetically Silences Crossover Hot Spots and Controls Chromosomal Domains of Meiotic Recombination in Arabidopsis. Genes Dev. 2015, 29, 2183–2202. [Google Scholar] [CrossRef] [Green Version]
- Underwood, C.J.; Choi, K.; Lambing, C.; Zhao, X.; Serra, H.; Borges, F.; Simorowski, J.; Ernst, E.; Jacob, Y.; Henderson, I.R.; et al. Epigenetic Activation of Meiotic Recombination near Arabidopsis Thaliana Centromeres via Loss of H3K9me2 and Non-CG DNA Methylation. Genome Res. 2018, 28, 519–531. [Google Scholar] [CrossRef] [Green Version]
- Rowan, B.A.; Heavens, D.; Feuerborn, T.R.; Tock, A.J.; Henderson, I.R.; Weigel, D. An Ultra High-Density Arabidopsis Thaliana Crossover Map That Refines the Influences of Structural Variation and Epigenetic Features. Genetics 2019, 213, 771–787. [Google Scholar] [CrossRef] [Green Version]
- Smukowski, C.S.; Noor, M.A.F. Recombination Rate Variation in Closely Related Species. Heredity 2011, 107. [Google Scholar] [CrossRef] [Green Version]
- Haenel, Q.; Laurentino, T.G.; Roesti, M.; Berner, D. Meta-Analysis of Chromosome-Scale Crossover Rate Variation in Eukaryotes and Its Significance to Evolutionary Genomics. Mol. Ecol. 2018, 27, 2477–2497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nei, M. Modification of Linkage Intensity by Natural Selection. Genetics 1967, 57, 625–641. [Google Scholar] [CrossRef]
- Feldman, M.; Liu, B.; Segal, G.; Abbo, S.; Levy, A.A.; Vega, J.M. Rapid Elimination of Low-Copy DNA Sequences in Polyploid Wheat: A Possible Mechanism for Differentiation of Homoeologous Chromosomes. Genetics 1997, 147, 1381–1387. [Google Scholar] [CrossRef]
- Coop, G.; Przeworski, M. An Evolutionary View of Human Recombination. Nat. Rev. Genet. 2007, 8, 23–34. [Google Scholar] [CrossRef] [PubMed]
- Blary, A.; Jenczewski, E. Manipulation of Crossover Frequency and Distribution for Plant Breeding. Theor. Appl. Genet. 2019, 132, 575–592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bomblies, K.; Higgins, J.D.; Yant, L. Meiosis Evolves: Adaptation to External and Internal Environments. New Phytol. 2015, 208, 306–323. [Google Scholar] [CrossRef]
- Drouaud, J.; Camilleri, C.; Bourguignon, P.-Y.; Canaguier, A.; Bérard, A.; Vezon, D.; Giancola, S.; Brunel, D.; Colot, V.; Prum, B.; et al. Variation in Crossing-over Rates across Chromosome 4 of Arabidopsis Thaliana Reveals the Presence of Meiotic Recombination “Hot Spots”. Genome Res. 2006, 16, 106–114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giraut, L.; Falque, M.; Drouaud, J.; Pereira, L.; Martin, O.C.; Mézard, C. Genome-Wide Crossover Distribution in Arabidopsis Thaliana Meiosis Reveals Sex-Specific Patterns along Chromosomes. PLoS Genet. 2011, 7, e1002354. [Google Scholar] [CrossRef] [Green Version]
- Bauer, E.; Falque, M.; Walter, H.; Bauland, C.; Camisan, C.; Campo, L.; Meyer, N.; Ranc, N.; Rincent, R.; Schipprack, W.; et al. Intraspecific Variation of Recombination Rate in Maize. Genome Biol. 2013, 14, R103. [Google Scholar] [CrossRef] [PubMed]
- Ziolkowski, P.A.; Underwood, C.J.; Lambing, C.; Martinez-Garcia, M.; Lawrence, E.J.; Ziolkowska, L.; Griffin, C.; Choi, K.; Franklin, F.C.H.; Martienssen, R.A.; et al. Natural Variation and Dosage of the HEI10 Meiotic E3 Ligase Control Arabidopsis Crossover Recombination. Genes Dev. 2017, 31, 306–317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lawrence, E.J.; Gao, H.; Tock, A.J.; Lambing, C.; Blackwell, A.R.; Feng, X.; Henderson, I.R. Natural Variation in TBP-Associated Factor 4b Controls Meiotic Crossover and Germline Transcription in Arabidopsis. Curr. Biol. 2019, 29, 2676–2686.e3. [Google Scholar] [CrossRef] [PubMed]
- Pelé, A.; Rousseau-Gueutin, M.; Chèvre, A.-M. Speciation Success of Polyploid Plants Closely Relates to the Regulation of Meiotic Recombination. Front. Plant Sci. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Leflon, M.; Grandont, L.; Eber, F.; Huteau, V.; Coriton, O.; Chelysheva, L.; Jenczewski, E.; Chèvre, A.-M. Crossovers Get a Boost in Brassica Allotriploid and Allotetraploid Hybrids. Plant Cell 2010, 22, 2253–2264. [Google Scholar] [CrossRef] [Green Version]
- Pelé, A.; Falque, M.; Trotoux, G.; Eber, F.; Nègre, S.; Gilet, M.; Huteau, V.; Lodé, M.; Jousseaume, T.; Dechaumet, S.; et al. Amplifying Recombination Genome-Wide and Reshaping Crossover Landscapes in Brassicas. PLoS Genet. 2017, 13, e1006794. [Google Scholar] [CrossRef] [PubMed]
- Leflon, M.; Eber, F.; Letanneur, J.C.; Chelysheva, L.; Coriton, O.; Huteau, V.; Ryder, C.D.; Barker, G.; Jenczewski, E.; Chèvre, A.M. Pairing and Recombination at Meiosis of Brassica Rapa (AA) × Brassica Napus (AACC) Hybrids. Theor. Appl. Genet. Theor. Angew. Genet. 2006, 113, 1467–1480. [Google Scholar] [CrossRef]
- Snowdon, R.J.; Abbadi, A.; Kox, T.; Schmutzer, T.; Leckband, G. Heterotic Haplotype Capture: Precision Breeding for Hybrid Performance. Trends Plant Sci. 2015, 20, 410–413. [Google Scholar] [CrossRef]
- Tourrette, E.; Bernardo, R.; Falque, M.; Martin, O.C. Assessing by Modeling the Consequences of Increased Recombination in Recurrent Selection of Oryza Sativa and Brassica Rapa. G3 Genes Genomes Genet. 2019, 9, 4169–4181. [Google Scholar] [CrossRef] [Green Version]
- Tourrette, E.; Falque, M.; Martin, O.C. Enhancing Backcross Programs through Increased Recombination. Genet. Sel. Evol. 2021, 53, 25. [Google Scholar] [CrossRef]
- Kumar, V.; Paillard, S.; Fopa-Fomeju, B.; Falentin, C.; Deniot, G.; Baron, C.; Vallée, P.; Manzanares-Dauleux, M.J.; Delourme, R. Multi-Year Linkage and Association Mapping Confirm the High Number of Genomic Regions Involved in Oilseed Rape Quantitative Resistance to Blackleg. Theor. Appl. Genet. 2018, 131, 1627–1643. [Google Scholar] [CrossRef] [PubMed]
- Pelé, A.; Trotoux, G.; Eber, F.; Lodé, M.; Gilet, M.; Deniot, G.; Falentin, C.; Nègre, S.; Morice, J.; Rousseau-Gueutin, M.; et al. The Poor Lonesome A Subgenome of Brassica Napus Var. Darmor (AACC) May Not Survive without Its Mate. New Phytol. 2017, 213, 1886–1897. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szadkowski, E.; Eber, F.; Huteau, V.; Lodé, M.; Huneau, C.; Belcram, H.; Coriton, O.; Manzanares-Dauleux, M.J.; Delourme, R.; King, G.J.; et al. The First Meiosis of Resynthesized Brassica Napus, a Genome Blender. New Phytol. 2010, 186, 102–112. [Google Scholar] [CrossRef]
- Howell, E.C.; Barker, G.C.; Jones, G.H.; Kearsey, M.J.; King, G.J.; Kop, E.P.; Ryder, C.D.; Teakle, G.R.; Vicente, J.G.; Armstrong, S.J. Integration of the Cytogenetic and Genetic Linkage Maps of Brassica Oleracea. Genetics 2002, 161, 1225–1234. [Google Scholar] [CrossRef]
- Wang, J.; Wu, J.; Liu, S.; Bai, Y.; Mun, J.-H.; Bancroft, I.; Cheng, F.; Huang, S.; Li, X.; Hua, W.; et al. The Genome of the Mesopolyploid Crop Species Brassica Rapa. Nat. Genet. 2011, 43, 1035–1039. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rousseau-Gueutin, M.; Belser, C.; Da Silva, C.; Richard, G.; Istace, B.; Cruaud, C.; Falentin, C.; Boideau, F.; Boutte, J.; Delourme, R.; et al. Long-Read Assembly of the Brassica Napus Reference Genome Darmor-Bzh. GigaScience 2020, 9. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- De Givry, S.; Bouchez, M.; Chabrier, P.; Milan, D.; Schiex, T. Carhta Gene: Multipopulation Integrated Genetic and Radiation Hybrid Mapping. Bioinformatics 2005, 21, 1703–1704. [Google Scholar] [CrossRef] [Green Version]
- Kosambi, D.D. The Estimation of Map Distances from Recombination Values. Ann. Eugen. 1943, 12, 172–175. [Google Scholar] [CrossRef]
- Lim, K.-B.; de Jong, H.; Yang, T.-J.; Park, J.-Y.; Kwon, S.-J.; Kim, J.S.; Lim, M.-H.; Kim, J.A.; Jin, M.; Jin, Y.-M.; et al. Characterization of RDNAs and Tandem Repeats in the Heterochromatin of Brassica Rapa. Mol. Cells 2005, 19, 436–444. [Google Scholar]
- Lim, K.-B.; Yang, T.-J.; Hwang, Y.-J.; Kim, J.S.; Park, J.-Y.; Kwon, S.-J.; Kim, J.; Choi, B.-S.; Lim, M.-H.; Jin, M.; et al. Characterization of the Centromere and Peri-Centromere Retrotransposons in Brassica Rapa and Their Distribution in Related Brassica Species. Plant J. 2007, 49, 173–183. [Google Scholar] [CrossRef] [PubMed]
- Yelina, N.E.; Choi, K.; Chelysheva, L.; Macaulay, M.; de Snoo, B.; Wijnker, E.; Miller, N.; Drouaud, J.; Grelon, M.; Copenhaver, G.P.; et al. Epigenetic Remodeling of Meiotic Crossover Frequency in Arabidopsis Thaliana DNA Methyltransferase Mutants. PLoS Genet. 2012, 8, e1002844. [Google Scholar] [CrossRef] [Green Version]
- Mason, A.S.; Rousseau-Gueutin, M.; Morice, J.; Bayer, P.E.; Besharat, N.; Cousin, A.; Pradhan, A.; Parkin, I.A.P.; Chevre, A.-M.; Batley, J.; et al. Centromere Locations in Brassica A and C Genomes Revealed Through Half-Tetrad Analysis. Genetics 2016, 202, 513–523. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiong, Z.; Pires, J.C. Karyotype and Identification of All Homoeologous Chromosomes of Allopolyploid Brassica Napus and Its Diploid Progenitors. Genetics 2011, 187. [Google Scholar] [CrossRef] [Green Version]
- Song, K.; Lu, P.; Tang, K.; Osborn, T.C. Rapid Genome Change in Synthetic Polyploids of Brassica and Its Implications for Polyploid Evolution. Proc. Natl. Acad. Sci. USA 1995, 92, 7719–7723. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gaeta, R.T.; Pires, J.C.; Iniguez-Luy, F.; Leon, E.; Osborn, T.C. Genomic Changes in Resynthesized Brassica Napus and Their Effect on Gene Expression and Phenotype. Plant Cell 2007, 19. [Google Scholar] [CrossRef] [Green Version]
- Xiong, Z.; Gaeta, R.T.; Pires, J.C. Homoeologous Shuffling and Chromosome Compensation Maintain Genome Balance in Resynthesized Allopolyploid Brassica Napus. Proc. Natl. Acad. Sci. USA 2011, 108, 7908–7913. [Google Scholar] [CrossRef] [Green Version]
- Xiong, Z.; Gaeta, R.T.; Edger, P.P.; Cao, Y.; Zhao, K.; Zhang, S.; Pires, J.C. Chromosome Inheritance and Meiotic Stability in Allopolyploid Brassica Napus. G3 Genes Genomes Genet. 2021, 11. [Google Scholar] [CrossRef]
- Rousseau-Gueutin, M.; Morice, J.; Coriton, O.; Huteau, V.; Trotoux, G.; Nègre, S.; Falentin, C.; Deniot, G.; Gilet, M.; Eber, F.; et al. The Impact of Open Pollination on the Structural Evolutionary Dynamics, Meiotic Behavior, and Fertility of Resynthesized Allotetraploid Brassica Napus L. G3 Genes Genomes Genet. 2017, 7. [Google Scholar] [CrossRef] [Green Version]
- Ferreira De Carvalho, J.; Stoeckel, S.; Eber, F.; Lodé-Taburel, M.; Gilet, M.-M.; Trotoux, G.; Morice, J.; Falentin, C.; Chèvre, A.-M.; Rousseau-Gueutin, M. Untangling Structural Factors Driving Genome Stabilization in Nascent Brassica Napus Allopolyploids. New Phytol. 2021, 230, 2072–2084. [Google Scholar] [CrossRef]
- Mason, A.S.; Snowdon, R.J. Oilseed Rape: Learning about Ancient and Recent Polyploid Evolution from a Recent Crop Species. Plant Biol. 2016, 18, 883–892. [Google Scholar] [CrossRef] [PubMed]
- Kittipol, V.; He, Z.; Wang, L.; Doheny-Adams, T.; Langer, S.; Bancroft, I. Genetic Architecture of Glucosinolate Variation in Brassica Napus. J. Plant Physiol. 2019, 240. [Google Scholar] [CrossRef] [PubMed]
- Pires, J.C.; Zhao, J.; Schranz, M.E.; Leon, E.J.; Quijada, P.A.; Lukens, L.N.; Osborn, T.C. Flowering Time Divergence and Genomic Rearrangements in Resynthesized Brassica Polyploids (Brassicaceae). Biol. J. Linn. Soc. 2004, 82, 675–688. [Google Scholar] [CrossRef]
- Zhao, J.; Udall, J.A.; Quijada, P.A.; Grau, C.R.; Meng, J.; Osborn, T.C. Quantitative Trait Loci for Resistance to Sclerotinia Sclerotiorum and Its Association with a Homeologous Non-Reciprocal Transposition in Brassica Napus L. Theor. Appl. Genet. 2006, 112, 509–516. [Google Scholar] [CrossRef]
- Stein, A.; Coriton, O.; Rousseau-Gueutin, M.; Samans, B.; Schiessl, S.V.; Obermeier, C.; Parkin, I.A.P.; Chèvre, A.-M.; Snowdon, R.J. Mapping of Homoeologous Chromosome Exchanges Influencing Quantitative Trait Variation in Brassica Napus. Plant Biotechnol. J. 2017, 15, 1478–1489. [Google Scholar] [CrossRef] [Green Version]
- Diederichsen, E.; Sacristan, M.D. Disease Response of Resynthesized Brassica Napus L. Lines Carrying Different Combinations of Resistance to Plasmodiophora Brassicae Wor. Plant Breed. 1996, 115, 5–10. [Google Scholar] [CrossRef]
- Diederichsen, E.; Beckmann, J.; Schondelmeier, J.; Dreyer, F. Genetics of Clubroot Resistance in Brassica Napus “Mendel”. Acta Hortic. 2006, 706, 307–311. [Google Scholar] [CrossRef]
- Qian, W.; Chen, X.; Fu, D.; Zou, J.; Meng, J. Intersubgenomic Heterosis in Seed Yield Potential Observed in a New Type of Brassica Napus Introgressed with Partial Brassica Rapa Genome. Theor. Appl. Genet. Theor. Angew. Genet. 2005, 110. [Google Scholar] [CrossRef] [PubMed]
- Attri, R.; Rahman, H. Introgression of Allelic Diversity from Genetically Distinct Variants of Brassica Rapa into Brassica Napus Canola and Inheritance of the B. Rapa Alleles. Crop Pasture Sci. 2017, 69, 94–106. [Google Scholar] [CrossRef]
- Pan, Q.; Zhu, B.; Zhang, D.; Tong, C.; Ge, X.; Liu, S.; Li, Z. Gene Expression Changes During the Allo-/Deallopolyploidization Process of Brassica Napus. Front. Genet. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.; Mao, L.; Qiu, J.; Wang, M.; Jia, L.; Wu, D.; He, Z.; Chen, M.; Shen, Y.; Shen, E.; et al. Genome-Wide Selection Footprints and Deleterious Variations in Young Asian Allotetraploid Rapeseed. Plant Biotechnol. J. 2019, 17, 1998–2010. [Google Scholar] [CrossRef]
- Liu, Y.; Xu, A.; Liang, F.; Yao, X.; Wang, Y.; Liu, X.; Zhang, Y.; Dalelhan, J.; Zhang, B.; Qin, M.; et al. Screening of Clubroot-Resistant Varieties and Transfer of Clubroot Resistance Genes to Brassica Napus Using Distant Hybridization. Breed. Sci. 2018, 68, 258–267. [Google Scholar] [CrossRef] [Green Version]
- Balesdent, M.; Fudal, I.; Ollivier, B.; Bally, P.; Grandaubert, J.; Eber, F.; Chèvre, A.; Leflon, M.; Rouxel, T. The Dispensable Chromosome of Leptosphaeria Maculans Shelters an Effector Gene Conferring Avirulence towards Brassica Rapa. New Phytol. 2013, 198, 887–898. [Google Scholar] [CrossRef]
- Delourme, R.; Laperche, A.; Bouchet, A.-S.; Jubault, M.; Paillard, S.; Manzanares-Dauleux, M.-J.; Nesi, N. Genes and quantitative trait loci mapping for major agronomic traits in Brassica napus L. In The Brassica napus Genome; Liu, S., Snowdon, R., Chalhoub, B., Eds.; Springer International Publishing: Cham, Switzerland, 2018; pp. 41–85. ISBN 978-3-319-43692-0. [Google Scholar]
- Song, J.-M.; Guan, Z.; Hu, J.; Guo, C.; Yang, Z.; Wang, S.; Liu, D.; Wang, B.; Lu, S.; Zhou, R.; et al. Eight High-Quality Genomes Reveal Pan-Genome Architecture and Ecotype Differentiation of Brassica Napus. Nat. Plants 2020, 6, 34–45. [Google Scholar] [CrossRef] [PubMed]
- Downey, R.K.; Klassen, A.J.; Stringam, G.R. Rapeseed and mustard. In Hybridization of Crop Plants; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 1980; pp. 495–509. ISBN 978-0-89118-566-6. [Google Scholar]
- Nikzad, A.; Kebede, B.; Pinzon, J.; Bhavikkumar, J.; Wang, X.; Yang, R.-C.; Rahman, H. Potential of the C Genome of the Different Variants of Brassica Oleracea for Heterosis in Spring B. Napus Canola. Front. Plant Sci. 2020, 10. [Google Scholar] [CrossRef] [PubMed]
- Chiang, M.S.; Chiang, B.Y.; Grant, W.F. Transfer of Resistance to Race 2 of Plasmodiophora Brassicae from Brassica Napus to Cabbage (B. Oleracea Var. Capitata). I. Interspecific Hybridization between B. Napus and B. Oleracea Var. Capitata. Euphytica 1977, 26, 319–336. [Google Scholar] [CrossRef]
- Yu, H.; Li, Z.; Ren, W.; Han, F.; Yang, L.; Zhuang, M.; Lv, H.; Liu, Y.; Fang, Z.; Zhang, Y. Creation of Fertility-Restored Materials for Ogura CMS in Brassica Oleracea by Introducing Rfo Gene from Brassica Napus via an Allotriploid Strategy. Theor. Appl. Genet. 2020, 133, 2825–2837. [Google Scholar] [CrossRef] [PubMed]
- Nagaharu, U. Genome Analysis in Brassica with Special Reference to the Experimental Formation of B. Napus and Peculiar Mode of Fertilization. Jpn. J. Bot. 1935, 7, 389–452. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).