Role of Persistent Organic Pollutants in Breast Cancer Progression and Identification of Estrogen Receptor Alpha Inhibitors Using In-Silico Mining and Drug-Drug Interaction Network Approaches
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Disease Selection
2.2. Identification of the Mutated Gene
2.3. Selection and Preparation of Targeted Protein
2.4. Validation of Virtual Screening (VS) Protocol
2.5. Screening and Toxicity Detection of Pollutants
2.6. Preparation of Ligand and Molecular Docking
2.7. Collection and Mining of Natural and Synthetic Compounds
2.8. Cluster Formation
2.9. Drug-Drug Interaction (DDI) Network
3. Results and Discussion
3.1. Validation of VS and Reliability of MD
3.2. Interactions between POPs and ERα
3.3. Natural and Synthetic Compounds Collection, Mining, and Clustering
3.4. DDI Network
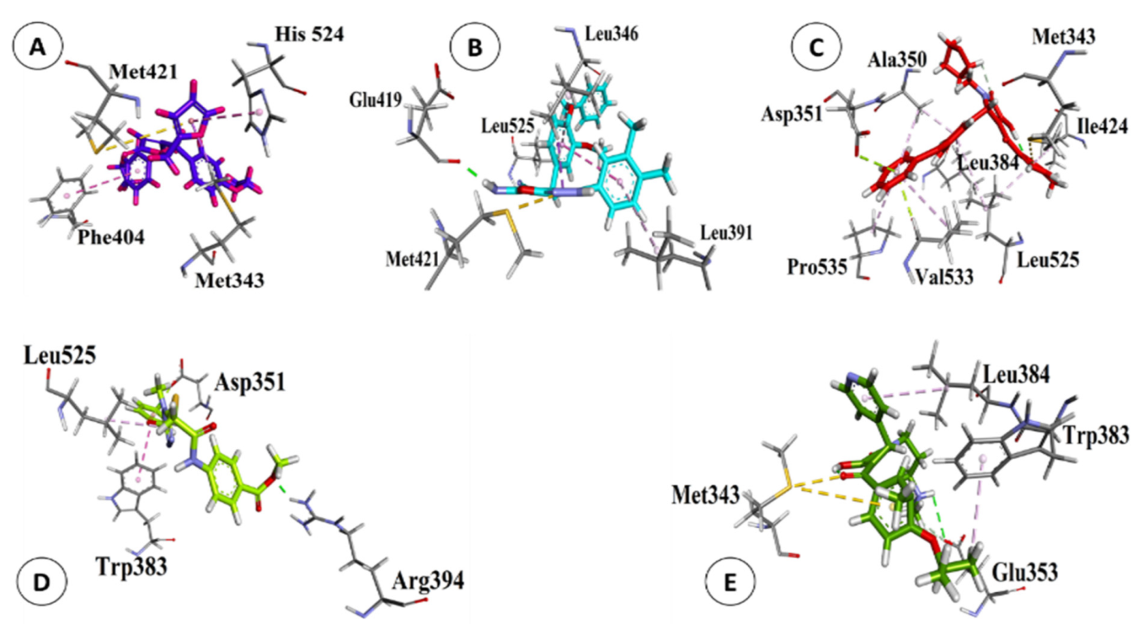
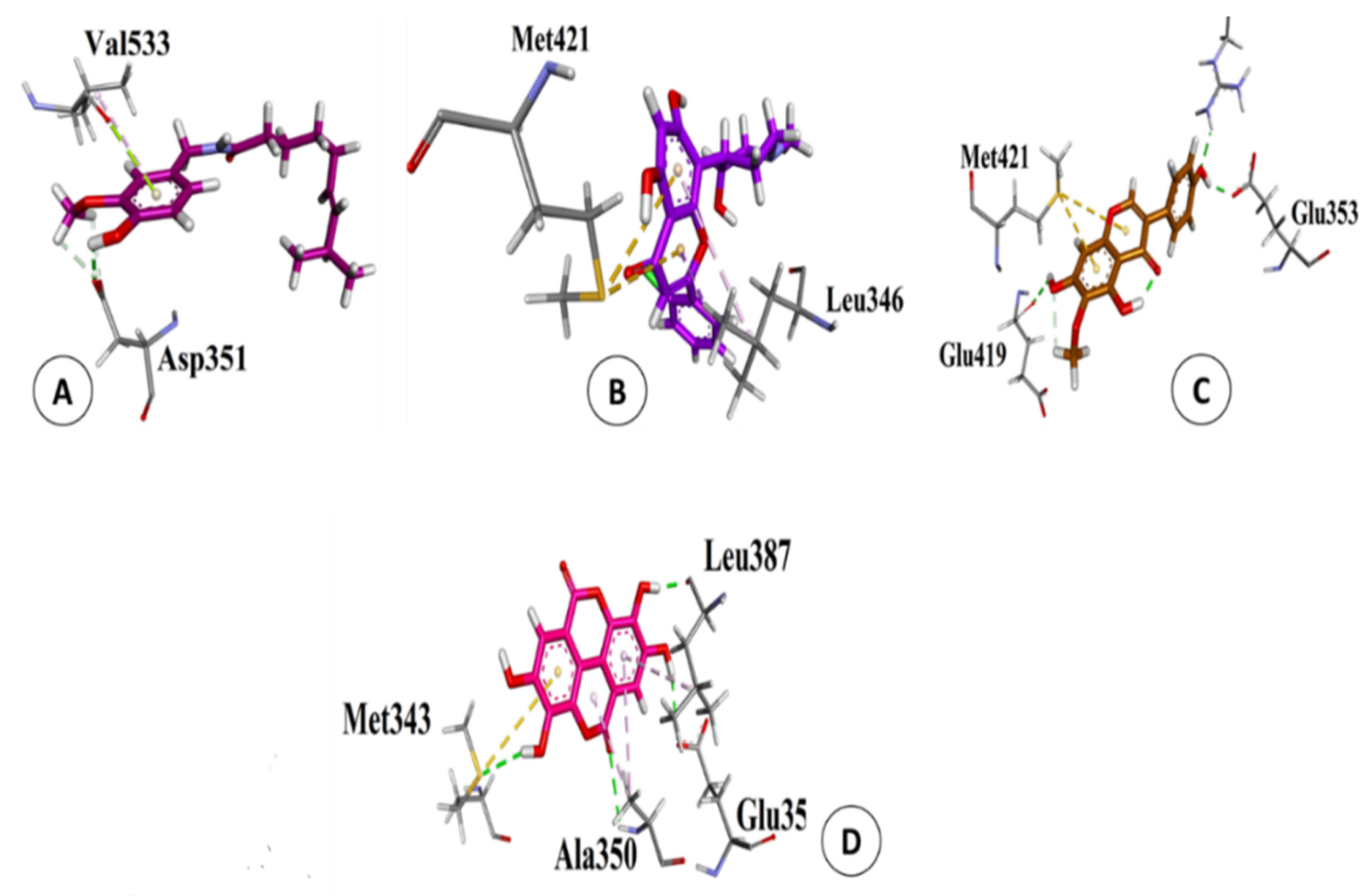
3.5. Validation of Natural and Synthetic Compounds
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- El-Shahawi, M.S.; Hamza, A.; Bashammakh, A.S.; Al-Saggaf, W.T. An overview on the accumulation, distribution, transformations, toxicity and analytical methods for the monitoring of persistent organic pollutants. Talanta 2010, 80, 1587–1597. [Google Scholar] [CrossRef]
- Hinkebein, T. Desalination: Limitations and Challenges; National Academies Press (US): Washington, DC, USA, 2004; ISBN 0-309-53173-X. [Google Scholar]
- Blanchard, M.; Teil, M.J.; Ollivon, D.; Legenti, L.; Chevreuil, M. Polycyclic aromatic hydrocarbons and polychlorobiphenyls in wastewaters and sewage sludges from the Paris area (France). Environ. Res. 2004, 95, 184–197. [Google Scholar] [CrossRef]
- Bräuner, E.V.; Raaschou-Nielsen, O.; Gaudreau, E.; Leblanc, A.; Tjønneland, A.; Overvad, K.; Sørensen, M. Predictors of adipose tissue concentrations of organochlorine pesticides in a general Danish population. J. Expo. Sci. Environ. Epidemiol. 2012, 22, 52–59. [Google Scholar] [CrossRef]
- Jones, K.C.; de Voogt, P. Persistent organic pollutants (POPs): State of the science. Environ. Pollut. 1999, 100, 209–221. [Google Scholar] [CrossRef]
- Bonefeld-Jorgensen, E.C.; Long, M.; Bossi, R.; Ayotte, P.; Asmund, G.; Krüger, T.; Ghisari, M.; Mulvad, G.; Kern, P.; Nzulumiki, P.; et al. Perfluorinated compounds are related to breast cancer risk in greenlandic inuit: A case control study. Environ. Health A Glob. Access Sci. Source 2011, 10, 1–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghosh, S.; Murinova, L.; Trnovec, T.; Christopher, A.L.; Washington, K.; Partha, S.M.; Sisir, K.D. Biomarkers linking PCB exposure and obesity. Curr. Pharm. Biotechnol. 2014, 15, 1058–1068. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soto, A.M.; Chung, K.L.; Sonnenschein, C. The pesticides endosulfan, toxaphene, and dieldrin have estrogenic effects on human estrogen-sensitive cells. Environ. Health Perspect. 1994, 102, 380–383. [Google Scholar] [CrossRef]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA. Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Subramanian, A.; Salhab, M.; Mokbel, K. Oestrogen producing enzymes and mammary carcinogenesis: A review. Breast Cancer Res. Treat. 2008, 111, 191–202. [Google Scholar] [CrossRef]
- Nagel, A.; Szade, J.; Iliszko, M.; Elzanowska, J.; Welnicka-Jaskiewicz, M.; Skokowski, J.; Stasilojc, G.; Bigda, J.; Sadej, R.; Zaczek, A.; et al. Clinical and biological significance of ESR1 gene alteration and estrogen receptors isoforms expression in breast cancer patients. Int. J. Mol. Sci. 2019, 20, 1881. [Google Scholar] [CrossRef] [Green Version]
- Ennour-Idrissi, K.; Ayotte, P.; Diorio, C. Persistent organic pollutants and breast cancer: A systematic review and critical appraisal of the literature. Cancers 2019, 11, 1063. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tong, C.W.S.; Wu, M.; Cho, W.C.S.; To, K.K.W. Recent advances in the treatment of breast cancer. Front. Oncol. 2018, 8. [Google Scholar] [CrossRef] [Green Version]
- Munir, A.; Elahi, S.; Masood, N. Clustering based drug-drug interaction networks for possible repositioning of drugs against EGFR mutations: Clustering based DDI networks for EGFR mutations. Comput. Biol. Chem. 2018, 75, 24–31. [Google Scholar] [CrossRef] [PubMed]
- Zainab, B.; Ayaz, Z.; Munir, A.; Hossam Mahmoud, A.; Soliman Elsheikh, M.; Mehmood, A.; Khan, S.; Rizwan, M.; Jahangir, K.; Mehmood Abbasi, A. Repositioning of strongly integrated drugs against achromatopsia (CNGB3). J. King Saud Univ. Sci. 2020, 32, 1793–1811. [Google Scholar] [CrossRef]
- Cronin, M.T.D.T.D.D.; Jaworska, J.S.; Walker, J.D.; Comber, M.H.I.; Watts, C.D.; Worth, A.P.P.; Basketter, D.; Casati, S.; Gerberick, G.F.; Griem, P.; et al. Report of the EPAA-ECVAM workshop on the validation of Integrated Testing Strategies (ITS). Altern. Lab. Anim. 2008, 27, 258–284. [Google Scholar] [CrossRef]
- Edelman, L.B.; Eddy, J.A.; Price, N.D. In silico models of cancer. Wiley Interdiscip. Rev. Syst. Biol. Med. 2010, 2, 438–459. [Google Scholar] [CrossRef] [PubMed]
- Nath, O.; Singh, A.; Singh, I.K. In-Silico drug discovery approach targeting receptor tyrosine kinase-like orphan receptor 1 for cancer treatment. Sci. Rep. 2017, 7, 1–10. [Google Scholar] [CrossRef]
- Cassidy, C.E.; Setzer, W.N. Cancer-relevant biochemical targets of cytotoxic Lonchocarpus flavonoids: A molecular docking analysis. J. Mol. Model. 2010, 16, 311–326. [Google Scholar] [CrossRef] [PubMed]
- Majeed, A.I.; Ullah, A.; Jadoon, M.; Ahmad, W.; Riazuddin, S. Screening, diagnosis and genetic study of breast cancer patients in Pakistan. Pakistan J. Med. Sci. 2020, 36, 16–20. [Google Scholar] [CrossRef] [Green Version]
- Safran, M.; Dalah, I.; Alexander, J.; Rosen, N.; Iny Stein, T.; Shmoish, M.; Nativ, N.; Bahir, I.; Doniger, T.; Krug, H.; et al. Gene Cards Version 3: The human gene integrator. Database 2010, 2010, 1–16. [Google Scholar] [CrossRef]
- Rose, P.W.; Prlić, A.; Altunkaya, A.; Bi, C.; Bradley, A.R.; Christie, C.H.; Di Costanzo, L.; Duarte, J.M.; Dutta, S.; Feng, Z.; et al. The RCSB protein data bank: Integrative view of protein, gene and 3D structural information. Nucleic Acids Res. 2017, 45, D271–D281. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Lou, C.; Sun, L.; Li, J.; Cai, Y.; Wang, Z.; Li, W.; Liu, G.; Tang, Y. AdmetSAR 2.0: Web-service for prediction and optimization of chemical ADMET properties. Bioinformatics 2019, 35, 1067–1069. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Bryant, S.H.; Cheng, T.; Wang, J.; Gindulyte, A.; Shoemaker, B.A.; Thiessen, P.A.; He, S.; Zhang, J. PubChem BioAssay: 2017 update. Nucleic Acids Res. 2017, 45, D955–D963. [Google Scholar] [CrossRef] [PubMed]
- Wadood, A.; Ahmed, N.; Shah, L.; Ahmad, A.; Hassan, H.; Shams, S. In-silico drug design: An approach which revolutionarised the drug discovery process. OA Drug Des. Deliv. 2013, 1. [Google Scholar] [CrossRef]
- Sapundzhi, F.I.; Dzimbova, T.A. Computer modelling of the CB1 receptor by Molecular Operating Environment. Bulg. Chem. Commun. 2018, 50, 15–19. [Google Scholar]
- Hall, M.; Frank, E.; Holmes, G.; Pfahringer, B.; Reutemann, P.; Witten, I.H. The WEKA data mining software: An update. ACM SIGKDD Explor. Newsl. 2009, 11, 10–18. [Google Scholar] [CrossRef]
- Bastian, M.; Heymann, S.; Jacomy, M. Gephi: An open source software for exploring and manipulating networks. BT-International AAAI Conference on Weblogs and Social. In Proceedings of the 3rd international AAAI conference on weblogs and social media, San Jose, CA, USA, 17–20 May 2009; pp. 361–362. [Google Scholar]
- Réau, M.; Langenfeld, F.; Zagury, J.F.; Lagarde, N.; Montes, M. Decoys selection in benchmarking datasets: Overview and perspectives. Front. Pharmacol. 2018, 9. [Google Scholar] [CrossRef]
- Prieto-Martínez, F.D.; Arciniega, M.; Medina-Franco, J.L. Acoplamiento molecular: Avances recientes y retos. TIP Rev. Espec. Cienc. Químico Biológicas 2018, 21, 65–87. [Google Scholar] [CrossRef] [Green Version]
- Hathout, R.M.; Abdelhamid, S.G.; El-Housseiny, G.S.; Metwally, A.A. Comparing cefotaxime and ceftriaxone in combating meningitis through nose-to-brain delivery using bio/chemoinformatics tools. Sci. Rep. 2020, 10, 1–7. [Google Scholar] [CrossRef]
- Belpomme, D.; Irigaray, P.; Hardell, L.; Clapp, R.; Montagnier, L.; Epstein, S.; Sasco, A.J. The multitude and diversity of environmental carcinogens. Environ. Res. 2007, 105, 414–429. [Google Scholar] [CrossRef]
- Arrebola, J.P.; Fernández-Rodríguez, M.; Artacho-Cordón, F.; Garde, C.; Perez-Carrascosa, F.; Linares, I.; Tovar, I.; González-Alzaga, B.; Expósito, J.; Torne, P.; et al. Associations of persistent organic pollutants in serum and adipose tissue with breast cancer prognostic markers. Sci. Total Environ. 2016, 566–567, 41–49. [Google Scholar] [CrossRef] [PubMed]
- Aubé, M.; Larochelle, C.; Ayotte, P. Differential effects of a complex organochlorine mixture on the proliferation of breast cancer cell lines. Environ. Res. 2011, 111, 337–347. [Google Scholar] [CrossRef] [PubMed]
- Fitriah, A.; Holil, K.; Syarifah, U.; Fitriyah, F.; Utomo, D.H. In silico approach for revealing the anti-breast cancer and estrogen receptor alpha inhibitory activity of Artocarpus altilis. AIP Conf. Proc. 2018, 2021, 1–7. [Google Scholar] [CrossRef]
- Science, E.; Ballschmiter, K. Persistent organic pollutants 274 review articles review articles: Persistent Organic Pollutants Man-made Chemicals Found in Remote Areas of the World. Environ. Sci. Pollut. Res. 2019. [Google Scholar] [CrossRef]
- Zheng, X.; Sun, Q.; Wang, S.; Li, X.; Liu, P.; Yan, Z.; Kong, X.; Fan, J. Advances in studies on toxic effects of short-chain chlorinated paraffins (SCCPs) and characterization of environmental pollution in china. Arch. Environ. Contam. Toxicol. 2020, 78, 501–512. [Google Scholar] [CrossRef]
- UNEP. Stockholm Convention on Persistent Organic Pollutants (POPs): Text and Annexesas Amended in 2009.Secretariat of the Stockholm Convention on Persistent Organic Pollutants; United Nations Environment Programme (UNEP): Nairobi, Kenya; International Environment House: Geneva, Switzerland, 2009. [Google Scholar]
- Fiedler, H. Short-chain chlorinated paraffins: Production, use and international regulations. In Chlorinated Paraffins; Springer: Berlin, Heidelberg, 2010; pp. 1–40. [Google Scholar] [CrossRef]
- Gong, Y.; Zhang, H.; Geng, N.; Xing, L.; Fan, J.; Luo, Y.; Song, X.; Ren, X.; Wang, F.; Chen, J. Short-chain chlorinated paraffins (SCCPs) induced thyroid disruption by enhancement of hepatic thyroid hormone influx and degradation in male Sprague Dawley rats. Sci. Total Environ. 2018, 625, 657–666. [Google Scholar] [CrossRef] [PubMed]
- Harno, E.; Ramamoorthy, T.G.; Coll, A.P.; White, A. POMC: The physiological power of hormone processing. Physiol. Rev. 2018, 98, 2381–2430. [Google Scholar] [CrossRef]
- Jiang, Y.; Yang, S.; Liu, J.; Ren, T.; Zhang, Y.; Sun, X. Degradation of hexabromocyclododecane (HBCD) by nanoscale zero-valent aluminum (nZVAl). Chemosphere 2020, 244, 125536. [Google Scholar] [CrossRef]
- Szabo, D.T. Hexabromocyclododecane. Encycl. Toxicol. Third Ed. 2014, 2, 864–868. [Google Scholar] [CrossRef]
- Maras, M.; Vanparys, C.; Muylle, F.; Robbens, J.; Berger, U.; Barber, J.L.; Blust, R.; De Coen, W. Estrogen-like properties of fluorotelomer alcohols as revealed by MCF-7 breast cancer cell proliferation. Environ. Health Perspect. 2006, 114, 100–105. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Ng, S.; Adesanya-Famuiya, O.; Anderson, K.; Bondy, C.A. Testosterone inhibits estrogen-induced mammary epithelial proliferation and suppresses estrogen receptor expression. FASEB J. 2000, 14, 1725–1730. [Google Scholar] [CrossRef] [Green Version]
- Folkerd, E.; Dowsett, M. Sex hormones and breast cancer risk and prognosis. Breast 2013, 22, S38–S43. [Google Scholar] [CrossRef] [PubMed]
- Somboonporn, W.; Davis, S.R. Testosterone effects on the breast: Implications for testosterone therapy for women. Endocr. Rev. 2004, 25, 374–388. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pasqualini, J.R.; Chetrite, G.S.; Pasqualini, J.R. Biological responses of progestogen metabolites in normal and cancerous human breast. Horm. Mol. Biol. Clin. Investig. 2010, 3, 427–435. [Google Scholar] [CrossRef] [PubMed]
- Trabert, B.; Sherman, M.E.; Kannan, N.; Stanczyk, F.Z. Progesterone and breast cancer. Endocr. Rev. 2019, 41, 320–344. [Google Scholar] [CrossRef] [PubMed]
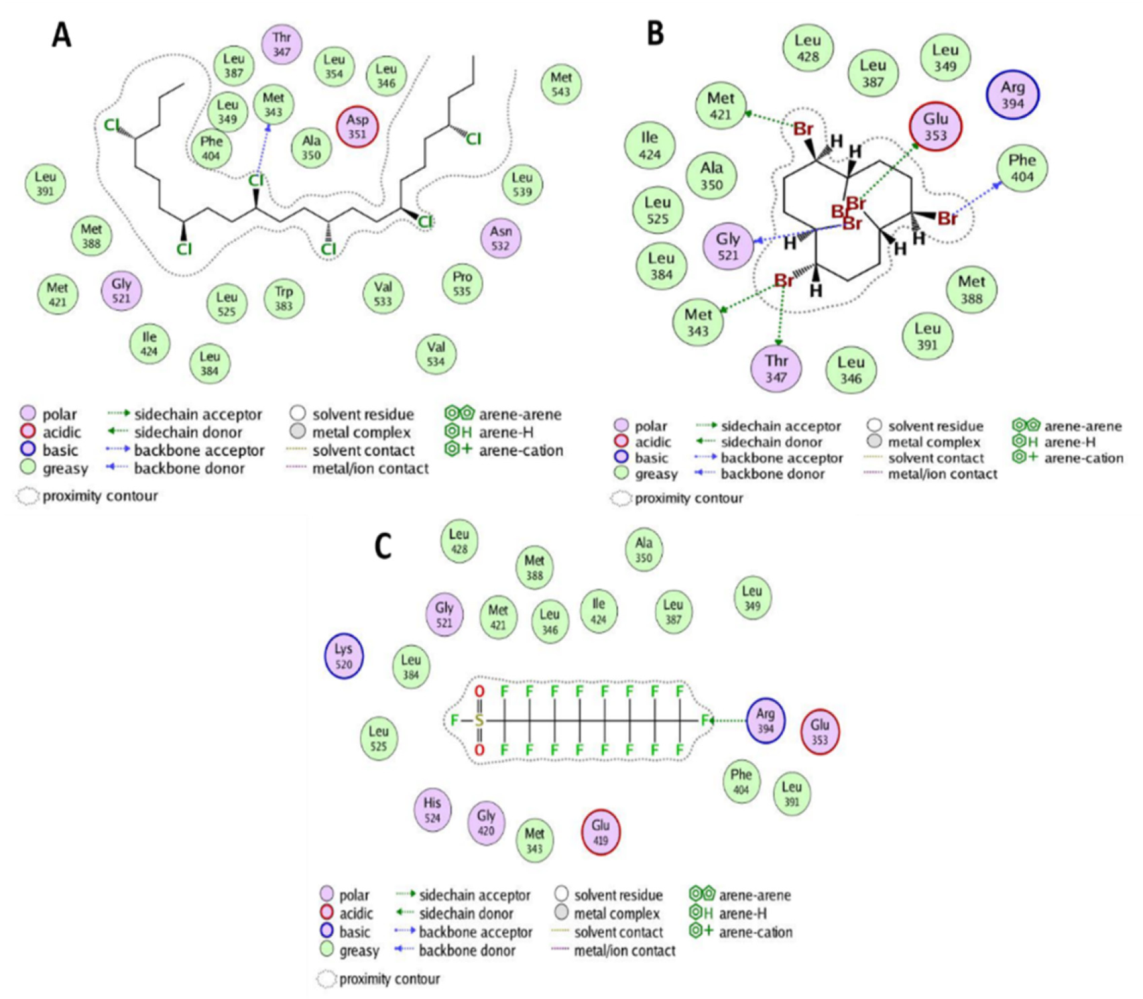

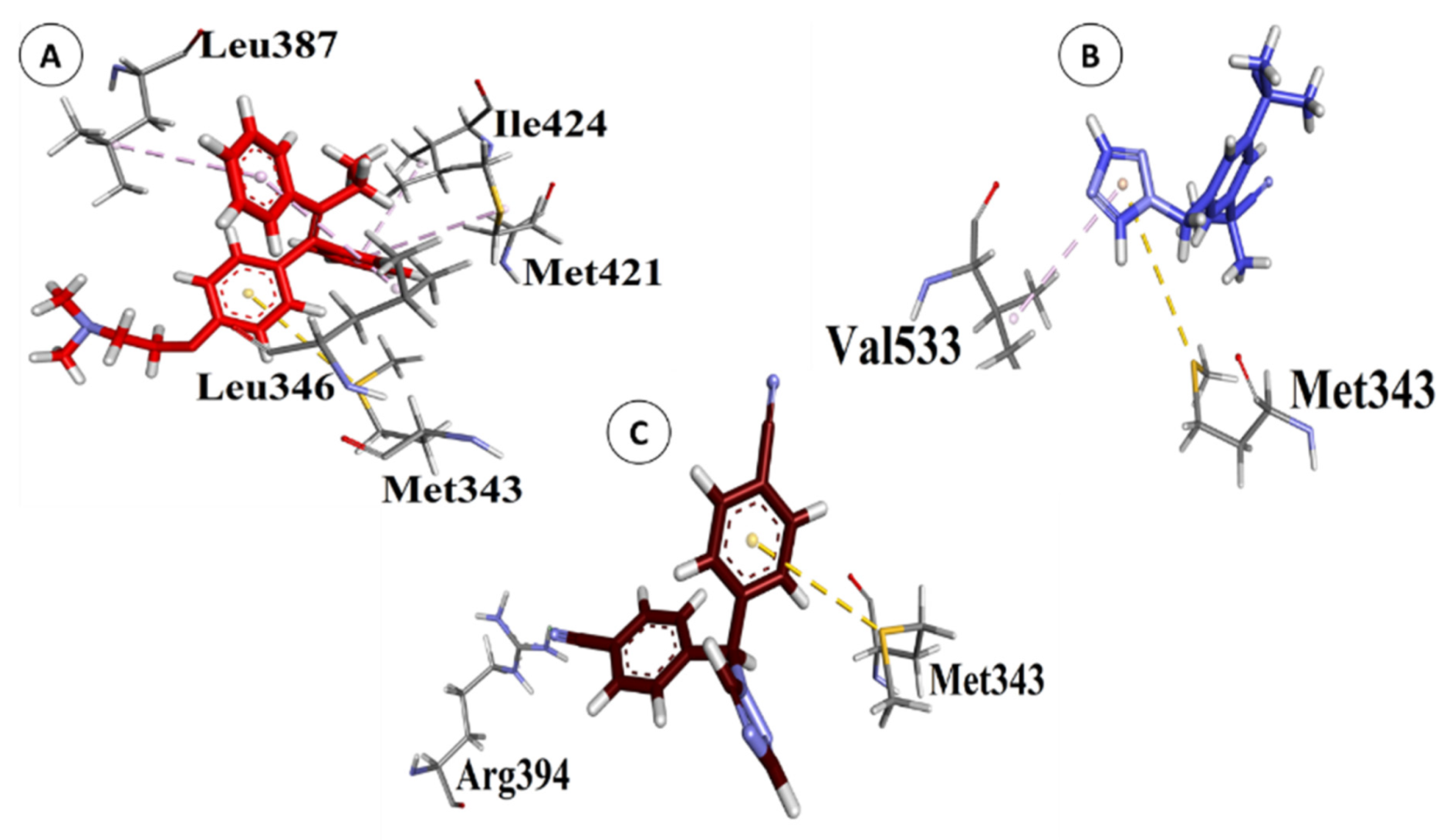
| S. # | Chemical Name | Structures | B.E. (kcal/mol) | RMSD (Å) |
|---|---|---|---|---|
| 1. | Short-chained chlorinated paraffin’s | 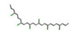 | −31.95 | 1.975 |
| 2. | HBCD (Hexabromocyclododecane) |  | −18.92 | 0.831 |
| 3. | PFOSF (Perfluorooctanesulfonyl fluoride) |  | −17.58 | 1.920 |
| 4. | Dieldrin | 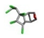 | −17.22 | 0.635 |
| 5. | DDT (dichloro-diphenyl-trichloroethane) | 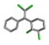 | −17.15 | 1.123 |
| 6. | PFOS (perfluorooctanesulfonic acid) |  | −17.13 | 0.884 |
| 7. | Endrin | 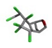 | −17.01 | 0.979 |
| 8. | Aldrin |  | −16.19 | 1.312 |
| 9. | Hexa bromodiphenyl ethers |  | −15.72 | 1.099 |
| 10. | Hexabromobiphenyl | 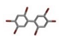 | −15.67 | 1.541 |
| 11. | Penta-bromodiphenyl ethers | 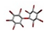 | −15.39 | 1.261 |
| 12. | PCDDs (Polychlorinated dibenzodioxins) | 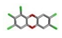 | −15.01 | 1.345 |
| 13. | Chlordane | 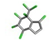 | −14.85 | 0.937 |
| 14. | Toxaphene | 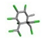 | −14.55 | 0.818 |
| 15. | PCDFs (polychlorinated dibenzofurans) | 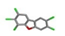 | −14.39 | 1.885 |
| 16. | Beta endosulfans | 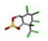 | −14.26 | 1.791 |
| 17. | PCBs (Polychlorinated biphenyls) | 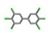 | −14.16 | 1.233 |
| 18. | α-endosulfans | 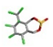 | −14.09 | 0.978 |
| 19. | Heptachlor | 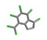 | −13.70 | 0.999 |
| 20. | Lindane | 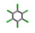 | −12.91 | 0.631 |
| 21. | Chlorinated_naphthalenes | 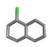 | −12.58 | 0.903 |
| 22. | Mirex | 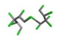 | −12.39 | 1.508 |
| 23. | Chlordecone | 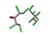 | −11.87 | 1.207 |
| 24. | Pentachlorophenol |  | −9.566 | 1.215 |
| 25. | Hexachlorobenzene | 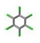 | −9.501 | 1.212 |
| 26. | Pentachlorobenzen | 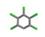 | −9.500 | 1.596 |
| 27. | Hexachlorobutadiene | 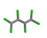 | −8.650 | 1.090 |
| Networks | AD | AWD | ND | GD | Ml | APL | N | E |
|---|---|---|---|---|---|---|---|---|
| 1. | 2.306 | 2.860 | 1.000 | 0.005 | 0.518 | 1.000 | 507.0 | 1169 |
| 2. | 2.120 | 2.519 | 1.000 | 0.008 | 0.524 | 1.000 | 266.0 | 564.0 |
| 3. | 2.070 | 2.649 | 1.000 | 0.011 | 0.538 | 1.000 | 185.0 | 383.0 |
| 4. | 1.531 | 2.266 | 1.000 | 0.024 | 0.613 | 1.000 | 64.00 | 98.00 |
| 5. | 2.325 | 2.476 | 1.000 | 0.003 | 0.509 | 1.000 | 923.0 | 2146 |
| 6. | 2.101 | 3.005 | 1.000 | 0.011 | 0.571 | 1.000 | 188.0 | 395.0 |
| 7. | 2.275 | 3.028 | 1.000 | 0.005 | 0.525 | 1.000 | 469.0 | 1067 |
| 8. | 1.629 | 2.056 | 1.000 | 0.001 | 0.552 | 1.000 | 107.0 | 181.0 |
| 9. | 2.371 | 2.792 | 1.000 | 0.006 | 0.500 | 1.000 | 367.0 | 870.0 |
| 10. | 2.155 | 2.662 | 1.000 | 0.004 | 0.501 | 1.000 | 541.0 | 1166 |
| 11. | 1.518 | 1.789 | 1.000 | 0.007 | 0.528 | 1.000 | 218.0 | 331.0 |
| 12. | 2.220 | 2.967 | 1.000 | 0.007 | 0.500 | 1.000 | 300.0 | 666.0 |
| FSIN | 2.325 | 2.476 | 1.000 | 0.003 | 0.503 | 1.000 | 923.0 | 2146 |
| S. # | Names | Structures | B.E. (kcal/mol) | RMSD (Å) |
|---|---|---|---|---|
| Positive control | ||||
| 1 | Tamoxifen | 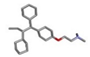 | −31.26 | 1.006 |
| 2 | Arimidex | 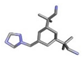 | −22.46 | 1.130 |
| 3 | Letrozole | 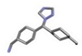 | −20.97 | 1.385 |
| Negative control | ||||
| 1 | Progestrone | 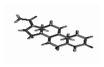 | −23.53 | 1.080 |
| 2 | Testosterone |  | −23.95 | 0.859 |
| Synthetic compounds | ||||
| 1 | 5-(4-butoxyphenyl)-4-(2,3-dihydro-1,4-benzodioxin-6-ylcarbonyl)-1-(2-furylmethyl)-3-hydroxy-1,5-dihydro-2H-pyrrol-2-one [ZINC08441573] |  | −32.47 | 1.521 |
| 2 | 2-(4-tert-butylphenyl)-3H-quinazolin-4-one [ZINC00664754] | 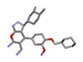 | −31.38 | 1.575 |
| 3 | (2S)-4-hydroxy-3-(5-methylfuran-2-carbonyl)-1-[[(2R)-oxolan-2-yl]methyl]-2-(3-phenoxyphenyl)-2H-pyrrol-5-one [ZINC00702695] | 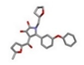 | −30.35 | 1.820 |
| 4 | (1R,6S)-6-[[2-[4-(4-methylphenyl) piperazine-1-carbonyl]phenyl]carbamoyl]cyclohex-3-ene-1-carboxylic acid [ZINC00627464] | 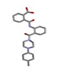 | −30.31 | 1.650 |
| 5 | (5S)-1-[3-(dimethylamino)propyl]-4-[hydroxy-(3-methyl-4-propan-2-yloxyphenyl)methylidene]-5-pyridin-4-ylpyrrolidine-2,3-dione [ZINC08440501] | 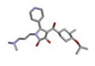 | −28.76 | 1.686 |
| Natural compounds | ||||
| 1 | Capsaicin [ZINC01530575] | 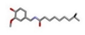 | −24.90 | 0.905 |
| 2 | Flavopiridol [ZINC21288966] | 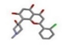 | −24.76 | 1.875 |
| 3 | Tectorigenin [ZINC00899915] | 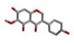 | −21.71 | 0.994 |
| 4 | Ellagic acid [ZINC03872446] | 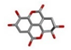 | −16.36 | 0.996 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zainab, B.; Ayaz, Z.; Rashid, U.; Al Farraj, D.A.; Alkufeidy, R.M.; AlQahtany, F.S.; Aljowaie, R.M.; Abbasi, A.M. Role of Persistent Organic Pollutants in Breast Cancer Progression and Identification of Estrogen Receptor Alpha Inhibitors Using In-Silico Mining and Drug-Drug Interaction Network Approaches. Biology 2021, 10, 681. https://doi.org/10.3390/biology10070681
Zainab B, Ayaz Z, Rashid U, Al Farraj DA, Alkufeidy RM, AlQahtany FS, Aljowaie RM, Abbasi AM. Role of Persistent Organic Pollutants in Breast Cancer Progression and Identification of Estrogen Receptor Alpha Inhibitors Using In-Silico Mining and Drug-Drug Interaction Network Approaches. Biology. 2021; 10(7):681. https://doi.org/10.3390/biology10070681
Chicago/Turabian StyleZainab, Bibi, Zainab Ayaz, Umer Rashid, Dunia A. Al Farraj, Roua M. Alkufeidy, Fatmah S. AlQahtany, Reem M. Aljowaie, and Arshad Mehmood Abbasi. 2021. "Role of Persistent Organic Pollutants in Breast Cancer Progression and Identification of Estrogen Receptor Alpha Inhibitors Using In-Silico Mining and Drug-Drug Interaction Network Approaches" Biology 10, no. 7: 681. https://doi.org/10.3390/biology10070681
APA StyleZainab, B., Ayaz, Z., Rashid, U., Al Farraj, D. A., Alkufeidy, R. M., AlQahtany, F. S., Aljowaie, R. M., & Abbasi, A. M. (2021). Role of Persistent Organic Pollutants in Breast Cancer Progression and Identification of Estrogen Receptor Alpha Inhibitors Using In-Silico Mining and Drug-Drug Interaction Network Approaches. Biology, 10(7), 681. https://doi.org/10.3390/biology10070681






