Unveiling the Fecal Microbiota in Two Captive Mexican Wolf (Canis lupus baileyi) Populations Receiving Different Type of Diets
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. General
2.2. Location, Experimental Sites, Environmental Conditions and Animal Diets
2.3. Sampling According to Gender, Age, and Type of Animal Grouping
2.4. DNA Extraction and Visualization
2.5. 16S rRNA Gene Amplicon Sequencing
2.6. Bio-Informatic Analyses
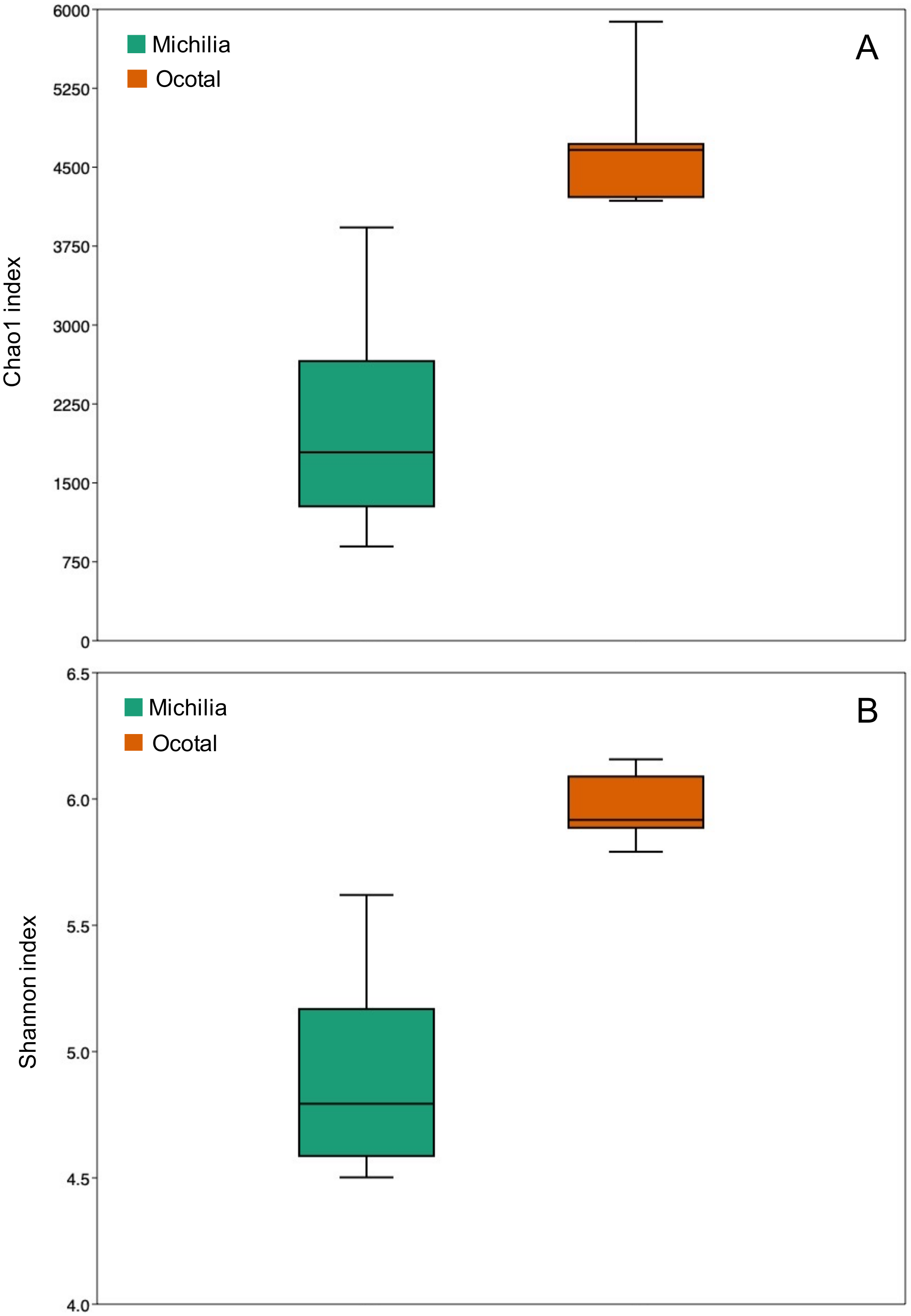
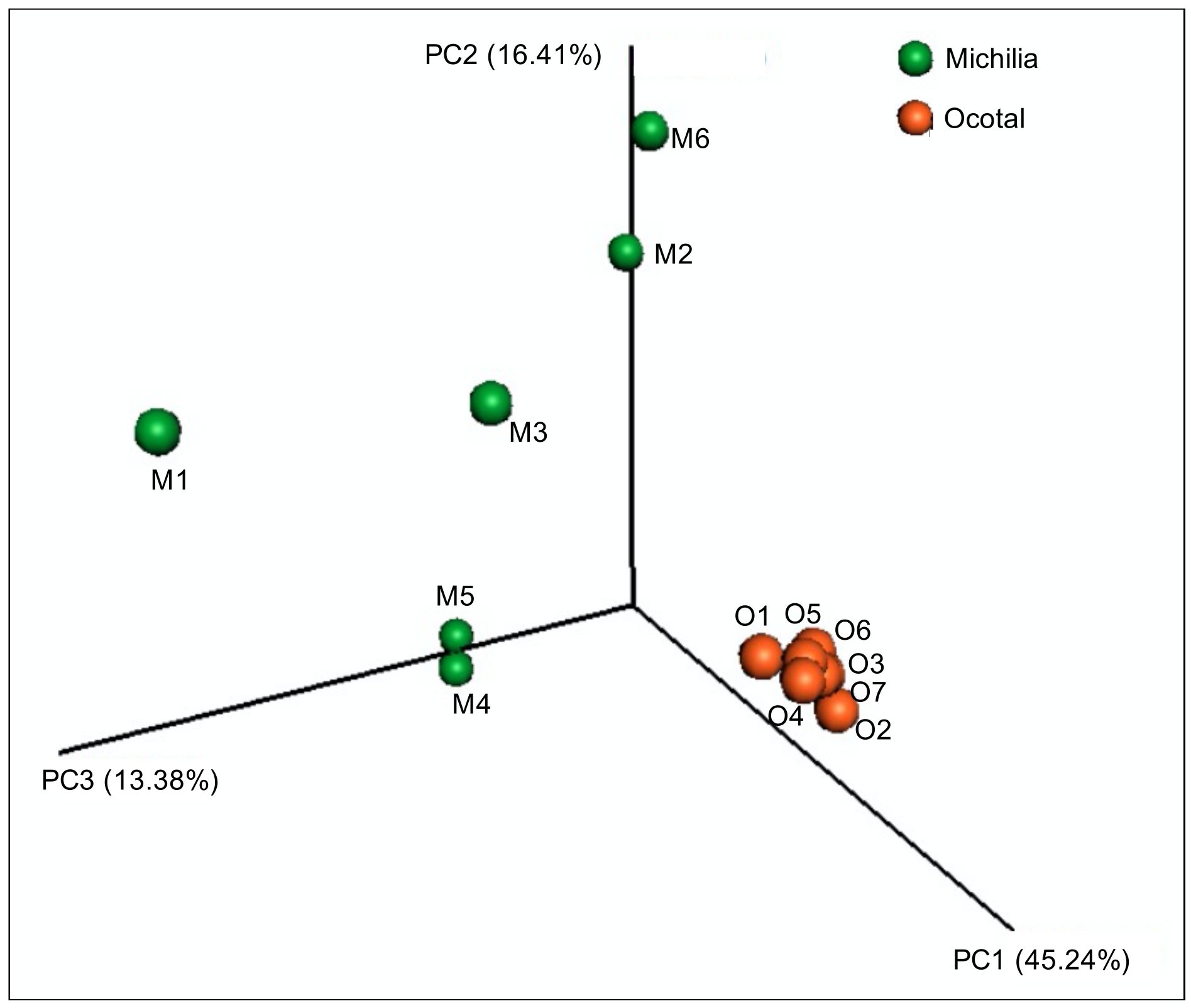
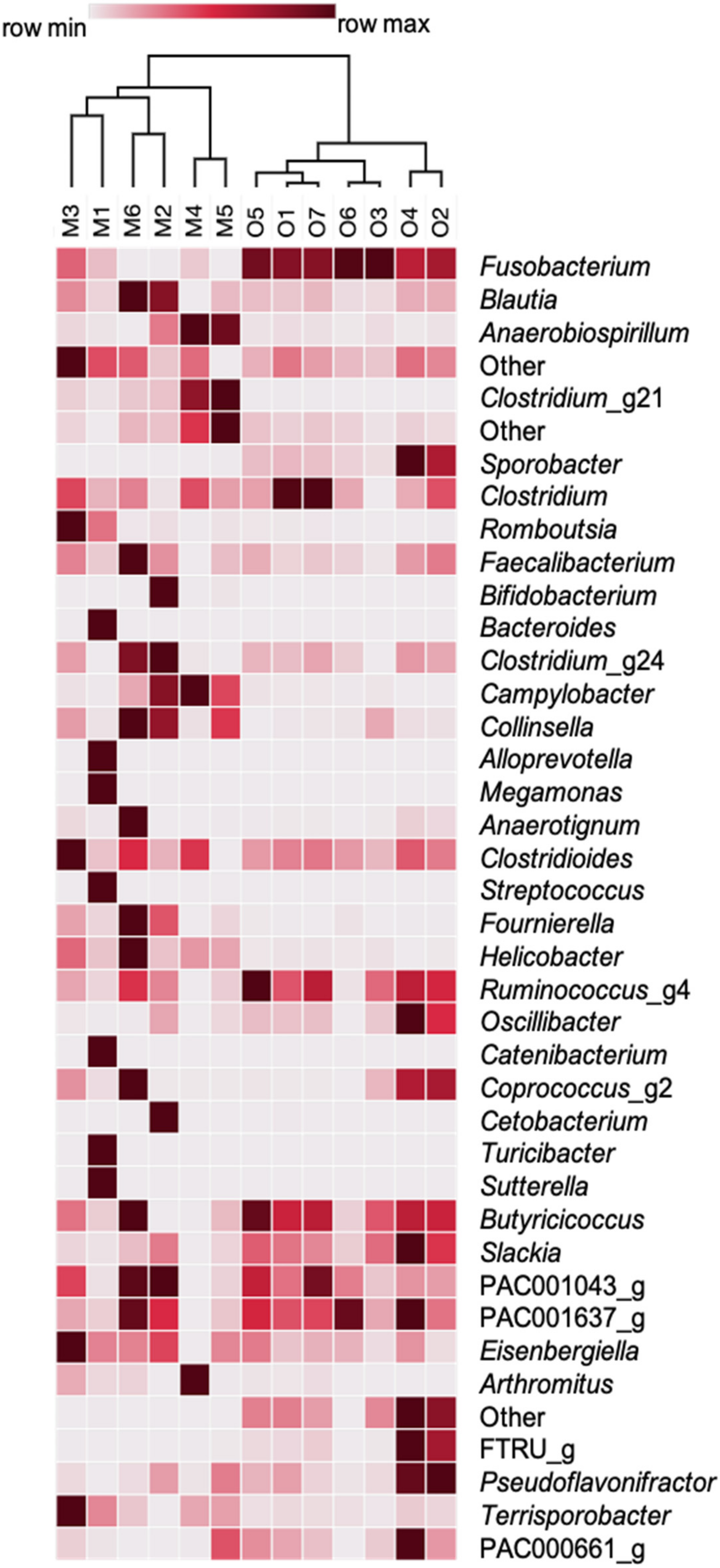
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Ethics Statement
References
- Hale, V.L.; Tan, C.L.; Knight, R.; Amato, K.R. Effect of preservation method on spider monkey (Ateles geoffroyi) fecal microbiota over 8 weeks. J. Microbiol. Methods 2015, 113, 16–26. [Google Scholar] [CrossRef] [PubMed]
- Borbón-García, A.; Reyes, A.; Vives-Flórez, M.; Caballero, S. Captivity shapes the gut microbiota of andean bears: Insights into health surveillance. Front. Microbiol. 2017, 8, 1316. [Google Scholar] [CrossRef] [Green Version]
- Stumpf, R.M.; Gomez, A.; Amato, K.R.; Yeoman, C.J.; Polk, J.D.; Wilson, B.A.; Nelson, K.E.; White, B.A.; Leigh, S.R. Microbiomes, metagenomics, and primate conservation: New strategies, tools, and applications. Biol. Conserv. 2016, 199, 56–66. [Google Scholar] [CrossRef] [Green Version]
- Cunningham, J.G.; Klein, B.G. Veterinary Physiology; Saunders Elsevier: Philadelphia, PA, USA, 2007. [Google Scholar]
- Pagliari, D.; Piccirillo, C.A.; Larbi, A.; Cianci, R. The interactions between innate immunity and microbiota in gastrointestinal diseases. J. Immunol. Res. 2015, 898297. [Google Scholar] [CrossRef]
- West, A.; Waite, D.; Deines, P.; Bourne, D.; Digby, A.; McKenzie, V.; Taylor, M. The microbiome in threatened species conservation. Biol. Conserv. 2019, 229, 85–98. [Google Scholar] [CrossRef]
- Tag, A.C. Large Canid (Canidae) Care Manual; Association of Zoos and Aquariums: Silver Spring, MD, USA, 2012. [Google Scholar]
- Bragg, M.; Freeman, E.W.; Lim, H.C.; Songsasen, N.; Muletz-Wolz, C.R. Gut Microbiomes Differ Among Dietary Types and Stool Consistency in the Captive Red Wolf (Canis rufus). Front. Microbiol. 2020, 11, 2777. [Google Scholar] [CrossRef]
- Wu, X.; Zhang, H.; Chen, J.; Shang, S.; Yan, J.; Chen, Y.; Tang, X.; Zhang, H. Analysis and comparison of the wolf microbiome under different environmental factors using three different data of Next Generation Sequencing. Sci. Rep. 2017, 7, 11332. [Google Scholar] [CrossRef] [Green Version]
- Lyu, T.; Liu, G.; Zhang, H.; Wang, L.; Zhou, S.; Dou, H.; Pang, B.; Sha, W.; Zhang, H. Changes in feeding habits promoted the differentiation of the composition and function of gut microbiotas between domestic dogs (Canis lupus familiaris) and gray wolves (Canis lupus). AMB Express 2018, 8, 1–12. [Google Scholar] [CrossRef]
- Carciofi, A.; Sakomura, N.; Kawauchi, I.; Vasconcellos, R. Digestibility and metabolizable energy of some carbohydrate sources for dogs. Anim. Feed Sci. Technol. 2010, 156, 121–125. [Google Scholar]
- Hang, I.; Rinttila, T.; Zentek, J.; Kettunen, A.; Alaja, S.; Apajalahti, J.; Harmoinen, J.; de Vos, W.M.; Spillmann, T. Effect of high contents of dietary animal-derived protein or carbohydrates on canine faecal microbiota. BMC Vet. Res. 2012, 8, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Fitak, R.R.; Rinkevich, S.E.; Culver, M. Genome-wide analysis of SNPs is consistent with no domestic dog ancestry in the endangered Mexican wolf (Canis lupus baileyi). J. Hered. 2018, 109, 372–383. [Google Scholar] [CrossRef]
- Servin, J. El periodo de apareamiento, nacimiento y crecimiento del lobo mexicano (Canis lupus baileyi). Acta Zoológica Mex. Nueva Ser. 1997, 71, 45–56. [Google Scholar]
- Sinding, M.-H.S.; Gopalakrishan, S.; Vieira, F.G.; Samaniego Castruita, J.A.; Raundrup, K.; Heide Jørgensen, M.P.; Meldgaard, M.; Petersen, B.; Sicheritz-Ponten, T.; Mikkelsen, J.B. Population genomics of grey wolves and wolf-like canids in North America. PLoS Genet. 2018, 14, e1007745. [Google Scholar] [CrossRef] [Green Version]
- Galindo, C. Recuperación del lobo Mexicano. In Patrimonio Natural de México. Cien Casos de Exito; Carabias, J., Sarukhán, J., de la Maza y, J., Galindo, C., Eds.; Comisión Nacional para el Conocimiento y Uso de la Biodiversidad: México, Mexico, D.F., 2010; pp. 80–81. [Google Scholar]
- SEGOB (Secretaría de Gobernación). NORMA Oficial Mexicana NOM-059-SEMARNAT-2010, Protección Ambiental-Especies Nativas de México de Flora y Fauna Silvestres-Categorías de Riesgo y Especificaciones para su Inclusión, Exclusión o Cambio-Lista de Especies en Riesgo; Diario Oficial de la Federación: Ciudad de México, Mexico, 2010. [Google Scholar]
- USFWS (U.S. Fish and Wildlife Service). Mexican Wolf Recovery Plan, First Revision. Region 2, Albuquerque, New Mexico, USA. 2017. Available online: https://www.fws.gov/southwest/es/mexicanwolf/pdf/2017MexicanWolfRecoveryPlanRevision1Final.pdf (accessed on 15 February 2021).
- SEMARNAT. Programa de Acción para la Conservación de la Especie, Lobo Gris Mexicano (Canis lupus baileyi). 2009. Available online: https://www.gob.mx/cms/uploads/attachment/file/251983/PACE_Lobo_Mexicano_2009.pdf (accessed on 16 February 2021).
- FASS. Guide for the Care and Use of Agricultural Animals in Agricultural Research and Teaching, 3rd ed.; Animal Science Journal: Champaing, IL, USA, 2010; p. 177. [Google Scholar]
- Gonzalez-Elizondo, S.; Gonzalez-Elizondo, M.; Cortes-Ortiz, A. Vegetación de la reserva de la biosfera La Michilia, Durango, México. Acta Botánica Mex. 1993, 22, 1–104. [Google Scholar] [CrossRef] [Green Version]
- Aguilar-Campos, P. Diagnóstico Ambiental de la Zona Norte del Parque Estatal El Ocotal, en Timilpan, Estado de México. Bachelor’s Thesis, Universidad Nacional Autónoma de México, México City, Mexico, 2009. [Google Scholar]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glockner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef]
- Illumina. 16S Metagenomic Sequencing Library Preparation, Preparing 16S Ribosomal RNA Gene Amplicons for the Illumina MiSeq System. 2020. Available online: https://support.illumina.com/documents/documentation/chemistry_documentation/16s/16s-metagenomic-library-prep-guide-15044223-b.pdf (accessed on 11 May 2021).
- Illumina. Nextera XT DNA Library Prep Kit Reference Guide. Available online: https://support.illumina.com/content/dam/illumina-support/documents/documentation/chemistry_documentation/samplepreps_nextera/nextera-xt/nextera-xt-library-prep-reference-guide-15031942-05.pdf (accessed on 11 May 2021).
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Pena, A.G.; Goodrich, J.K.; Gordon, J.I. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Kobert, K.; Flouri, T.; Stamatakis, A. PEAR: A fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics 2014, 30, 614–620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoon, S.-H.; Ha, S.-M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613. [Google Scholar] [CrossRef] [PubMed]
- Beals, E.W. Bray-Curtis ordination: An effective strategy for analysis of multivariate ecological data. Adv. Ecol. Res. 1984, 14, 1–55. [Google Scholar]
- Vazquez-Baeza, Y.; Pirrung, M.; Gonzalez, A.; Knight, R. EMPeror: A tool for visualizing high-throughput microbial community data. GigaScience 2013, 2, 16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clarke, K.R. Non-parametric multivariate analyses of changes in community structure. Aust. J. Ecol. 1993, 18, 117–143. [Google Scholar] [CrossRef]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmidt, M.; Unterer, S.; Suchodolski, J.S.; Honneffer, J.B.; Guard, B.C.; Lidbury, J.A.; Steiner, J.M.; Fritz, J.; Kölle, P. The fecal microbiome and metabolome differs between dogs fed Bones and Raw Food (BARF) diets and dogs fed commercial diets. PLoS ONE 2018, 13, e0201279. [Google Scholar] [CrossRef] [Green Version]
- Isaiah, A.; Parambeth, J.C.; Steiner, J.M.; Lidbury, J.A.; Suchodolski, J.S. The fecal microbiome of dogs with exocrine pancreatic insufficiency. Anaerobe 2017, 45, 50–58. [Google Scholar] [CrossRef]
- Trinh, P.; Zaneveld, J.R.; Safranek, S.; Rabinowitz, P.M. One health relationships between human, animal, and environmental microbiomes: A mini-review. Front. Public Health 2018, 6, 235. [Google Scholar] [CrossRef]
- Garcia-Mazcorro, J.F.; Dowd, S.E.; Poulsen, J.; Steiner, J.M.; Suchodolski, J.S. Abundance and short-term temporal variability of fecal microbiota in healthy dogs. Microbiol. Open 2012, 1, 340–347. [Google Scholar] [CrossRef]
- Hand, D.; Wallis, C.; Colyer, A.; Penn, C.W. Pyrosequencing the canine faecal microbiota: Breadth and depth of biodiversity. PLoS ONE 2013, 8, e53115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Honneffer, J.B.; Steiner, J.M.; Lidbury, J.A.; Suchodolski, J.S. Variation of the microbiota and metabolome along the canine gastrointestinal tract. Metabolomics 2017, 13, 26. [Google Scholar] [CrossRef]
- Bermingham, E.N.; Maclean, P.; Thomas, D.G.; Cave, N.J.; Young, W. Key bacterial families (Clostridiaceae, Erysipelotrichaceae and Bacteroidaceae) are related to the digestion of protein and energy in dogs. PeerJ 2017, 5, e3019. [Google Scholar] [CrossRef] [PubMed]
- Song, S.J.; Lauber, C.; Costello, E.K.; Lozupone, C.A.; Humphrey, G.; Berg-Lyons, D.; Caporaso, J.G.; Knights, D.; Clemente, J.C.; Nakielny, S.; et al. Cohabiting family members share microbiota with one another and with their dogs. eLife 2013, 2, e00458. [Google Scholar] [CrossRef]
- Pilla, R.; Suchodolski, J.S. The role of the canine gut microbiome and metabolome in health and gastrointestinal disease. Front. Vet. Sci. 2020, 6, 498. [Google Scholar] [CrossRef] [Green Version]
- Reed, J.E.; Ballard, W.B.; Gipson, P.S.; Kelly, B.T.; Krausman, P.R.; Wallace, M.C.; Wester, D.B. Diets of free-ranging Mexican gray wolves in Arizona and New Mexico. Wildl. Soc. Bull. 2006, 34, 1127–1133. [Google Scholar] [CrossRef]
- Merkle, J.; Stahler, D.; Smith, D. Interference competition between gray wolves and coyotes in Yellowstone National Park. Can. J. Zool. 2009, 87, 56–63. [Google Scholar] [CrossRef]
- Zhu, L.; Wu, Q.; Deng, C.; Zhang, M.; Zhang, C.; Chen, H.; Lu, G.; Wei, F. Adaptive evolution to a high purine and fat diet of carnivorans revealed by gut microbiomes and host genomes. Environ. Microbiol. 2018, 20, 1711–1722. [Google Scholar] [CrossRef]
- USFWS (U.S. Fish and Wildlife Service). Mexican Gray Wolf Husbandry Manual: Guidelines for Captive Management. 2009. Available online: https://www.fws.gov/southwest/es/mexicanwolf/pdf/Mexican_Wolf_Husbandry_Manual_2009.pdf (accessed on 15 February 2021).
- Henson, L.H.; Songsasen, N.; Waddell, W.; Wolf, K.N.; Emmons, L.; Gonzalez, S.; Freeman, E.; Maldonado, J. Characterization of genetic variation and basis of inflammatory bowel disease in the Toll-like receptor 5 gene of the red wolf and the maned wolf. Endanger. Species Res. 2017, 32, 135–144. [Google Scholar] [CrossRef] [Green Version]
- Alessandri, G.; Milani, C.; Mancabelli, L.; Mangifesta, M.; Lugli, G.A.; Viappiani, A.; Duranti, S.; Turroni, F.; Ossiprandi, M.C.; van Sinderen, D. The impact of human-facilitated selection on the gut microbiota of domesticated mammals. FEMS Microbiol. Ecol. 2019, 95, fiz121. [Google Scholar] [CrossRef] [PubMed]
- Moxham, G. Waltham feces scoring system-A tool for veterinarians and pet owners: How does your pet rate. Walth. Focus 2001, 11, 24–25. [Google Scholar]
- Herstad, K.M.; Gajardo, K.; Bakke, A.M.; Moe, L.; Ludvigsen, J.; Rudi, K.; Rud, I.; Sekelja, M.; Skancke, E. A diet change from dry food to beef induces reversible changes on the faecal microbiota in healthy, adult client-owned dogs. BMC Vet. Res. 2017, 13, 1–13. [Google Scholar] [CrossRef]
- Kim, J.; An, J.U.; Kim, W.; Lee, S.; Cho, S. Differences in the gut microbiota of dogs (Canis lupus familiaris) fed a natural diet or a commercial feed revealed by the Illumina MiSeq platform. Gut Pathog. 2017, 9, 68. [Google Scholar] [CrossRef] [Green Version]
- Xenoulis, P.G.; Palculict, B.; Allenspach, K.; Steiner, J.M.; Van House, A.M.; Suchodolski, J.S. Molecular-phylogenetic characterization of microbial communities imbalances in the small intestine of dogs with inflammatory bowel disease. FEMS Microbiol. Ecol. 2008, 66, 579–589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Minamoto, Y.; Otoni, C.C.; Steelman, S.M.; Büyükleblebici, O.; Steiner, J.M.; Jergens, A.E.; Suchodolski, J.S. Alteration of the fecal microbiota and serum metabolite profiles in dogs with idiopathic inflammatory bowel disease. Gut Microbes 2015, 6, 33–47. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Location | Sex | Age (Years) | Grouping |
|---|---|---|---|
| M1 | Female | 3 | Cohabitating with M2 |
| M2 | Female | 3 | Cohabitating with M1 |
| M3 | Male | 12 | Alone |
| M4 | Male | 2 | Cohabitating with M5 and M6 |
| M5 | Male | 2 | Cohabitating with M4 AND M6 |
| M6 | Male | 2 | Cohabitating with M4 AND M5 |
| O1 | Male | 3 | Pack |
| O2 | Male | 3 | Pack |
| O3 | Male | 1 | Pack |
| O4 | Female | 5 | Pack |
| O5 | Female | 5 | Pack |
| O6 | Female | 1 | Pack |
| O7 | Female | 1 | Pack |
| Location | Total Reads | Assembled Reads | QB 1 | OTUs 2 |
|---|---|---|---|---|
| M1 | 142,533 | 42,013 | 35,441 | 735 |
| M2 | 190,819 | 20,205 | 13,051 | 1074 |
| M3 | 168,432 | 49,194 | 31,220 | 2537 |
| M4 | 150,884 | 49,018 | 33,674 | 1836 |
| M5 | 132,288 | 41,866 | 25,773 | 1631 |
| M6 | 181,892 | 28,113 | 19,643 | 1038 |
| Mean | 161,141 | 38,402 | 26,467 | 1475 |
| O1 | 119,488 | 46,215 | 28,913 | 3085 |
| O2 | 141,153 | 61,695 | 43,526 | 3506 |
| O3 | 140,534 | 64,251 | 44,318 | 3512 |
| O4 | 151,679 | 76,461 | 54,132 | 3891 |
| O5 | 119,588 | 51,454 | 34,252 | 3430 |
| O6 | 128,670 | 57,787 | 35,222 | 3554 |
| O7 | 89,352 | 38,777 | 18,936 | 3165 |
| Mean | 127,209 | 56,663 | 37,043 | 3449 |
| Taxon | AVD | Contribution % | Cumulative % | Mean M | Mean O | U | p-Value |
|---|---|---|---|---|---|---|---|
| Phylum | |||||||
| Fusobacteria | 32.93 | 49.87 | 49.87 | 0.085 | 0.744 | 0 | 0.001 |
| Firmicutes | 17.63 | 26.69 | 76.56 | 0.589 | 0.238 | 1 | 0.001 |
| Proteobacteria | 9.58 | 14.51 | 91.07 | 0.202 | 0.011 | 0 | 0.001 |
| Bacteroidetes | 3.07 | 4.64 | 95.72 | 0.061 | 0.000 | 14 | 0.353 |
| Actinobacteria | 2.81 | 4.26 | 99.98 | 0.062 | 0.007 | 13 | 0.283 |
| Family | |||||||
| Fusobacteriaceae | 32.93 | 44.01 | 44.01 | 0.085 | 0.744 | 0 | 0.001 |
| Lachnospiraceae | 14.09 | 18.83 | 62.84 | 0.350 | 0.082 | 6 | 0.038 |
| Succinivibrionaceae | 7.75 | 10.36 | 73.20 | 0.160 | 0.009 | 12 | 0.224 |
| Peptostreptococcaceae | 4.92 | 6.57 | 79.77 | 0.126 | 0.049 | 13 | 0.283 |
| Ruminococcaceae | 3.30 | 4.41 | 84.18 | 0.046 | 0.075 | 15 | 0.432 |
| Bifidobacteriaceae | 1.94 | 2.59 | 86.77 | 0.039 | 0.000 | - | - |
| Bacteroidaceae | 1.89 | 2.52 | 89.29 | 0.038 | 0.000 | 14 | 0.350 |
| Campylobacteraceae | 1.43 | 1.90 | 91.19 | 0.029 | 0.001 | 5 | 0.026 |
| Prevotellaceae | 1.18 | 1.58 | 92.77 | 0.024 | 0.000 | - | - |
| Clostridiaceae | 1.14 | 1.52 | 94.29 | 0.019 | 0.027 | 20 | 0.943 |
| Selenomonadaceae | 1.13 | 1.51 | 95.80 | 0.023 | 0.000 | - | - |
| Coriobacteriaceae | 0.89 | 1.19 | 96.99 | 0.023 | 0.007 | 13 | 0.283 |
| Streptococcaceae | 0.77 | 1.03 | 98.02 | 0.015 | 0.000 | - | - |
| Helicobacteraceae | 0.46 | 0.62 | 98.64 | 0.010 | 0.000 | 0 | 0.003 |
| Genus | |||||||
| Fusobacterium | 33.14 | 41.80 | 41.80 | 0.081 | 0.744 | 0 | 0.001 |
| Anaerobiospirillum | 7.75 | 9.78 | 51.58 | 0.160 | 0.008 | 12 | 0.224 |
| Blautia | 6.71 | 8.47 | 60.05 | 0.157 | 0.045 | 15 | 0.432 |
| Clostridium_g21 | 4.50 | 5.68 | 65.73 | 0.091 | 0.001 | 0 | 0.001 |
| Other | 2.51 | 3.16 | 68.89 | 0.069 | 0.039 | 13 | 0.283 |
| Sporobacter | 2.31 | 2.91 | 71.80 | 0.000 | 0.046 | 0 | 0.001 |
| Romboutsia | 2.10 | 2.65 | 74.45 | 0.043 | 0.001 | 5 | 0.026 |
| Other | 2.06 | 2.59 | 77.04 | 0.048 | 0.011 | 13 | 0.283 |
| Bifidobacterium | 1.94 | 2.44 | 79.48 | 0.039 | 0.000 | - | - |
| Bacteroides | 1.89 | 2.38 | 81.86 | 0.038 | 0.000 | 14 | 0.350 |
| Campylobacter | 1.43 | 1.80 | 83.66 | 0.029 | 0.001 | 5 | 0.026 |
| Faecalibacterium | 1.20 | 1.52 | 85.17 | 0.029 | 0.013 | 16 | 0.520 |
| Alloprevotella | 1.18 | 1.49 | 86.66 | 0.024 | 0.000 | - | - |
| Megamonas | 1.13 | 1.43 | 88.09 | 0.023 | 0.000 | - | - |
| Clostridium | 1.08 | 1.37 | 89.45 | 0.016 | 0.027 | 20 | 0.943 |
| Anaerotignum | 1.01 | 1.28 | 90.73 | 0.020 | 0.002 | 19 | 0.829 |
| Collinsella | 0.90 | 1.14 | 91.87 | 0.022 | 0.005 | 6 | 0.038 |
| Clostridium_g24 | 0.84 | 1.06 | 92.93 | 0.021 | 0.010 | 19 | 0.830 |
| Streptococcus | 0.76 | 0.96 | 93.90 | 0.015 | 0.000 | - | - |
| Fournierella | 0.62 | 0.78 | 94.68 | 0.013 | 0.000 | 6.5 | 0.045 |
| Helicobacter | 0.46 | 0.58 | 95.26 | 0.010 | 0.000 | 0 | 0.001 |
| Clostridioides | 0.45 | 0.56 | 95.83 | 0.012 | 0.008 | 20 | 0.943 |
| Oscillibacter | 0.20 | 0.26 | 96.35 | 0.001 | 0.004 | 9 | 0.100 |
| Turicibacter | 0.17 | 0.22 | 96.81 | 0.003 | 0.000 | - | - |
| Core Microbiota | ||
|---|---|---|
| Genera/Species | Michilia | Ocotal |
| Agathobaculum | - | * |
| Anaerobiospirillum | - | * |
| Blautia | * | * |
| Butyricicoccus | - | * |
| Campylobacter | * | * |
| Clostridium | * | * |
| Clostridium_g21 | * | * |
| Clostridium_g24 | * | * |
| Collinsella | * | * |
| Eisenbergiella | * | * |
| Faecalibacterium | - | * |
| Faecalimonas | - | * |
| Fusobacterium | - | * |
| Fusobacterium necrophorum | - | * |
| GU302778_g | - | * |
| Helicobacter | * | * |
| PAC001043_g | * | * |
| PAC001200_g | - | * |
| PAC001637_g | - | * |
| Romboutsia | * | - |
| Roseburia | - | * |
| Ruminococcus_g4 | * | * |
| Slackia | - | * |
| Sporobacter | - | * |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barraza-Guerrero, S.I.; Meza-Herrera, C.A.; García-De la Peña, C.; Ávila-Rodríguez, V.; Vaca-Paniagua, F.; Díaz-Velásquez, C.E.; Pacheco-Torres, I.; Valdez-Solana, M.A.; Siller-Rodríguez, Q.K.; Valenzuela-Núñez, L.M.; et al. Unveiling the Fecal Microbiota in Two Captive Mexican Wolf (Canis lupus baileyi) Populations Receiving Different Type of Diets. Biology 2021, 10, 637. https://doi.org/10.3390/biology10070637
Barraza-Guerrero SI, Meza-Herrera CA, García-De la Peña C, Ávila-Rodríguez V, Vaca-Paniagua F, Díaz-Velásquez CE, Pacheco-Torres I, Valdez-Solana MA, Siller-Rodríguez QK, Valenzuela-Núñez LM, et al. Unveiling the Fecal Microbiota in Two Captive Mexican Wolf (Canis lupus baileyi) Populations Receiving Different Type of Diets. Biology. 2021; 10(7):637. https://doi.org/10.3390/biology10070637
Chicago/Turabian StyleBarraza-Guerrero, Sergio I., César A. Meza-Herrera, Cristina García-De la Peña, Verónica Ávila-Rodríguez, Felipe Vaca-Paniagua, Clara E. Díaz-Velásquez, Irene Pacheco-Torres, Mónica A. Valdez-Solana, Quetzaly K. Siller-Rodríguez, Luis M. Valenzuela-Núñez, and et al. 2021. "Unveiling the Fecal Microbiota in Two Captive Mexican Wolf (Canis lupus baileyi) Populations Receiving Different Type of Diets" Biology 10, no. 7: 637. https://doi.org/10.3390/biology10070637
APA StyleBarraza-Guerrero, S. I., Meza-Herrera, C. A., García-De la Peña, C., Ávila-Rodríguez, V., Vaca-Paniagua, F., Díaz-Velásquez, C. E., Pacheco-Torres, I., Valdez-Solana, M. A., Siller-Rodríguez, Q. K., Valenzuela-Núñez, L. M., & Herrera-Salazar, J. C. (2021). Unveiling the Fecal Microbiota in Two Captive Mexican Wolf (Canis lupus baileyi) Populations Receiving Different Type of Diets. Biology, 10(7), 637. https://doi.org/10.3390/biology10070637






