Origin and Potential Expansion of the Invasive Longan Lanternfly, Pyrops candelaria (Hemiptera: Fulgoridae) in Taiwan
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Genetic Structure and Phylogenetic Reconstruction of P. candelaria in Taiwan and Related Regions
2.1.1. Sample Preparation, PCR Amplification and Sequencing
2.1.2. Sequence Analyses and Phylogenetic Reconstruction
2.2. Prediction of Habitat Suitability of P. candelaria
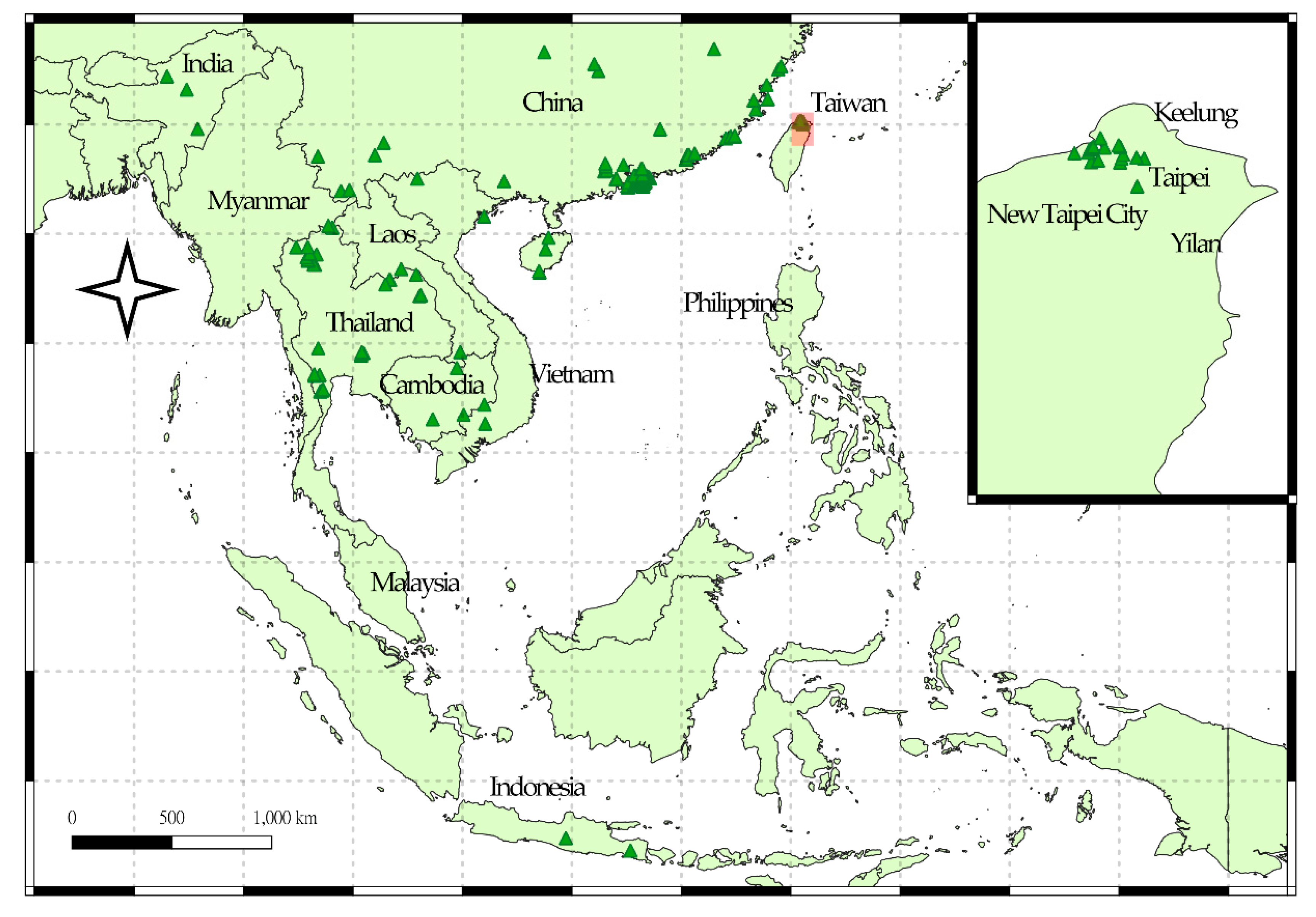
2.2.1. Species Occurrence Dataset
2.2.2. Environmental Variables
2.2.3. Distribution Modeling
2.3. Comparisons of Habitat Suitability of P. candelaria and Crop Cultivation Areas
3. Results
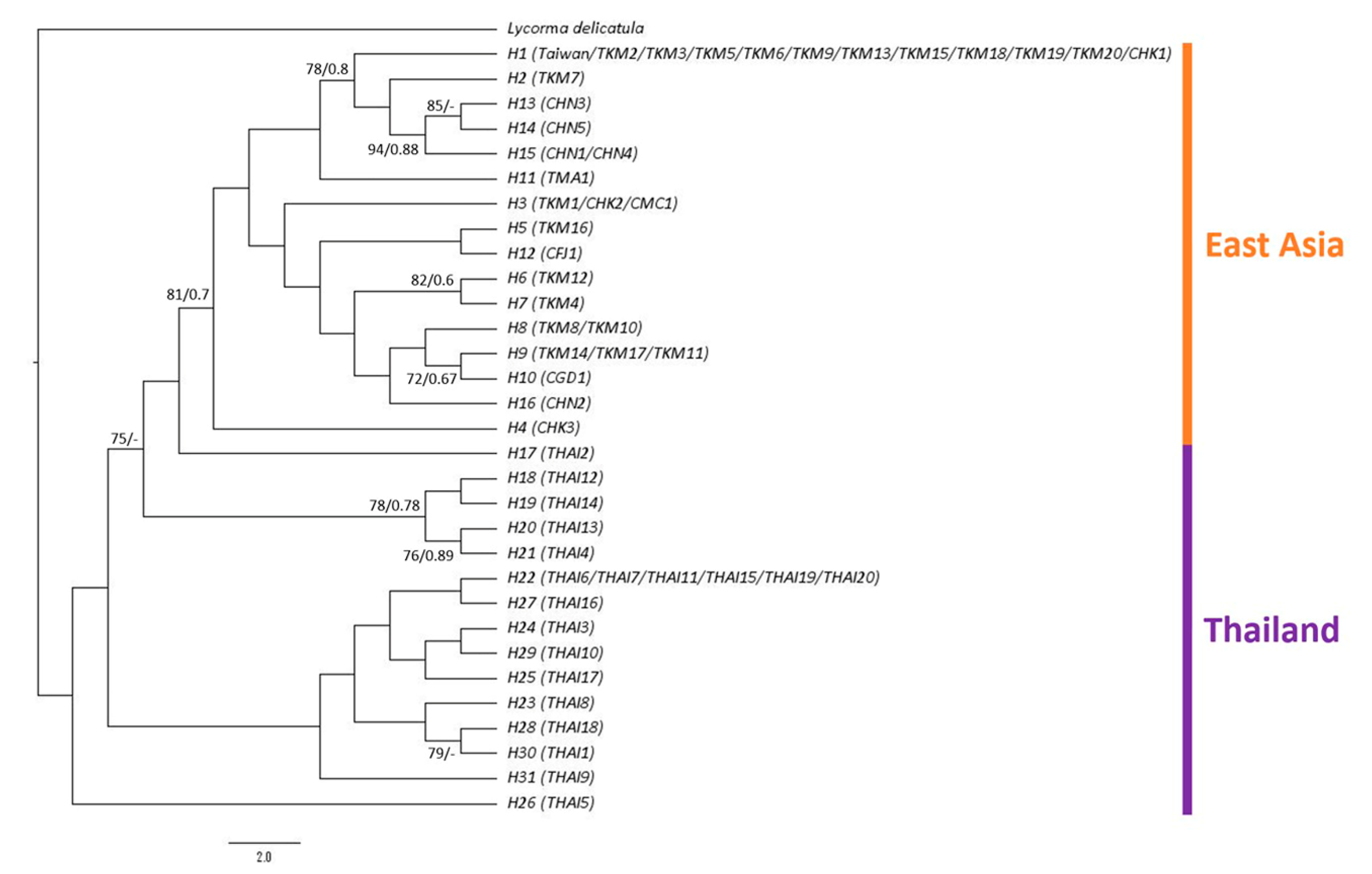
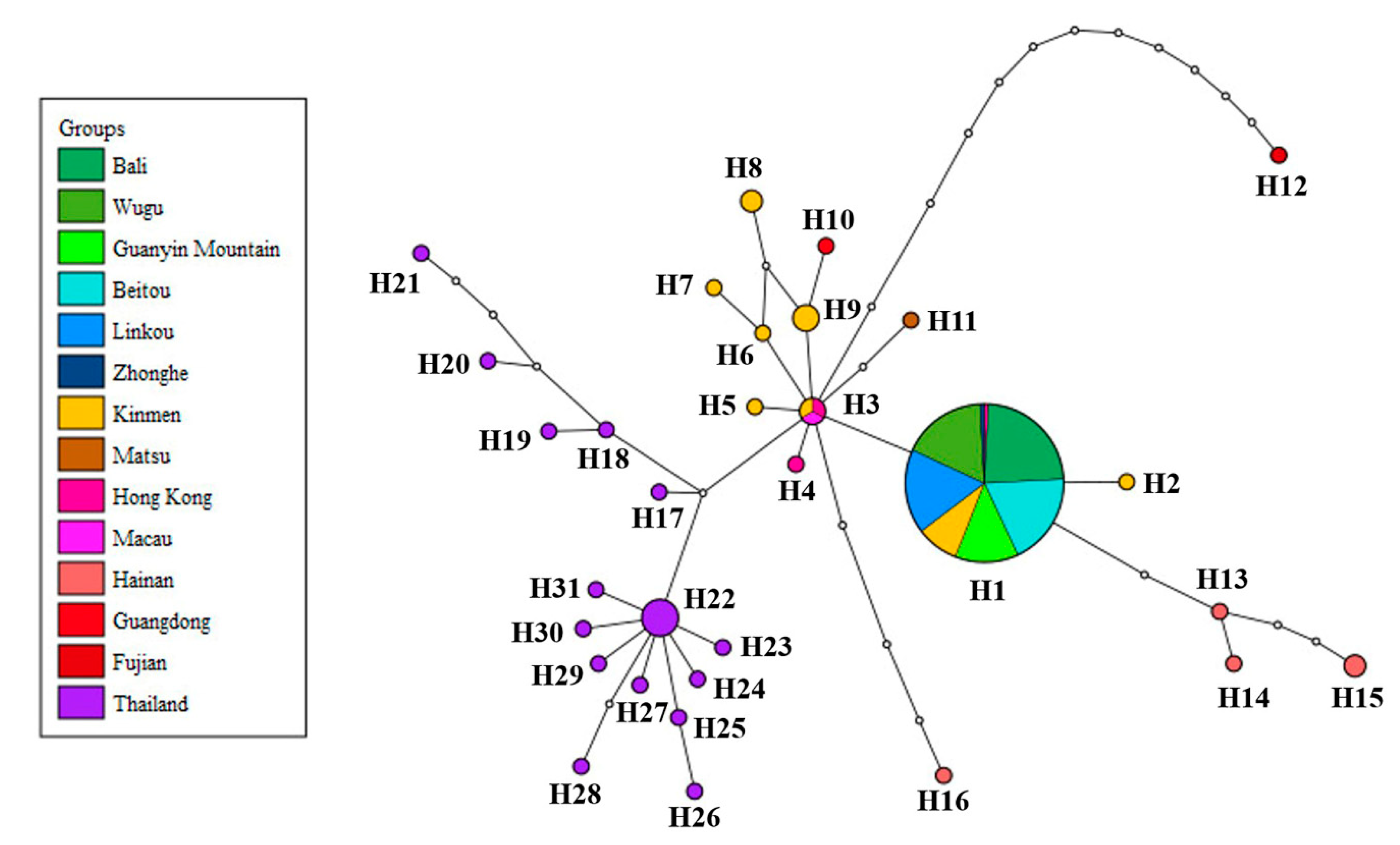
3.1. Phylogeny, Haplotype Network and Genetic Distance of P. candelaria in Taiwan and Related Regions
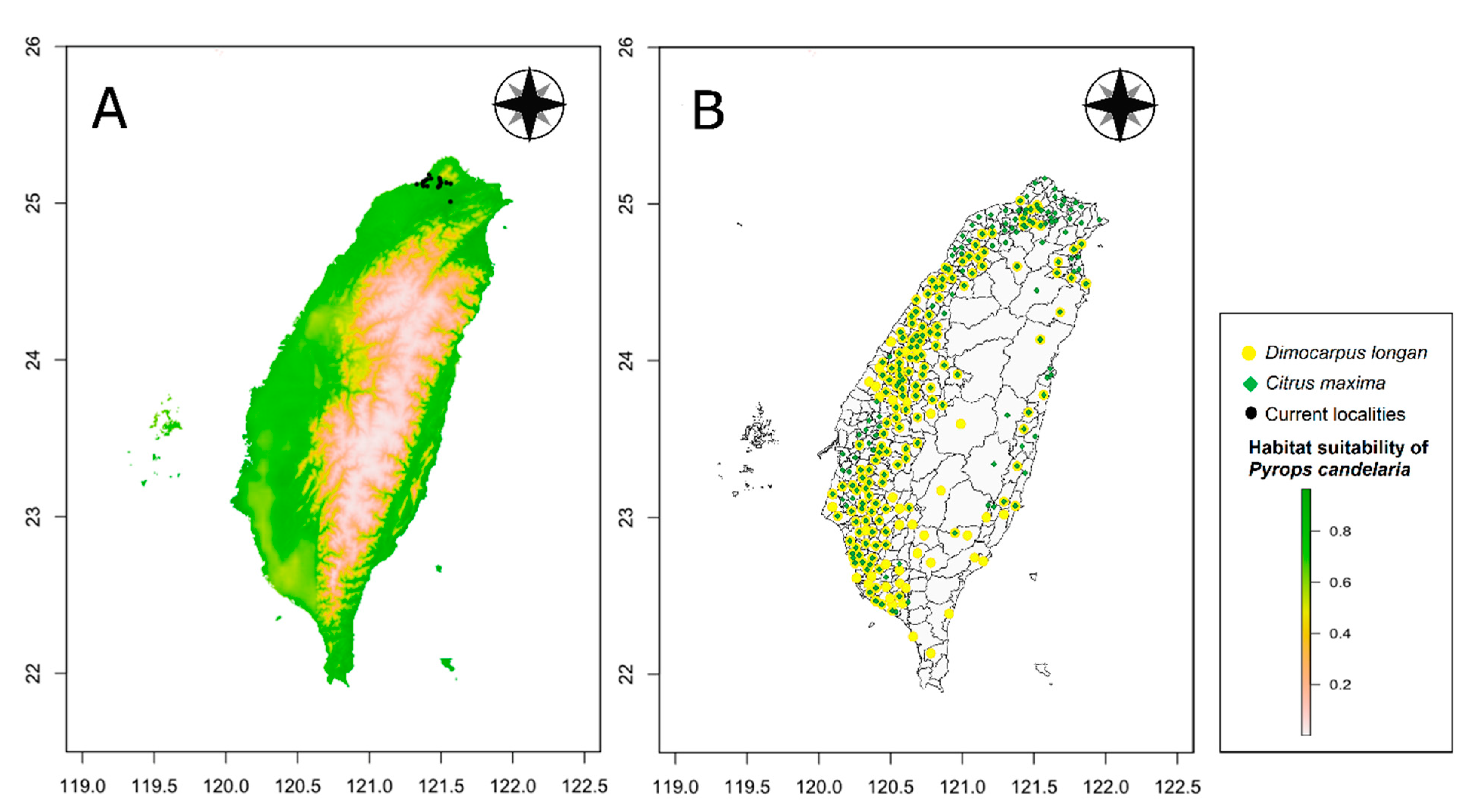
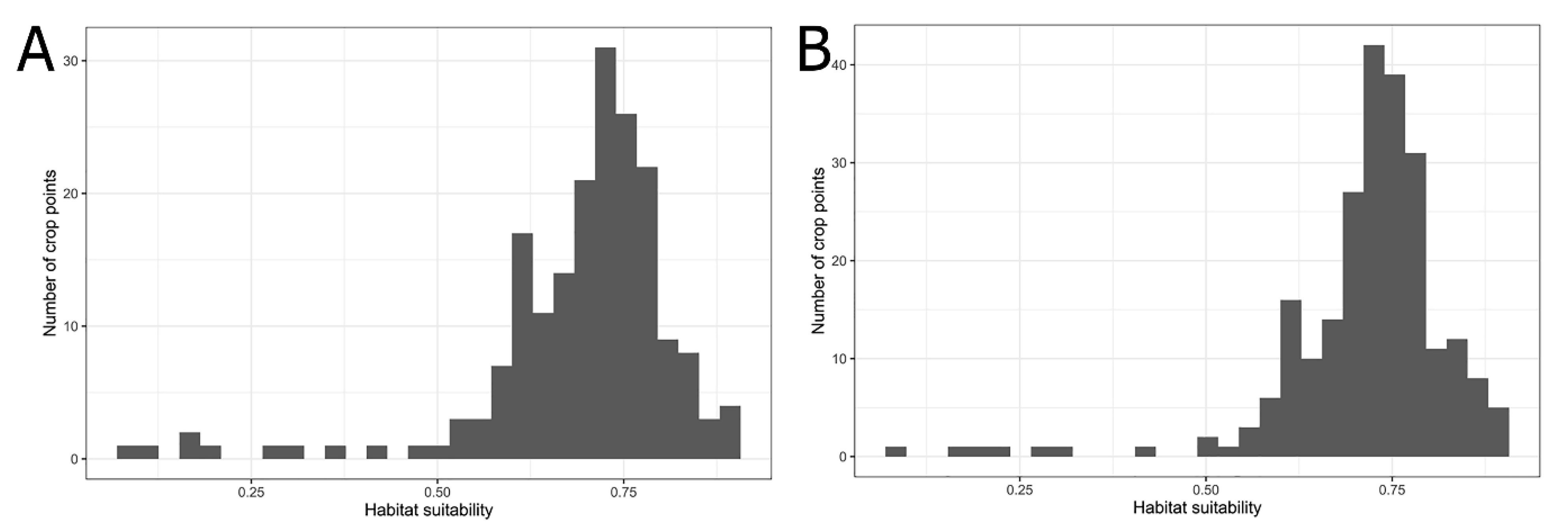
3.2. Comparison of Potential Distributions of P. candelaria and Cultivation Areas in Taiwan
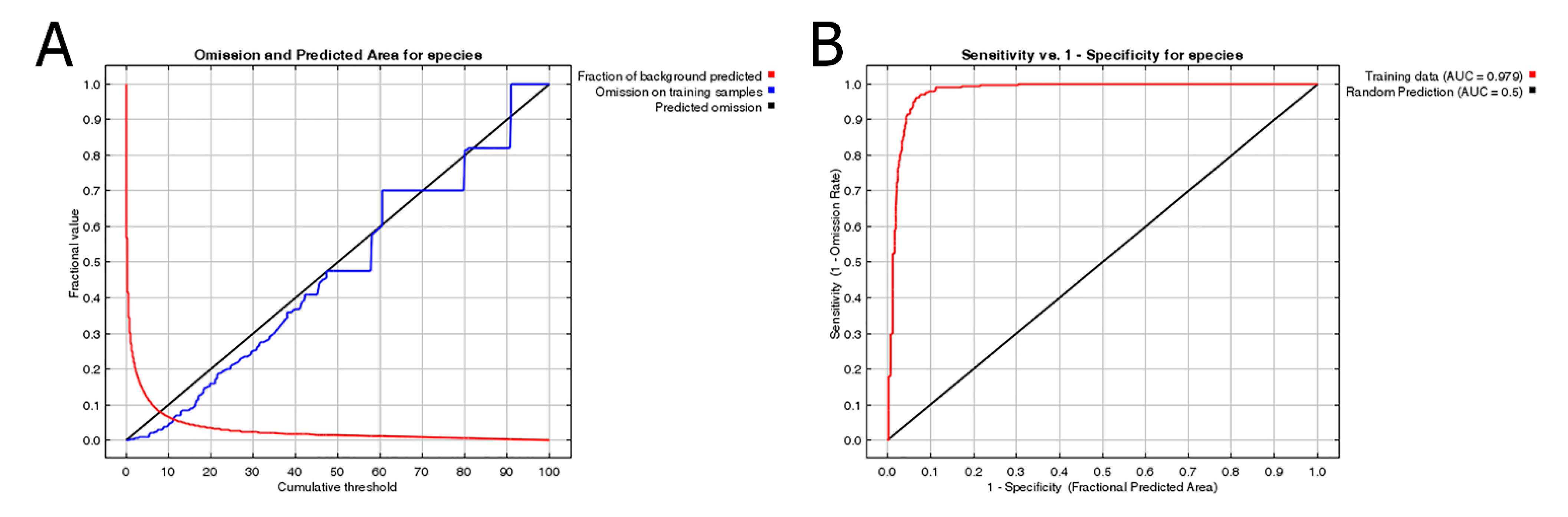
3.3. Environmental Variables Influencing the Distribution of P. candelaria
4. Discussion
4.1. Invasion History of P. candelaria in Taiwan
4.2. Habitat Suitability, Potential Expansion and Comparison with Crop Cultivation Areas
4.3. Environmental Variables Affect the Establishment of P. candelaria
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hulme, P.E. Trade, transport and trouble: Managing invasive species pathways in an era of globalization. J. Appl. Ecol. 2009, 46, 10–18. [Google Scholar] [CrossRef]
- Pimentel, D.; Zuniga, R.; Morrison, D. Update on the environmental and economic costs associated with alien-invasive species in the United States. Ecol. Econ. 2005, 52, 273–288. [Google Scholar] [CrossRef]
- Bellard, C.; Cassey, P.; Blackburn, T.M. Alien species as a driver of recent extinctions. Biol. Lett. 2016, 12, 20150623. [Google Scholar] [CrossRef]
- Lockwood, J.L.; Hoopes, M.F.; Marchetti, M.P. Invasion Ecology, 2nd ed.; Wiley-Blackwell: Oxford, UK, 2013. [Google Scholar]
- Lockwood, J.; Welbourne, D.J.; Romagosa, C.M.; Cassey, P.; Mandrak, N.E.; Strecker, A.; Leung, B.; Stringham, O.C.; Udell, B.; Episcopio-Sturgeon, D.J.; et al. When pets become pests: The role of the exotic pet trade in producing invasive vertebrate animals. Front. Ecol. Environ. 2019, 17, 323–330. [Google Scholar] [CrossRef] [Green Version]
- Du, Z.; Wu, Y.; Chen, Z.; Cao, L.; Ishikawa, T.; Kamitani, S.; Sota, T.; Song, F.; Tian, L.; Cai, W.; et al. Global phylogeography and invasion history of the spotted lanternfly revealed by mitochondrial phylogenomics. Evol. Appl. 2021, 14, 915–930. [Google Scholar] [CrossRef]
- Kim, H.; Kim, M.; Kwon, D.H.; Park, S.; Lee, Y.; Huang, J.; Kai, S.; Lee, H.-S.; Hong, K.-J.; Jang, Y.; et al. Molecular comparison of Lycorma delicatula (Hemiptera: Fulgoridae) isolates in Korea, China, and Japan. J. Asia-Pacific Ѐntomol. 2013, 16, 503–506. [Google Scholar] [CrossRef]
- Wu, Y.-H.; Kamiyama, M.T.; Chung, C.-C.; Tzeng, H.-Y.; Hsieh, C.-H.; Yang, C.-C.S. Population Monitoring, Egg Parasitoids, and Genetic Structure of the Invasive Litchi Stink Bug, Tessaratoma papillosa in Taiwan. Insects 2020, 11, 690. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.-S.; Takahashi, J.; Yeh, W.-C.; Lu, M.-L.; Huang, J.-Y.; Lin, Y.-J.; Sung, I.-H. Evidence for Range Expansion and Origins of an Invasive Hornet Vespa bicolor (Hymenoptera, Vespidae) in Taiwan, with Notes on Its Natural Status. Insects 2021, 12, 320. [Google Scholar] [CrossRef]
- Phillips, S.J.; Anderson, R.P.; Schapire, R.E. Maximum entropy modeling of species geographic distributions. Ecol. Model. 2006, 190, 231–259. [Google Scholar] [CrossRef] [Green Version]
- Yeh, H.-T.; Cheah, H.-Y.; Chiu, M.-C.; Liao, J.-R.; Ko, C.-C. Assessment of potential invasion for six phytophagous quarantine pests in Taiwan. Sci. Rep. 2021, 11, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Liao, J.-R.; Ho, C.-C.; Chiu, M.-C.; Ko, C.-C. Niche Modeling May Explain the Historical Population Failure of Phytoseiulus persimilis in Taiwan: Implications of Biocontrol Strategies. Insects 2021, 12, 418. [Google Scholar] [CrossRef]
- Xu, D.; Li, X.; Jin, Y.; Zhuo, Z.; Yang, H.; Hu, J.; Wang, R. Influence of climatic factors on the potential distribution of pest Heortia vitessoides Moore in China. Glob. Ecol. Conserv. 2020, 23, e01107. [Google Scholar] [CrossRef]
- Lallemand, V. Révision des Fulgoridae (Homoptera). Deuxième partie. faunes asiatique et Australienne. Mém. Inst. R. Sci. Nat. Belg. 1963, 75, 1–99. [Google Scholar]
- Wang, G.Y.; Huang, J.; Huang, B.K. Studies on the biology of Fulgora candelaria (L.) (Homoptera: Fulgoridae). Entomol. J. East China 2000, 9, 61–65. [Google Scholar]
- Pham, H.T. A checklist of the family Fulgoridae (Homoptera: Auchenorrhyncha: Fulgoroidea) from Vietnam. In Proceedings of the 3rd National Scientific Conference On Ecology and Biological Resources, Hanoi, Vietnam, 22 October 2009; pp. 317–321. [Google Scholar]
- Constant, J.; Phauk, S.; Bourgoin, T. Updating lanternflies biodiversity knowledge in Cambodia (Hemiptera: Fulgoromorpha: Fulgoridae) by optimizing field work surveys with citizen science involvement through Facebook networking and data access in FLOW website. Belg. J. Entomol. 2016, 37, 1–16. [Google Scholar]
- Kershaw, J.C.W.; Kirkaldy, G.W. A memoir to the anatomy and life-history of the Homopterous insect Pyrops candelaria (or “candle-fly”). Zool. Jb. Syst. 1910, 29, 105–124. [Google Scholar]
- Chou, I.; Lu, J.S.; Huang, J.; Wang, S.Z. Economic Insect Fauna of China Fascicle 36. Homoptera, Fulgoroidea; Science Press: Beijing, China, 1985; pp. 116–117. [Google Scholar]
- Wang, W.Q.; Xu, S.L.; Qin, D.Z. The lanternfly genus Pyrops Spinola (Hemiptera: Fulgoridae) from China with description of a new species. Entomotaxonomia 2018, 40, 296–309. [Google Scholar]
- Folmer, O.; Black, M.; Hoeh, W.; Lutz, R.; Vrijenhoek, R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 1994, 3, 294–299. [Google Scholar] [PubMed]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef] [Green Version]
- Templeton, A.; Crandall, K.; Sing, C. A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping and DNA sequence data. III. Cladogram estimation. Genetics 1992, 132, 619–633. [Google Scholar] [CrossRef]
- Clement, M.; Posada, D.; Crandall, K.A. TCS: A computer program to estimate gene genealogies. Mol. Ecol. 2000, 9, 1657–1659. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Nylander, A.A. MrModeltest, v2. Program Distributed by the Author. 2004. Available online: http://www.abc.se/nylander/mrmodeltest2/mrmodeltest2.html (accessed on 20 April 2021).
- Trifinopoulos, J.; Nguyen, L.-T.; Von Haeseler, A.; Minh, B.Q. W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016, 44, W232–W235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Hoehna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice Across a Large Model Space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ambaut, A. Computer Program Distributed by the Author. 2009. Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 4 January 2011).
- Global Biodiversity Information Facility. Available online: www.gbif.org (accessed on 3 November 2010).
- QGIS Development Team. QGIS Geographic Information System. Open Source Geospatial Foundation Project. 2020. Available online: http://qgis.osgeo.org (accessed on 15 April 2020).
- Phillips, S.J. A Brief Tutorial on MAXENT. Available online: http://biodiversityinformatics.amnh.org/open_source/maxent/ (accessed on 20 December 2020).
- Fick, S.E.; Hijmans, R.J. WorldClim 2: New 1-km Spatial Resolution Climate Surfaces for Global Land Areas. Int. J. Climatol. 2017, 37, 4302–4315. [Google Scholar] [CrossRef]
- Booth, T.H.; Nix, H.A.; Busby, J.R.; Hutchinson, M.F. Bioclim: The first species distribution modelling package, its early applications and relevance to most currentMaxEntstudies. Divers. Distrib. 2014, 20, 1–9. [Google Scholar] [CrossRef]
- De Meyer, M.; Robertson, M.P.; Mansell, M.W.; Ekesi, S.; Tsuruta, K.; Mwaiko, W.; Vayssieres, J.-F.; Peterson, A.T. Ecological niche and potential geographic distribution of the invasive fruit fly Bactrocera invadens (Diptera, Tephritidae). Bull. Ѐntomol. Res. 2009, 100, 35–48. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hijmans, R. Package “Raster”. Available online: http://raster.r-forge.r-project.org/ (accessed on 22 December 2020).
- Bivand, R.; Keitt, T.; Rowlingson, B. Package “Rgdal”. Available online: https://CRAN.R-project.org/package=rgdal (accessed on 22 December 2020).
- Phillips, S.J.; Dudik, M.; Schapire, R.E. Maxent Software for Modeling Species Niches and Distribution (Version 3.4.1). Available online: https://biodiversityinformatics.amnh.org/open_source/maxent/ (accessed on 20 December 2020).
- Hijmans, R.J.; Phillips, S.; Leathwick, J.; Elith, J. An {R} Package for Species Distribution Modelling. 2012. Available online: http://cran.r-project.org/web/packages/dismo/index.html (accessed on 4 June 2013).
- Muscarella, R.; Galante, P.J.; Soley-Guardia, M.; Boria, R.A.; Kass, J.; Uriarte, M.; Anderson, R.P. ENMeval: An R package for conducting spatially independent evaluations and estimating optimal model complexity forMaxentecological niche models. Methods Ecol. Evol. 2014, 5, 1198–1205. [Google Scholar] [CrossRef]
- Hernandez, P.A.; Graham, C.; Master, L.L.; Albert, D.L. The effect of sample size and species characteristics on performance of different species distribution modeling methods. Ecography 2006, 29, 773–785. [Google Scholar] [CrossRef]
- Peterson, A.T.; Papeş, M.; Soberon, J. Rethinking receiver operating characteristic analysis applications in ecological niche modeling. Ecol. Model. 2008, 213, 63–72. [Google Scholar] [CrossRef]
- Liu, C.; White, M.T.; Newell, G. Selecting thresholds for the prediction of species occurrence with presence-only data. J. Biogeogr. 2013, 40, 778–789. [Google Scholar] [CrossRef]
- Kumar, S.; Neven, L.G.; Zhu, H.; Zhang, R. Assessing the Global Risk of Establishment of Cydia pomonella(Lepidoptera: Tortricidae) using CLIMEX and MaxEnt Niche Models. J. Econ. Ѐntomol. 2015, 108, 1708–1719. [Google Scholar] [CrossRef] [PubMed]
- Peterson, A.T.; Soberón, J.; Anderson, R.P.; Pearson, R.G.; Martínez-Meyer, E.; Nakamura, M.; Araújo, M.B. Ecological Niches and Geographic Distributions: A Modeling Perspective; Princeton University Press: Princeton, NJ, USA, 2011. [Google Scholar]
- Boria, R.A.; Olson, L.E.; Goodman, S.M.; Anderson, R.P. Spatial filtering to reduce sampling bias can improve the performance of ecological niche models. Ecol. Model. 2014, 275, 73–77. [Google Scholar] [CrossRef]
- Velasco, J.A.; González-Salazar, C. Akaike information criterion should not be a “test” of geographical prediction accuracy in ecological niche modelling. Ecol. Inform. 2019, 51, 25–32. [Google Scholar] [CrossRef]
- Shih, H.T.; Ho, C.C.; Wu, W.J.; Yang, J.T.; Tsai, M.Y.; Fang, S.J.; Yang, C.J.; Wang, D.T.; Chou, H.C. A review and present status of entomological research in Kinmen Islands and Matsu Islands, Fujian. Formosan Entomol. Spec. Publ. 2004, 6, 187–210. [Google Scholar]

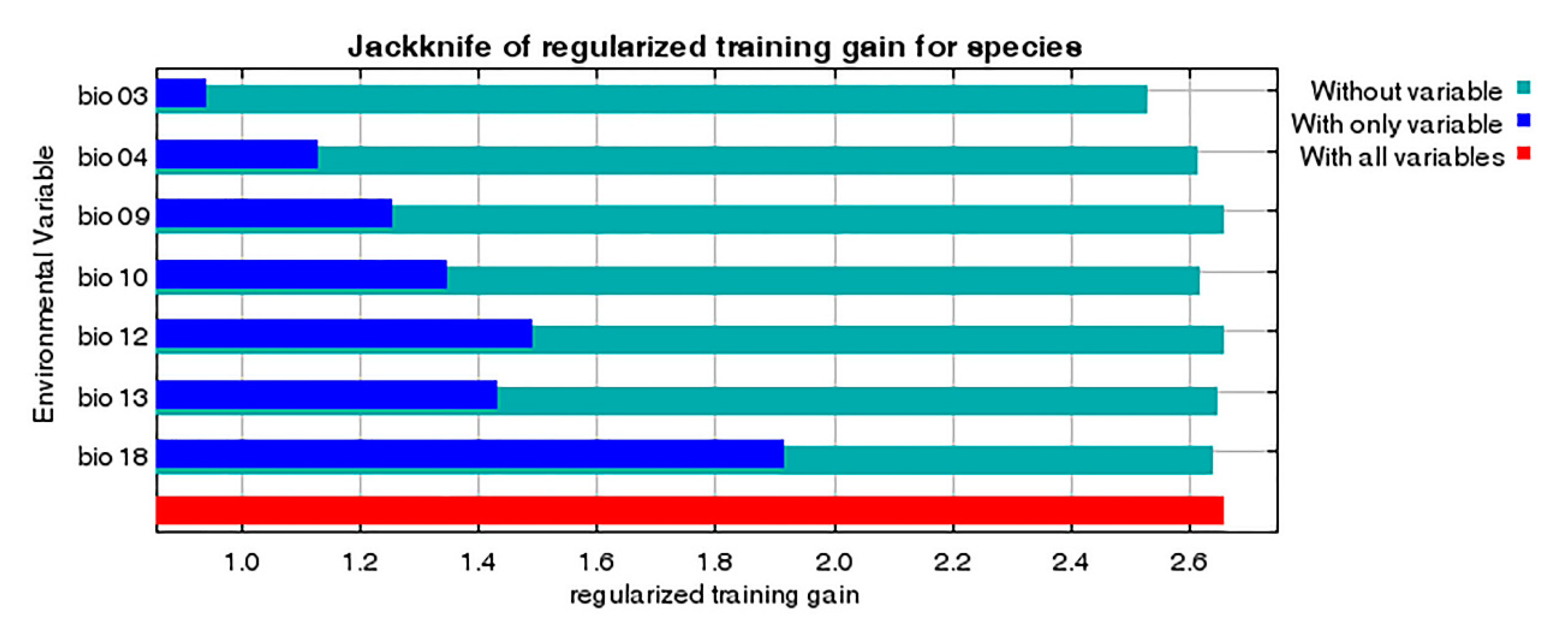
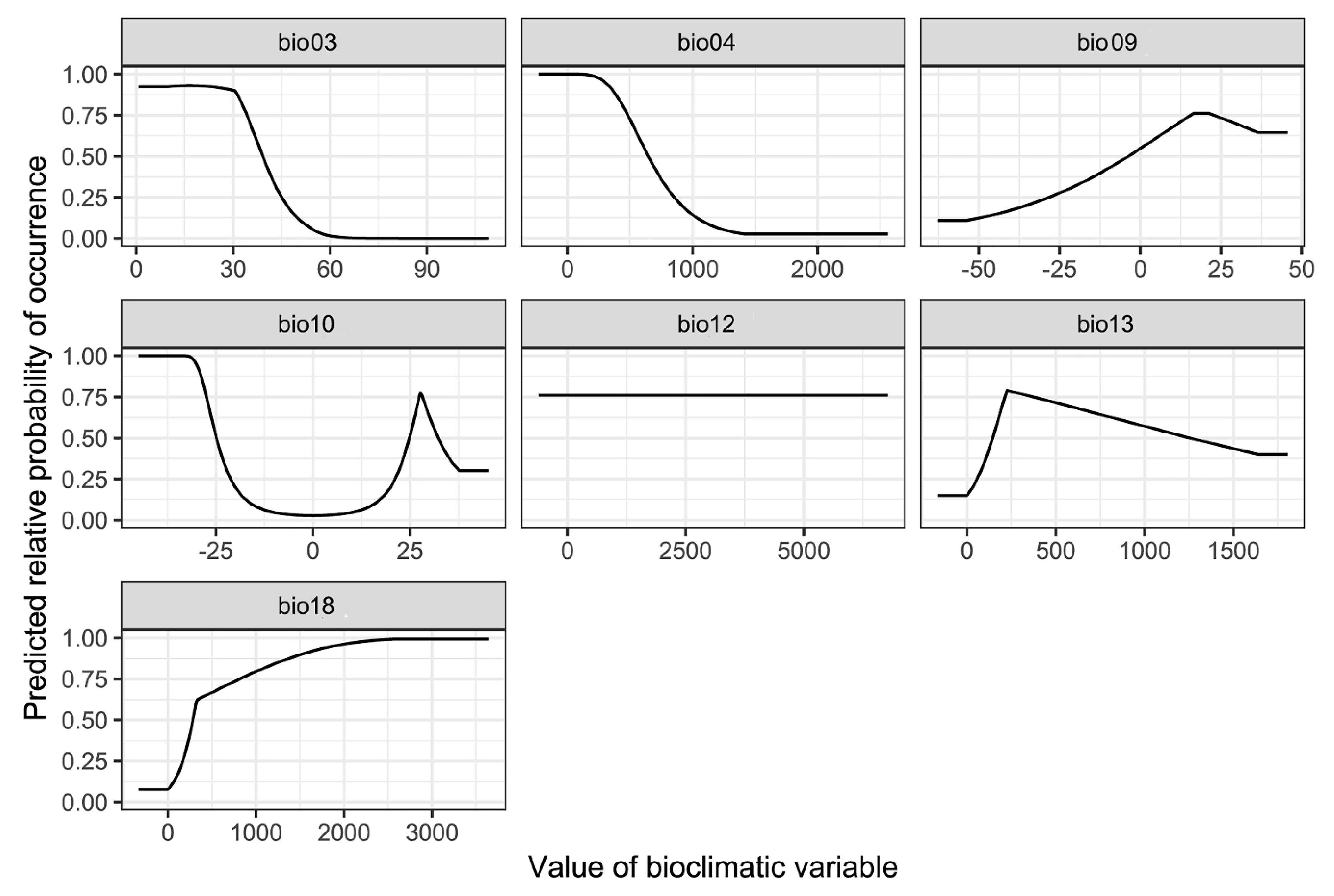
| Variable Code | Environmental Variables | Contribution Rate % | Permutation Importance % |
|---|---|---|---|
| bio18 | Precipitation of warmest quarter (mm) | 59.1 | 5 |
| bio03 | Isothermality (°C) | 18.5 | 66.8 |
| bio13 | Precipitation of wettest month (mm) | 15.7 | 0.9 |
| bio09 | Mean temperature of driest quarter (°C) | 4.5 | 1 |
| bio10 | Mean temperature of warmest quarter (°C) | 1.9 | 9.1 |
| bio04 | Temperature seasonality (°C) | 0.4 | 17.2 |
| bio12 | Annual precipitation (mm) | 0 | 0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, Y.-S.; Liao, J.-R.; Shiao, S.-F.; Ko, C.-C. Origin and Potential Expansion of the Invasive Longan Lanternfly, Pyrops candelaria (Hemiptera: Fulgoridae) in Taiwan. Biology 2021, 10, 678. https://doi.org/10.3390/biology10070678
Lin Y-S, Liao J-R, Shiao S-F, Ko C-C. Origin and Potential Expansion of the Invasive Longan Lanternfly, Pyrops candelaria (Hemiptera: Fulgoridae) in Taiwan. Biology. 2021; 10(7):678. https://doi.org/10.3390/biology10070678
Chicago/Turabian StyleLin, You-Sheng, Jhih-Rong Liao, Shiuh-Feng Shiao, and Chiun-Cheng Ko. 2021. "Origin and Potential Expansion of the Invasive Longan Lanternfly, Pyrops candelaria (Hemiptera: Fulgoridae) in Taiwan" Biology 10, no. 7: 678. https://doi.org/10.3390/biology10070678
APA StyleLin, Y.-S., Liao, J.-R., Shiao, S.-F., & Ko, C.-C. (2021). Origin and Potential Expansion of the Invasive Longan Lanternfly, Pyrops candelaria (Hemiptera: Fulgoridae) in Taiwan. Biology, 10(7), 678. https://doi.org/10.3390/biology10070678






