Morphological and Molecular Characterization of Five Species Including Three New Species of Golden Gorgonians (Cnidaria: Octocorallia) from Seamounts in the Western Pacific
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Specimen Collection and Morphological Examination
2.2. DNA Extraction and Sequencing
2.3. Genetic Distance and Phylogenetic Analyses
| Species | Voucher Number | Locatiom | References | GenBank Accession Numbers | ||
|---|---|---|---|---|---|---|
| mtMutS | COI | 28S rDNA | ||||
| Iridogorgia densispiralis sp. nov. | MBM286454 | 10.37° N, 140.30° E | present study | MW841033 | MW841036 | MW841043 |
| Iridogorgia flexilis sp. nov. | MBM286453 | 10.40° N, 140.90° E | present study | MW841031 | MW841035 | MW841041 |
| Iridogorgia verrucosa sp. nov. | MBM286455 | 10.37° N, 140.04° E | present study | MW841034 | MW840138 | MW841044 |
| Iridogorgia densispicula | MBM286538 | 10.35° N, 140.07° E | [7] | MK431864 | MW841037 | MW841040 |
| Iridogorgia magnispiralis | MBM286450 | 10.50° N, 140.11° E | present study | MW841032 | MW841039 | MW841042 |
| Iridogorgia squarrosa | MBM286539 | 11.16° N, 139.25° E | [7] | MK431865 | – | – |
| Iridogorgia fontinalis | YPM 38584 | 34.81° N, 50.50° W | [6] | EU293802 | GQ868321 | – |
| Iridogorgia magnispiralis | YPM: IZ: 38580 | – | France, unpublished | DQ860108 | – | – |
| Iridogorgia magnispiralis | MNHN-Oct.0000-0576 | 38.47° N, 27.9° W | [6] | GQ353316 | – | – |
| Iridogorgia magnispiralis | – | 38.78° N, 63.96° W | [6] | EU268055 | – | – |
| Iridogorgia magnispiralis | – | unknown | France & Pante, unpublished | – | FJ268639 | – |
| Iridogorgia magnispiralis | YPM 38580 | 38.78° N, 63.96° W | [6] | JN227997 | – | – |
| Iridogorgia magnispiralis | – | 35.19° N, 47.68° W | [6] | GQ223116 | GQ868318 | – |
| Iridogorgia magnispiralis | YPM 38581 | 38.26° N, 60.55° W | [6] | GQ180141 | – | – |
| Iridogorgia magnispiralis | USNM 1092265 | 34.58° N, 56.84° W | [6] | GQ180142 | – | – |
| Iridogorgia magnispiralis | YPM 38582 | 38.86° N, 63.91° W | [6] | GQ180140 | – | – |
| Iridogorgia magnispiralis | – | Gulf of Mexico | [28,29] | KC788263 | KC788237 | KX890214 |
| Iridogorgia sp. | – | 21.32° N, 157.02° W | [6] | GQ868342 | GQ868323 | – |
| Iridogorgia sp. | YPM 28866 | 33.79° N, 62.59° W | [6] | DQ297422 | – | – |
| Iridogorgia sp. type C | – | 23.05° N, 163.16° W | [6] | JN227919 | – | – |
| Iridogorgia sp. type A | MNHN-IC.2009-0001 | 9.15° S, 158.27° E | [6] | GQ180145 | – | – |
| Iridogorgia splendens | YPM 35397, 38586 | 38.85° N, 63.76° W | [6] | DQ860109 | – | – |
| Iridogorgia splendens | USNM 1092267 | 38.79° N, 64.13° W | [6] | JN227996 | GQ868313 | – |
| Iridogorgia splendens | YPM 38585 | 37.46° N, 59.95° W | [6] | JN228005 | GQ868330 | – |
| Iridogorgia splendens | – | Gulf of Mexico | [28] | KC788271 | KC788229 | – |
| Iridogorgia splendens | – | Gulf of Mexico | [29] | – | – | KX890215 |
| Iridogorgia splendens | USNM 1092267 | Kelvin Seamount, NW Atlantic | [30] | GQ180143 | – | – |
| Iridogorgia splendens | YPM: IZ: 38585 | 37.46° N, 59.95° W | [30] | GQ180144 | – | – |
| Rhodaniridogorgia fragilis | YPM 38588 | 34.46° N, 56.73° W | [6] | JN228000 | JN227954 | – |
| Chrysogorgia averta | – | Gulf of Mexico | [28] | KC788265 | KC788235 | KC788258 |
| Chrysogorgia sp. | – | Gulf of Mexico | [28] | KC788268 | KC788223 | KC788240 |
| Chrysogorgia sp. | – | Gulf of Mexico | [29] | – | – | KX890212 |
| Pseudochrysogorgia bellona | MNHN-IC.2008-007 Paratype | 21.12° S, 158.5° E | [6] | GQ868332 | GQ868310 | – |
| Pseudochrysogorgia sp. | XMUB7697 | 15.52° N, 110.96° E | [31] | – | – | MW336977 |
| Metallogorgia melanotrichos | – | 34.53° N, 47.79° W | [6] | GQ180158 | FJ268633 | – |
| Metallogorgia macrospina | NIWA15642 | 37.21° S, 177.24° E | [6] | JN228001 | JN227952 | – |
| Radicipes stonei | USNM: IZ:1418007 | Alaska | [32] | MG986912 | MG986961 | MG980134 |
| Stephanogorgia faulkneri | NTM C014927 | 7.19° N, 134.32° E | [6] | GQ342485 | GQ342406 | JX203718 |
| Chrysogorgiidae sp. | – | 41.37° S, 42.85° E | [33] | KP324387 | KP678004 | KP324611 |
| Plexaura kuna | RMNH Coel.40836 | South Africa | [34] | JX203807 | JX203866 | JX203748 |
3. Results
3.1. Systematics
3.2. Genetic Distance and Phylogenetic Analyses
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Roberts, J.M.; Wheeler, A.J.; Freiwald, A.; Cairns, S.D. Cold-Water Corals: The Biology and Geology of Deep-Sea Coral Habitats; Cambridge University Press: Cambridge, UK, 2009; pp. 1–368. [Google Scholar]
- Dautova, T.N. Octocorallia as a key taxon in the vulnerable marine ecosystems of the Emperor Chain (Northwest Pacific): Diversity, distribution and biogeographical boundary. In Marine Biodiversity for a Healthy Ocean—Biodiversity, Functional Groups and Ocean Health, Proceedings of the Russia-China Bilateral Workshop, Vladivostok, Russia, 10–11 October 2019; Lutaenko, K.A., Ed.; Publishing House of the Far Eastern Federal University: Vladivostok, Russia, 2019; pp. 68–80. [Google Scholar]
- Pérez, C.D.; de Moura Neves, B.; Cordeiro, R.T.S.; Williams, G.C.; Cairns, S.D. Diversity and distribution of Octocorallia. In The Cnidaria, Past, Present and Future; Goffredo, S., Dubinsky, Z., Eds.; Springer: Cham, Switzerland, 2016; pp. 109–123. [Google Scholar] [CrossRef]
- Watling, L. A review of the genus Iridogorgia (Octocorallia: Chrysogorgiidae) and its relatives, chiefly from the North Atlantic Ocean. J. Mar. Biol. Assoc. UK 2007, 87, 393–402. [Google Scholar] [CrossRef]
- Watling, L.; France, S.C.; Pante, E.; Simpson, A. Biology of deep-water octocorals. In Advances in Marine Biology; Lesser, M., Ed.; Elsevier Academic Press: Burlington, NJ, USA, 2011; pp. 41–122. [Google Scholar]
- Pante, E.; France, S.C.; Couloux, A.; Cruaud, C.; McFadden, C.S.; Samadi, S.; Watling, L. Deep-sea origin and in-situ diversification of chrysogorgiid octocorals. PLoS ONE 2012, 7, e38357. [Google Scholar] [CrossRef]
- Xu, Y.; Zhan, Z.; Li, Y.; Xu, K. Morphology and phylogenetic analysis of two new species of deep-sea golden gorgonians (Cnidaria: Octocorallia: Chrysogorgiidae) from seamounts in the Western Pacific Ocean. Zootaxa 2020, 4731, 249–262. [Google Scholar] [CrossRef]
- Watling, L.; Rowley, S.; Guinotte, J. The world’s largest known gorgonian. Zootaxa 2013, 3630, 198–199. [Google Scholar] [CrossRef]
- Cordeiro, R.; McFadden, C.; van Ofwegen, L.; Williams, G. World List of Octocorallia. Iridogorgia Verrill. 1883. World Register of Marine Species. Available online: http://www.marinespecies.org/aphia.php?p=taxdetails&id=125295 (accessed on 10 June 2021).
- Verrill, A.E. Report on the Anthozoa, and on some additional species dredged by the “Blake” in 1877–1879, and by the U.S. Fish Commission steamer “Fish Hawk” in 1880–1882. Bull. Mus. Comp. Zool. Harv. Coll. 1883, 11, 1–72. [Google Scholar]
- Cairns, S.D.; Hourigan, T.F. A Comprehensive List of Known Deep-Sea Corals Occurring in the EEZ of the United States and its Possessions. 2017. Available online: https://deepseacoraldata.noaa.gov/ (accessed on 28 April 2021).
- Nutting, C.C. Descriptions of the Alcyonaria collected by the U.S. Bureau of Fisheries steamer Albatross in the vicinity of the Hawaiian Islands in 1902. Proc. U.S. Nat. Mus. 1908, 34, 543–601. [Google Scholar] [CrossRef][Green Version]
- Parrish, F.A.; Baco-Tayor, A.R.; Kelley, C.; Cairns, S.D.; Hourigan, T.F. Deep-Sea Coral Taxa in the Hawaiian Archipelago and other U.S. Pacific Islands: Depth and Geographical Distribution. 2017. Available online: https://deepseacoraldata.noaa.gov/ (accessed on 28 April 2021).
- Auscavitch, S.R.; Deere, M.C.; Keller, A.G.; Rotjan, R.D.; Shank, T.M.; Cordes, E.E. Oceanographic Drivers of Deep-Sea Coral Species Distribution and Community Assembly on Seamounts, Islands, Atolls, and Reefs Within the Phoenix Islands Protected Area. Front. Mar. Sci. 2020, 7, 42. [Google Scholar] [CrossRef]
- Bayer, F.M.; Grasshoff, M.; Verseveldt, J. Illustrated Trilingual Glossary of Morphological and Anatomical Terms Applied to Octocorallia; E. J. Brill Publishers: Leiden, The Netherlands, 1983; p. 75. [Google Scholar]
- Herrera, S.; Baco, A.; Sánchez, J.A. Molecular systematics of the bubblegum coral genera (Paragorgiidae, Octocorallia) and description of a new deep-sea species. Mol. Phylogenet. Evol. 2010, 55, 123–135. [Google Scholar] [CrossRef]
- Sánchez, J.A.; Lasker, H.R.; Taylor, D.J. Phylogenetic analyses among octocorals (Cnidaria): Mitochondrial and nuclear DNA sequences (lsu-rRNA, 16S and ssu-rRNA, 18S) support two convergent clades of branching gorgonians. Mol. Biol. Evol. 2003, 29, 31–42. [Google Scholar] [CrossRef]
- Folmer, O.; Black, M.; Hoeh, W.; Lutz, R.; Vrijenhoek, R. DNA primers for amplification of mitochondrial cytochrome coxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotech. 1994, 3, 294–299. [Google Scholar]
- McFadden, C.S.; van Ofwegen, L.P. A second, cryptic species of the soft coral genus Incrustatus (Anthozoa: Octocorallia: Clavulariidae) from Tierra del Fuego, Argentina revealed by DNA barcoding. Helgol. Mar. Res. 2013, 67, 137–147. [Google Scholar] [CrossRef][Green Version]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids Sym. Ser. 1999, 41, 95–98. [Google Scholar]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef]
- Hillis, D.M.; Bull, J.J. An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Syst. Biol. 1993, 42, 182–192. [Google Scholar] [CrossRef]
- Ronquist, F.R.; Huelsenbeck, J.P. Mrbayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef]
- Rambaut, A.; Drummond, A.J. Tracer v1.4. Available online: http://beast.bio.ed.ac.uk/Tracer (accessed on 28 April 2021).
- Alfaro, M.E.; Zoller, S.; Lutzoni, F. Bayes or Bootstrap? A simulation study comparing the performance of Bayesian Markov chain Monte Carlo sampling and bootstrapping in assessing phylogenetic confidence. Mol. Biol. Evol. 2003, 20, 255–266. [Google Scholar] [CrossRef] [PubMed]
- Quattrini, A.M.; Georgian, S.E.; Byrnes, L.; Stevens, A.; Falco, R.; Cordes, E.E. Niche divergence by deep-sea octocorals in the genus Callogorgia across the continental slope of the Gulf of Mexico. Mol. Ecol. 2013, 22, 4123–4140. [Google Scholar] [CrossRef]
- Quattrini, A.M.; Gómez, C.E.; Cordes, E.E. Environmental filtering and neutral processes shape octocoral community assembly in the deep sea. Oecologia 2017, 183, 221–236. [Google Scholar] [CrossRef]
- Thoma, J.N.; Pante, E.; Brugler, M.R.; France, S.C. Deep-sea octocorals and antipatharians show no evidence of seamount-scale endemism in the NW Atlantic. Mar. Ecol. Prog. Ser. 2009, 397, 25–35. [Google Scholar] [CrossRef]
- Song, X.; Lyu, M.; Zhang, X.; Ruthensteiner, B.; Ahn, I.-Y.; Pastorino, G.; Wang, Y.; Gu, Y.; Ta, K.; Sun, J.; et al. Large plastic debris dumps: New biodiversity hot spots emerging on the deep-sea floor. Environ. Sci. Technol. Lett. 2021, 8, 148–154. [Google Scholar] [CrossRef]
- Cairns, S.D.; Wirshing, H.H. A phylogenetic analysis of the Primnoidae (Anthozoa: Octocorallia: Calcaxonia) with analyses of character evolution and a key to the genera and subgenera. BMC Evol. Biol. 2018, 18, 1–20. [Google Scholar] [CrossRef]
- Taylor, M.L.; Rogers, A.D. Evolutionary dynamics of a common sub-Antarctic octocoral family. Mol. Phylogenet. Evol. 2014, 84, 185–204. [Google Scholar] [CrossRef]
- McFadden, C.S.; van Ofwegen, L.P. Stoloniferous octocorals (Anthozoa, Octocorallia) from South Africa, with descriptions of a new family of Alcyonacea, a new genus of Clavulariidae, and a new species of Cornularia (Cornulariidae). Invertebr. Syst. 2012, 26, 331–356. [Google Scholar] [CrossRef]
- Ehrenberg, C.G. Beiträge zur physiologischen Kenntniss der Corallenthiere im allgemeinen, und besonders des rothen Meeres, nebst einem Versuche zur physiologischen Systematik derselben. Abh. Königlichen Akad. Wiss. Berl. 1834, 1, 225–380. [Google Scholar]
- Haeckel, E. Generelle Morphologie der Organismen; Verlag von Georg Reimer: Berlin, Germany, 1866; Volume 2, p. 652. [Google Scholar] [CrossRef]
- Lamouroux, J.V.F. Extrait d’un mémoire sur la classification des polypiers coralligènes non entièrement piérreux. Nouv. Bull. Sci. Société Philomath. Paris 1812, 3, 181–188. [Google Scholar]
- Grasshoff, M. The shallow-water gorgonians of New Caledonia and adjacent islands (Coelenterata, Octocorallia). Senckenbergiana Biol. 1999, 78, 1–121. [Google Scholar]
- MacIntosh, H.; Althaus, F.; Williams, A.; Tanner, J.E.; Alderslade, P.; Ahyong, S.T.; Bax, N.; Criscione, F.; Crowther, A.L.; Farrelly, C.A.; et al. Invertebrate diversity in the deep Great Australian Bight (200–5000 m). Mar. Biodivers. Rec. 2018, 11, 23. [Google Scholar] [CrossRef]
- Cairns, S.D.; Gerhswin, L.A.; Brook, F.; Pugh, P.R.; Dawson, E.W.; Ocaña, V.O.; Vervoort, W.; Williams, G.; Watson, J.; Opresko, D.M.; et al. Phylum Cnidaria: Corals, medusae, hydroids, myxozoans. In New Zealand Inventory of Biodiversity: Volume 1. Kingdom Animalia: Radiata, Lophotrochozoa, Deuterostomia; Gordon, D.P., Ed.; Canterbury University Press: Christchurch, New Zealand, 2009; p. 566. [Google Scholar]
- McFadden, C.S.; Benayahu, Y.; Pante, E.; Thoma, J.N.; Nevarez, P.A.; France, S.C. Limitations of mitochondrial gene barcoding in Octocorallia. Mol. Ecol. Res. 2011, 11, 19–31. [Google Scholar] [CrossRef]
- McFadden, C.S.; Brown, A.S.; Brayton, C.; Hunt, C.B.; van Ofwegen, L.P. Application of DNA barcoding in biodiversity studies of shallow-water octocorals: Molecular proxies agree with morphological estimates of species richness in Palau. Coral Reefs 2014, 33, 275–286. [Google Scholar] [CrossRef]
- Dong, D.; Li, X.; Yang, M.; Gong, L.; Li, Y.; Sui, J.; Gan, Z.; Kou, Q.; Xiao, N.; Zhang, J. Report of epibenthic macrofauna found from Haima cold seeps and adjacent deep-sea habitats, South China Sea. Mar. Life Sci. Technol. 2020, 3, 1–2. [Google Scholar] [CrossRef]
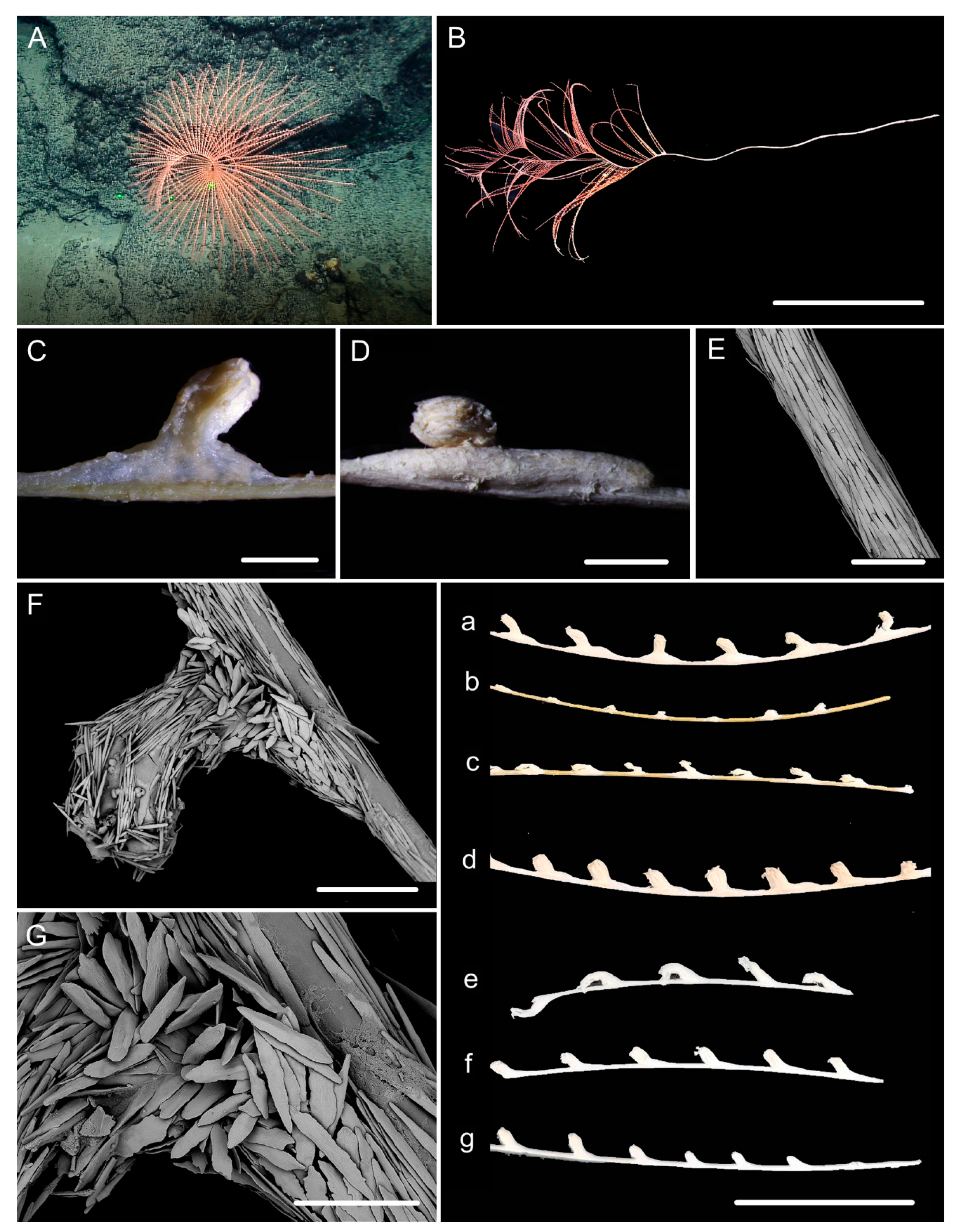



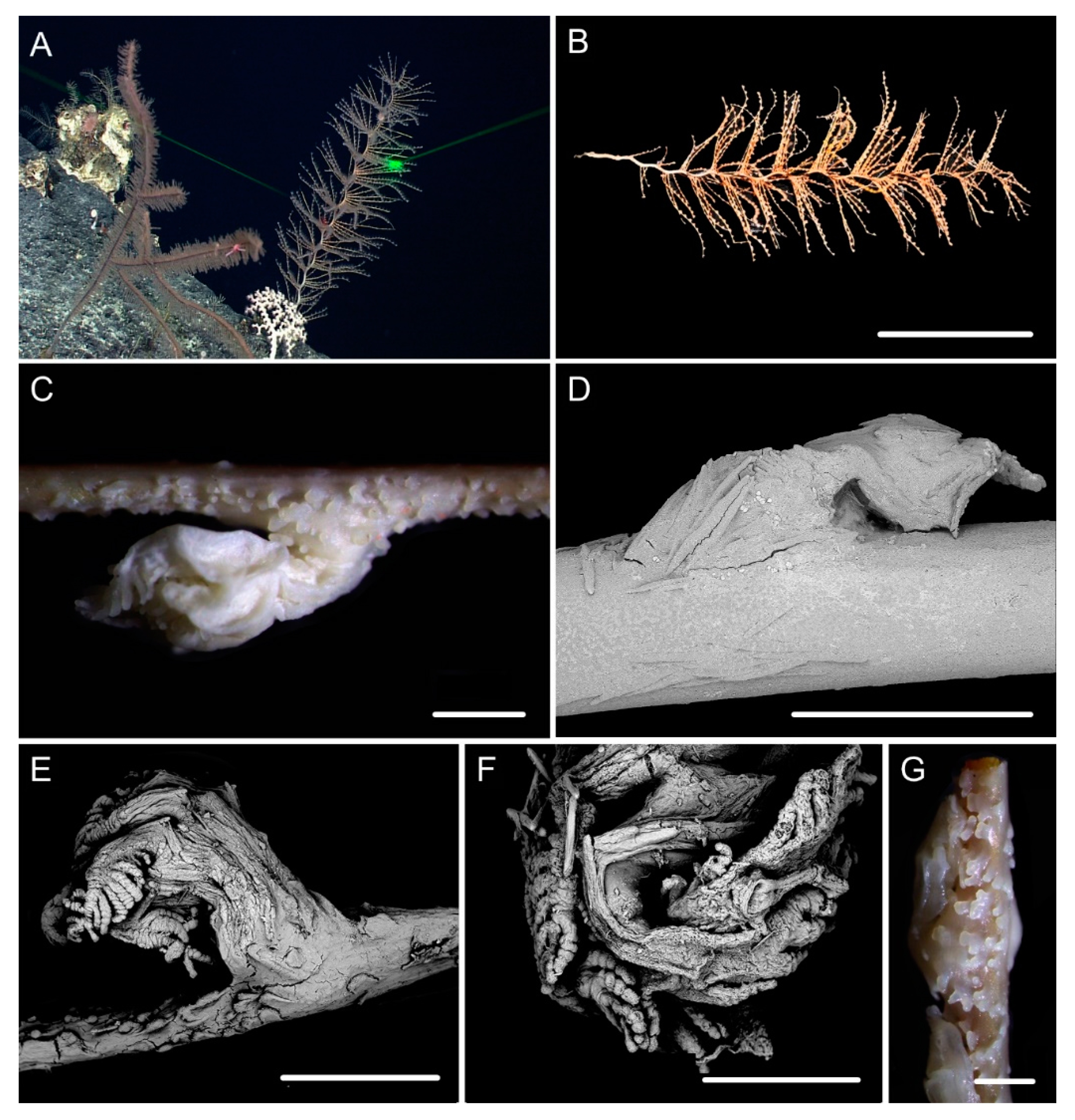

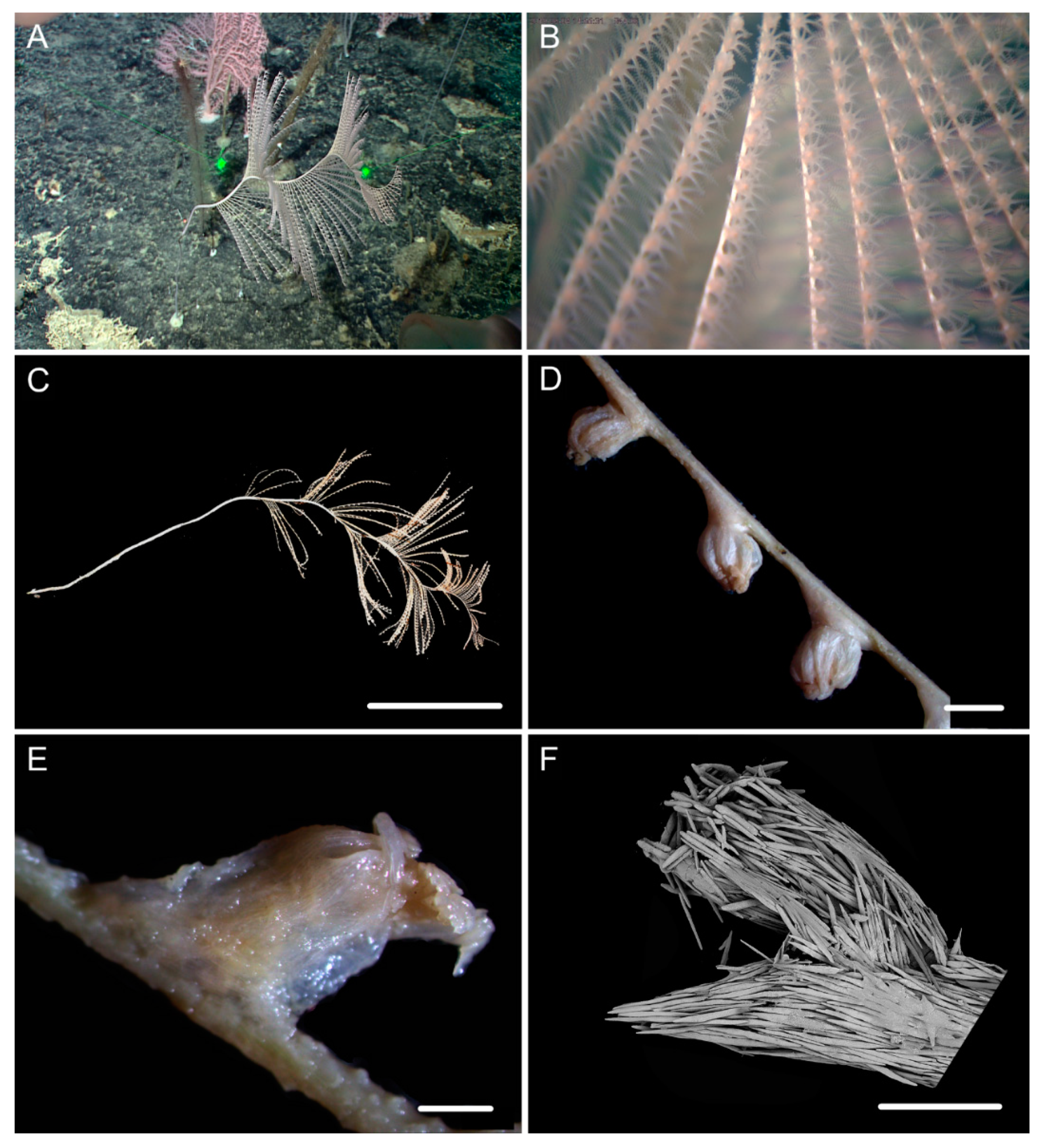

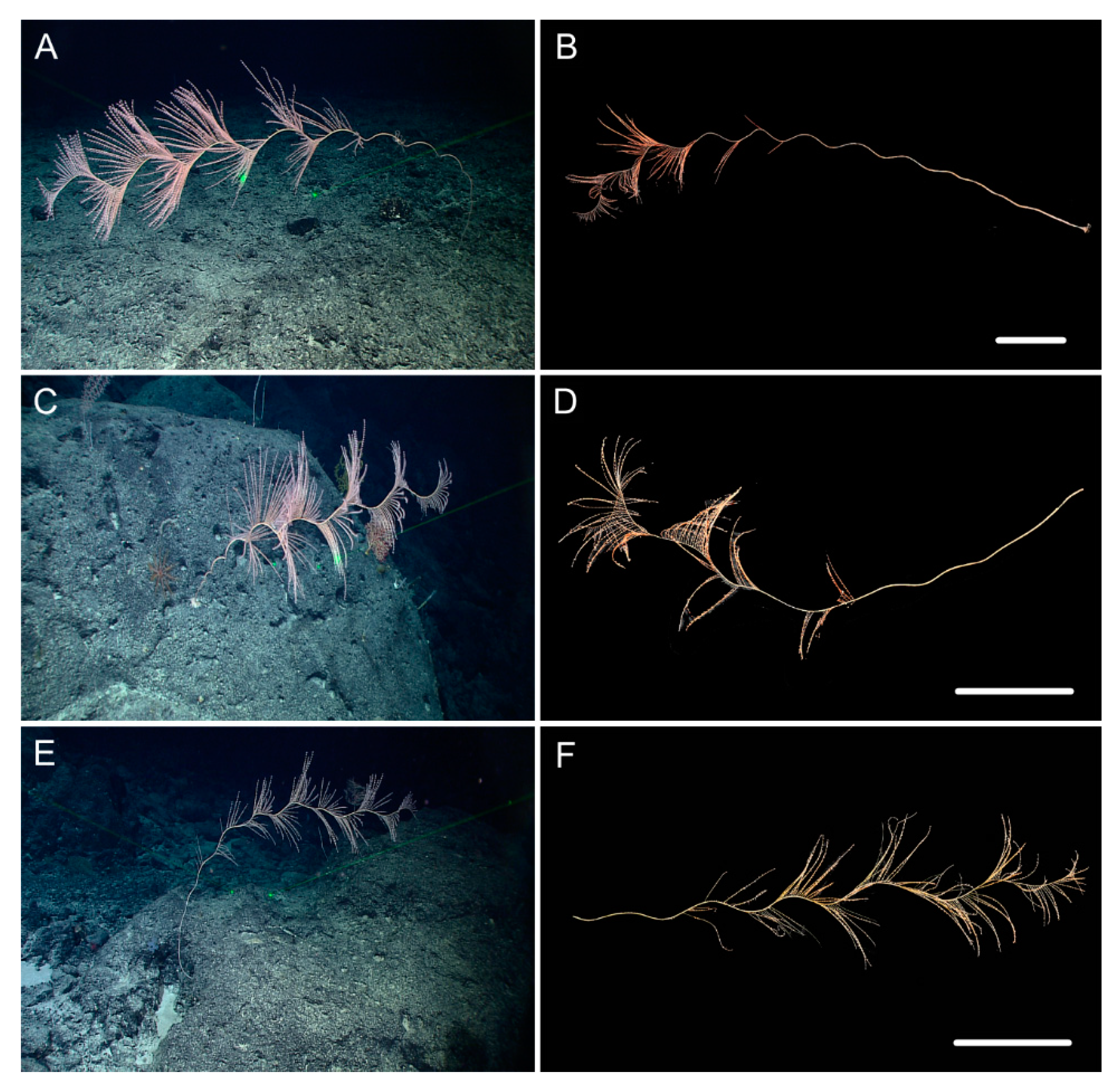
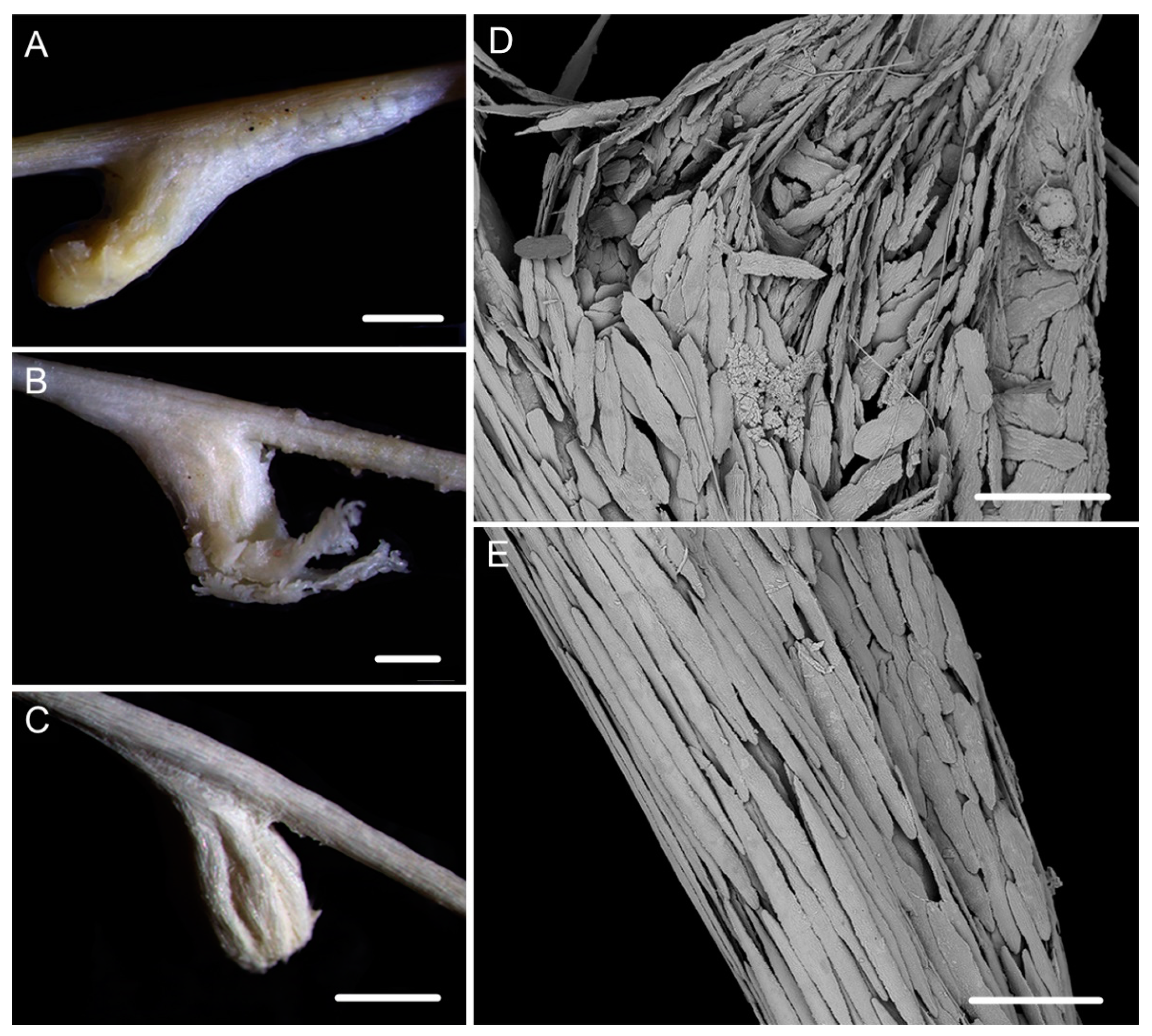
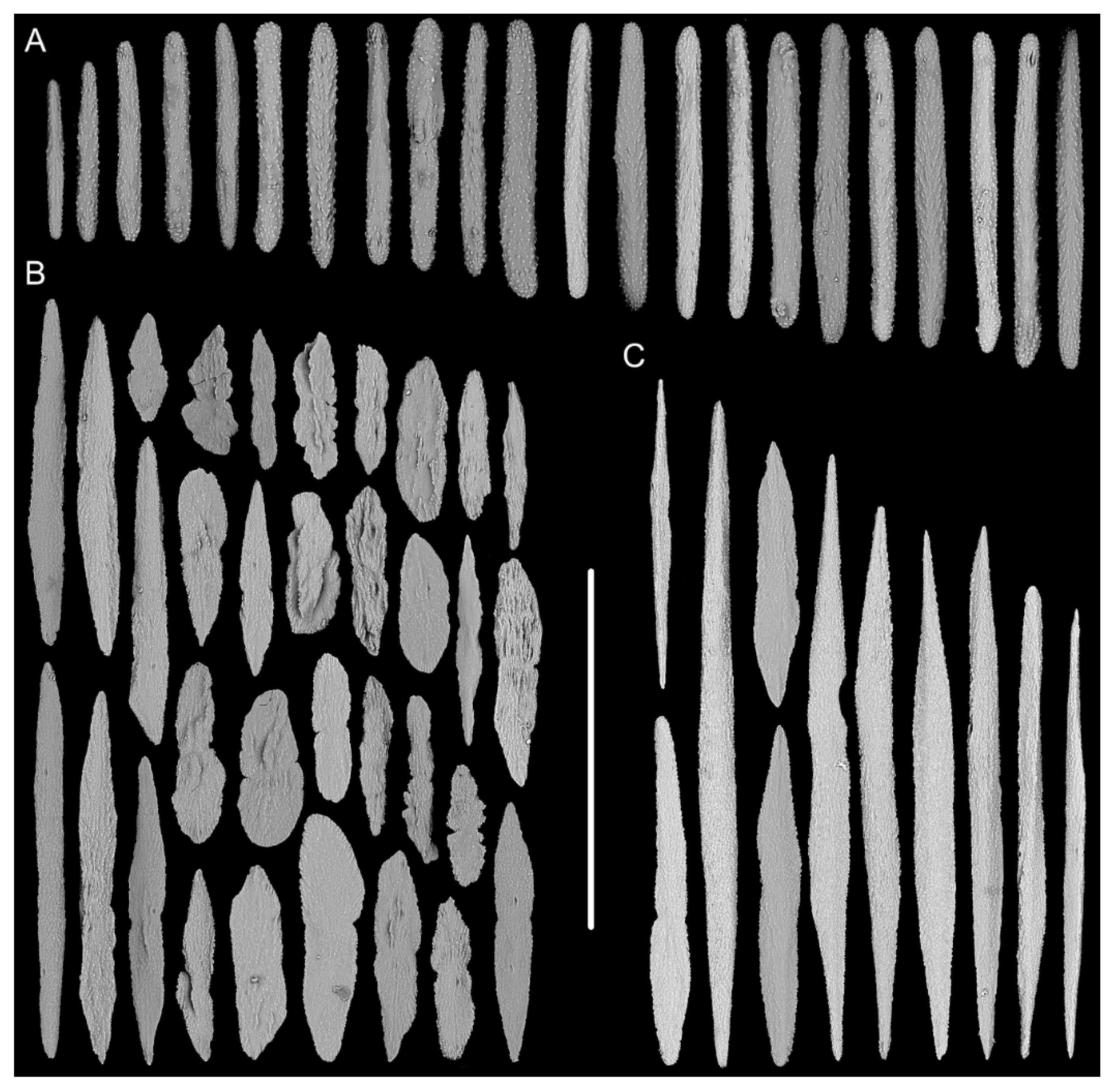





| Characters/Species | I. densispicula | I. squarrosa | I. bella | I. fontinalis | I. magnispiralis |
| Each turn height (cm) | 13–18 | 13–15 | – | 4 | 12–20 |
| Helical diameter (cm) | 3–4 | 4–5 | – | 2.5 | 1–9 |
| Branch intervals (mm) | 3–4 | 3–4 | 4 | 1–2 | 2–5 |
| Polyp height and width (mm) | 2–4/2–4 | 2–4/1–2 | – | 1 mm from branch to tentacle base | 1–3/2–3 |
| Polyp intervals (mm) | 2–7 | 2–6 | 7 | 5–9 | 2–7 |
| Verrucae | rare | a few | sparse | abundant | abundant |
| Sclerites in coenenchyme (μm) | spindles usually with two sharp ends: 203–967 × 11–65 | spindles usually with two sharp ends: 180–840 × 23–60 | – | spindles and a few scales: 397–1024 × 25–74 | spindles usually with two sharp ends: 322–1032 × 21–81 |
| Sclerites in polyp bodies (μm) | spindles and scales coarse, usually lobed with irregular shape and rugged and ridged surface in the upper part, and regular and flat in the basal part: 80–627 × 14–110 | scales with coarse surface and various shape: 72–569 × 24–150 | needle-like or bar-shaped | rods: 175–482 × 27–50 | spindles thick and slender, nearly smooth or with many fine warts: 224–383 × 23–57 |
| Sclerites in tentacles (μm) | rods: 137–945 × 11–98 | rods: 325–433 × 26–43 scales: 467–805 × 48–66 | needle-like or bar-shaped | shorter rods than bodies | rods: 157–548 × 18–78 |
| Distribution | Western Pacific | Western Pacific | near Hawaii | North Atlantic | North Atlantic, near Hawaii, Western Pacific |
| References | [7], present study | [7] | [6,7,11,13] | [4] | [4,8,11,14], present study |
| continued | |||||
| Characters/Species | I. pourtalesii | I. splendens | I. flexilis sp. nov. | I. densispiralis sp. nov. | I. verrucosa sp. nov. |
| Each turn height (cm) | – | 5 | 13–15 | 2.5–3.0 | 3–4 |
| Helical diameter (cm) | – | 1–2 | 3–4 | 0.5 | 1 |
| Branch intervals (mm) | 3–6 | 3–4 | 3 | 2–3 | 1–3 |
| Polyp height and width (mm) | – | less than 1 mm from branch to tentacle base | 1–3/2–4 | 1–2/1–3 | 2/1–2 |
| Polyp intervals (mm) | 5 | 6–8.5 | 4–6 | 3–9 | 4–6 |
| Verrucae | numerous | numerous | rare | a few | numerous |
| Sclerites in coenenchyme (μm) | lacking in the inter- polyps | scales and spindles under polyps; absent to rare in the inter-polyps: 274–592 × 24–51 | spindles usually with two rounded ends: 322–892 × 25–72 | rods and spindles usually with two rounded ends, sometimes sparse in the inter-polyps: 90–523 × 19–65 | spindles same as the body wall, usually with two rounded ends: 169–438 × 19–62 |
| Sclerites in polyp bodies (μm) | spindles smooth: average 400 | scales with a constriction midway: 143–268 × 29–45 | spindles and scales stout and thick with nearly smooth surface: 172–679 × 27–219 | spindles, rods and a few elongated scales often thick with sparse and fine warts: 84–347 × 18–60 | spindles and elongated scales with sparse and fine warts and irregular edges: 116–450 × 21–65 |
| Sclerites in tentacles (μm) | sparse rods: 200–900 | few rods: 169–274 × 27–39 | rods: 261–532 × 19–58 | rods: 95–442 × 11–52 | rods: 160–459 × 17–73 |
| Distribution | North Atlantic | North Atlantic | Western Pacific | Western Pacific | Western Pacific |
| References | [4,10,11] | [4,6,11] | present study | present study | present study |
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | ||
|---|---|---|---|---|---|---|---|---|
| 1 | Iridogorgia densispiralis sp. nov. MW841043 | - | 1.17% | 0.31% | 3.48% | 1.64% | - | - |
| 2 | Iridogorgia flexilis sp. nov. MW841041 | 2.30% | - | 1.09% | 2.76% | 0.46% | - | - |
| 3 | Iridogorgia verrucosa sp. nov. MW841044 | 0.65% | 2.13% | - | 3.40% | 1.56% | - | - |
| 4 | Iridogorgia densispicula MW841040 | 6.08% | 4.68% | 5.90% | - | 3.24% | - | - |
| 5 | Iridogorgia magnispiralis MW841042 | 2.97% | 0.65% | 2.80% | 5.37% | - | - | - |
| 6 | Iridogorgia magnispiralis KX890214 | 3.30% | 0.98% | 3.13% | 5.54% | 0.32% | - | - |
| 7 | Iridogorgia splendens KX890215 | 0.32% | 2.30% | 0.98% | 5.72% | 2.97% | 3.30% | - |
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Iridogorgia densispiralissp. nov. MW841033 | - | ||||||||||||
| 2 | I. verrucosasp. nov. MW841034 | 0.00% | - | |||||||||||
| 3 | I. densispicula MK431864 | 1.11% | 1.11% | - | ||||||||||
| 4 | I. flexilissp. nov. MW841031 | 0.15% | 0.15% | 0.95% | - | |||||||||
| 5 | I. splendens DQ860109, JN227996, JN228005, KC788271, GQ180143 | 0.16% | 0.16% | 0.95% | 0.00% | 0.00% | ||||||||
| 6 | I. splendens GQ180144 | 0.47% | 0.47% | 1.33% | 0.32% | 0.33% | - | |||||||
| 7 | I. squarrosa MK431865 | 0.32% | 0.32% | 1.11% | 0.16% | 0.16% | 0.49% | - | ||||||
| 8 | I. fontinalis EU293802 | 0.48% | 0.48% | 1.27% | 0.32% | 0.32% | 0.66% | 0.48% | - | |||||
| 9 | I. magnispiralisMW841032 DQ860108, GQ353316, EU268055, JN227997, GQ223116, GQ180141, GQ180142, GQ180140, KC788263 | 0.47% | 0.47% | 1.27% | 0.32% | 0.32% | 0.66% | 0.47% | 0.00% | 0.00% | ||||
| 10 | Iridogorgia sp. JN227919 | 0.00% | 0.00% | 1.11% | 0.15% | 0.16% | 0.47% | 0.32% | 0.48% | 0.47% | - | |||
| 11 | Iridogorgia sp. GQ868342 | 0.30% | 0.30% | 1.11% | 0.15% | 0.16% | 0.47% | 0.00% | 0.48% | 0.47% | 0.30% | - | ||
| 12 | Iridogorgia sp. DQ297422, GQ180145 | 0.46% | 0.46% | 1.27% | 0.30% | 0.32% | 0.63% | 0.47% | 0.00% | 0.00% | 0.46% | 0.46% | 0.00% | |
| 13 | Rhodaniridogorgia fragilis JN228000 | 0.70% | 0.70% | 1.41% | 0.47% | 0.47% | 0.70% | 0.93% | 0.93% | 0.93% | 0.70% | 0.70% | 0.93% | - |
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | ||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Iridogorgia densispiralis sp. nov. MW841036 | - | ||||||||
| 2 | Iridogorgia verrucosa sp. nov. MW840138 | 0 | - | |||||||
| 3 | Iridogorgia flexilis sp. nov. MW841035 | 0 | 0 | - | ||||||
| 4 | Iridogorgia splendens GQ868313, GQ868330, KC788229 | 0 | 0 | 0 | 0 | |||||
| 5 | Iridogorgia sp. GQ868323 | 0 | 0 | 0 | 0 | - | ||||
| 6 | Iridogorgia densispicula MW841037 | 0.20% | 0.20% | 0.20% | 0.20% | 0.20% | - | |||
| 7 | Iridogorgia fontinalis GQ868321 | 0.19% | 0.19% | 0.19% | 0.19% | 0.19% | 0.39% | - | ||
| 8 | Iridogorgia magnispiralis MW841039, FJ268639, GQ868318, KC788237 | 0.19% | 0.19% | 0.19% | 0.19% | 0.19% | 0.39% | 0 | 0 | |
| 9 | Rhodaniridogorgia fragilis JN227954 | 0.20% | 0.20% | 0.20% | 0.20% | 0.20% | 0.39% | 0 | 0 | 0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, Y.; Zhan, Z.; Xu, K. Morphological and Molecular Characterization of Five Species Including Three New Species of Golden Gorgonians (Cnidaria: Octocorallia) from Seamounts in the Western Pacific. Biology 2021, 10, 588. https://doi.org/10.3390/biology10070588
Xu Y, Zhan Z, Xu K. Morphological and Molecular Characterization of Five Species Including Three New Species of Golden Gorgonians (Cnidaria: Octocorallia) from Seamounts in the Western Pacific. Biology. 2021; 10(7):588. https://doi.org/10.3390/biology10070588
Chicago/Turabian StyleXu, Yu, Zifeng Zhan, and Kuidong Xu. 2021. "Morphological and Molecular Characterization of Five Species Including Three New Species of Golden Gorgonians (Cnidaria: Octocorallia) from Seamounts in the Western Pacific" Biology 10, no. 7: 588. https://doi.org/10.3390/biology10070588
APA StyleXu, Y., Zhan, Z., & Xu, K. (2021). Morphological and Molecular Characterization of Five Species Including Three New Species of Golden Gorgonians (Cnidaria: Octocorallia) from Seamounts in the Western Pacific. Biology, 10(7), 588. https://doi.org/10.3390/biology10070588






