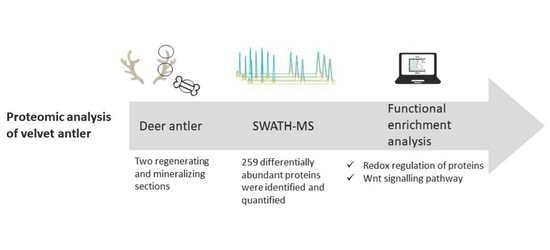
SWATH-MS Quantitative Proteomic Analysis of Deer Antler from Two Regenerating and Mineralizing Sections
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection of the Deer Antler
2.2. Protein Extraction and Tryptic Digestion
2.3. Protein Identification and Reference Spectral Library Building
2.4. Protein Quantification by SWATH-MS
2.5. ELISA Procedure for IGF-1 and IFN-γ Determination
3. Results
3.1. Comparisons of Deer Proteomes Tip vs. Middle Antler, and Middle Antler vs. Ribs
3.1.1. Comparison Tip vs. Middle Antler
3.1.2. Comparison Middle Section of Antler vs. Rib
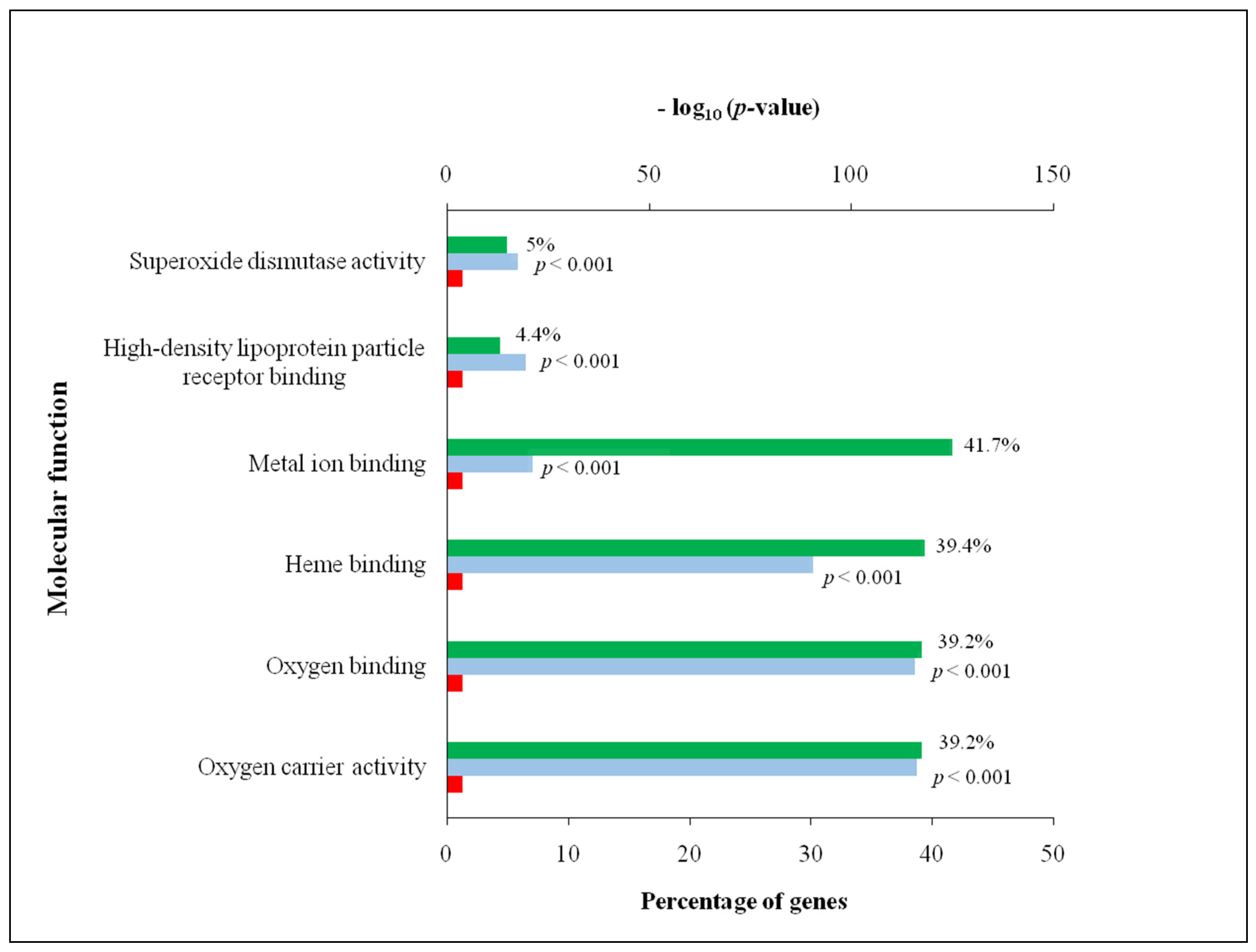
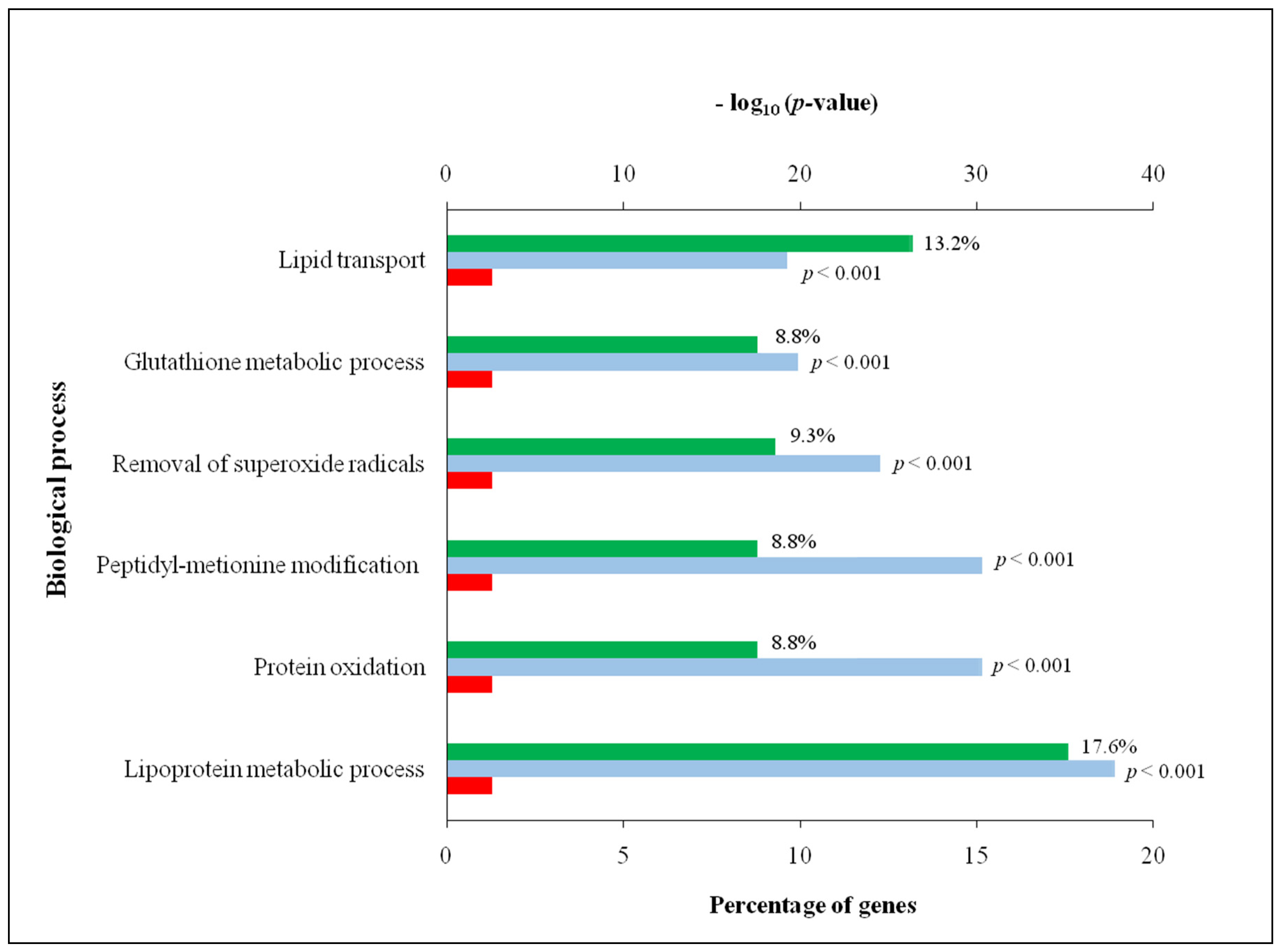
3.2. Comprehensive Analysis of Deer Antler Proteome
4. Discussion
4.1. Comparisons of Deer Proteomes Tip vs. Middle Antler, and Middle Antler vs. Ribs
4.1.1. Comparison Tip vs. Middle Antler
4.1.2. Comparison Middle Section of Antler vs. Rib
4.2. Comprehensive Analysis of Deer Antler Proteome
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Ethic Statement
References
- Gaspar-López, E.; Landete-Castillejos, T.; Estevez, J.A.; Ceacero, F.; Gallego, L.; García, A.J. Biometrics, testosterone, cortisol and antler growth cycle in iberian red deer stags (Cervus elaphus hispanicus). Reprod. Domest. Anim. 2010, 45, 243–249. [Google Scholar] [CrossRef]
- Shi, H.; Yu, T.; Li, Z.; Lu, W.; Zhang, M.; Ye, J. Bone regeneration strategy inspired by the study of calcification behavior in deer antler. Mater. Sci. Eng. C 2015, 57, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Landete-Castillejos, T.; Kierdorf, H.; Gomez, S.; Luna, S.; García, A.J.; Cappelli, J.; Pérez-Serrano, M.; Pérez-Barbería, J.; Gallego, L.; Kierdorf, U. Antlers—Evolution, development, structure, composition, and biomechanics of an outstanding type of bone. Bone 2019, 128, 115046. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, C.; Wang, N.; Li, Z.; Heller, R.; Liu, R.; Zhao, Y.; Han, J.; Pan, X.; Zheng, Z.; et al. Genetic basis of ruminant headgear and rapid antler regeneration. Science 2019, 364, eaav6335. [Google Scholar] [CrossRef] [PubMed]
- Chonco, L.; Landete-Castillejos, T.; Serrano-Heras, G.; Serrano, M.P.; Pérez-Barbería, F.J.; González-Armesto, C.; García, A.; de Cabo, C.; Lorenzo, J.M.; Li, C.; et al. Anti-tumour activity of deer growing antlers and its potential applications in the treatment of malignant gliomas. Sci. Rep. 2021, 11, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.J.; Jeon, B.T.; Wang, Y.; Choi, E.J.; Kim, Y.S.; Hwang, J.W.; Park, P.J.; Moon, S.H.; Kim, E.K. First evidence that sika deer (Cervus nippon) velvet antler extract suppresses migration of human prostate cancer cells. Korean J. Food Sci. Anim. Resour. 2015, 35, 507–514. [Google Scholar] [CrossRef] [Green Version]
- Yang, H.; Wang, L.; Sun, H.; He, X.; Zhang, J.; Liu, F. Anticancer activity in vitro and biological safety evaluation in vivo of Sika deer antler protein. J. Food Biochem. 2017, 41, 1–8. [Google Scholar] [CrossRef]
- Fraser, A.; Haines, S.R.; Stuart, E.C.; Scandlyn, M.J.; Alexander, A.; Somers-Edgar, T.J.; Rosengren, R.J. Deer velvet supplementation decreases the grade and metastasis of azoxymethane-induced colon cancer in the male rat. Food Chem. Toxicol. 2010, 48, 1288–1292. [Google Scholar] [CrossRef]
- Hu, W.; Qi, L.; Tian, Y.H.; Hu, R.; Wu, L.; Meng, X.Y. Studies on the purification of polypeptide from sika antler plate and activities of antitumor. BMC Complement. Altern. Med. 2015, 15, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Li, H.; Jin, L.; Li, X.; Ma, Y.; You, J.; Li, S.; Xu, Y. Deer antler base as a traditional Chinese medicine: A review of its traditional uses, chemistry and pharmacology. J. Ethnopharmacol. 2013, 145, 403–415. [Google Scholar] [CrossRef]
- Sui, Z.; Sun, H.; Weng, Y.; Zhang, X.; Sun, M.; Sun, R.; Zhao, B.; Liang, Z.; Zhang, Y.; Li, C.; et al. Quantitative proteomics analysis of deer antlerogenic periosteal cells reveals potential bioactive factors in velvet antlers. J. Chromatogr. A 2020, 1609, 460496. [Google Scholar] [CrossRef] [PubMed]
- Su, H.; Tang, X.; Zhang, X.; Liu, L.; Jing, L.; Pan, D.; Sun, W.; He, H.; Yang, C.; Zhao, D.; et al. Comparative proteomics analysis reveals the difference during antler regeneration stage between red deer and sika deer. PeerJ 2019, 2019, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Ko, S.C.; Moon, S.H.; Lee, S.H. Protective effects of novel antioxidant peptide purified from alcalase hydrolysate of velvet antler against oxidative stress in chang liver cells in vitro and in a zebrafish model in vivo. Int. J. Mol. Sci. 2019, 20, 5187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dong, Z.; Ba, H.; Zhang, W.; Coates, D.; Li, C. iTRAQ-based quantitative proteomic analysis of the potentiated and dormant antler stem cells. Int. J. Mol. Sci. 2016, 17, 1778. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, D.; Berg, D.; Ba, H.; Sun, H.; Wang, Z.; Li, C. Deer antler stem cells are a novel type of cells that sustain full regeneration of a mammalian organ—Deer antler. Cell Death Dis. 2019, 10, 443. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, D.; Ba, H.; Li, C.; Zhao, Q.; Li, C. Proteomic analysis of plasma membrane proteins of antler stem cells using label-free LC-MS/MS. Int. J. Mol. Sci. 2018, 19, 3477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dong, Z.; Haines, S.; Coates, D. Proteomic Profiling of Stem Cell Tissues during Regeneration of Deer Antler: A Model of Mammalian Organ Regeneration. J. Proteome Res. 2020, 19, 1760–1775. [Google Scholar] [CrossRef]
- Boletín Oficial del Estado. Ley 2/2018, de 15 de Marzo, por la que se Modifican la Ley 3/2015, de 5 de Marzo, de Caza de Castilla-La Mancha y Otras Normas en Materia Medioambiental y Fiscal; Agencia Estatal Boletín Oficial del Estado: Madrid, Spain, 2018. [Google Scholar]
- Pathan, M.; Keerthikumar, S.; Ang, C.S.; Gangoda, L.; Quek, C.Y.J.; Williamson, N.A.; Mouradov, D.; Sieber, O.M.; Simpson, R.J.; Salim, A.; et al. FunRich: An open access standalone functional enrichment and interaction network analysis tool. Proteomics 2015, 15, 2597–2601. [Google Scholar] [CrossRef] [PubMed]
- López-pedrouso, M.; Franco, D.; Serrano, M.P.; Maggiolino, A. A proteomic-based approach for the search of biomarkers in Iberian wild deer ( Cervus elaphus ) as indicators of meat quality. J. Proteomics 2019, 205, 103422. [Google Scholar] [CrossRef]
- Li, C.; Suttie, J.M.; Clark, D.E. Histological examination of antler regeneration in red deer (Cervus elaphus). Anat. Rec. Part A Discov. Mol. Cell. Evol. Biol. 2005, 282, 163–174. [Google Scholar] [CrossRef]
- Ba, H.; Wang, D.; Yau, T.O.; Shang, Y.; Li, C. Transcriptomic analysis of different tissue layers in antler growth Center in Sika Deer (Cervus nippon). BMC Genom. 2019, 20, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Finkel, T.; Holbrook, N.J. Oxidants, Oxidative Stress and Biology of Ageing. Insight Rev. Artic. 2000, 408, 239–247. [Google Scholar] [CrossRef]
- Balogh, E.; Paragh, G.; Jeney, V. Influence of iron on bone homeostasis. Pharmaceuticals 2018, 11, 107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cesari, M.; Pahor, M.; Lauretani, F.; Penninx, B.W.H.J.; Bartali, B.; Russo, R.; Cherubini, A.; Woodman, R.; Bandinelli, S.; Guralnik, J.M.; et al. Bone density and hemoglobin levels in older persons: Results from the InCHIANTI study. Osteoporos. Int. 2005, 16, 691–699. [Google Scholar] [CrossRef] [Green Version]
- Golizeh, M.; Lee, K.; Ilchenko, S.; Ösme, A.; Bena, J.; Sadygov, R.G.; Kashyap, S.; Kasumov, T. Increased serotransferrin and ceruloplasmin turnover in diet-controlled patients with type 2 diabetes. Free Radic. Biol. Med. 2017, 113, 461–469. [Google Scholar] [CrossRef]
- Deng, M.; Zhang, W.; Tang, H.; Ye, Q.; Liao, Q.; Zhou, Y.; Wu, M.; Xiong, W.; Zheng, Y.; Guo, X.; et al. Lactotransferrin acts as a tumor suppressor in nasopharyngeal carcinoma by repressing AKT through multiple mechanisms. Oncogene 2013, 32, 4273–4283. [Google Scholar] [CrossRef] [Green Version]
- Mihara, E.; Hirai, H.; Yamamoto, H.; Tamura-Kawakami, K.; Matano, M.; Kikuchi, A.; Sato, T.; Takagi, J. Active and water-soluble form of lipidated wnt protein is maintained by a serum glycoprotein afamin/α-albumin. elife 2016, 5, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Steger, D.J.; Grant, G.R.; Schupp, M.; Tomaru, T.; Lefterova, M.I.; Schug, J.; Manduchi, E.; Stoeckert, C.J.; Lazar, M.A. Propagation of adipogenic signals through an epigenomic transition state. Genes Dev. 2010, 24, 1035–1044. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baker, J.; Liu, J.P.; Robertson, E.J.; Efstratiadis, A. Role of insulin-like growth factors in embryonic and postnatal growth. Cell 1993, 75, 73–82. [Google Scholar] [CrossRef]
- Sun, D.; Zhao, Y.Y.; Dai, S.P.; Fang, K.; Dong, L.Y.; Ding, Q. Cloning and analysis of human α-1B glycoprotein precursor gene: A novel member of human immunoglobulin superfamily. Acta Genet. Sin. 2002, 29, 299–302. [Google Scholar] [PubMed]
- Kreunin, P.; Zhao, J.; Rosser, C.; Urquidi, V.; Lubman, D.M.; Goodison, S. Bladder cancer associated glycoprotein signatures revealed by urinary proteomic profiling. J. Proteome Res. 2007, 6, 2631–2639. [Google Scholar] [CrossRef] [Green Version]
- Landete-Castillejos, T.; Estevez, J.A.; Cearero, F.; García, A.J.; Gallego, L. A review of factors affecting antler composition and mechanics Tomas. Front. Biosci. 2012, 4, 2328–2339. [Google Scholar] [CrossRef]
- Lee, K.S.; Cao, Y.; Witwicka, H.E.; Tom, S.; Tapscott, S.J.; Wang, E.H. RNA-binding protein muscleblind-like 3 (MBNL3) disrupts myocyte enhancer factor 2 (Mef2) β-exon splicing. J. Biol. Chem. 2010, 285, 33779–33787. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, H.W.; Kim, H.D.; Kim, T.S.; Kim, J. Senescent Cells Differentially Translate Senescence-Related mRNAs Via Ribosome Heterogeneity. J. Gerontol. Ser. A Biol. Sci. Med. Sci. 2019, 74, 1015–1024. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.; Grafe, I.; Alexander, S.; Lee, B. Genetic causes and mechanisms of Osteogenesis Imperfecta. Bone 2017, 102, 40–49. [Google Scholar] [CrossRef] [PubMed]
- Hu, P.; Wang, T.; Liu, H.; Xu, J.; Wang, L.; Zhao, P.; Xing, X. Full-length transcriptome and microRNA sequencing reveal the specific gene-regulation network of velvet antler in sika deer with extremely different velvet antler weight. Mol. Genet. Genom. 2019, 294, 431–443. [Google Scholar] [CrossRef] [PubMed]
- Gomez, S.; Garcia, A.J.; Luna, S.; Kierdorf, U.; Kierdorf, H.; Gallego, L.; Landete-Castillejos, T. Labeling studies on cortical bone formation in the antlers of red deer (Cervus elaphus). Bone 2013, 52, 506–515. [Google Scholar] [CrossRef] [PubMed]
- Borsy, A.; Podani, J.; Stéger, V.; Balla, B.; Horváth, A.; Kósa, J.P.; Gyurján, I.; Molnár, A.; Szabolcsi, Z.; Szabó, L.; et al. Identifying novel genes involved in both deer physiological and human pathological osteoporosis. Mol. Genet. Genom. 2009, 281, 301–313. [Google Scholar] [CrossRef] [PubMed]
- Baxter, B.J.; Andrews, R.N.; Barrell, G.K. Bone turnover associated with antler growth in red deer (Cervus elaphus). Anat. Rec. 1999, 256, 14–19. [Google Scholar] [CrossRef]
- Urs, S.; Harrington, A.; Liaw, L.; Small, D. Selective expression of an aP2/Fatty Acid Binding Protein4-Cre transgene in non-adipogenic tissues during embryonic development. Transgenic Res. 2006, 15, 647–653. [Google Scholar] [CrossRef]
- Wergedal, J.E.; Mohan, S.; Lundy, M.; Baylink, D.J. Skeletal growth factor and other growth factors known to be present in bone matrix stimulate proliferation and protein synthesis in human bone cells. J. Bone Miner. Res. 1990, 5, 179–186. [Google Scholar] [CrossRef] [PubMed]
- Wauquier, F.; Leotoing, L.; Coxam, V.; Guicheux, J.; Wittrant, Y. Oxidative stress in bone remodelling and disease. Trends Mol. Med. 2009, 15, 468–477. [Google Scholar] [CrossRef] [PubMed]
- Bartelt, A.; Koehne, T.; Tödter, K.; Reimer, R.; Müller, B.; Behler-Janbeck, F.; Heeren, J.; Scheja, L.; Niemeier, A. Quantification of bone fatty acid metabolism and its regulation by adipocyte lipoprotein lipase. Int. J. Mol. Sci. 2017, 18, 1264. [Google Scholar] [CrossRef] [PubMed]
- Poprac, P.; Jomova, K.; Simunkova, M.; Kollar, V.; Rhodes, C.J.; Valko, M. Targeting Free Radicals in Oxidative Stress-Related Human Diseases. Trends Pharmacol. Sci. 2017, 38, 592–607. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Fang, Y.Z.; Yang, S.; Lupton, J.R.; Turner, N.D. Glutathione Metabolism and Its Implications for Health. J. Nutr. 2004, 134, 489–492. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruiz-Romero, C.; Calamia, V.; Mateos, J.; Carreira, V.; Martínez-Gomariz, M.; Fernández, M.; Blanco, F.J. Mitochondrial dysregulation of osteoarthritic human articular chondrocytes analyzed by proteomics: A decrease in mitochondrial superoxide dismutase points to a redox imbalance. Mol. Cell. Proteom. 2009, 8, 172–189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kiang, J.G.; Tsokos, G.C. Heat shock protein 70 kDa: Molecular biology, biochemistry, and physiology. Pharmacol. Ther. 1998, 80, 183–201. [Google Scholar] [CrossRef]
- Chunjuan, L.; Shouqin, Z.; Dongsheng, L.; Jianpeng, D. Detection and quantitation of insulin-like growth factor-1 in velvet antler by an enzyme-linked immunosorbent assay. In Proceedings of the 2011 International Conference on Consumer Electronics, Communications and Networks (CECNet), Xianning, China, 16–18 April 2011; pp. 1162–1168. [Google Scholar] [CrossRef]


| Items | Protein Name | Gene Names | Middle Section | Tip | FC |
|---|---|---|---|---|---|
| Mean ± SE | Mean ± SE | ||||
| Hemoglobin subunit alpha | HBA/SCN2A | 2,563,475.0 ± 869,235.6 | 155,785.3 ± 39,216.0 | 16.46 | |
| Adult beta-globin 1 | HBB | 45,176,500.0 ± 16,757,107.7 | 3,050,606.7 ± 1,088,464.0 | 14.81 | |
| Adult beta-globin 2 | HBB | 13,515,916.7 ± 4,092,463.0 | 1,083,311.7 ± 331,202.8 | 12.48 | |
| Creatine kinase B-type | CKB | 311,250.0 ± 37,004.7 | 40,426.8 ± 3039.0 | 7.70 | |
| Alpha-2-macroglobulin (A2MG) | A2M | 316,528.3 ± 77,031.0 | 52,790.2 ± 5947.1 | 6.00 | |
| Inter-alpha-trypsin inhibitor heavy chain H1 | ITIH1 | 165,838.3 ± 53,177.3 | 30,431.5 ± 1875.7 | 5.45 | |
| Amiloride-sensitive amine oxidase | AOC1 | 39,327.2 ± 6441.8 | 9712.9 ± 1135.1 | 4.05 | |
| Carbonic anhydrase (CAH3) | CA3 | 22,310.6 ± 5010.6 | 5560.7 ± 1178.0 | 4.01 | |
| Alkaline phosphatase (PPBN) | ALPG | 15,993.0 ± 1618.5 | 4000.0 ± 467.9 | 4.00 | |
| Inter-alpha-trypsin inhibitor heavy chain H2 | ITIH2 | 87,390.0 ± 17,580.9 | 24,580.3 ± 2302.8 | 3.56 | |
| Peroxiredoxin-2 | PRDX2 | 37,583.5 ± 4204.3 | 11,079.8 ± 510.7 | 3.39 | |
| Apolipoprotein A-II | APOA2 | 60,995.7 ± 16,710.0 | 18,302.5 ± 1743.9 | 3.33 | |
| Alpha-amylase 1A | AMY1A | 22,315.3 ± 4160.5 | 6740.0 ± 955.1 | 3.31 | |
| Plasminogen (PLMN) | PLG | 37,045.7 ± 9978.9 | 11,666.0 ± 644.5 | 3.18 | |
| Glutathione S-transferase Mu 1 | GSTM1 | 94,826.5 ± 25,325.8 | 30,432.3 ± 2409.7 | 3.12 | |
| Complement C3 | C3 | 712,043.3 ± 109,393.3 | 252,156.7 ± 27,537.1 | 2.82 | |
| Afamin (AFAM) | AFM | 25,198.7 ± 5103.3 | 9541.6 ± 1423.6 | 2.64 | |
| Serpin 3-6 | Serpin | 378,931.7 ± 87,373.6 | 145,784.2 ± 24,405.9 | 2.60 | |
| Mimecan (MIME) | OGN | 55,036.5 ± 9328.6 | 21,511.8 ± 2205.4 | 2.56 | |
| Alpha-1B-glycoprotein | A1BG | 2,213,076.7 ± 441,457.5 | 866,086.7 ± 63,588.4 | 2.56 | |
| Fetuin-B (FETUB) | Fetuin-B | 56,124.7 ± 8673.8 | 22,155.8 ± 2673.3 | 2.53 | |
| Alpha-2-antiplasmin (A2AP) | SERPINF2 | 24,572.3 ± 4954.9 | 9877.7 ± 579.0 | 2.49 | |
| Complement factor B (CFAB) | CFB | 90,554.5 ± 16,717.9 | 36,896.7 ± 2915.5 | 2.45 | |
| Heparin cofactor 2 (HEP2) | SERPIND1 | 19,863.8 ± 3310.1 | 8067.4 ± 843.7 | 2.46 | |
| Retinol-binding protein 4 (RET4) | RBP4 | 20,127.3 ± 4240.2 | 8274.5 ± 783.4 | 2.43 | |
| Annexin A2 | ANXA2 | 52,405.0 ± 7452.6 | 22,504.7 ± 1287.2 | 2.33 | |
| Beta-N-acetylhexosaminidase | HEXB | 4104.9 ± 607.5 | 1851.1 ± 250.1 | 2.22 | |
| Lactotransferrin | LTF | 1444,530.0 ± 231,686.6 | 659,118.3 ± 34,072.7 | 2.19 | |
| Hemopexin (fragment) | HPX | 173,172.2 ± 38,801.3 | 80,237.7 ± 9734.1 | 2.16 | |
| Antithrombin-III (ANT3) | SERPINC1 | 64,056.0 ± 10,572.3 | 30,005.0 ± 1717.2 | 2.13 | |
| 40S ribosomal protein S27a (RS27A) | RPS27A | 78,794.3 ± 10,336.1 | 38,137.8 ± 4586.8 | 2.07 | |
| Apolipoprotein A-I | APOA1 | 4,144,766.7 ± 798,136.1 | 2,013,133.3 ± 167,607.1 | 2.06 | |
| Alpha-1-acid glycoprotein 1 (A1AG1) | ORM1 | 95,798.0 ± 18,143.9 | 47,375.3 ± 3783.3 | 2.02 | |
| Hemopexin (HEMO) | HPX | 346,240.0 ± 72,507.6 | 172,841.7 ± 15,826.5 | 2.00 | |
| Actin-depolymerizing factor | GSN | 78,246.3 ± 13,025.7 | 39,946.8 ± 1462.5 | 1.96 | |
| Serotransferrin (TRFE) | TF | 2,125,016.7 ± 347,685.8 | 1,096,858.3 ± 76,391.0 | 1.94 | |
| Adenosylhomocysteinase (SAHH) | AHCY | 9665.6 ± 765.9 | 5004.9 ± 1092.1 | 1.93 | |
| Lumican | LUM | 206,390.0 ± 30,250.1 | 109,842.0 ± 11,393.3 | 1.88 | |
| ATP synthase subunit beta (ATPB) | ATP5F1B | 163,660.0 ± 11,615.0 | 87,713.2 ± 3889.4 | 1.87 | |
| Cu/Zn superoxide dismutase (SODC) | SOD1 | 69,312.8 ± 7322.5 | 37,554.7 ± 3418.6 | 1.85 | |
| Peroxiredoxin-6 | PRDX6 | 88,053.3 ± 12,976.1 | 47,894.5 ± 2267.1 | 1.84 | |
| Cystatin-B (CSTB) | Cystatin-B | 26,072.0 ± 4123.7 | 14,697.6 ± 1446.9 | 1.77 | |
| Transaldolase (TALDO) | TALDO1 | 12,790.6 ± 1895.0 | 7283.0 ± 1154.7 | 1.76 | |
| Plastin-3 | PLS3 | 32,357.3 ± 3587.1 | 18,817.3 ± 869.4 | 1.72 | |
| Pyruvate kinase (KPYM) | PKM | 171,291.7 ± 19,236.7 | 264,250.0 ± 9462.0 | 0.65 | |
| L-lactate dehydrogenase | LDHB | 100,577.0 ± 9101.6 | 155,815.0 ± 8110.5 | 0.65 | |
| Polyadenylate-binding protein (PABP2) | PABPN1 | 5875.5 ± 1150.7 | 9190.6 ± 365.0 | 0.64 | |
| Chloride intracellular channel protein 1 | CLIC1 | 9348.2 ± 1694.4 | 14,520.7 ± 453.0 | 0.64 | |
| Heat shock 70kDa protein 5 | HSPA5 | 186,508.3 ± 33,958.7 | 301,630.0 ± 5751.1 | 0.62 | |
| Heterogeneous nuclear ribonucleoprotein M | HNRNPM | 10,044.3 ± 1370.7 | 16,531.0 ± 530.7 | 0.61 | |
| Eukaryotic translation initiation factor 5A (IF5A1) | EIF5A | 19,182.3 ± 3339.0 | 32,350.0 ± 3133.9 | 0.59 | |
| Aggrecan core protein (PGCA) | ACAN | 49,556.0 ± 4794.4 | 85,088.7 ± 5985.1 | 0.58 | |
| Glutathione S-transferase P | GSTP1 | 24,142.5 ± 2739.0 | 44,152.7 ± 8435.2 | 0.55 | |
| Peptidylprolyl isomerase | PPWD1 | 3916.0 ± 800.8 | 7129.1 ± 562.9 | 0.55 | |
| Proteasome endopeptidase complex/PSB | PSMB | 2137.7 ± 470.8 | 3933.2 ± 136.7 | 0.54 | |
| Olfactomedin-like protein 3 (OLFL3) | OLFML3 | 9038.2 ± 1023.4 | 17,887.7 ± 2661.1 | 0.51 | |
| Putative ATP-dependent RNA helicase | DHX57 | 3944.2 ± 1136.7 | 7865.7 ± 679.4 | 0.50 | |
| Protein disulfide-isomerase | PDIA6 | 178,383.3 ± 22,517.8 | 390,683.3 ± 12,154.9 | 0.46 | |
| Calponin | CNN2 | 3543.7 ± 896.7 | 8039.1 ± 1697.6 | 0.44 | |
| Septin-7 (SEPT7) | SEPTIN7 | 3874.0 ± 1053.9 | 9256.4 ± 506.5 | 0.42 | |
| Thrombospondin-1 (TSP1) | THBS1 | 8282.2 ± 2059.1 | 20,655.8 ± 2272.9 | 0.40 | |
| Heterogeneous nuclear ribonucleoprotein (HNRPK) | HNRNPK | 9548.3 ± 1725.5 | 24,761.2 ± 969.2 | 0.39 | |
| Collagen alpha-1(II) chain (CO2A1) | COL2A1 | 26,222.3 ± 7333.0 | 67,945.7 ± 7406.7 | 0.39 | |
| Glucosidase 2 subunit beta (GLU2B) | PRKCSH | 2971.2 ± 894.4 | 8639.6 ± 737.5 | 0.34 | |
| Protein disulfide-isomerase (fragment) | PDIA6 | 105,751.3 ± 16,292.9 | 334,988.3 ± 8084.2 | 0.32 | |
| Heterogeneous nuclear ribonucleoproteins C1/C2 | HNRNPC | 1700.7 ± 405.7 | 6226.1 ± 350.6 | 0.27 | |
| Hyaluronan and proteoglycan link protein 1 (HPLN1) | HAPLN1 | 63,678.3 ± 6588.1 | 235,995.0 ± 14,162.8 | 0.27 | |
| Elongation factor 1-gamma (EF1G) | EEF1G | 6767.1 ± 1803.8 | 25,447.8 ± 1352.1 | 0.27 | |
| 60S acidic ribosomal protein P1 (RLA1) | RPLP1 | 3699.7 ± 955.8 | 15,261.1 ± 1570.3 | 0.24 | |
| 60S acidic ribosomal protein P2 (RLA2) | RPLP2 | 8009.4 ± 1102.0 | 35,171.8 ± 3258.9 | 0.23 | |
| Ribosome-associated molecular chaperone SSB1 | SSB1 | 2193.3 ± 778.7 | 10,186.9 ± 346.4 | 0.22 | |
| Elongation factor 1-delta (EF1D) | EEF1D | 6809.5 ± 1064.5 | 32,352.3 ± 928.9 | 0.21 | |
| Protein disulfide-isomerase A4 | PDIA4 | 15,204.7 ± 2296.0 | 77,964.3 ± 3747.7 | 0.20 | |
| Peptidyl-prolyl cis-trans isomerase (PIN4) | PIN4 | 25,527.3 ± 9262.3 | 132,480.8 ± 21,660.1 | 0.19 | |
| Hsc70-interacting protein (F10A1) | ST13 | 2395.8 ± 725.3 | 14,398.3 ± 804.9 | 0.17 | |
| Y-box-binding protein 1 (YBOX1) | YBX1 | 1189.9 ± 396.4 | 7032.0 ± 683.7 | 0.17 | |
| 60S acidic ribosomal protein P0 (RLA0) | RPLP0 | 1920.2 ± 265.3 | 12,717.7 ± 779.5 | 0.15 | |
| Endoplasmin (ENPL) | HSP90B1 | 27,822.7 ± 4283.1 | 243,358.3 ± 8040.5 | 0.11 |
| Protein Name | Gene Names | Rib | Middle Section | ||
|---|---|---|---|---|---|
| Mean ± SE | Mean ± SE | FC | |||
| Collagen alpha-2(I) chain (CO1Aa) | COL1A2 | 23,664,457.4 ± 4,097,670.2 | 481,201.8 ± 110,505.6 | 49.17 | |
| Fatty acid-binding protein, adipocyte | FABP4 | 477,239.0 ± 58,119.2 | 40,306.7 ± 5080.5 | 11.84 | |
| 40S ribosomal protein S12 | 29,511.2 ± 7966.9 | 2719.0 ± 682.7 | 10.85 | ||
| Keratin, type II cytoskeletal 6A | KRT6A | 340,515.4 ± 68,869.3 | 53,010.4 ± 16,401.4 | 6.42 | |
| Fructose-bisphosphate aldolase | ALDOA | 515,078.4 ± 93,413.7 | 81,491.0 ± 7750.3 | 6.32 | |
| Carbonic anhydrase (CAH3) | CA3 | 128,113.8 ± 21,821.4 | 22,310.6 ± 5010.6 | 5.74 | |
| Cellular retinoic acid-binding protein 2 | CRABP2 | 14,697.1 ± 3304.0 | 2776.5 ± 616.7 | 5.29 | |
| Hsc70-interacting protein (F10A1) | ST13 | 12,672.4 ± 1992.2 | 2395.8 ± 725.3 | 5.28 | |
| Keratin, type I cytoskeletal 10 | KRT10 | 360,962.0 ± 76,985.9 | 70,201.7 ± 19,902.1 | 5.14 | |
| Transaldolase (TALDO) | TALDO1 | 60,475.7 ± 6149.7 | 12,790.6 ± 1895.0 | 4.72 | |
| Y-box-binding protein 1 (YBOX1) | YBX1 | 5441.0 ± 1179.4 | 1189.9 ± 396.4 | 4.57 | |
| Annexin A2 (fragment) | ANXA2 | 5372.1 ± 764.6 | 1182.3 ± 309.8 | 4.54 | |
| Heterogeneous nuclear ribonucleoproteins C1/C2 | HNRNPC | 7177.4 ± 1158.7 | 1700.7 ± 405.7 | 4.22 | |
| Alpha-amylase 1A | AMY1A | 92,107.7 ± 12,024.1 | 22,315.3 ± 4160.5 | 4.13 | |
| Protein disulfide-isomerase A4 | PDIA4 | 61,477.6 ± 5982.7 | 15,204.7 ± 2296.0 | 4.04 | |
| 40S ribosomal protein S27a (RS27A) | RPS27A | 308,538.6 ± 23,001.6 | 78,794.3 ± 10,336.1 | 3.92 | |
| Glutathione S-transferase Mu 1 | GSTM1 | 349,396.0 ± 39,083.3 | 94,826.5 ± 25,325.8 | 3.68 | |
| Glucose-6-phosphate isomerase | GPI | 56,018.1 ± 7541.4 | 15,638.1 ± 1216.3 | 3.58 | |
| Heterogeneous nuclear ribonucleoprotein K(HNRPK) | HNRNPK | 33,963.0 ± 5938.7 | 9548.3 ± 1725.5 | 3.56 | |
| L-lactate dehydrogenase | LDHB | 339,487.1 ± 68,674.4 | 100,577.0 ± 9101.6 | 3.38 | |
| Polyadenylate-binding protein (PABP2) (fragment) | PABPN1 | 14,151.0 ± 3025.1 | 4320.8 ± 740.8 | 3.28 | |
| Olfactomedin-like protein 3 (OLFL3) | OLFML3 | 28,187.8 ± 2177.8 | 9038.2 ± 1023.4 | 3.12 | |
| Polypyrimidine tract-binding protein 1 | PTBP1 | 12,884.6 ± 1449.8 | 4236.4 ± 382.5 | 3.04 | |
| Putative ATP-dependent RNA helicase | DHX57 | 11,623.6 ± 2252.4 | 3944.2 ± 1136.7 | 2.95 | |
| Tetranectin | CLEC3B | 31,409.1 ± 5411.8 | 10,775.4 ± 2341.5 | 2.91 | |
| 60S acidic ribosomal protein P0 (RLA0) | RPLP0 | 5587.1 ± 730.0 | 1920.2 ± 265.3 | 2.91 | |
| Triosephosphate isomerase | TPI1 | 548,662.6 ± 54,009.9 | 192,349.8 ± 28.673,1 | 2.85 | |
| Ubiquitin-conjugating enzyme E2 variant 2 | UBE2V2 | 14,901.8 ± 2303.7 | 5305.7 ± 1703.4 | 2.81 | |
| 6-phosphogluconate dehydrogenase decarboxylating | PGD | 30,659.1 ± 3472.5 | 10,972.5 ± 2533.0 | 2.79 | |
| Lupus La protein | SSB | 6042.3 ± 964.5 | 2193.3 ± 781.8 | 2.75 | |
| UDP-glucose 6-dehydrogenase | UGDH | 8707.4 ± 1631.5 | 3215.2 ± 674.1 | 2.71 | |
| Malate dehydrogenase, cytoplasmic | MDH1 | 11,937.7 ± 2189.3 | 4467.4 ± 1815.3 | 2.67 | |
| Decorin | DCN | 203,457.1 ± 19,766.6 | 78,507.4 ± 9628.3 | 2.59 | |
| Phosphoglycerate kinase 1 | PGK1 | 102,757.0 ± 15,094.9 | 39,944.3 ± 3442.8 | 2.57 | |
| Peroxiredoxin-6 | PRDX6 | 226,238.3 ± 16,359.3 | 88,053.3 ± 12,976.1 | 2.57 | |
| Cu/Zn superoxide dismutase (SODC) | SOD1 | 173,733.6 ± 23,314.9 | 69,312.8 ± 7322.5 | 2.51 | |
| Biglycan | BGN | 100,093.6 ± 13,826.5 | 41,794.6 ± 5319,2 | 2.39 | |
| Elongation factor 1-gamma (EF1G) | EEF1G | 15,255.3 ± 2163.9 | 6767.1 ± 1803.8 | 2.25 | |
| Glutathione S-transferase P | GSTP1 | 54,208.3 ± 5030.5 | 24,142.5 ± 2739.0 | 2.25 | |
| Glucosidase 2 subunit beta (GLU2B) | PRKCSH | 6611.3 ± 939.7 | 2971.2 ± 894.4 | 2.23 | |
| Chloride intracellular channel protein 1 | CLIC1 | 20,704.72 ± 3379.9 | 9348.2 ± 1694.4 | 2.21 | |
| Glutathione S-transferase P (fragment) | GSTP1 | 42,414.8 ± 7016.7 | 19,194.5 ± 5.535.9 | 2.21 | |
| 60S acidic ribosomal protein P2 (RLA2) | RPLP2 | 17,289.1 ± 1634.2 | 8009.4 ± 1102.0 | 2.16 | |
| Alpha-2-HS-glycoprotein | AHSG | 274,143.4 ± 36,049.6 | 129,638.6 ± 29,231.7 | 2.11 | |
| Transgelin | TAGLN | 84,419.6 ± 10,442.4 | 40,987.1 ± 5652.8 | 2.06 | |
| Transgelin (fragment) | TAGLN | 37,823.7 ± 3877.1 | 18,390.6 ± 2628.0 | 2.06 | |
| Calponin | CNN2 | 7285.9 ± 849.1 | 3543.7 ± 896.7 | 2.06 | |
| Polyadenylate-binding protein (PABP2) | PABPN1 | 11,923.6 ± 887.0 | 5875.5 ± 1150.7 | 2.03 | |
| Phosphatidylethanolamine-binding protein 1 | PEBP1 | 17,399.2 ± 2682.0 | 8613.4 ± 1756.9 | 2.02 | |
| Fatty acid-binding protein 5 | FABP5 | 7163.9 ± 843.8 | 3625.2 ± 699.1 | 1.98 | |
| Heterogeneous nuclear ribonucleoprotein H | HNRNPH1 | 31,298.3 ± 2793.2 | 16,102.7 ± 1632.7 | 1.94 | |
| Endoplasmin (ENPL) | HSP90B1 | 51,370.1 ± 3589.4 | 27,822.7 ± 4283.1 | 1.85 | |
| Lumican | LUM | 380,040.3 ± 43,044.1 | 206,390.0 ± 30,250.1 | 1.84 | |
| Transitional endoplasmic reticulum ATPase | VCP | 68,467.5 ± 8692.5 | 39,195.5 ± 4453.9 | 1.75 | |
| Phosphatidylethanolamine-binding protein 1 | PEBP1 | 42,253.9 ± 5040.5 | 24,291.3 ± 4633.2 | 1.74 | |
| Peptidyl-prolyl cis-trans isomerase (PIN4) | PIN4 | 375,942.8 ± 29,644.9 | 25,527.3 ± 9262.3 | 1.61 | |
| UMP-CMP kinase | CMPK1 | 6599.5 ± 647.2 | 4105.2 ± 500.7 | 1.61 | |
| Pyruvate kinase (KPYM) | PKM | 261,337.6 ± 27,813.7 | 171,291.7 ± 19,236.7 | 1.53 | |
| Galectin 1 | LGALS1 | 49,839.4 ± 5621.5 | 76,707.49 ± 10,793.0 | 0.65 | |
| Nucleobindin-1 | NUCB1 | 24,434.2 ± 3012.5 | 38,920.5 ± 4924.8 | 0.63 | |
| Heparin cofactor 2 (HEP2) | SERPIND1 | 11,873.5 ± 1969.7 | 19,863.8 ± 3310.1 | 0.60 | |
| Elongation factor 2 | EEF2 | 51,547.7 ± 4948.4 | 86,627.3 ± 12,626.1 | 0.60 | |
| Plastin-3 | PLS3 | 18,711.7 ± 2677.2 | 32,357.3 ± 3587.1 | 0.58 | |
| Heat shock protein 70 1A | HSPA1A | 65,543.3 ± 8657.8 | 114,598.05 ± 8998.6 | 0.57 | |
| Serotransferrin (TRFE) | TF | 1,215,178.8 ± 193,911.2 | 2,125,016.7 ± 347,685.8 | 0.57 | |
| Inter-alpha-trypsin inhibitor heavy chain H2 | ITIH2 | 47,937.9 ± 5216.0 | 87,390.0 ± 17,580.9 | 0.55 | |
| Apolipoprotein A-IV | APOA4 | 45,231.5 ± 4775.7 | 85,435.1 ± 9548.5 | 0.53 | |
| Nucleoside diphosphate kinase mitochondrial | NME4 | 67,760.2 ± 5520.7 | 129,968.3 ± 14,280.8 | 0.52 | |
| Fibrinogen alpha chain | FGA | 135,860.8 ± 21,986.1 | 291,686.2 ± 55,713.2 | 0.47 | |
| Vimentin | VIM | 156,778.7 ± 17,997.7 | 341,121.7 ± 67,935.6 | 0.46 | |
| Hemopexin | HPX | 79,373.2 ± 11,927.3 | 173,172.2 ± 38,801.3 | 0.46 | |
| Afamin (AFAM) | AFM | 10,596.6 ± 1972.3 | 25,198.7 ± 5103.3 | 0.42 | |
| Alpha-1B-glycoprotein | A1BG | 878,594.0 ± 101,549.5 | 2,213,076.7 ± 441,457.5 | 0.40 | |
| Thioredoxin domain-containing protein 5 | TXNDC5 | 47,903.8 ± 6878.6 | 121,210.10 ± 10,595.2 | 0.40 | |
| Eukaryotic translation initiation factor 5A (IF5A1) | EIF5A | 7520.5 ± 1315.2 | 19,182.3 ± 3339.0 | 0.39 | |
| Transthyretin | TTR | 98,470.6 ± 19,719.4 | 251,207.1 ± 73,458.0 | 0.39 | |
| Apolipoprotein A-I | APOA1 | 1,599,381.1 ± 272,199.7 | 4,144,766.7 ± 798,136.1 | 0.39 | |
| Creatine kinase B-type | CKB | 119,883.7 ± 15,308.5 | 311,250.0 ± 37,004.7 | 0.39 | |
| Amiloride-sensitive amine oxidase | AOC1 | 14,519.1 ± 3374.7 | 39,327.2 ± 6441.8 | 0.37 | |
| Hemoglobin subunit alpha | HBA/SCN2A | 905,083.4 ± 296,103.9 | 2,563,475.0 ± 869.235.6 | 0.35 | |
| Antithrombin-III (ANT3) | SERPINC1 | 22,535.4 ± 2287.5 | 64,056.0 ± 10,572.3 | 0.35 | |
| Apolipoprotein A-II | APOA2 | 20,745.2 ± 5114.2 | 60,995.7 ± 16,710.0 | 0.34 | |
| Collagen alpha-1(I) chain | COL1A1 | 87,968.2 ± 11,426.9 | 275,756.5 ± 54,450.2 | 0.32 | |
| Complement factor B (CFAB) | CFB | 27,628.1 ± 3354.1 | 90,554.5 ± 16,717.9 | 0.31 | |
| Histidine-rich glycoprotein | HRG | 46,388.9 ± 4259.7 | 159,018.1 ± 45,430.2 | 0.29 | |
| Albumin | ALB | 4,096,318.3 ± 713,885.6 | 19,510,270.2 ± 3,628,081.0 | 0.21 | |
| Plasminogen (PLMN) | PLG | 7232.5 ± 2021.8 | 37,045.7 ± 9978.9 | 0.20 | |
| Complement C3 | C3 | 131,933.9 ± 14,398.5 | 712,043.3 ± 109,393.3 | 0.19 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
López-Pedrouso, M.; Lorenzo, J.M.; Landete-Castillejos, T.; Chonco, L.; Pérez-Barbería, F.J.; García, A.; López-Garrido, M.-P.; Franco, D. SWATH-MS Quantitative Proteomic Analysis of Deer Antler from Two Regenerating and Mineralizing Sections. Biology 2021, 10, 679. https://doi.org/10.3390/biology10070679
López-Pedrouso M, Lorenzo JM, Landete-Castillejos T, Chonco L, Pérez-Barbería FJ, García A, López-Garrido M-P, Franco D. SWATH-MS Quantitative Proteomic Analysis of Deer Antler from Two Regenerating and Mineralizing Sections. Biology. 2021; 10(7):679. https://doi.org/10.3390/biology10070679
Chicago/Turabian StyleLópez-Pedrouso, María, José M. Lorenzo, Tomás Landete-Castillejos, Louis Chonco, Francisco Javier Pérez-Barbería, Andrés García, María-Pilar López-Garrido, and Daniel Franco. 2021. "SWATH-MS Quantitative Proteomic Analysis of Deer Antler from Two Regenerating and Mineralizing Sections" Biology 10, no. 7: 679. https://doi.org/10.3390/biology10070679
APA StyleLópez-Pedrouso, M., Lorenzo, J. M., Landete-Castillejos, T., Chonco, L., Pérez-Barbería, F. J., García, A., López-Garrido, M.-P., & Franco, D. (2021). SWATH-MS Quantitative Proteomic Analysis of Deer Antler from Two Regenerating and Mineralizing Sections. Biology, 10(7), 679. https://doi.org/10.3390/biology10070679