Mitochondrial Protein Abundance Gradients Require the Distribution of Separated Mitochondria
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. RNA Interference
2.3. Sample Preparation for Fluorescence Microscopy
2.4. Fluorescence Microscopy
2.5. Quantification of the Distribution of Mitochondrial Proteins
2.6. Statistical Analysis
3. Results and Discussion
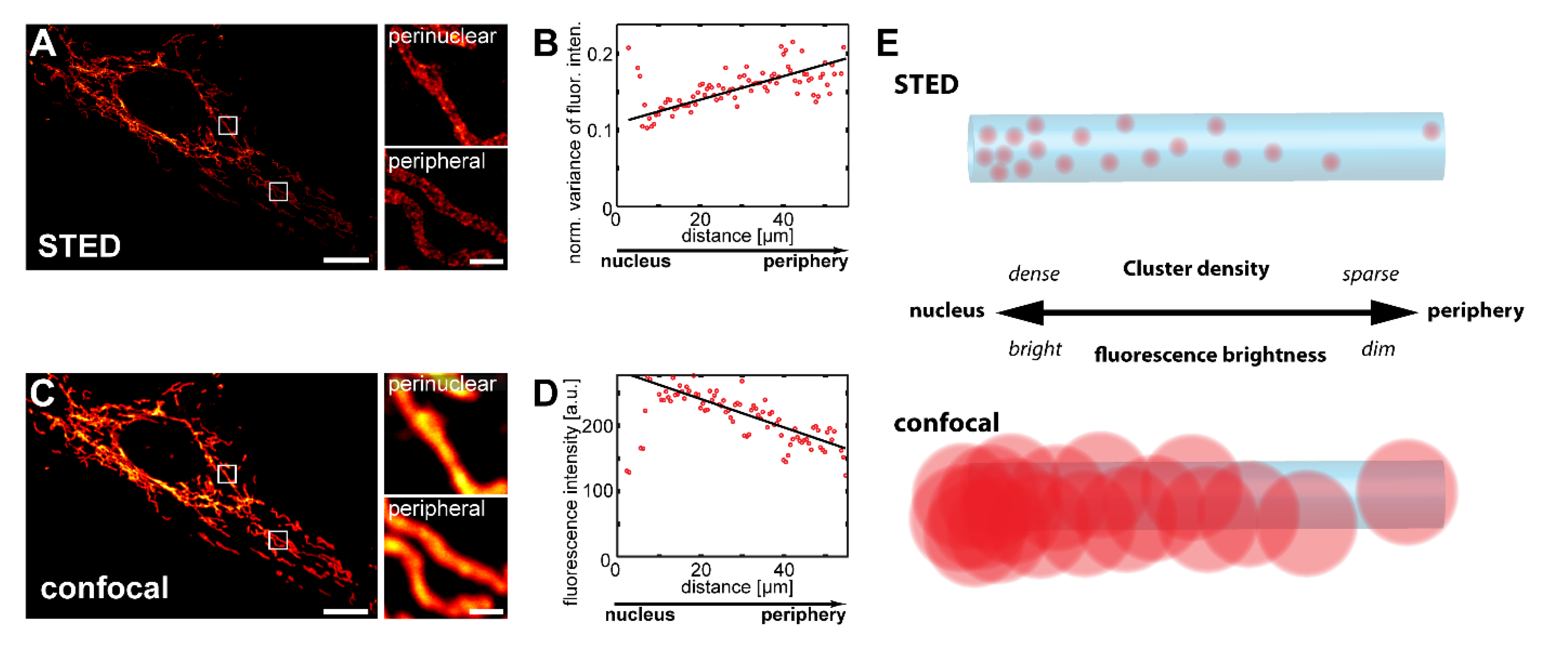
3.1. Confocal Microscopy Enables the Visualization of Inner-Cellular Mitochondrial Protein Abundance Gradients
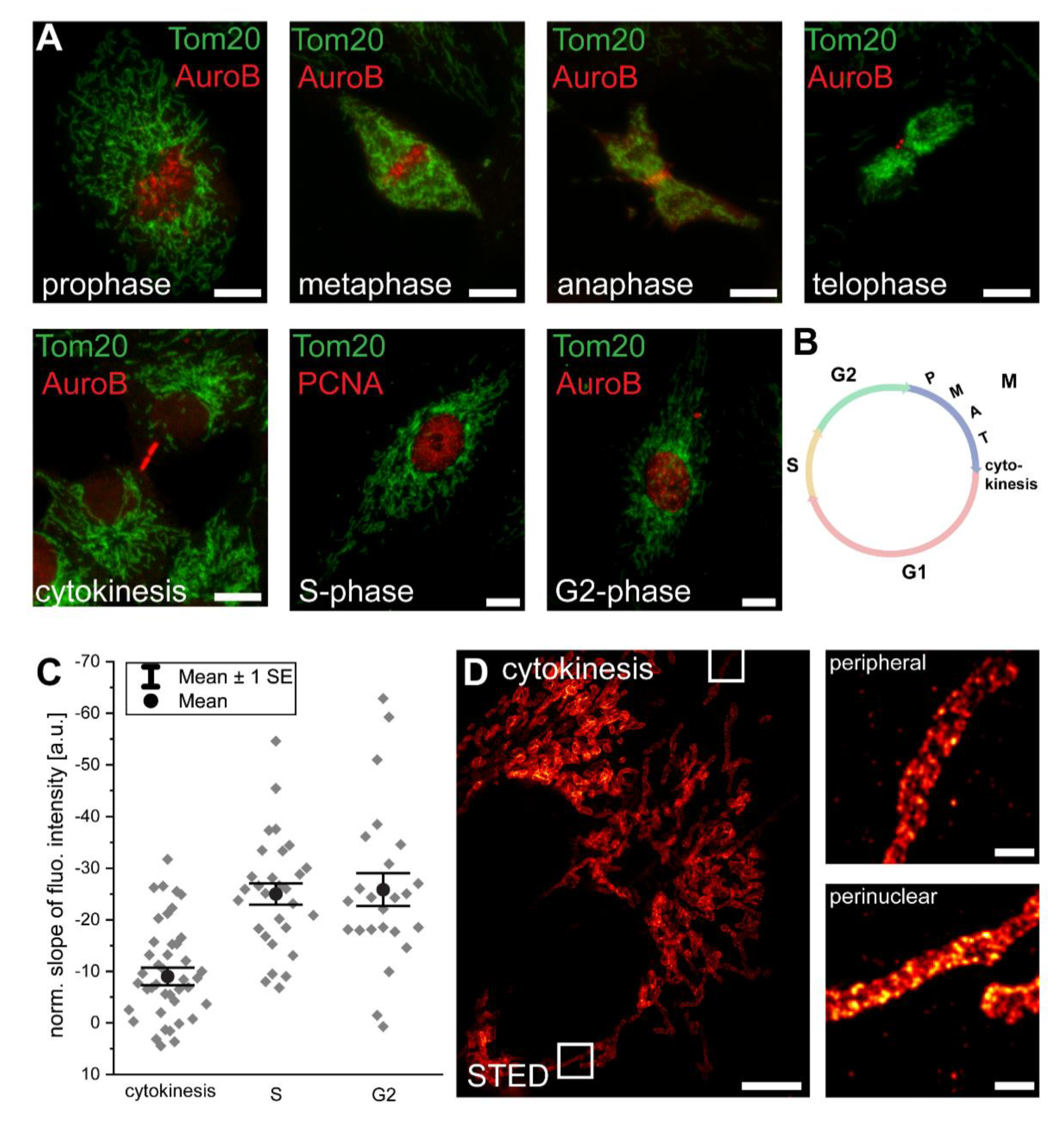
3.2. Mitochondrial Protein Abundance Gradients Are Established Immediately after Cell Division
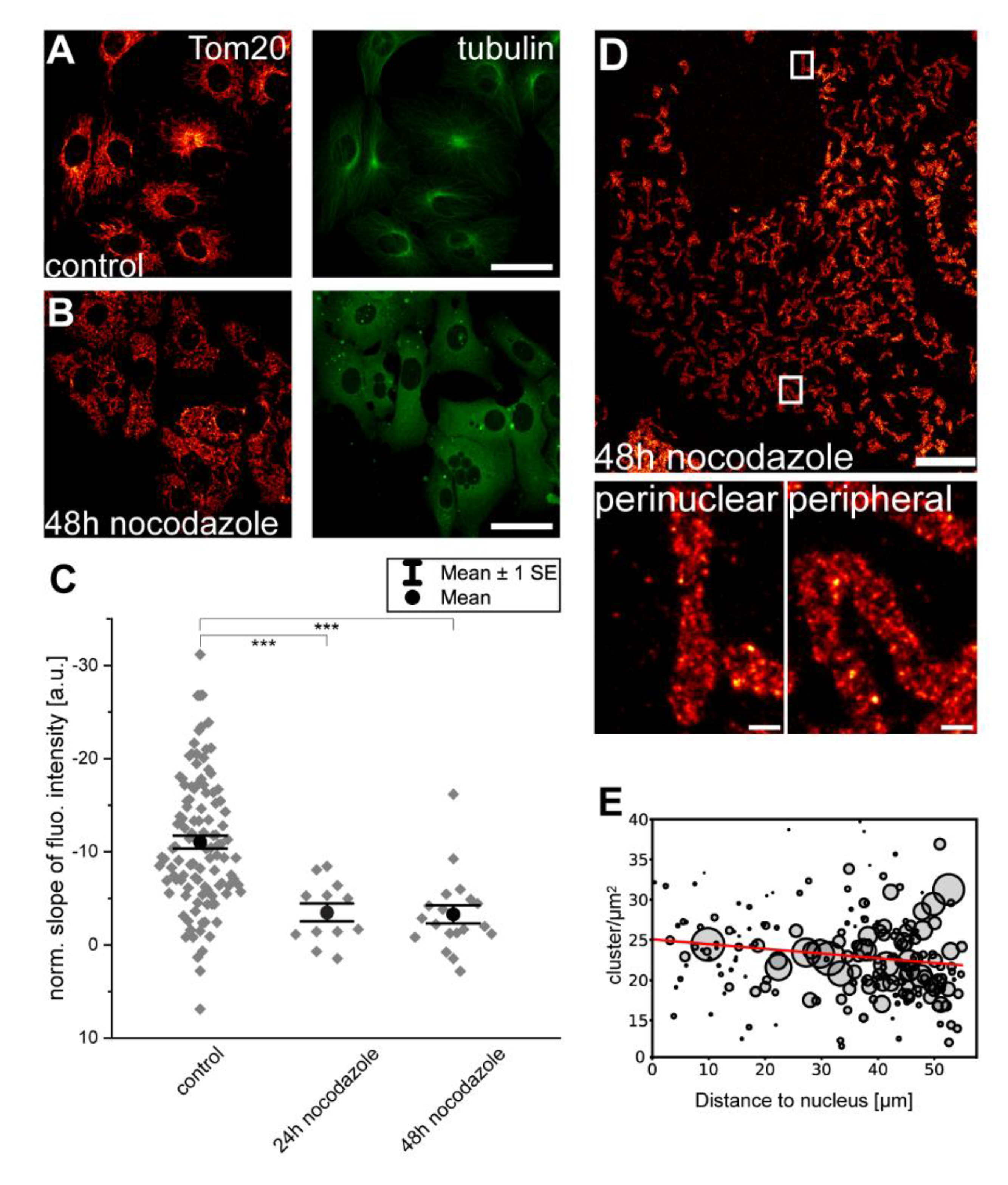
3.3. Disruption of Microtubules Reduces Inner-Cellular Mitochondrial Protein Abundance Gradients
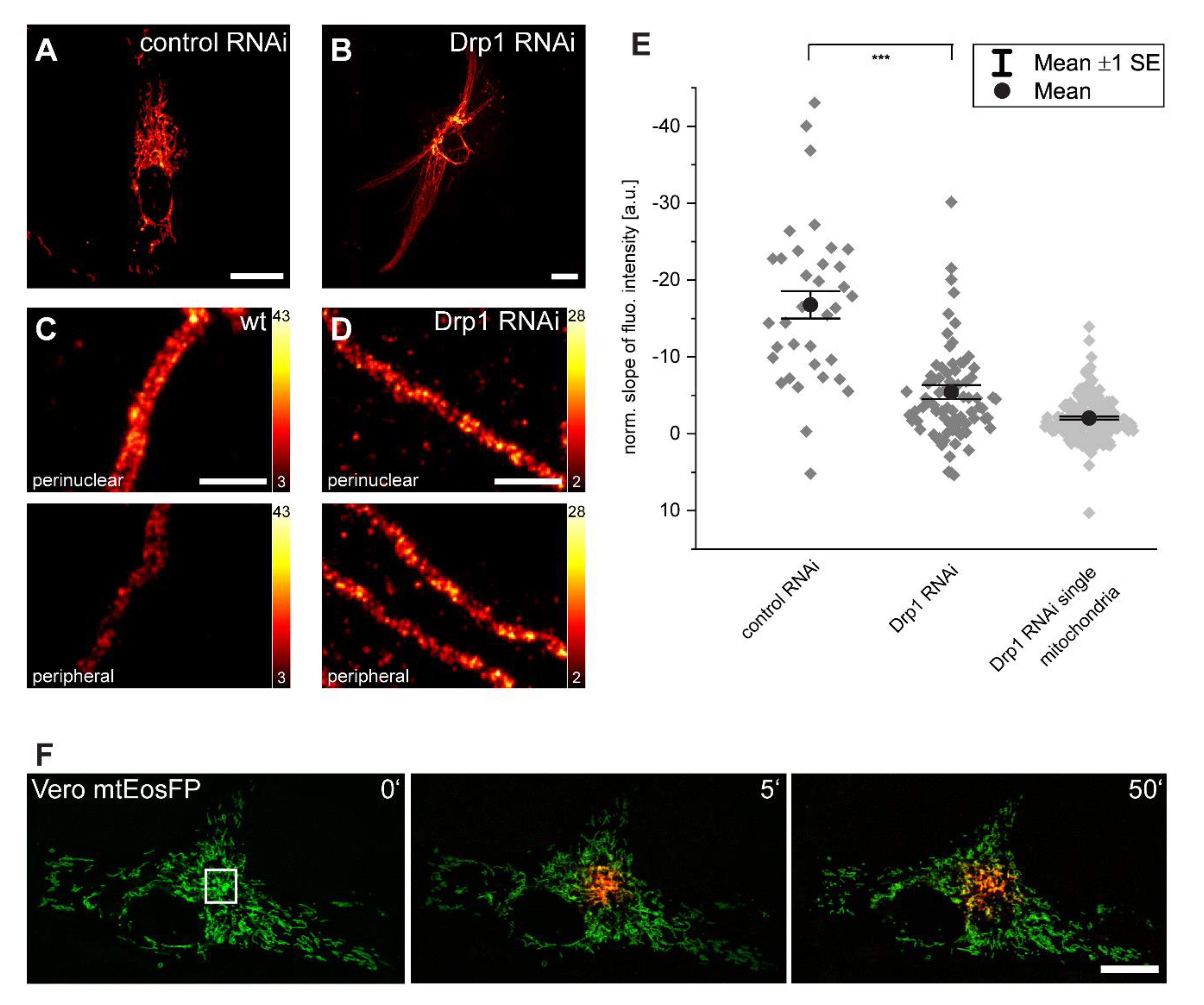
3.4. Inner-Cellular Mitochondrial Protein Gradients Result from Protein Abundance Differences between Separated Mitochondria
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tait, S.; Green, D. Mitochondria and cell death: Outer membrane permeabilization and beyond. Nat. Rev. Mol. Cell Biol. 2010, 11, 621–632. [Google Scholar] [CrossRef]
- Nunnari, J.; Suomalainen, A. Mitochondria: In Sickness and in Health. Cell 2012, 148, 1145–1159. [Google Scholar] [CrossRef]
- Rizzuto, R.; De Stefani, D.; Raffaello, A.; Mammucari, C. Mitochondria as sensors and regulators of calcium signalling. Nat. Rev. Mol. Cell Biol. 2012, 13, 566–578. [Google Scholar] [CrossRef] [PubMed]
- Horvath, S.E.; Daum, G. Lipids of mitochondria. Prog. Lipid Res. 2013, 52, 590–614. [Google Scholar] [CrossRef] [PubMed]
- Labbé, K.; Murley, A.; Nunnari, J. Determinants and Functions of Mitochondrial Behavior. Annu. Rev. Cell Dev. Biol. 2014, 30, 357–391. [Google Scholar] [CrossRef] [PubMed]
- Wurm, C.A.; Neumann, D.; Lauterbach, M.A.; Harke, B.; Egner, A.; Hell, S.W.; Jakobs, S.; Wurm, C.A.; Neumann, D.; Lauterbach, M.A.; et al. Nanoscale distribution of mitochondrial import receptor Tom20 is adjusted to cellular conditions and exhibits an inner-cellular gradient. Proc. Natl. Acad. Sci. USA 2011, 108, 13546–13551. [Google Scholar] [CrossRef]
- Jans, D.C.; Wurm, C.A.; Riedel, D.; Wenzel, D.; Stagge, F.; Deckers, M.; Rehling, P.; Jakobs, S. STED super-resolution microscopy reveals an array of MINOS clusters along human mitochondria. Proc. Natl. Acad. Sci. USA 2013, 110, 8936–8941. [Google Scholar] [CrossRef]
- Klotzsch, E.; Smorodchenko, A.; Löfler, L.; Moldzio, R.; Parkinson, E.; Schütz, G.J.; Pohl, E.E. Superresolution microscopy reveals spatial separation of UCP4 and F0F1-ATP synthase in neuronal mitochondria. Proc. Natl. Acad. Sci. USA 2015, 112, 130–135. [Google Scholar] [CrossRef] [PubMed]
- Cogliati, S.; Enriquez, J.A.; Scorrano, L. Mitochondrial Cristae: Where Beauty Meets Functionality. Trends Biochem. Sci. 2016, 41, 261–273. [Google Scholar] [CrossRef]
- Stoldt, S.; Stephan, T.; Jans, D.C.; Brüser, C.; Lange, F.; Keller-Findeisen, J.; Riedel, D.; Hell, S.W.; Jakobs, S. Mic60 exhibits a coordinated clustered distribution along and across yeast and mammalian mitochondria. Proc. Natl. Acad. Sci. USA 2019, 116, 9853–9858. [Google Scholar] [CrossRef] [PubMed]
- Collins, T.J.; Berridge, M.J.; Lipp, P.; Bootman, M.D. Mitochondria are morphologically and functionally heterogeneous within cells. EMBO J. 2002, 21, 1616–1627. [Google Scholar] [CrossRef]
- Kuznetsov, A.V.; Troppmair, J.; Sucher, R.; Hermann, M.; Saks, V.; Margreiter, R. Mitochondrial subpopulations and heterogeneity revealed by confocal imaging: Possible physiological role? Biochim. Biophys. Acta (BBA) Bioenerg. 2006, 1757, 686–691. [Google Scholar] [CrossRef]
- Jakobs, S.; Stoldt, S.; Neumann, D. Light Microscopic Analysis of Mitochondrial Heterogeneity in Cell Populations and Within Single Cells. Blue Biotechnol. 2010, 124, 1–19. [Google Scholar]
- Wolf, D.M.; Segawa, M.; Kondadi, A.K.; Anand, R.; Bailey, S.T.; Reichert, A.S.; Van Der Bliek, A.M.; Shackelford, D.B.; Liesa, M.; Shirihai, O.S. Individual cristae within the same mitochondrion display different membrane potentials and are functionally independent. EMBO J. 2019, 38, e101056. [Google Scholar] [CrossRef] [PubMed]
- Wikstrom, J.D.; Twig, G.; Shirihai, O.S. What can mitochondrial heterogeneity tell us about mitochondrial dynamics and autophagy? Int. J. Biochem. Cell Biol. 2009, 41, 1914–1927. [Google Scholar] [CrossRef] [PubMed]
- Bereiter-Hahn, J.; Vöth, M. Dynamics of mitochondria in living cells: Shape changes, dislocations, fusion, and fission of mitochondria. Microsc. Res. Tech. 1994, 27, 198–219. [Google Scholar] [CrossRef] [PubMed]
- Nunnari, J.; Marshall, W.F.; Straight, A.; Murray, A.; Sedat, J.W.; Walter, P. Mitochondrial transmission during mating in Saccharomyces cerevisiae is determined by mitochondrial fusion and fission and the intramitochondrial segregation of mitochondrial DNA. Mol. Biol. Cell 1997, 8, 1233–1242. [Google Scholar] [CrossRef]
- Chan, D.C. Fusion and Fission: Interlinked Processes Critical for Mitochondrial Health. Annu. Rev. Genet. 2012, 46, 265–287. [Google Scholar] [CrossRef]
- Buchholz, T.-O.; Prakash, M.; Krull, A.; Jug, F. DenoiSeg: Joint Denoising and Segmentation. arXiv 2020, arXiv:2005.02987v2. [Google Scholar]
- Jakobs, S.; Stephan, T.; Ilgen, P.; Brüser, C. Light Microscopy of Mitochondria at the Nanoscale. Annu. Rev. Biophys. 2020, 49, 289–308. [Google Scholar] [CrossRef]
- Arakaki, N.; Nishihama, T.; Owaki, H.; Kuramoto, Y.; Suenaga, M.; Miyoshi, E.; Emoto, Y.; Shibata, H.; Shono, M.; Higuti, T. Dynamics of Mitochondria during the Cell Cycle. Biol. Pharm. Bull. 2006, 29, 1962–1965. [Google Scholar] [CrossRef]
- Mitra, K.; Wunder, C.; Roysam, B.; Lin, G.; Lippincott-Schwartz, J. A hyperfused mitochondrial state achieved at G1-S regulates cyclin E buildup and entry into S phase. Proc. Natl. Acad. Sci. USA 2009, 106, 11960–11965. [Google Scholar] [CrossRef] [PubMed]
- Kanfer, G.; Kornmann, B. Dynamics of the mitochondrial network during mitosis. Biochem. Soc. Trans. 2016, 44, 510–516. [Google Scholar] [CrossRef] [PubMed]
- Moore, A.S.; Coscia, S.M.; Simpson, C.L.; Ortega, F.E.; Wait, E.C.; Heddleston, J.M.; Nirschl, J.J.; Obara, C.J.; Guedes-Dias, P.; Boecker, C.A.; et al. Actin cables and comet tails organize mitochondrial networks in mitosis. Nature 2021, 591, 659–664. [Google Scholar] [CrossRef] [PubMed]
- Saxton, W.M.; Hollenbeck, P.J. The axonal transport of mitochondria. J. Cell Sci. 2012, 125 Pt 9, 2095–2104. [Google Scholar] [CrossRef]
- Twig, G.; Graf, S.A.; Wikstrom, J.D.; Mohamed, H.; Haigh, S.E.; Elorza, A.; Deutsch, M.; Zurgil, N.; Reynolds, N.; Shirihai, O.S. Tagging and tracking individual networks within a complex mitochondrial web with photoactivatable GFP. Am. J. Physiol. Physiol. 2006, 291, C176–C184. [Google Scholar] [CrossRef]
- Lee, Y.-J.; Jeong, S.-Y.; Karbowski, M.; Smith, C.L.; Youle, R.J. Roles of the Mammalian Mitochondrial Fission and Fusion Mediators Fis1, Drp1, and Opa1 in Apoptosis. Mol. Biol. Cell 2004, 15, 5001–5011. [Google Scholar] [CrossRef] [PubMed]
- Partikian, A.; Ölveczky, B.; Swaminathan, R.; Li, Y.; Verkman, A.S. Rapid Diffusion of Green Fluorescent Protein in the Mitochondrial Matrix. J. Cell Biol. 1998, 140, 821–829. [Google Scholar] [CrossRef]
- Jakobs, S.; Schauss, A.C.; Hell, S.W. Photoconversion of matrix targeted GFP enables analysis of continuity and intermixing of the mitochondrial lumen. FEBS Lett. 2003, 554, 194–200. [Google Scholar] [CrossRef]
- Ivanchenko, S.; Röcker, C.; Oswald, F.; Wiedenmann, J.; Nienhaus, G.U. Targeted Green-Red Photoconversion of EosFP, a Fluorescent Marker Protein. J. Biol. Phys. 2005, 31, 249–259. [Google Scholar] [CrossRef][Green Version]
- Karbowski, M.; Arnoult, D.; Chen, H.; Chan, D.C.; Smith, C.L.; Youle, R.J. Quantitation of mitochondrial dynamics by photolabeling of individual organelles shows that mitochondrial fusion is blocked during the Bax activation phase of apoptosis. J. Cell Biol. 2004, 164, 493–499. [Google Scholar] [CrossRef] [PubMed]
- Schuler, M.-H.; Lewandowska, A.; Di Caprio, G.; Skillern, W.; Upadhyayula, S.; Kirchhausen, T.; Shaw, J.M.; Cunniff, B. Miro1-mediated mitochondrial positioning shapes intracellular energy gradients required for cell migration. Mol. Biol. Cell 2017, 28, 2159–2169. [Google Scholar] [CrossRef]
- Chan, D.C. Mitochondrial Dynamics and Its Involvement in Disease. Annu. Rev. Pathol. Mech. Dis. 2020, 15, 235–259. [Google Scholar] [CrossRef] [PubMed]
- Friedman, J.; Nunnari, J. Mitochondrial form and function. Nature 2014, 505, 335–343. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bollmann, F.; Dohrke, J.-N.; Wurm, C.A.; Jans, D.C.; Jakobs, S. Mitochondrial Protein Abundance Gradients Require the Distribution of Separated Mitochondria. Biology 2021, 10, 572. https://doi.org/10.3390/biology10070572
Bollmann F, Dohrke J-N, Wurm CA, Jans DC, Jakobs S. Mitochondrial Protein Abundance Gradients Require the Distribution of Separated Mitochondria. Biology. 2021; 10(7):572. https://doi.org/10.3390/biology10070572
Chicago/Turabian StyleBollmann, Franziska, Jan-Niklas Dohrke, Christian A. Wurm, Daniel C. Jans, and Stefan Jakobs. 2021. "Mitochondrial Protein Abundance Gradients Require the Distribution of Separated Mitochondria" Biology 10, no. 7: 572. https://doi.org/10.3390/biology10070572
APA StyleBollmann, F., Dohrke, J.-N., Wurm, C. A., Jans, D. C., & Jakobs, S. (2021). Mitochondrial Protein Abundance Gradients Require the Distribution of Separated Mitochondria. Biology, 10(7), 572. https://doi.org/10.3390/biology10070572





