DNA Damage Repair Gene Set as a Potential Biomarker for Stratifying Patients with High Tumor Mutational Burden
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Mutation Data
2.2. Mutation Matrix Construction
2.3. Examination of the Association between the Mutation Status of DDR Genes and TMB
2.4. Identification of a DDR Gene Set as a Potential Biomarker
2.4.1. Effect Size Calculation
2.4.2. Stepwise Selection Method
3. Results
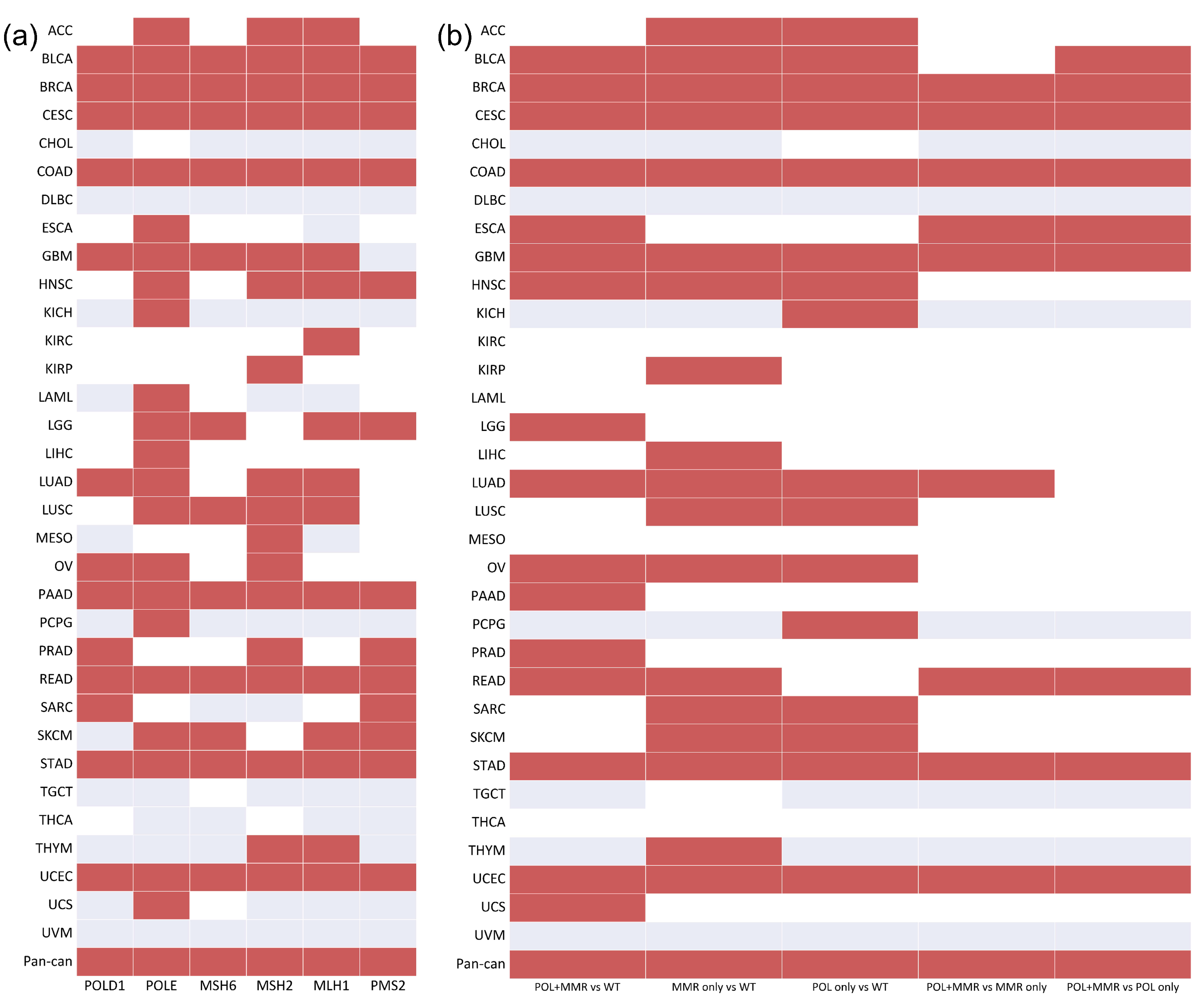
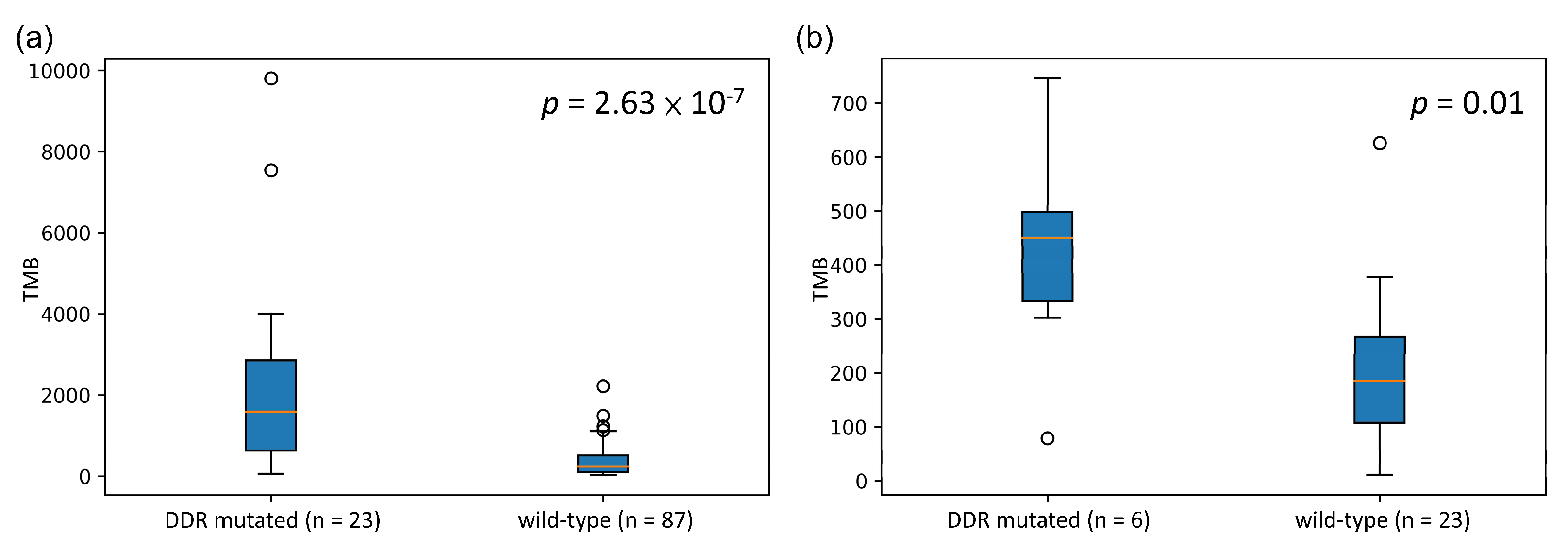
3.1. Association between the Mutation Status of the DDR Genes and TMB
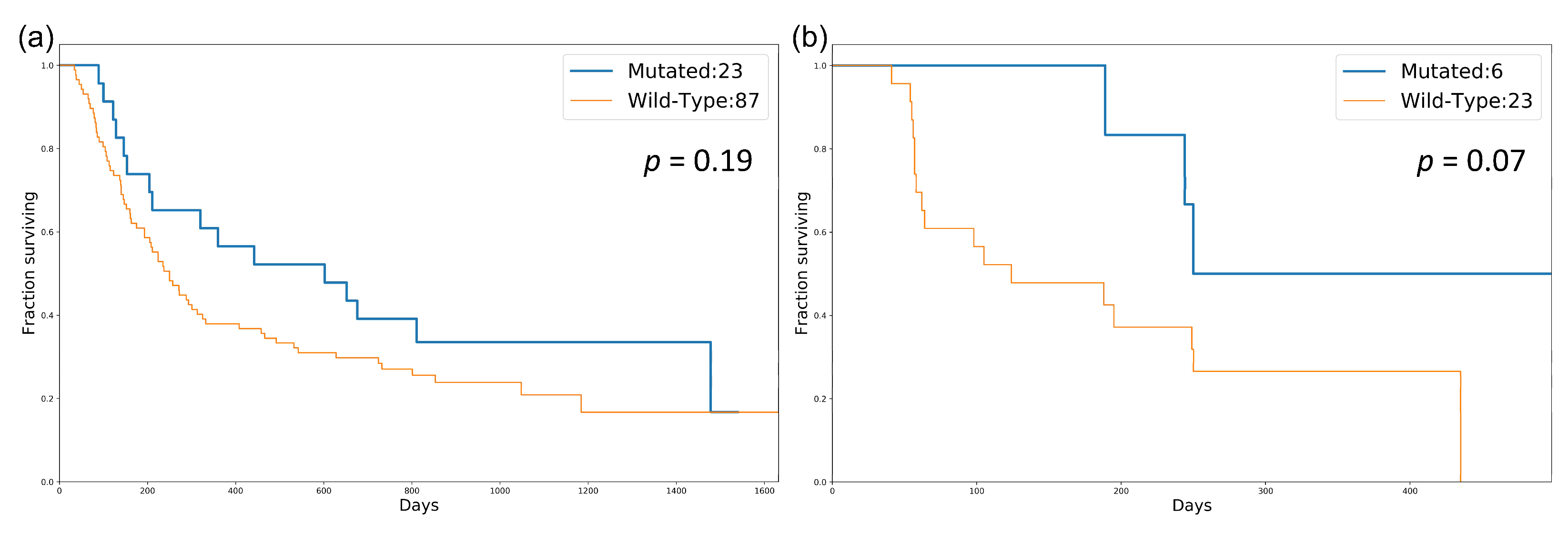
3.2. Identification of a DDR Gene Set as a Potential Biomarker
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Havel, J.J.; Chowell, D.; Chan, T.A. The evolving landscape of biomarkers for checkpoint inhibitor immunotherapy. Nat. Rev. Cancer 2019, 19, 133–150. [Google Scholar] [CrossRef] [PubMed]
- Legrand, F.A.; Gandara, D.R.; Mariathasan, S.; Powles, T.; He, X.; Zhang, W.; Jhunjhunwala, S.; Nickles, D.; Bourgon, R.; Schleifman, E.; et al. Association of high tissue TMB and atezolizumab efficacy across multiple tumor types. J. Clin. Oncol. 2018, 36, 12000. [Google Scholar] [CrossRef]
- Goodman, A.M.; Kato, S.; Bazhenova, L.; Patel, S.P.; Frampton, G.M.; Miller, V.; Stephens, P.J.; Daniels, G.A.; Kurzrock, R. Tumor Mutational Burden as an Independent Predictor of Response to Immunotherapy in Diverse Cancers. Mol. Cancer Ther. 2017, 16, 2598–2608. [Google Scholar] [CrossRef] [PubMed]
- Gibney, G.T.; Weiner, L.M.; Atkins, M.B. Predictive biomarkers for checkpoint inhibitor-based immunotherapy. Lancet Oncol. 2016, 17, e542–e551. [Google Scholar] [CrossRef]
- Wu, H.-X.; Wang, Z.-X.; Zhao, Q.; Wang, F.; Xu, R.-H. Designing gene panels for tumor mutational burden estimation: The need to shift from ‘correlation’ to ‘accuracy’. J. Immunother. Cancer 2019, 7, 206. [Google Scholar] [CrossRef] [PubMed]
- Allgäuer, M.; Budczies, J.; Christopoulos, P.; Endris, V.; Lier, A.; Rempel, E.; Volckmar, A.-L.; Kirchner, M.; Von Winterfeld, M.; Leichsenring, J.; et al. Implementing tumor mutational burden (TMB) analysis in routine diagnostics—A primer for molecular pathologists and clinicians. Transl. Lung Cancer Res. 2018, 7, 703–715. [Google Scholar] [CrossRef] [PubMed]
- Chalmers, Z.R.; Connelly, C.F.; Fabrizio, D.; Gay, L.; Ali, S.M.; Ennis, R.; Schrock, A.; Campbell, B.; Shlien, A.; Chmielecki, J.; et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med. 2017, 9, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Budczies, J.; Allgäuer, M.; Litchfield, K.; Rempel, E.; Christopoulos, P.; Kazdal, D.; Endris, V.; Thomas, M.; Fröhling, S.; Peters, S.; et al. Optimizing panel-based tumor mutational burden (TMB) measurement. Ann. Oncol. 2019, 30, 1496–1506. [Google Scholar] [CrossRef] [PubMed]
- De Velasco, G.; Wankowicz, S.A.; Madison, R.; Ali, S.M.; Norton, C.; Duquette, A.; Ross, J.S.; Bossé, D.; Lalani, A.-K.A.; Miller, V.A.; et al. Targeted genomic landscape of metastases compared to primary tumours in clear cell metastatic renal cell carcinoma. Br. J. Cancer 2018, 118, 1238–1242. [Google Scholar] [CrossRef] [PubMed]
- Garofalo, A.; Sholl, L.; Reardon, B.; Taylor-Weiner, A.; Amin-Mansour, A.; Miao, D.; Liu, D.; Oliver, N.; MacConaill, L.; Ducar, M.; et al. The impact of tumor profiling approaches and genomic data strategies for cancer precision medicine. Genome Med. 2016, 8, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Zehir, A.; Benayed, R.; Shah, R.; Syed, A.; Middha, S.; Kim, H.R.; Srinivasan, P.; Gao, J.; Chakravarty, D.; Devlin, S.M.; et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat. Med. 2017, 23, 703–713. [Google Scholar] [CrossRef] [PubMed]
- Yao, L.; Fu, Y.; Mohiyuddin, M.; Lam, H.Y.K. ecTMB: A robust method to estimate and classify tumor mutational burden. Sci. Rep. 2020, 10, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Büttner, R.; Longshore, J.W.; López-Ríos, F.; Merkelbach-Bruse, S.; Normanno, N.; Rouleau, E.; Penault-Llorca, F. Implementing TMB measurement in clinical practice: Considerations on assay requirements. ESMO Open 2019, 4, e000442. [Google Scholar] [CrossRef]
- Wu, C.-E.; Yeh, D.-W.; Pan, Y.-R.; Huang, W.-K.; Chen, M.-H.; Chang, J.W.-C.; Chen, J.-S.; Wang, Y.-C.; Yeh, C.-N. Chromosomal Instability May Not Be a Predictor for Immune Checkpoint Inhibitors from a Comprehensive Bioinformatics Analysis. Life 2020, 10, 276. [Google Scholar] [CrossRef] [PubMed]
- Wiesmüller, L.; Ford, J.M.; Schiestl, R.H. DNA Damage, Repair, and Diseases. J. Biomed. Biotechnol. 2002, 2, 45. [Google Scholar] [CrossRef] [PubMed]
- Knijnenburg, T.A.; Wang, L.; Zimmermann, M.T.; Chambwe, N.; Gao, G.; Cherniack, A.D.; Fan, H.; Shen, H.; Way, G.P.; Greene, C.S.; et al. Genomic and Molecular Landscape of DNA Damage Repair Deficiency across The Cancer Genome Atlas. Cell Rep. 2018, 23, 239–254.e6. [Google Scholar] [CrossRef]
- Jeggo, P.A.; Pearl, L.H.; Carr, A.M. DNA repair, genome stability and cancer: A historical perspective. Nat. Rev. Cancer 2016, 16, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Wang, C.; Zhang, Y.; Xu, L.; Fang, W.; Zhu, Y.; Zheng, Y.; Chen, X.; Xie, X.; Hu, X.; et al. Genomic signatures reveal DNA damage response deficiency in colorectal cancer brain metastases. Nat. Commun. 2019, 10, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Parikh, A.R.; He, Y.; Hong, T.S.; Corcoran, R.B.; Clark, J.W.; Ryan, D.P.; Zou, L.; Ting, D.; Catenacci, D.V.; Chao, J.; et al. Analysis of DNA Damage Response Gene Alterations and Tumor Mutational Burden Across 17,486 Tubular Gastrointestinal Carcinomas: Implications for Therapy. Oncology 2019, 24, 1340–1347. [Google Scholar] [CrossRef] [PubMed]
- Chae, Y.K.; Davis, A.A.; Raparia, K.; Agte, S.; Pan, A.; Mohindra, N.; Villaflor, V.; Giles, F. Association of Tumor Mutational Burden with DNA Repair Mutations and Response to Anti–PD-1/PD-L1 Therapy in Non–Small-Cell Lung Cancer. Clin. Lung Cancer 2019, 20, 88.e6–96.e6. [Google Scholar] [CrossRef] [PubMed]
- Mei, P.; Freitag, C.E.; Wei, L.; Zhang, Y.; Parwani, A.V.; Li, Z. High tumor mutation burden is associated with DNA damage repair gene mutation in breast carcinomas. Diagn. Pathol. 2020, 15, 1–7. [Google Scholar] [CrossRef]
- Ma, J.; Setton, J.; Lee, N.Y.; Riaz, N.; Powell, S.N. The therapeutic significance of mutational signatures from DNA repair deficiency in cancer. Nat. Commun. 2018, 9, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Campbell, B.B.; Light, N.; Fabrizio, D.; Zatzman, M.; Fuligni, F.; De Borja, R.; Davidson, S.; Edwards, M.; Elvin, J.A.; Hodel, K.P.; et al. Comprehensive Analysis of Hypermutation in Human Cancer. Cell 2017, 171, 1042–1056.e10. [Google Scholar] [CrossRef] [PubMed]
- Bonneville, R.; Krook, M.A.; Kautto, E.A.; Miya, J.; Wing, M.R.; Chen, H.-Z.; Reeser, J.W.; Yu, L.; Roychowdhury, S. Landscape of Microsatellite Instability Across 39 Cancer Types. JCO Precis. Oncol. 2017, 1, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Ellrott, K.; Bailey, M.H.; Saksena, G.; Covington, K.R.; Kandoth, C.; Stewart, C.; Hess, J.; Ma, S.; Chiotti, K.E.; McLellan, M.; et al. Scalable Open Science Approach for Mutation Calling of Tumor Exomes Using Multiple Genomic Pipelines. Cell Syst. 2018, 6, 271.e7–281.e7. [Google Scholar] [CrossRef] [PubMed]
- Durlak, J.A. How to Select, Calculate, and Interpret Effect Sizes. J. Pediatr. Psychol. 2009, 34, 917–928. [Google Scholar] [CrossRef] [PubMed]
- Ascierto, P.A.; Agarwala, S.S.; Botti, G.; Budillon, A.; Davies, M.A.; Dummer, R.; Ernstoff, M.; Ferrone, S.; Formenti, S.; Gajewski, T.F.; et al. Perspectives in melanoma: Meeting report from the Melanoma Bridge (November 29th–1 December 1st, 2018, Naples, Italy). J. Transl. Med. 2019, 17, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Auslander, N.; Zhang, G.; Lee, J.S.; Frederick, D.T.; Miao, B.; Moll, T.; Tian, T.; Wei, Z.; Madan, S.; Sullivan, R.J.; et al. Robust prediction of response to immune checkpoint blockade therapy in metastatic melanoma. Nat. Med. 2018, 24, 1545–1549. [Google Scholar] [CrossRef]
- Borcoman, E.; Nandikolla, A.; Long, G.; Goel, S.; Le Tourneau, C. Patterns of Response and Progression to Immunotherapy. Am. Soc. Clin. Oncol. Educ. Book 2018, 38, 169–178. [Google Scholar] [CrossRef] [PubMed]
- Pavan, A.; Attili, I.; Pasello, G.; Guarneri, V.; Conte, P.; Bonanno, L. Immunotherapy in small-cell lung cancer: From molecular promises to clinical challenges. J. Immunother. Cancer 2019, 7, 205. [Google Scholar] [CrossRef] [PubMed]
- Rizvi, N.A.; Hellmann, M.D.; Snyder, A.; Kvistborg, P.; Makarov, V.; Havel, J.J.; Lee, W.; Yuan, J.; Wong, P.; Ho, T.S.; et al. Mutational landscape determines sensitivity to PD-1 blockade in non–small cell lung cancer. Science 2015, 348, 124–128. [Google Scholar] [CrossRef] [PubMed]
- Van Allen, E.M.; Miao, D.; Schilling, B.; Shukla, S.A.; Blank, C.; Zimmer, L.; Sucker, A.; Hillen, U.; Foppen, M.H.G.; Goldinger, S.M.; et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science 2015, 350, 207–211. [Google Scholar] [CrossRef] [PubMed]
- Martin, S.A.; Lord, C.J.; Ashworth, A. Therapeutic Targeting of the DNA Mismatch Repair Pathway. Clin. Cancer Res. 2010, 16, 5107–5113. [Google Scholar] [CrossRef]
- Li, X.; Heyer, W.-D. Homologous recombination in DNA repair and DNA damage tolerance. Cell Res. 2008, 18, 99–113. [Google Scholar] [CrossRef] [PubMed]
- Piérard, G.E. Cell Proliferation in Cutaneous Malignant Melanoma: Relationship with Neoplastic Progression. ISRN Dermatol. 2012, 2012, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Giannakis, M.; Mu, X.J.; Shukla, S.A.; Qian, Z.R.; Cohen, O.; Nishihara, R.; Bahl, S.; Cao, Y.; Amin-Mansour, A.; Yamauchi, M.; et al. Genomic Correlates of Immune-Cell Infiltrates in Colorectal Carcinoma. Cell Rep. 2016, 15, 857–865. [Google Scholar] [CrossRef] [PubMed]
- Mankor, J.; CPCT Consortium; Paats, M.S.; Groenendijk, F.H.; Roepman, P.; Dinjens, W.N.M.; Dubbink, H.J.; Sleijfer, S.; Cuppen, E.; Lolkema, M.P.J.K. Impact of panel design and cut-off on tumour mutational burden assessment in metastatic solid tumour samples. Br. J. Cancer 2020, 122, 953–956. [Google Scholar] [CrossRef]

| SKCM | ||||
|---|---|---|---|---|
| NBN | LIG4 | MLH1 | RAD50 | PMS2 |
| FANCA | MRE11A | PMS1 | MSH3 | |
| LUAD | ||||
| UBE2T | MGMT | XPC | ALKBH3 | TDG |
| XRCC2 | CUL5 | NBN | FANCC | BARD1 |
| ERCC4 | MSH2 | XRCC4 | UNG | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chiu, T.-Y.; Lin, R.W.; Huang, C.-J.; Yeh, D.-W.; Wang, Y.-C. DNA Damage Repair Gene Set as a Potential Biomarker for Stratifying Patients with High Tumor Mutational Burden. Biology 2021, 10, 528. https://doi.org/10.3390/biology10060528
Chiu T-Y, Lin RW, Huang C-J, Yeh D-W, Wang Y-C. DNA Damage Repair Gene Set as a Potential Biomarker for Stratifying Patients with High Tumor Mutational Burden. Biology. 2021; 10(6):528. https://doi.org/10.3390/biology10060528
Chicago/Turabian StyleChiu, To-Yuan, Ryan Weihsiang Lin, Chien-Jung Huang, Da-Wei Yeh, and Yu-Chao Wang. 2021. "DNA Damage Repair Gene Set as a Potential Biomarker for Stratifying Patients with High Tumor Mutational Burden" Biology 10, no. 6: 528. https://doi.org/10.3390/biology10060528
APA StyleChiu, T.-Y., Lin, R. W., Huang, C.-J., Yeh, D.-W., & Wang, Y.-C. (2021). DNA Damage Repair Gene Set as a Potential Biomarker for Stratifying Patients with High Tumor Mutational Burden. Biology, 10(6), 528. https://doi.org/10.3390/biology10060528





