Construction of a Three-Color Prism-Based TIRF Microscope to Study the Interactions and Dynamics of Macromolecules
Abstract
Simple Summary
Abstract
1. Introduction
2. Design
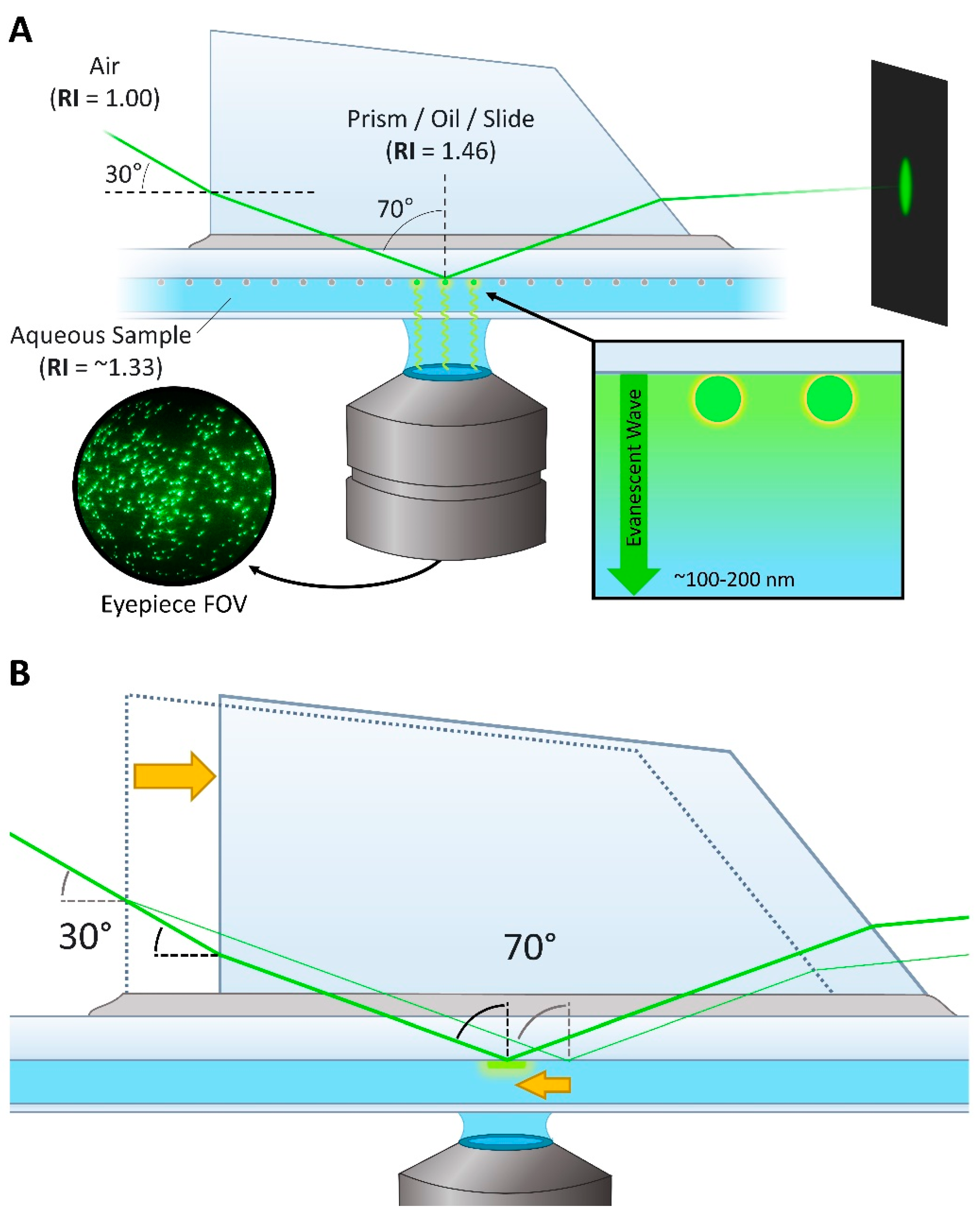
2.1. Prism-Based vs. Objective-Based TIRF
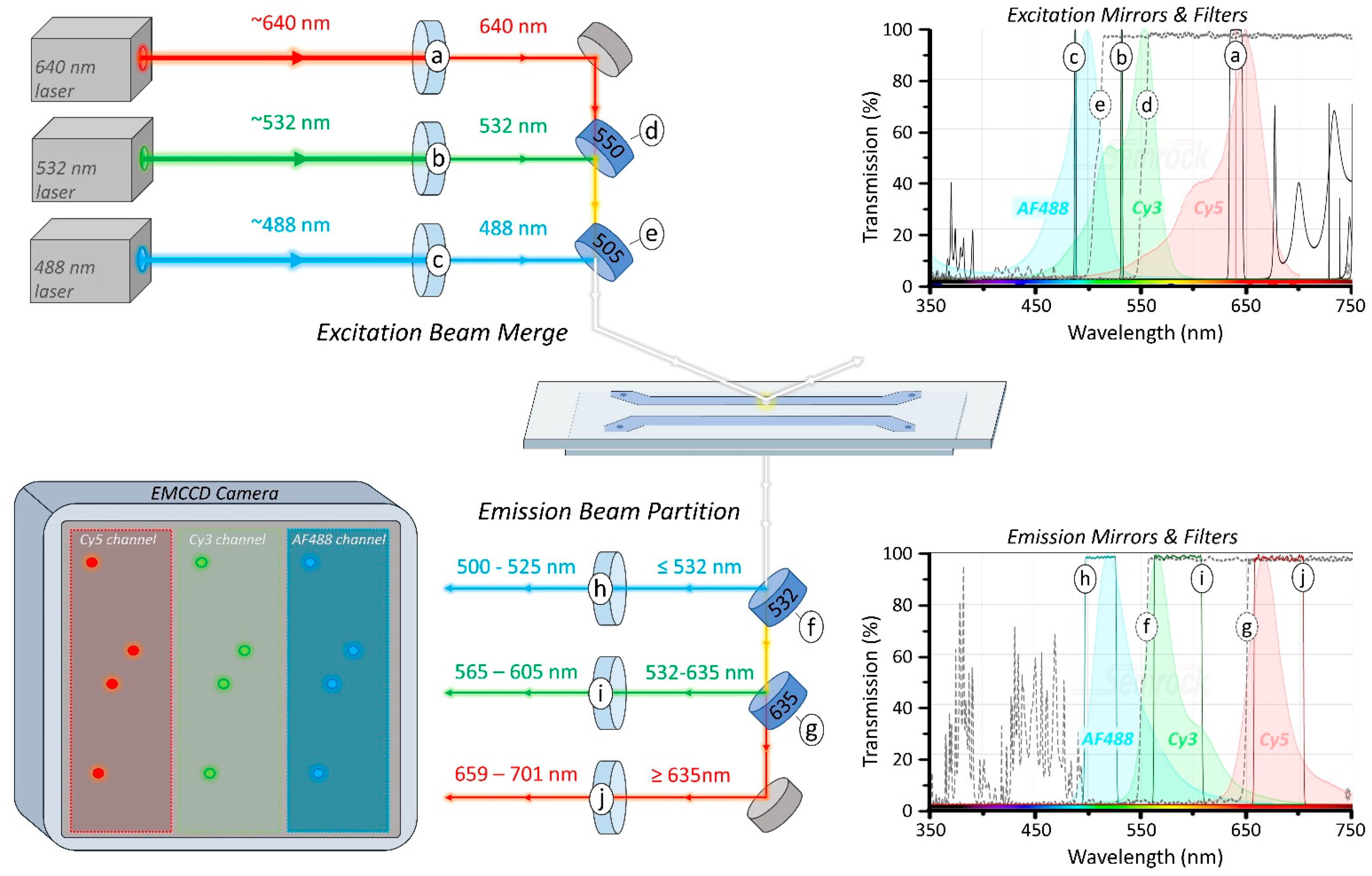
2.2. Fluorescent Labels and Excitation Lasers
2.3. Longpass Dichroic Mirrors
2.4. Bandpass Filters
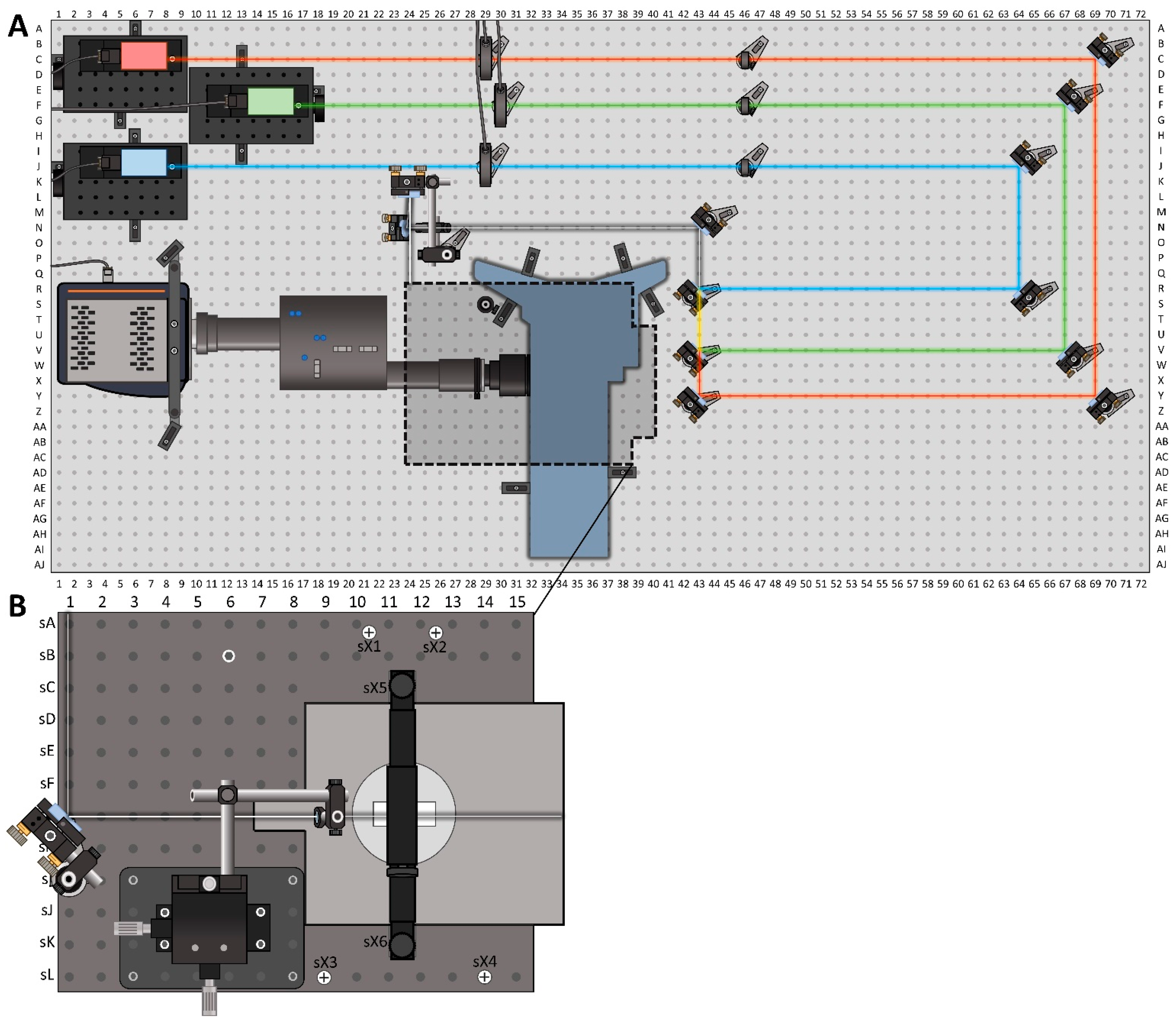
3. Construction
3.1. Excitation Beam Path
3.1.1. Safety Considerations
3.1.2. Optical Table and Operating Space
3.1.3. Excitation Lasers, Shutters and Clean-Up Filters
3.1.4. Leveling and Merging the Laser Beam Paths
3.1.5. Beam to Stage Path
3.2. Microscope Stage
3.2.1. Inverted Microscope and Stage Breadboard
3.2.2. TIR Angle and the Stage Mirror
3.2.3. Focusing Lens and Prism Assemblies
3.3. Emission Path
3.3.1. Optosplit III Emission Splitting
3.3.2. EMCCD Camera
4. Operation
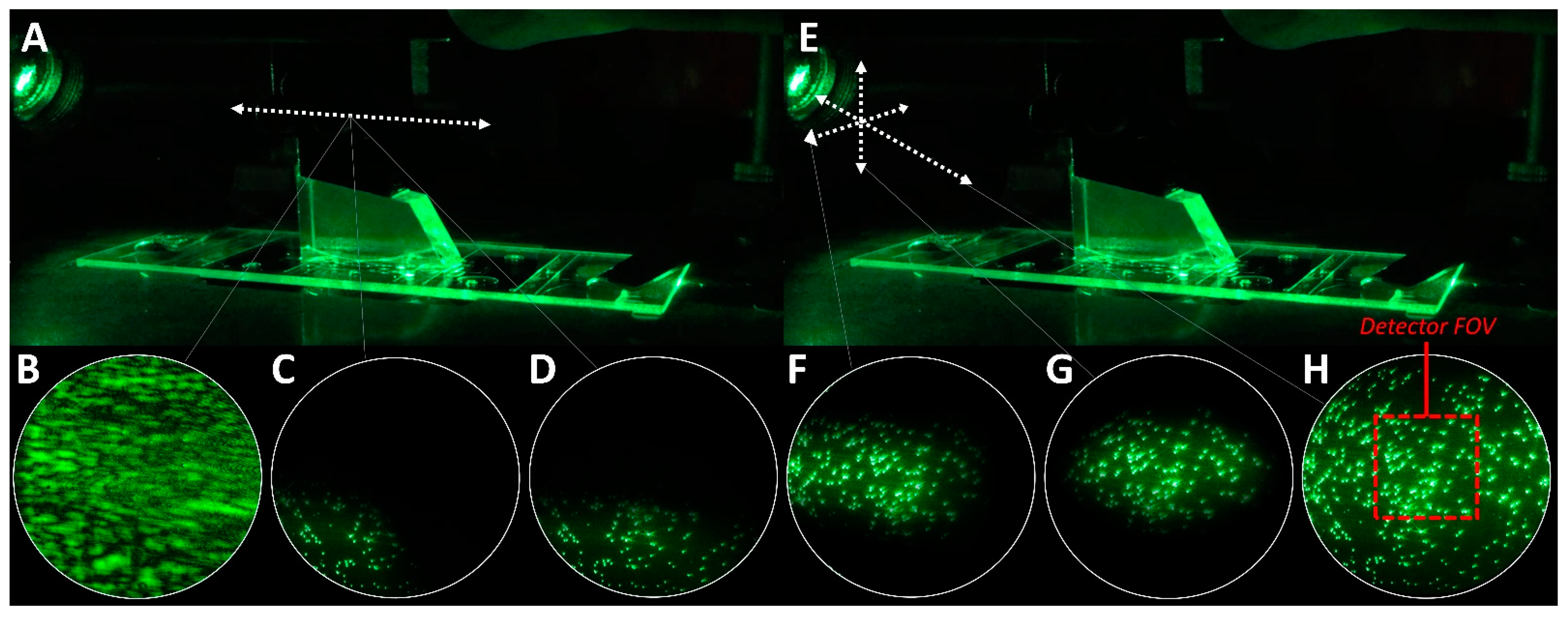
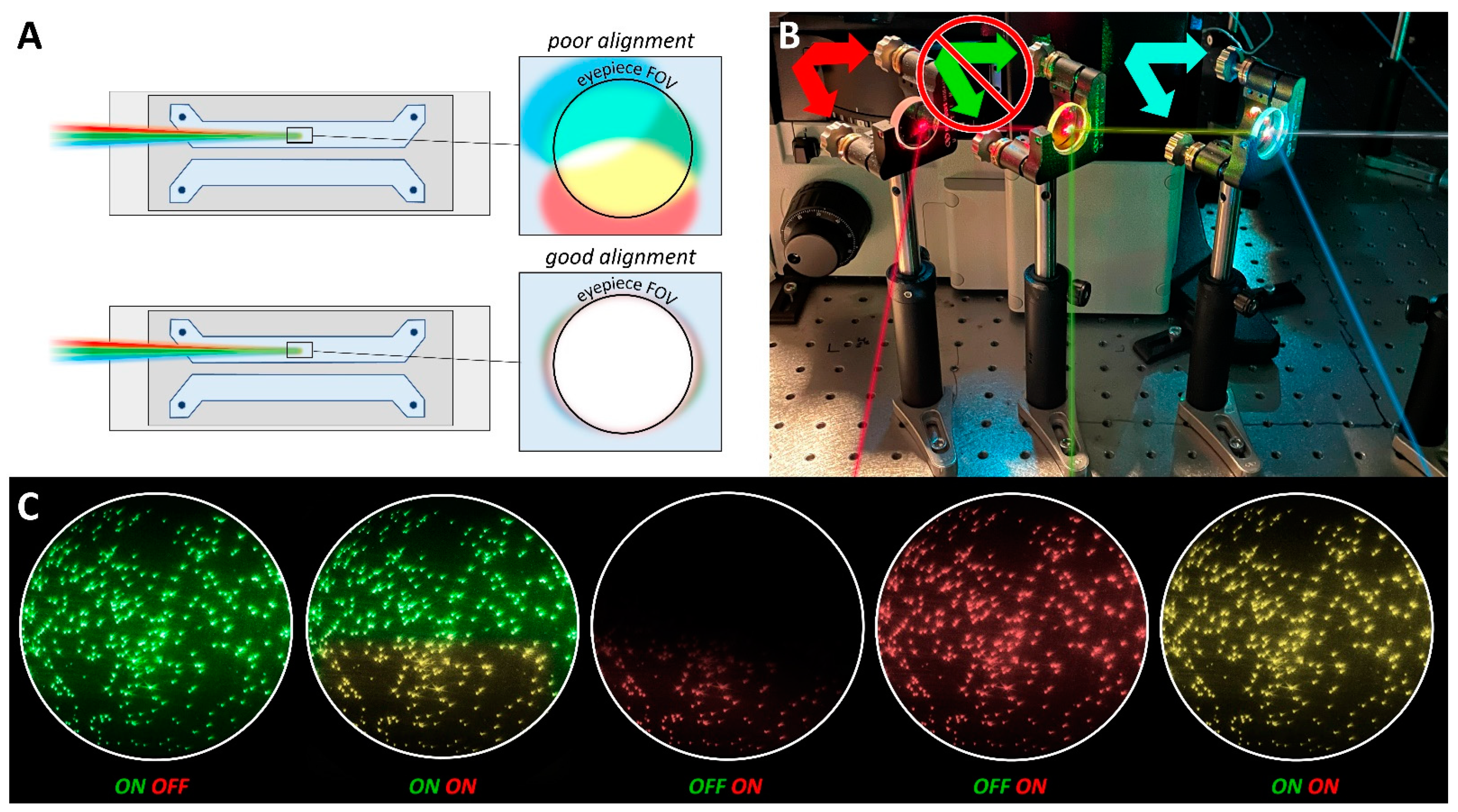
4.1. Bead Slides and TIR Acquisition
4.2. Excitation Beam Alignment
4.3. Emission Channels and Camera Settings
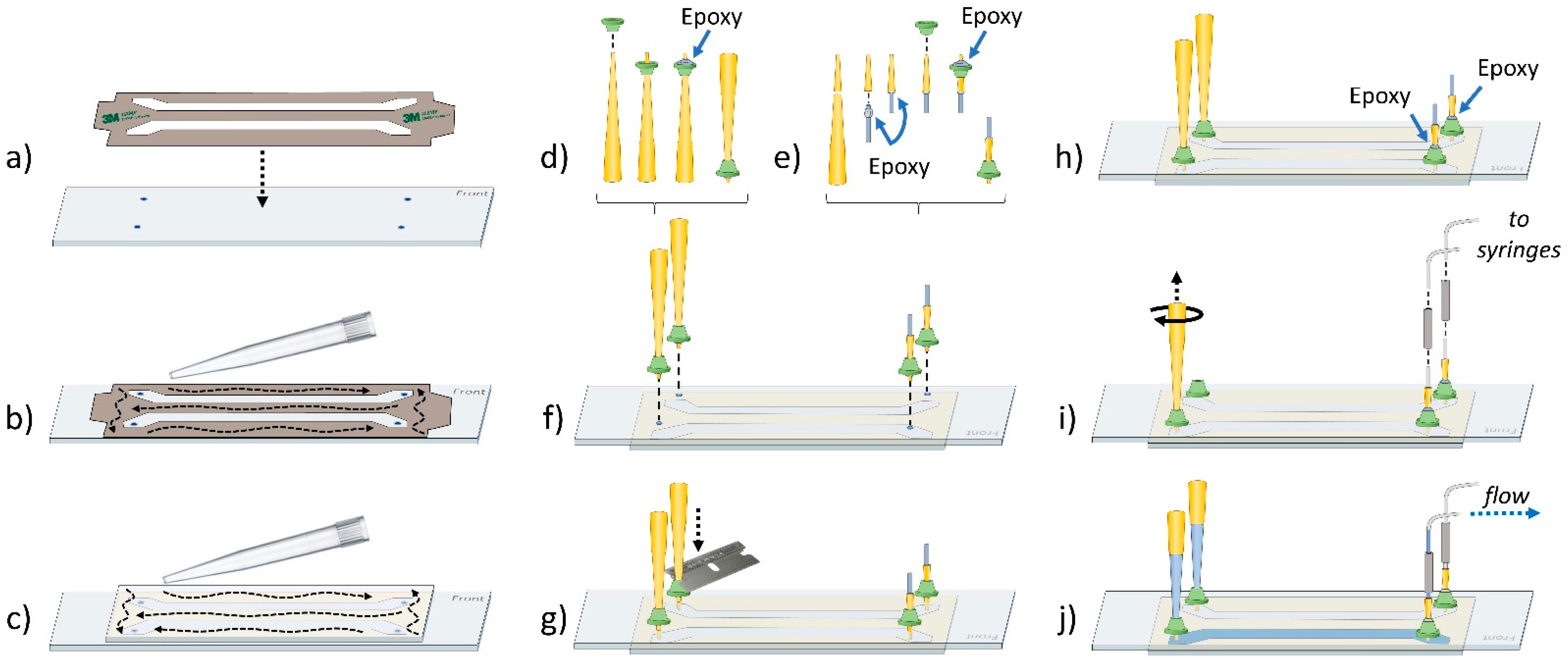
4.4. Sample Chamber Assembly
4.5. Data Collection
5. Applications
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Selvin, P.R.; Ha, T. Single-Molecule Techniques: A Laboratory Manual; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2008. [Google Scholar]
- Kudalkar, E.M.; Davis, T.N.; Asbury, C.L. Single-Molecule Total Internal Reflection Fluorescence Microscopy. Cold Spring Harb. Protoc. 2016, 2016. [Google Scholar] [CrossRef]
- Roy, R.; Hohng, S.; Ha, T. A practical guide to single-molecule FRET. Nat. Methods 2008, 5, 507–516. [Google Scholar] [CrossRef]
- Kapanidis, A.N.; Weiss, S. Fluorescent probes and bioconjugation chemistries for single-molecule fluorescence analysis of biomolecules. J. Chem. Phys. 2002, 117, 10953–10964. [Google Scholar] [CrossRef]
- Hohng, S.; Joo, C.; Ha, T. Single-molecule three-color FRET. Biophys. J. 2004, 87, 1328–1337. [Google Scholar] [CrossRef] [PubMed]
- Gibbs, D.R.; Kaur, A.; Megalathan, A.; Sapkota, K.; Dhakal, S. Build Your Own Microscope: Step-By-Step Guide for Building a Prism-Based TIRF Microscope. Methods Protoc. 2018, 1, 40. [Google Scholar] [CrossRef]
- Johnson, D.S.; Jaiswal, J.K.; Simon, S. Total internal reflection fluorescence (TIRF) microscopy illuminator for improved imaging of cell surface events. Curr. Protoc. Cytom. 2012, 12, 12.29.11–12.29.19. [Google Scholar] [CrossRef]
- Ambrose, B.; Baxter, J.M.; Cully, J.; Willmott, M.; Steele, E.M.; Bateman, B.C.; Martin-Fernandez, M.L.; Cadby, A.; Shewring, J.; Aaldering, M.; et al. The smfBox is an open-source platform for single-molecule FRET. Nat. Commun. 2020, 11, 5641. [Google Scholar] [CrossRef]
- Boehm, E.M.; Subramanyam, S.; Ghoneim, M.; Washington, M.T.; Spies, M. Quantifying the Assembly of Multicomponent Molecular Machines by Single-Molecule Total Internal Reflection Fluorescence Microscopy. Methods Enzymol. 2016, 581, 105–145. [Google Scholar] [CrossRef]
- Ghoneim, M.; Spies, M. Direct correlation of DNA binding and single protein domain motion via dual illumination fluorescence microscopy. Nano Lett. 2014, 14, 5920–5931. [Google Scholar] [CrossRef]
- Kudalkar, E.M.; Davis, T.N.; Asbury, C.L. Preparation of Reactions for Imaging with Total Internal Reflection Fluorescence Microscopy. Cold Spring Harb. Protoc. 2016, 2016. [Google Scholar] [CrossRef][Green Version]
- Gotz, M.; Wortmann, P.; Schmid, S.; Hugel, T. Using Three-color Single-molecule FRET to Study the Correlation of Protein Interactions. J. Vis. Exp. 2018, 2018. [Google Scholar] [CrossRef]
- Axelrod, D. Total internal reflection fluorescence microscopy in cell biology. Methods Enzymol. 2003, 361, 1–33. [Google Scholar] [CrossRef]
- Fish, K.N. Total internal reflection fluorescence (TIRF) microscopy. Curr. Protoc. Cytom. 2009, 50, 12.18.11–12.18.13. [Google Scholar] [CrossRef] [PubMed]
- Joo, C.; Ha, T. Objective-type total internal reflection microscopy (emission) for single-molecule FRET. Cold Spring Harb Protoc 2012, 2012, 1192–1194. [Google Scholar] [CrossRef] [PubMed]
- Friedman, L.J.; Chung, J.; Gelles, J. Viewing dynamic assembly of molecular complexes by multi-wavelength single-molecule fluorescence. Biophys. J. 2006, 91, 1023–1031. [Google Scholar] [CrossRef]
- Peet, M.; Erdogan, T. Searchlight-Welcome. Available online: https://searchlight.semrock.com/ (accessed on 5 March 2021).
- Eggeling, C.; Widengren, J.; Brand, L.; Schaffer, J.; Felekyan, S.; Seidel, C.A. Analysis of photobleaching in single-molecule multicolor excitation and Forster resonance energy transfer measurements. J. Phys. Chem. A 2006, 110, 2979–2995. [Google Scholar] [CrossRef]
- Pokhrel, N.; Caldwell, C.C.; Corless, E.I.; Tillison, E.A.; Tibbs, J.; Jocic, N.; Tabei, S.M.A.; Wold, M.S.; Spies, M.; Antony, E. Dynamics and selective remodeling of the DNA-binding domains of RPA. Nat. Struct. Mol. Biol. 2019, 26, 129–136. [Google Scholar] [CrossRef]
- Reichman, J. Handbook of Optical Filters for Fluorescence Microscopy; Chroma Technology Corporation: Bellows Falls, VT, USA, 2000. [Google Scholar]
- Delay, M. Maximizing the Performance of Advanced Microscopes by Controlling Wavefront Error Using Optical Filters. 2018. Available online: https://www.semrock.com/Data/Sites/1/semrockpdfs/idx2678-smk-maxperf-wp.pdf (accessed on 5 March 2021).
- Joo, C.; Ha, T. Single-molecule FRET with total internal reflection microscopy. Cold Spring Harb. Protoc. 2012, 2012. [Google Scholar] [CrossRef]
- Hohlbein, J.; Craggs, T.D.; Cordes, T. Alternating-laser excitation: Single-molecule FRET and beyond. Chem. Soc. Rev. 2014, 43, 1156–1171. [Google Scholar] [CrossRef]
- Kapanidis, A.N.; Lee, N.K.; Laurence, T.A.; Doose, S.; Margeat, E.; Weiss, S. Fluorescence-aided molecule sorting: Analysis of structure and interactions by alternating-laser excitation of single molecules. Proc. Natl. Acad. Sci. USA 2004, 101, 8936–8941. [Google Scholar] [CrossRef]
- Axelrod, D. Chapter 7: Total internal reflection fluorescence microscopy. Methods Cell Biol. 2008, 89, 169–221. [Google Scholar] [CrossRef]
- Trache, A.; Meininger, G.A. Total internal reflection fluorescence (TIRF) microscopy. Curr. Protoc. Microbiol. 2008, 10, 2A.2.1–2A.2.22. [Google Scholar] [CrossRef] [PubMed]
- Oheim, M.; Salomon, A.; Weissman, A.; Brunstein, M.; Becherer, U. Calibrating Evanescent-Wave Penetration Depths for Biological TIRF Microscopy. Biophys. J. 2019, 117, 795–809. [Google Scholar] [CrossRef] [PubMed]
- Martin-Fernandez, M.L.; Tynan, C.J.; Webb, S.E. A ‘pocket guide’ to total internal reflection fluorescence. J. Microsc. 2013, 252, 16–22. [Google Scholar] [CrossRef]
- Daniel Axelrod, J.C.L.; Davidson, M.W. Evanescent Field Penetration Depth—Java Tutorial. Available online: https://www.olympus-lifescience.com/en/microscope-resource/primer/java/tirf/penetration/ (accessed on 5 March 2021).
- Millis, B.A. Evanescent-wave field imaging: An introduction to total internal reflection fluorescence microscopy. Methods Mol. Biol. 2012, 823, 295–309. [Google Scholar] [CrossRef]
- Saurabh, S.; Maji, S.; Bruchez, M.P. Evaluation of sCMOS cameras for detection and localization of single Cy5 molecules. Opt. Express 2012, 20, 7338–7349. [Google Scholar] [CrossRef] [PubMed]
- Bain, F.E.; Wu, C.G.; Spies, M. Single-molecule sorting of DNA helicases. Methods 2016, 108, 14–23. [Google Scholar] [CrossRef]
- Joo, C.; Ha, T. Preparing sample chambers for single-molecule FRET. Cold Spring Harb. Protoc. 2012, 2012, 1104–1108. [Google Scholar] [CrossRef] [PubMed]
- Chandradoss, S.D.; Haagsma, A.C.; Lee, Y.K.; Hwang, J.H.; Nam, J.M.; Joo, C. Surface passivation for single-molecule protein studies. J. Vis. Exp. 2014, 2014, e50549. [Google Scholar] [CrossRef]
- Jain, A.; Liu, R.; Xiang, Y.K.; Ha, T. Single-molecule pull-down for studying protein interactions. Nat. Protoc. 2012, 7, 445–452. [Google Scholar] [CrossRef]
- Tibbs, J.; Ghoneim, M.; Caldwell, C.C.; Buzynski, T.; Bowie, W.; Boehm, E.M.; Washington, M.T.; Tabei, S.M.A.; Spies, M. KERA: Analysis tool for multi-process, multi-state single-molecule data. Nucleic Acids Res. 2021, 49, e53. [Google Scholar] [CrossRef]
- Friedman, L.J.; Gelles, J. Multi-wavelength single-molecule fluorescence analysis of transcription mechanisms. Methods 2015, 86, 27–36. [Google Scholar] [CrossRef]
- Boehm, E.M.; Spies, M.; Washington, M.T. PCNA tool belts and polymerase bridges form during translesion synthesis. Nucleic Acids Res. 2016, 44, 8250–8260. [Google Scholar] [CrossRef]
- Honda, M.; Park, J.; Pugh, R.A.; Ha, T.; Spies, M. Single-molecule analysis reveals differential effect of ssDNA-binding proteins on DNA translocation by XPD helicase. Mol. Cell 2009, 35, 694–703. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.S.; Dagdas, Y.S.; Kleinstiver, B.P.; Welch, M.M.; Sousa, A.A.; Harrington, L.B.; Sternberg, S.H.; Joung, J.K.; Yildiz, A.; Doudna, J.A. Enhanced proofreading governs CRISPR-Cas9 targeting accuracy. Nature 2017, 550, 407–410. [Google Scholar] [CrossRef]
- Chen, R.; Subramanyam, S.; Elcock, A.H.; Spies, M.; Wold, M.S. Dynamic binding of replication protein a is required for DNA repair. Nucleic Acids Res. 2016, 44, 5758–5772. [Google Scholar] [CrossRef] [PubMed]
- Boehm, E.M.; Powers, K.T.; Kondratick, C.M.; Spies, M.; Houtman, J.C.; Washington, M.T. The Proliferating Cell Nuclear Antigen (PCNA)-interacting Protein (PIP) Motif of DNA Polymerase eta Mediates Its Interaction with the C-terminal Domain of Rev1. J. Biol. Chem. 2016, 291, 8735–8744. [Google Scholar] [CrossRef] [PubMed]
- Roy, U.; Greene, E.C. Single-Stranded DNA Curtains for Single-Molecule Visualization of Rad51-ssDNA Filament Dynamics. Methods Mol. Biol. 2021, 2281, 193–207. [Google Scholar] [CrossRef] [PubMed]
- Unciuleac, M.C.; Meir, A.; Xue, C.; Warren, G.M.; Greene, E.C.; Shuman, S. Clutch mechanism of chemomechanical coupling in a DNA resecting motor nuclease. Proc. Natl. Acad. Sci. USA 2021, 118, e2023955118. [Google Scholar] [CrossRef]
- Gallardo, I.F.; Pasupathy, P.; Brown, M.; Manhart, C.M.; Neikirk, D.P.; Alani, E.; Finkelstein, I.J. High-Throughput Universal DNA Curtain Arrays for Single-Molecule Fluorescence Imaging. Langmuir 2015, 31, 10310–10317. [Google Scholar] [CrossRef]
- Calcines-Cruz, C.; Finkelstein, I.J.; Hernandez-Garcia, A. CRISPR-Guided Programmable Self-Assembly of Artificial Virus-Like Nucleocapsids. Nano Lett. 2021, 21, 2752–2757. [Google Scholar] [CrossRef] [PubMed]
- Kong, M.; Beckwitt, E.C.; Springall, L.; Kad, N.M.; Van Houten, B. Single-Molecule Methods for Nucleotide Excision Repair: Building a System to Watch Repair in Real Time. Methods Enzymol. 2017, 592, 213–257. [Google Scholar] [CrossRef]
- Jang, S.; Kumar, N.; Beckwitt, E.C.; Kong, M.; Fouquerel, E.; Rapic-Otrin, V.; Prasad, R.; Watkins, S.C.; Khuu, C.; Majumdar, C.; et al. Damage sensor role of UV-DDB during base excision repair. Nat. Struct. Mol. Biol. 2019, 26, 695–703. [Google Scholar] [CrossRef] [PubMed]
- Singh, D.; Wang, Y.; Mallon, J.; Yang, O.; Fei, J.; Poddar, A.; Ceylan, D.; Bailey, S.; Ha, T. Mechanisms of improved specificity of engineered Cas9s revealed by single-molecule FRET analysis. Nat. Struct. Mol. Biol. 2018, 25, 347–354. [Google Scholar] [CrossRef]
- Dagdas, Y.S.; Chen, J.S.; Sternberg, S.H.; Doudna, J.A.; Yildiz, A. A conformational checkpoint between DNA binding and cleavage by CRISPR-Cas9. Sci. Adv. 2017, 3, eaao0027. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Lee, S.; Ragunathan, K.; Joo, C.; Ha, T.; Hohng, S. Single-molecule four-color FRET. Angew. Chem. Int. Ed. Engl. 2010, 49, 9922–9925. [Google Scholar] [CrossRef]
- Paudel, B.P.; Moye, A.L.; Abou Assi, H.; El-Khoury, R.; Cohen, S.B.; Holien, J.K.; Birrento, M.L.; Samosorn, S.; Intharapichai, K.; Tomlinson, C.G.; et al. A mechanism for the extension and unfolding of parallel telomeric G-quadruplexes by human telomerase at single-molecule resolution. eLife 2020, 9, e56428. [Google Scholar] [CrossRef] [PubMed]
- Subramanyam, S.; Kinz-Thompson, C.D.; Gonzalez, R.L., Jr.; Spies, M. Observation and Analysis of RAD51 Nucleation Dynamics at Single-Monomer Resolution. Methods Enzymol. 2018, 600, 201–232. [Google Scholar] [CrossRef]
- Mohapatra, S.; Lin, C.T.; Feng, X.A.; Basu, A.; Ha, T. Single-Molecule Analysis and Engineering of DNA Motors. Chem. Rev. 2020, 120, 36–78. [Google Scholar] [CrossRef]
- Brown, A.E.; Hategan, A.; Safer, D.; Goldman, Y.E.; Discher, D.E. Cross-correlated TIRF/AFM reveals asymmetric distribution of force-generating heads along self-assembled, “synthetic” myosin filaments. Biophys. J. 2009, 96, 1952–1960. [Google Scholar] [CrossRef]
- Maki, K.; Han, S.W.; Hirano, Y.; Yonemura, S.; Hakoshima, T.; Adachi, T. Real-time TIRF observation of vinculin recruitment to stretched alpha-catenin by AFM. Sci. Rep. 2018, 8, 1575. [Google Scholar] [CrossRef] [PubMed]
- Ha, T.; Enderle, T.; Ogletree, D.F.; Chemla, D.S.; Selvin, P.R.; Weiss, S. Probing the interaction between two single molecules: Fluorescence resonance energy transfer between a single donor and a single acceptor. Proc. Natl. Acad. Sci. USA 1996, 93, 6264–6268. [Google Scholar] [CrossRef]
- Hohng, S.; Zhou, R.; Nahas, M.K.; Yu, J.; Schulten, K.; Lilley, D.M.; Ha, T. Fluorescence-force spectroscopy maps two-dimensional reaction landscape of the holliday junction. Science 2007, 318, 279–283. [Google Scholar] [CrossRef]
- Roy, R.; Kozlov, A.G.; Lohman, T.M.; Ha, T. SSB protein diffusion on single-stranded DNA stimulates RecA filament formation. Nature 2009, 461, 1092–1097. [Google Scholar] [CrossRef]
- Zhou, R.; Kozlov, A.G.; Roy, R.; Zhang, J.; Korolev, S.; Lohman, T.M.; Ha, T. SSB functions as a sliding platform that migrates on DNA via reptation. Cell 2011, 146, 222–232. [Google Scholar] [CrossRef] [PubMed]
- Myong, S.; Bruno, M.M.; Pyle, A.M.; Ha, T. Spring-loaded mechanism of DNA unwinding by hepatitis C virus NS3 helicase. Science 2007, 317, 513–516. [Google Scholar] [CrossRef]
- Arslan, S.; Khafizov, R.; Thomas, C.D.; Chemla, Y.R.; Ha, T. Protein structure. Engineering of a superhelicase through conformational control. Science 2015, 348, 344–347. [Google Scholar] [CrossRef]
- Ngo, T.T.; Zhang, Q.; Zhou, R.; Yodh, J.G.; Ha, T. Asymmetric unwrapping of nucleosomes under tension directed by DNA local flexibility. Cell 2015, 160, 1135–1144. [Google Scholar] [CrossRef] [PubMed]
- Myong, S.; Rasnik, I.; Joo, C.; Lohman, T.M.; Ha, T. Repetitive shuttling of a motor protein on DNA. Nature 2005, 437, 1321–1325. [Google Scholar] [CrossRef]
- Park, J.; Myong, S.; Niedziela-Majka, A.; Lee, K.S.; Yu, J.; Lohman, T.M.; Ha, T. PcrA helicase dismantles RecA filaments by reeling in DNA in uniform steps. Cell 2010, 142, 544–555. [Google Scholar] [CrossRef]
- Ragunathan, K.; Joo, C.; Ha, T. Real-time observation of strand exchange reaction with high spatiotemporal resolution. Structure 2011, 19, 1064–1073. [Google Scholar] [CrossRef] [PubMed]
- Ragunathan, K.; Liu, C.; Ha, T. RecA filament sliding on DNA facilitates homology search. eLife 2012, 1, e00067. [Google Scholar] [CrossRef] [PubMed]
- Myler, L.R.; Gallardo, I.F.; Soniat, M.M.; Deshpande, R.A.; Gonzalez, X.B.; Kim, Y.; Paull, T.T.; Finkelstein, I.J. Single-Molecule Imaging Reveals How Mre11-Rad50-Nbs1 Initiates DNA Break Repair. Mol. Cell 2017, 67, 891–898.e4. [Google Scholar] [CrossRef] [PubMed]
- Soniat, M.M.; Myler, L.R.; Finkelstein, I.J. Assembling the Human Resectosome on DNA Curtains. Methods Mol. Biol. 2019, 1999, 225–244. [Google Scholar] [CrossRef] [PubMed]
- Deshpande, R.A.; Myler, L.R.; Soniat, M.M.; Makharashvili, N.; Lee, L.; Lees-Miller, S.P.; Finkelstein, I.J.; Paull, T.T. DNA-dependent protein kinase promotes DNA end processing by MRN and CtIP. Sci. Adv. 2020, 6, eaay0922. [Google Scholar] [CrossRef]
- Rothenberg, E.; Grimme, J.M.; Spies, M.; Ha, T. Human Rad52-mediated homology search and annealing occurs by continuous interactions between overlapping nucleoprotein complexes. Proc. Natl. Acad. Sci. USA 2008, 105, 20274–20279. [Google Scholar] [CrossRef]
- Hwang, H.; Myong, S. Protein induced fluorescence enhancement (PIFE) for probing protein-nucleic acid interactions. Chem. Soc. Rev. 2014, 43, 1221–1229. [Google Scholar] [CrossRef]
- Qiu, Y.; Antony, E.; Doganay, S.; Koh, H.R.; Lohman, T.M.; Myong, S. Srs2 prevents Rad51 filament formation by repetitive motion on DNA. Nat. Commun. 2013, 4, 2281. [Google Scholar] [CrossRef]
- Subramanyam, S.; Ismail, M.; Bhattacharya, I.; Spies, M. Tyrosine phosphorylation stimulates activity of human RAD51 recombinase through altered nucleoprotein filament dynamics. Proc. Natl. Acad. Sci. USA 2016, 113, E6045–E6054. [Google Scholar] [CrossRef]
- Joo, C.; McKinney, S.A.; Nakamura, M.; Rasnik, I.; Myong, S.; Ha, T. Real-time observation of RecA filament dynamics with single monomer resolution. Cell 2006, 126, 515–527. [Google Scholar] [CrossRef]
- Qi, Z.; Greene, E.C. Visualizing recombination intermediates with single-stranded DNA curtains. Methods 2016, 105, 62–74. [Google Scholar] [CrossRef]
- Yao, N.Y.; Georgescu, R.E.; Finkelstein, J.; O’Donnell, M.E. Single-molecule analysis reveals that the lagging strand increases replisome processivity but slows replication fork progression. Proc. Natl. Acad. Sci. USA 2009, 106, 13236–13241. [Google Scholar] [CrossRef]
- Pauszek, R.F., 3rd; Lamichhane, R.; Rajkarnikar Singh, A.; Millar, D.P. Single-molecule view of coordination in a multi-functional DNA polymerase. eLife 2021, 10. [Google Scholar] [CrossRef] [PubMed]
- Duderstadt, K.E.; Geertsema, H.J.; Stratmann, S.A.; Punter, C.M.; Kulczyk, A.W.; Richardson, C.C.; van Oijen, A.M. Simultaneous Real-Time Imaging of Leading and Lagging Strand Synthesis Reveals the Coordination Dynamics of Single Replisomes. Mol. Cell 2016, 64, 1035–1047. [Google Scholar] [CrossRef]
- Graham, J.E.; Marians, K.J.; Kowalczykowski, S.C. Independent and Stochastic Action of DNA Polymerases in the Replisome. Cell 2017, 169, 1201–1213e.17. [Google Scholar] [CrossRef]
- Zaher, M.S.; Rashid, F.; Song, B.; Joudeh, L.I.; Sobhy, M.A.; Tehseen, M.; Hingorani, M.M.; Hamdan, S.M. Missed cleavage opportunities by FEN1 lead to Okazaki fragment maturation via the long-flap pathway. Nucleic Acids Res. 2018, 46, 2956–2974. [Google Scholar] [CrossRef] [PubMed]
- Vrtis, K.B.; Dewar, J.M.; Chistol, G.; Wu, R.A.; Graham, T.G.W.; Walter, J.C. Single-strand DNA breaks cause replisome disassembly. Mol. Cell 2021, 81, 1309–1318.e6. [Google Scholar] [CrossRef]
- Yardimci, H.; Loveland, A.B.; Habuchi, S.; van Oijen, A.M.; Walter, J.C. Uncoupling of sister replisomes during eukaryotic DNA replication. Mol. Cell 2010, 40, 834–840. [Google Scholar] [CrossRef] [PubMed]
- van Oijen, A.M.; Loparo, J.J. Single-molecule studies of the replisome. Annu. Rev. Biophys. 2010, 39, 429–448. [Google Scholar] [CrossRef]
- Rashid, F.; Raducanu, V.S.; Zaher, M.S.; Tehseen, M.; Habuchi, S.; Hamdan, S.M. Initial state of DNA-Dye complex sets the stage for protein induced fluorescence modulation. Nat. Commun. 2019, 10, 2104. [Google Scholar] [CrossRef]
- Ray, S.; Qureshi, M.H.; Malcolm, D.W.; Budhathoki, J.B.; Celik, U.; Balci, H. RPA-mediated unfolding of systematically varying G-quadruplex structures. Biophys. J. 2013, 104, 2235–2245. [Google Scholar] [CrossRef]
- Qureshi, M.H.; Ray, S.; Sewell, A.L.; Basu, S.; Balci, H. Replication protein A unfolds G-quadruplex structures with varying degrees of efficiency. J. Phys. Chem. B 2012, 116, 5588–5594. [Google Scholar] [CrossRef] [PubMed]
- Donovan, B.T.; Huynh, A.; Ball, D.A.; Patel, H.P.; Poirier, M.G.; Larson, D.R.; Ferguson, M.L.; Lenstra, T.L. Live-cell imaging reveals the interplay between transcription factors, nucleosomes, and bursting. EMBO J. 2019, 38, e100809. [Google Scholar] [CrossRef]
- Kilic, S.; Felekyan, S.; Doroshenko, O.; Boichenko, I.; Dimura, M.; Vardanyan, H.; Bryan, L.C.; Arya, G.; Seidel, C.A.M.; Fierz, B. Single-molecule FRET reveals multiscale chromatin dynamics modulated by HP1alpha. Nat. Commun. 2018, 9, 235. [Google Scholar] [CrossRef] [PubMed]
- Wilson, M.D.; Renault, L.; Maskell, D.P.; Ghoneim, M.; Pye, V.E.; Nans, A.; Rueda, D.S.; Cherepanov, P.; Costa, A. Retroviral integration into nucleosomes through DNA looping and sliding along the histone octamer. Nat. Commun. 2019, 10, 4189. [Google Scholar] [CrossRef] [PubMed]
- Willhoft, O.; Ghoneim, M.; Lin, C.L.; Chua, E.Y.D.; Wilkinson, M.; Chaban, Y.; Ayala, R.; McCormack, E.A.; Ocloo, L.; Rueda, D.S.; et al. Structure and dynamics of the yeast SWR1-nucleosome complex. Science 2018, 362, eaat7716. [Google Scholar] [CrossRef]
- Senavirathne, G.; Bertram, J.G.; Jaszczur, M.; Chaurasiya, K.R.; Pham, P.; Mak, C.H.; Goodman, M.F.; Rueda, D. Activation-induced deoxycytidine deaminase (AID) co-transcriptional scanning at single-molecule resolution. Nat. Commun. 2015, 6, 10209. [Google Scholar] [CrossRef]
| Ref # | Item | Notes | Catalog ID | Vendor | # |
|---|---|---|---|---|---|
| Table | |||||
| #1 | Optical Table | 3’ × 6’ × 12”, 1/4”-20 threads | RPR-36-12 | Newport | 1 |
| #2 | Table Legs | SL-600 Series, 22” height, 2400 lb max. capacity | SL-600-422 | Newport | 4 |
| #3 | Overhead Table Shelf | Fits 6’ table, w/ electrical outlets | ATS-6 | Newport | 1 |
| Mounting Hardware, Clamps, and Posts | |||||
| #4 | 2" Post | 1/2” diameter, top tapped 8-32, bottom tapped 1/4”-20 | 9621 | Newport | 4 |
| #5 | 3” Post | 1/2” diameter, top tapped 8-32, bottom tapped 1/4”-20 | 9622 | Newport | 1 |
| #6 | 4” Post | 1/2” diameter, top tapped 8-32, bottom tapped 1/4”-20 | 9623 | Newport | 7 |
| #7 | 6” Post | 1/2” diameter, top tapped 8-32, bottom tapped 1/4”-20 | 9624 | Newport | 20 |
| #8 | 4” Adjustable Post Holder | Fits 1/2” diameter posts, bottom tapped 1/4”-20 | VPH-4PK | Newport | 20 |
| #9 | Pedestal Base Adaptor | 1-1/4” diameter, 0.19” height, top thread 1/4”-20 | PS-A-PK | Newport | 20 |
| #10 | Slotted Clamping Fork | Fits Pedestal Base Adaptor (#9), slot for 1/4”-20 bolt | PS-F-PK | Newport | 30 |
| #11 | L-shaped Table Clamp | (for mounting #20, #30, #46) | CL5-P5 | Thorlabs | 19 |
| #12 | Right-Angle Post Clamp | Fits 1/2” diameter posts (#4–7) | 9935 | Newport | 7 |
| #13 | 1” Ultima Clear Edge Mirror Mount | Fits 1” diameter mirrors (#27–#29), 2 knobs, left-handed | U100-A-LH-2K | Newport | 13 |
| #14 | 1/2” Lens Mount | Fits 1/2” diameter lenses (#24-26, #55), 8–32 Thread | LH-0.5A | Newport | 4 |
| #15 | 1” Adjustable Post Holder + Pedestal Base | Fits 1/2” diameter posts (for stage mirror assembly) | VPH-1-P | Newport | 1 |
| Excitation Lasers | |||||
| #16 | OBIS 488nm LS 100 mW Laser | CW, Diode, beam diameter 0.7 ± 0.05 mm, (to excite AF488) | 1226419 | Coherent | 1 |
| #17 | OBIS 532nm LS 100 mW Laser | CW, Diode, beam diameter 0.7 ± 0.05 mm, (to excite Cy3) | 1261781 | Coherent | 1 |
| #18 | OBIS 640nm LX 100 mW Laser | CW, Diode, beam diameter 0.8 ± 0.1 mm, (to excite Cy5) | 1185055 | Coherent | 1 |
| #19 | OBIS Laser Heat Sink | Fits LX/LS OBIS Lasers, 2.7” height (optional) | 1193289 | Coherent | 3 |
| #20 | Vertical Translation Stage | 84 mm–184 mm height, M6 thread holes (mounted via #11) | 860-0075 | Eksma | 3 |
| #21 | OBIS Laser Remote and Power Supply | Includes six 1-meter SDR cables | 1234466 | Coherent | 1 |
| Laser Shutters | |||||
| #22 | Laser Shutter | 3mm laser shutter, teflon coating, no electronic sync | LS3S2T0-100 | Uniblitz | 3 |
| #23 | Shutter Driver | four-channels, includes 710C shutter interconnect cables | VMM-D4 | Uniblitz | 1 |
| Excitation Dichroic Mirrors and Filters | |||||
| #24 | 488 nm Laser clean-up filter | 0.5” diameter, 488 nm, MaxLine® | LL01-488-12.5 | Semrock | 1 |
| #25 | 532 nm Laser clean-up filter | 0.5” diameter, 532 nm, MaxLine® | LL01-532-12.5 | Semrock | 1 |
| #26 | 640 nm Laser clean-up filter | 0.5” diameter, 640/8 nm, MaxDiode™ | LD01-640/8-12.5 | Semrock | 1 |
| #27 | Broadband Mirror | 1” diameter, 6.0 mm thick, RWE: λ/10 @ 633 nm | BB1-E02 | Thorlabs | 11 |
| #28 | 505 nm Cut-off Longpass Dichroic Mirror | 1” diameter, 3.2 mm thick, TWE: λ/4 @ 633 nm | DMLP505 | Thorlabs | 1 |
| #29 | 550 nm Cut-off Longpass Dichroic Mirror | 1” diameter, 3.2 mm thick, TWE: λ/4 @ 633 nm | DMLP550 | Thorlabs | 1 |
| Inverted Microscope Components | |||||
| #30 | IX73 Microscope Frame (1 deck) | Single deck | IX73P1F | Olympus | 1 |
| #31 | 60X Water Objective (NA 1.20) | UPLSAPO60XW; U Plan S-Apo, WD0.28,W/CC0.13-0.21 | 1-U2B893 | Olympus | 1 |
| #32 | C-Mount Camera Adapter | Centerable (U-TV1XC) (1x Mag) | U-V111C | Olympus | 1 |
| #33 | Fluorescent Turret | (IX3-RFACS-1-2 CODED) | 5-UR416-1 | Olympus | 1 |
| #34 | Binocular Observation Tube | GX/IX (U-BI90-1-2) | 3-U243 | Olympus | 1 |
| #35 | 10X Eyepiece | (FN:22 WHN10X-1-7) | 2-U1007 | Olympus | 1 |
| #36 | 10X Eyepiece (Adjustable Focus) | (FN:22 WHN10X-H-1-7) | 2-U100H6 | Olympus | 1 |
| #37 | Left Handle Stage with Short Stalk | (IX3-SVL) | 4-U222 | Olympus | 1 |
| #38 | Stage Clips | FOR IX STAGE (IX-SCL) (includes 2) | FV4-U291 | Olympus | 1 |
| #39 | 6-position Nosepiece | (IX3-D6RES CODED) | U-R380 | Olympus | 1 |
| Custom Components and Screws | |||||
| #40 | Custom Stage Adapter (Rear) | Schematics can be found at www.freudenthallab.com (accessed on 21 June 2021) | CAD file ID 40 | machine shop | 1 |
| #41 | Custom Stage Adapter (Front) | CAD file ID 41 | local shop | 1 | |
| #42 | Custom Stage Breadboard | CAD file ID 42 | local shop | 1 | |
| #43 | Custom Prism Adapter | CAD file ID 43 | local shop | 1 | |
| #44 | Custom Prism Connector Piece | CAD file ID 44 | local shop | 1 | |
| #45 | Custom Prism Overhead Arm | CAD file ID 45 | local shop | 1 | |
| #46 | Custom Camera Mount | CAD file ID 46 (A–C) | local shop | 1 | |
| #47 | Custom Slide Holder Insert | CAD file ID 47 | local shop | 1 | |
| #48 | Thumb Screws | 1/4”-20 × 3/4”, knurled head 3/4” diameter | 60746807 | mscdirect | 3 |
| #49 | Prism Connector Screws | 4-40 × 1/4”, Mini Socket Cap Screw | 43494 | Hillman | 3 |
| #50 | Camera Mount Screws | 10-32 × 3/4”, Zinc, Flat Head, Phillips | 101100 | Hillman | 4 |
| #51 | Front Stage Breadboard Screws | 1/4”-20 × 1-1/4”, Zinc, Flat Head, Phillips | 101140 | Hillman | 2 |
| #52 | Front Stage Breadboard Screws | M5—0.8 mm × 40 mm, Socket Cap Screw | 43103 | Hillman | 2 |
| #53 | Back Stage Breadboard Screws | 10-24 × 1”, Socket Cap Screw | 3197 | Hillman | 2 |
| #54 | Back Stage Breadboard Screws | M5—0.8 mm × 25 mm, Socket Cap Screw | 43102 | Hillman | 4 |
| Focusing Lens Micrometer Components | |||||
| #55 | Plano-Convex Lens | 1/2” diameter, 50.0 mm focal length, uncoated | LA1213-N-BK7 | Thorlabs | 1 |
| #56 | Optical Breadboard (4” x 6”) | 1/4”-20 thread on 1” grid, aluminum | SA2-04x06 | Newport | 1 |
| #57 | XYZ Quick-Mount Linear Stage | 1/2” travel, right-handed, 1/4”-20 thread | 460A-XYZ | Newport | 1 |
| #58 | Micrometer Actuator | 1 µm vernier, 13 mm travel, 50.8 TPI (fits #57) | SM-13 | Newport | 3 |
| Emission Dichroic Mirrors and Filters | |||||
| #59 | AF488 Bandpass Filter | 1” diameter, 512/25, BrightLine® | FF01-512/25-25 | Semrock | 1 |
| #60 | Cy3 Bandpass Filter | 1” diameter, 585/40, BrightLine® | FF01-585/40-25 | Semrock | 1 |
| #61 | Cy5 Bandpass Filter | 1” diameter, 680/42, BrightLine® | FF01-680/42-25 | Semrock | 1 |
| #62 | 532 nm Cutoff Longpass Dichroic Mirror | 25.2 × 35.6 × 1.1mm, RWE: 1λ P-V @ 632.8 nm, BrightLine® | DI03-R532-T1-25X36 | Semrock | 1 |
| #63 | 635 nm Cutoff Longpass Dichroic Mirror | 25.2 × 35.6 × 1.1mm, RWE: 1λ P-V @ 632.8 nm, BrightLine® | DI03-R635-T1-25X36 | Semrock | 1 |
| Camera and Emission Splitter | |||||
| #64 | IXON ULTRA 897 EMCCD | 56 FPS, 512 X 512, 16 UM, USB | OAT-DU-897U-CS0-#BV | Olympus | 1 |
| #65 | Optosplit III | Three-way image splitter,LS, 1X mag (contains #59–63) | O89-P280/310/0LS | Cairn | 1 |
| Tools and Screws | |||||
| #66 | Hex Driver Set (Imperial) | 20-Piece Balldriver and Hex Key Kit, w/Stand, Imperial | TC2 | Thorlabs | 1 |
| #67 | Hex Driver Set (Metric) | 15-Piece Balldriver and Hex Key Kit, w/Stand, Metric | TC3/M | Thorlabs | 1 |
| #68 | 1/2” Spanner Wrench | For threaded retaining rings (for 1/2” fixed lens mounts) | LT05-WR | Newport | 1 |
| #69 | Imperial Screw and Hardware Kit (1/4”-20) | 1/4”-20 Cap Screw and Hardware Kit | HW-KIT2 | Thorlabs | 1 |
| #70 | 1” Iris | 25.0 mm max aperture, TR3 Post (used for alignment) | ID25 | Thorlabs | 2 |
| #71 | Thin Slip-On Post Collar | Fits 0.5” diameter posts (maintains height of iris) | R2T | Thorlabs | 2 |
| #72 | Beam Height Measurement Tool | 12” tall, (used to measure height of beam and iris) | BHM4 | Thorlabs | 1 |
| Miscelaneous/Consumables | |||||
| #73 | Pellin-Broca Quartz Prism | Fused Silica | 325-1206 | Eksma | 1+ |
| #74 | 5 Minute Epoxy | Devcon, syringe | 12084 | Tap Plastics | 1+ |
| #75 | Quartz Microscope slide | 1” × 3” × 1 mm | 1X3X1MM | Finkenbeiner | 1+ |
| #76 | TetraSpeck Microspheres | 0.2 µm diameter, blue/green/orange/dark red | T7280 | Thermo Fisher | 1+ |
| #77 | Plain Glass Microscope Slides | Glass, 25 × 75 mm, 90° Ground Edges, Plain | 1301 | Globe Scientific | 1+ |
| #78 | Low Autofluorescence Immersion Oil | n = 1.518, Olympus Type F, 30 mL | MOIL-30 | Thorlabs | 1+ |
| #79 | Solvent Dropper Bottle | 2 oz. (60 mL) (to add water to objective) | LAB-14 | Newport | 1 |
| #80 | ETFE Tubing (ID 1.0 mm, OD 1/16”) | 3 m length, translucent | 18114238 | Cytiva | 1+ |
| #81 | Fisherbrand™ Redi-Tip™ 200 µL Pipet Tips | General purpose, yellow, 1000/PK | 02-707-500 | Fisher Scientific | 1+ |
| #82 | HEPA Air Purifier | Holmes small room 3-speed, w/optional ionizer | B0000DK35B | Holmes | 1 |
| #83 | 3M Double-Sided Adhesive Sheet | Clear, 5 MIL, double linered, 12 × 12” | 7955MP | Hisco | 1+ |
| #84 | Double Stick Tape | Scotch 665 Permanent 3M, Permanent, 1/2” × 250”, Clear | 917243 | Office Depot | 1+ |
| #85 | Cover Glass | 24 × 60 mm, No 1.5, 1 oz | 48393-251 | VWR | 1+ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fairlamb, M.S.; Whitaker, A.M.; Bain, F.E.; Spies, M.; Freudenthal, B.D. Construction of a Three-Color Prism-Based TIRF Microscope to Study the Interactions and Dynamics of Macromolecules. Biology 2021, 10, 571. https://doi.org/10.3390/biology10070571
Fairlamb MS, Whitaker AM, Bain FE, Spies M, Freudenthal BD. Construction of a Three-Color Prism-Based TIRF Microscope to Study the Interactions and Dynamics of Macromolecules. Biology. 2021; 10(7):571. https://doi.org/10.3390/biology10070571
Chicago/Turabian StyleFairlamb, Max S., Amy M. Whitaker, Fletcher E. Bain, Maria Spies, and Bret D. Freudenthal. 2021. "Construction of a Three-Color Prism-Based TIRF Microscope to Study the Interactions and Dynamics of Macromolecules" Biology 10, no. 7: 571. https://doi.org/10.3390/biology10070571
APA StyleFairlamb, M. S., Whitaker, A. M., Bain, F. E., Spies, M., & Freudenthal, B. D. (2021). Construction of a Three-Color Prism-Based TIRF Microscope to Study the Interactions and Dynamics of Macromolecules. Biology, 10(7), 571. https://doi.org/10.3390/biology10070571







