Cultivating Multidisciplinarity: Manufacturing and Sensing Challenges in Cultured Meat Production
Abstract
Simple Summary
Abstract
1. Cellular Agriculture
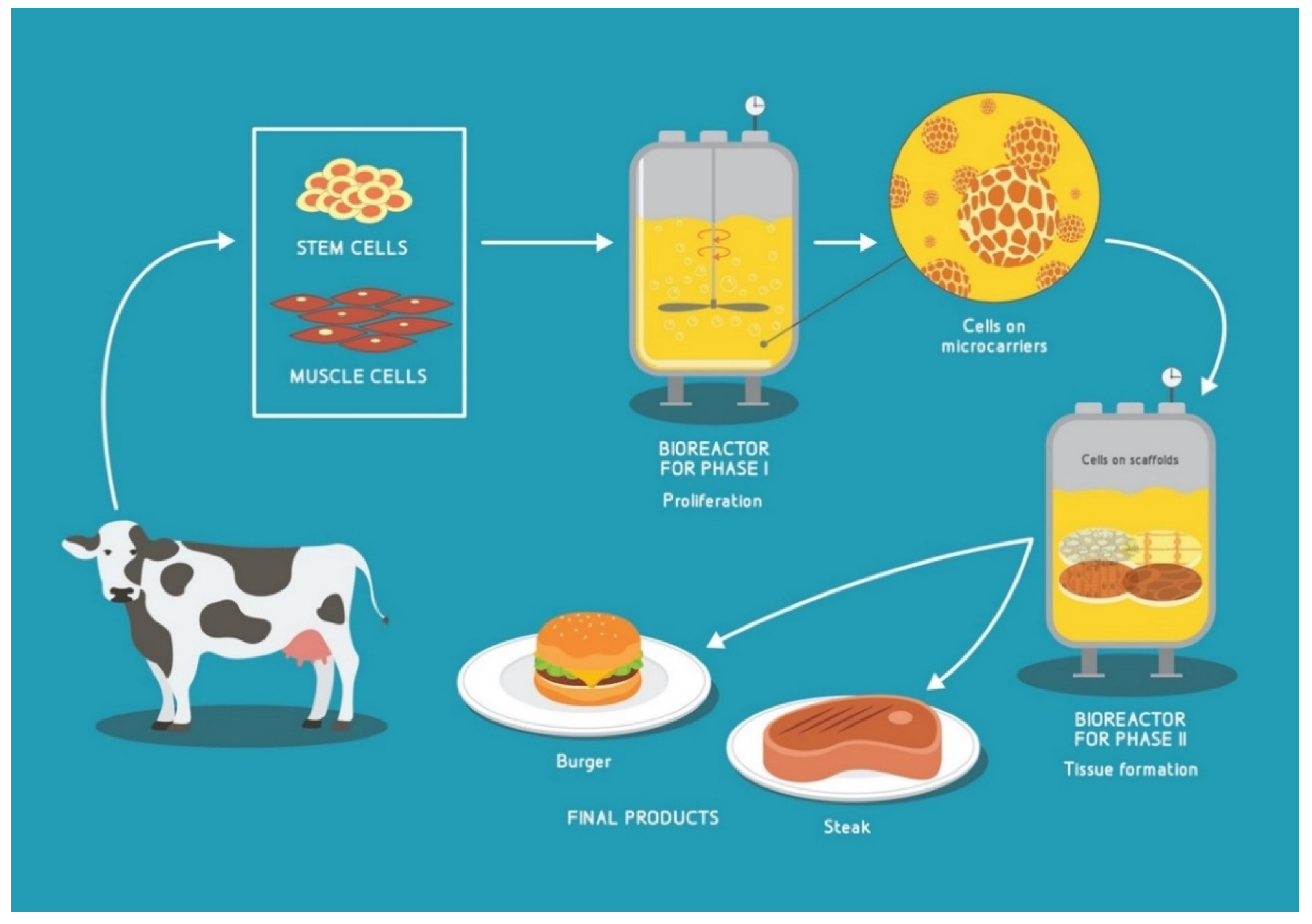
1.1. Cultured Meat (CM)
1.2. The Main Challenges Related to the Cultivated Meat Commercialization
1.3. Regulation
2. Bioreactors and Scaffolding
2.1. Types of Bioreactors
2.2. Microbioreactors
2.3. Microcarriers and Scaffolds
2.4. 3D Bioprinting for Cultured Meat
3. Mathematical Modeling and Computer Fluid Dynamics (CFDs)
3.1. Modeling for Stirred Tank Bioreactor
3.2. Modeling for Rocking/Wave Bioreactor
4. Sensors
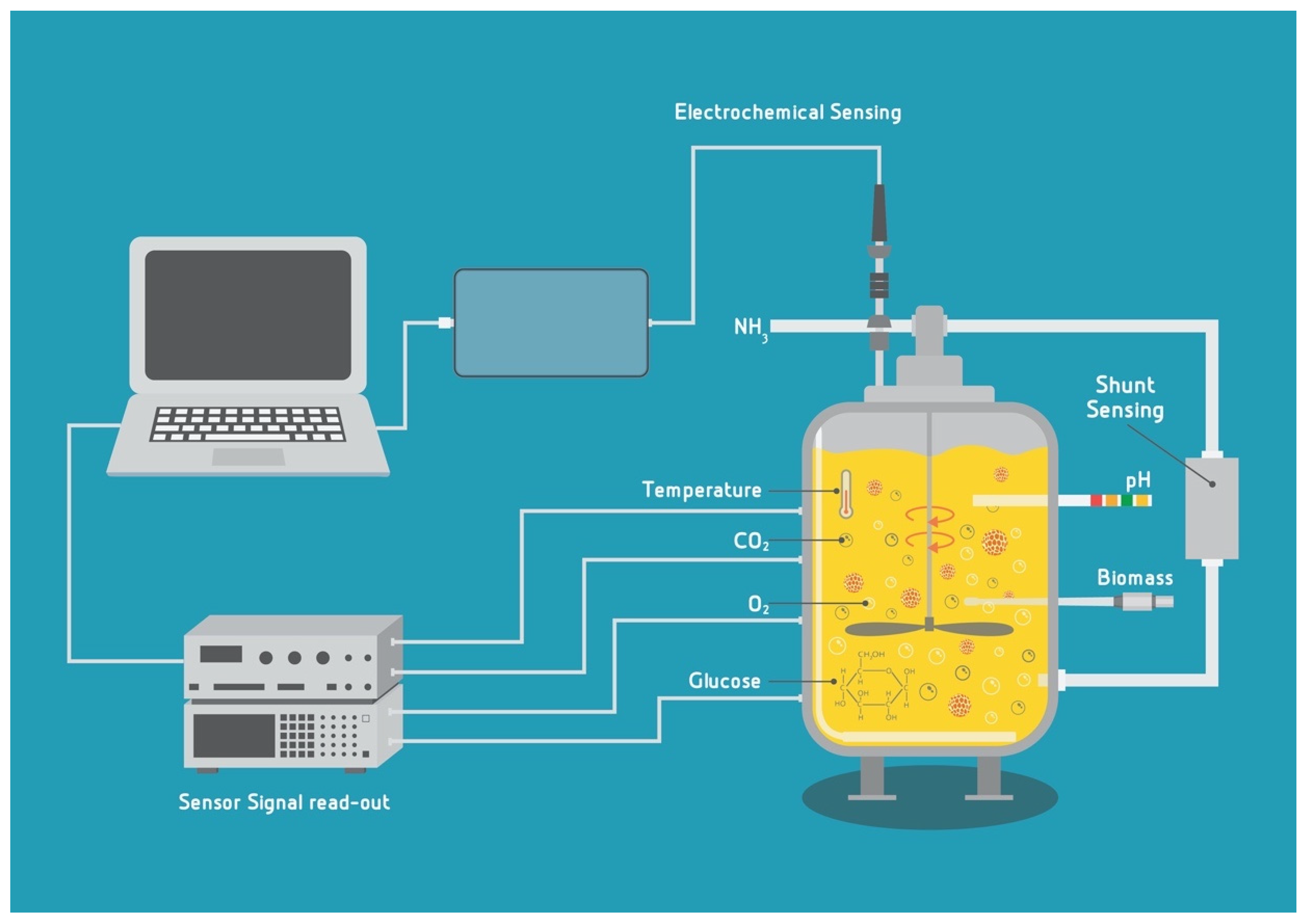
4.1. Sensing Options for pH, DO, CO2 and Temperature
4.2. Biomass Sensors in Bioreactors
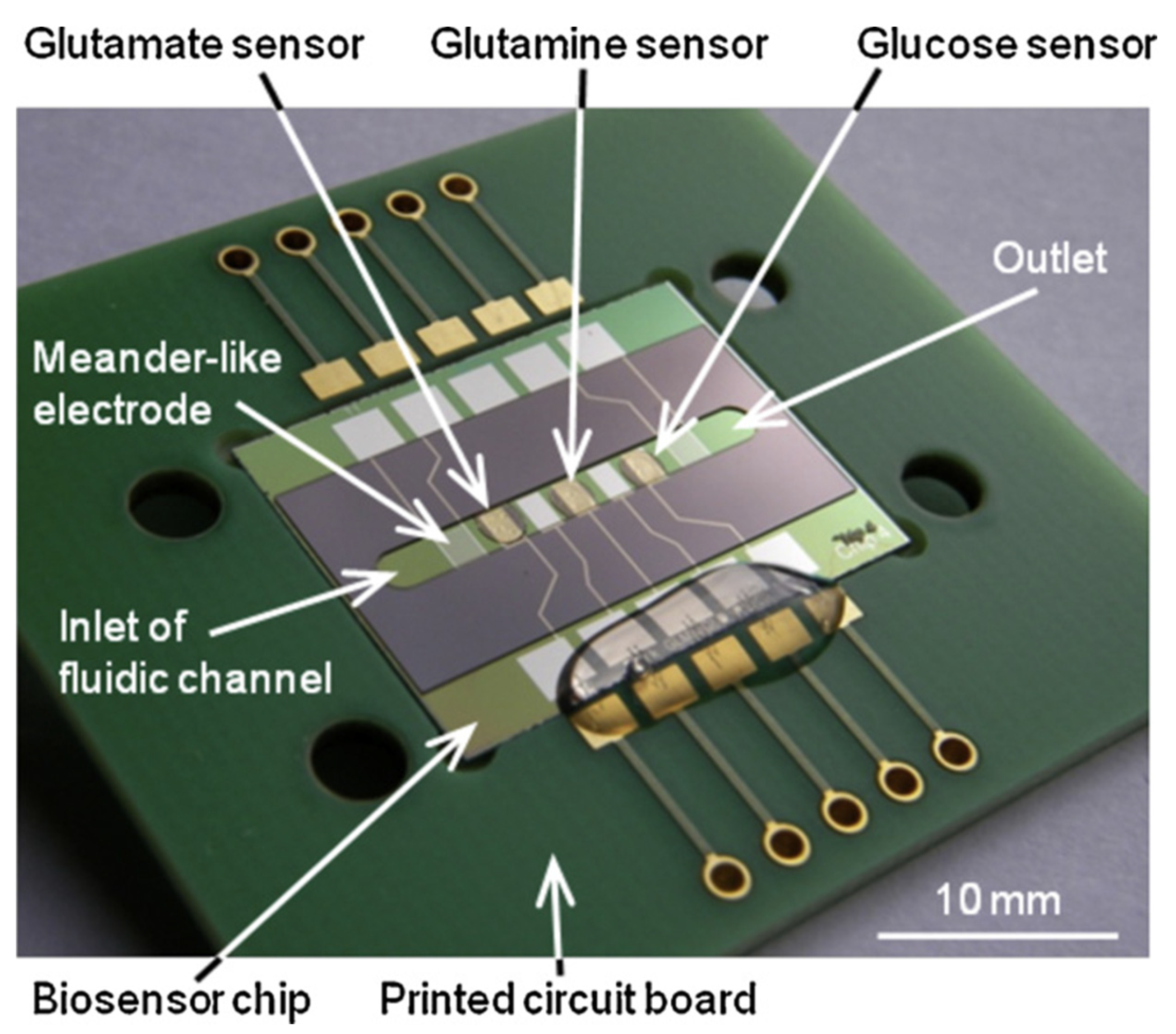
4.3. Electrochemical Biosensors for Nutrients and Metabolites
4.4. Photonic Sensors as Prospective Tool for Optical Monitoring of Cell Proliferation and Maturation
4.5. Longevity of the Sensing Elements in Real-Life Conditions
4.6. Image Detection and Recognition Techniques
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ABS | acrylonitrile butadiene styrene |
| AC | alternating current |
| ADA | alginate di-aldehyde |
| AMR | antimicrobial resistance |
| aMZI | asymmetric Mach-Zehnder interferometer |
| Au | gold |
| AWG | arrayed waveguide gratings |
| bASCs | bovine adipose-derived stem cells |
| BPH-1 | benign prostatic epithelial cells |
| BR | bioreactor |
| CA | cellular agriculture |
| CFD | computational fluid dynamics |
| CH4 | methane |
| CM | cultured meat |
| CMOS | complementary metal oxide semiconductor |
| CNCs | cellulose nanocrystals |
| COVID-19 | coronavirus disease-19 |
| CO2 | carbon dioxide |
| DNA | deoxyribonucleic acid |
| DO | dissolved oxygen |
| ECBs | electrochemical biosensors |
| ECM | extracellular matrix |
| EI | environmental impact |
| EU | European Union |
| FAO | Food and Agriculture Organization |
| FBS | fetal bovine serum |
| FDA | Food and Drug Administration |
| FIA | flow-injection analysis |
| FMIA | Federal Meat Inspection Act |
| FSANZ | Food Standards Australia New Zealand |
| GHG | greenhouse gas |
| GluOx | glutamate oxidase |
| GMO | genetically modified organism |
| GNPs-MPS | gold nanoparticles-mesoporous silica composite |
| GOx | glucose oxidase |
| HeLa | Henrietta Lacks |
| hMSCs | human mesenchymal stem cells |
| H2O2 | hydrogen peroxide |
| H5N1 | Hemagglutinin Type 5 and Neuraminidase Type 1 |
| H7N9 | Hemagglutinin Type 7 and Neuraminidase Type 9 |
| IO4− | periodate |
| iPSCs | induced pluripotent stem cells |
| IR | infra-red |
| KCl | potassium chloride |
| LCA | life cycle assessment |
| LED | light-emitting diode |
| LoD | limit of detection |
| LTCC | low-temperature co-fired ceramic |
| MCs | microcarriers |
| MSCs | mesenchymal stem cells |
| MSN-PtNP-GOx | mesoporous silica nanoparticles-platinum nanoparticles-glucose oxidase |
| MWCNT | multi-wall carbon nanotubes |
| µBRs | microbioreactors |
| N2O | nitrous oxide |
| NDIR | non-dispersive infrared |
| NH2 group | amino group |
| NH3 | ammonia |
| NIR | near-infrared |
| NPs | nanoparticles |
| NR | nanorods |
| PANI | polyaniline |
| PBE | population balance equations |
| PD | photodiodes |
| PDMS | polydimethylsiloxane |
| PEG | polyethylene glycol |
| PGA | polyglycolic acid |
| PLA | poly(L-lactic acid) |
| PLGA | poly(lactic-co-glycolic acid) |
| PMMA | poly(methyl methacrylate) |
| PPy | polypyrrole |
| PS | photonic sensors |
| Pt | platinum |
| PtNPs | platinum nanoparticles |
| PVC | polyvinyl chloride |
| RF | radio frequency |
| RIU | refractive index unit |
| SARS-CoV-2 | severe acute respiratory syndrome coronavirus 2 |
| SCs | satellite cells |
| SFA | Singapore Food Agency |
| Si | silicon |
| SiO2 | silicon dioxide |
| Si3N4 | silicon nitride |
| SMCs | single cardiac myocytes |
| SS | stainless steel |
| SU-8 | “Structured by Uv”-“8 epoxy groups” |
| SUBs | single use bioreactors |
| Ti | titanium |
| US | United States |
| USDA–FSIS | US Department of Agriculture Food Safety and Inspection Service |
| ZnO | zinc oxide |
| 2D | two-dimensional |
| 3D | three-dimensional |
| 3T3 cells | “3-day transfer, inoculum 3 × 105 cells” |
References
- Food and Agriculture Organization of the United Nations. The Future of Food and Agriculture: Trends and Challenges; Food and Agriculture Organization of the United Nations: Rome, Italy, 2017. [Google Scholar]
- Agovino, M.; Casaccia, M.; Ciommi, M.; Ferrara, M.; Marchesano, K. Agriculture, climate change and sustainability: The case of EU-28. Ecol. Indic. 2019, 105, 525–543. [Google Scholar] [CrossRef]
- Opio, C.; Gerber, P.; Mottet, A.; Falcucci, A.; Tempio, G.; MacLeod, M.; Vellinga, T.; Henderson, B.; Steinfeld, H. Greenhouse Gas Emissions from Ruminant Supply Chains—A Global Life Cycle Assessment. Greenhouse Gas: Emissions from Ruminant Supply Chains—A Global Life Cycle Assessment; Food and Agriculture Organization of the United Nations: Rome, Italy, 2013. [Google Scholar]
- Reisinger, A.; Clark, H. How much do direct livestock emissions actually contribute to global warming? Glob. Chang. Biol. 2018, 24, 1749–1761. [Google Scholar] [CrossRef] [PubMed]
- Poore, J.; Nemecek, T. Reducing food’s environmental impacts through producers and consumers. Science 2018, 360, 987–992. [Google Scholar] [CrossRef] [PubMed]
- Alexander, P.; Brown, C.; Arneth, A.; Dias, C.; Finnigan, J.; Moran, D.; Rounsevell, M.D.A. Could consumption of insects, cultured meat or imitation meat reduce global agricultural land use? Glob. Food Sec. 2017, 15, 22–32. [Google Scholar] [CrossRef]
- Tuomisto, H.L.; de Mattos, M.J.T. Environmental impacts of cultured meat production. Environ. Sci. Technol. 2011, 45, 6117–6123. [Google Scholar] [CrossRef]
- Tuomisto, H.L. The eco-friendly burger: Could cultured meat improve the environmental sustainability of meat products? Embo Rep. 2019, 20, e47395. [Google Scholar] [CrossRef] [PubMed]
- Lynch, J.; Pierrehumbert, R. Climate impacts of cultured meat and beef cattle. Front. Sustain. Food Syst. 2019, 3, 5. [Google Scholar] [CrossRef]
- Gibb, R.; Redding, D.W.; Chin, K.Q.; Donnelly, C.A.; Blackburn, T.M.; Newbold, T.; Jones, K.E. Zoonotic host diversity increases in human-dominated ecosystems. Nature 2020, 584, 398–402. [Google Scholar] [CrossRef] [PubMed]
- Anomaly, J. Cultured meat would prevent the next Covid crisis. Anim. Sentience 2020, 5, 5. [Google Scholar] [CrossRef]
- Verhagen, J.H.; Fouchier, R.A.M.; Lewis, N. Highly Pathogenic Avian Influenza Viruses at the Wild-Domestic Bird Interface in Europe: Future Directions for Research and Surveillance. Viruses 2021, 13, 212. [Google Scholar] [CrossRef] [PubMed]
- Van Boeckel, T.P.; Pires, J.; Silvester, R.; Zhao, C.; Song, J.; Criscuolo, N.G.; Gilbert, M.; Bonhoeffer, S.; Laxminarayan, R. Global trends in antimicrobial resistance in animals in low- and middle-income countries. Science 2019, 365, eaaw1944. [Google Scholar] [CrossRef]
- O’Neill, E.N.; Cosenza, Z.A.; Baar, K.; Block, D.E. Considerations for the development of cost-effective cell culture media for cultivated meat production. Comp. Rev. Food Sci. Food Saf. 2020, 20, 686–709. [Google Scholar] [CrossRef]
- Kolkmann, A.M.; Post, M.J.; Rutjens, M.A.M.; van Essen, A.L.M.; Moutsatsou, P. Serum-free media for the growth of primary bovine myoblasts. Cytotechnology 2020, 72, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Specht, L. An Analysis of Culture Medium Costs and Production Volumes for Cultivated Meat; The Good Food Institute: Washington, DC, USA, 2020. [Google Scholar]
- Sexton, A.E.; Garnett, T.; Lorimer, J. Framing the future of food: The contested promises of alternative proteins. Environ. Plan. E Nat. Space 2019, 2, 47–72. [Google Scholar] [CrossRef] [PubMed]
- Rischer, H.; Szilvay, G.R.; Oksman-Caldentey, K.-M. Cellular agriculture—Industrial biotechnology for food and materials. Curr. Opin. Biotechnol. 2020, 61, 128–134. [Google Scholar] [CrossRef] [PubMed]
- Ismail, B.P.; Senaratne-Lenagala, L.; Stube, A.; Brackenridge, A. Protein demand: Review of plant and animal proteins used in alternative protein product development and production. Anim. Front. 2020, 10, 53–63. [Google Scholar] [CrossRef] [PubMed]
- Ong, S.; Choudhury, D.; Naing, M.W. Cell-based meat: Current ambiguities with nomenclature. Trends Food Sci. Technol. 2020, 102, 223–231. [Google Scholar] [CrossRef]
- Fox, E.M.; Leonard, N.; Jordan, K. Molecular diversity of Listeria monocytogenes isolated from Irish dairy farms. Foodborne Pathog. Dis. 2011, 8, 635–641. [Google Scholar] [CrossRef] [PubMed]
- Mattick, C.S. Cellular agriculture: The coming revolution in food production. Bull. At. Sci. 2018, 74, 32–35. [Google Scholar] [CrossRef]
- Specht, E.A.; Welch, D.R.; Rees Clayton, E.M.; Lagally, C.D. Opportunities for applying biomedical production and manufacturing methods to the development of the clean meat industry. Biochem. Eng. J. 2018, 132, 161–168. [Google Scholar] [CrossRef]
- Bhat, Z.F.; Bhat, H.; Kumar, S. Cultured meat—A humane meat production system. In Principles of Tissue Engineering; Elsevier: Amsterdam, The Netherlands, 2020; pp. 1369–1388. [Google Scholar]
- Ben-Arye, T.; Levenberg, S. Tissue engineering for clean meat production. Front. Sustain. Food Syst. 2019, 3, 46. [Google Scholar] [CrossRef]
- Post, M.J. Cultured meat from stem cells: Challenges and prospects. Meat Sci. 2012, 92, 297–301. [Google Scholar] [CrossRef]
- Kadim, I.T.; Mahgoub, O.; Baqir, S.; Faye, B.; Purchas, R. Cultured meat from muscle stem cells: A review of challenges and prospects. J. Integr. Agric. 2015, 14, 222–233. [Google Scholar] [CrossRef]
- Ding, S.; Wang, F.; Liu, Y.; Li, S.; Zhou, G.; Hu, P. Characterization and isolation of highly purified porcine satellite cells. Cell Death Discov. 2017, 3, 17003. [Google Scholar] [CrossRef] [PubMed]
- Ding, S.; Swennen GN, M.; Messmer, T.; Gagliardi, M.; Molin DG, M.; Li, C.; Zhou, G.; Post, M.J. Maintaining bovine satellite cells stemness through p38 pathway. Sci. Rep. 2018, 8, 10808. [Google Scholar] [CrossRef] [PubMed]
- Fish, K.D.; Rubio, N.R.; Stout, A.J.; Yuen, J.S.K.; Kaplan, D.L. Prospects and challenges for cell-cultured fat as a novel food ingredient. Trends Food Sci. Technol. 2020, 98, 53–67. [Google Scholar] [CrossRef] [PubMed]
- Choudhury, D.; Tseng, T.W.; Swartz, E. The business of cultured meat. Trends Biotechnol. 2020, 38, 573–577. [Google Scholar] [CrossRef] [PubMed]
- Post, M.J. Cultured beef: Medical technology to produce food. J. Sci. Food Agric. 2014, 94, 1039–1041. [Google Scholar] [CrossRef]
- Post, M.J.; Levenberg, S.; Kaplan, D.L.; Genovese, N.; Fu, J.; Bryant, C.J.; Negowetti, N.; Verzijden, K.; Moutsatsou, P. Scientific, sustainability and regulatory challenges of cultured meat. Nat. Food 2020, 1, 403–415. [Google Scholar] [CrossRef]
- Zhang, G.; Zhao, X.; Li, X.; Du, G.; Zhou, J.; Chen, J. Challenges and possibilities for bio-manufacturing cultured meat. Trends Food Sci. Technol. 2020, 97, 443–450. [Google Scholar] [CrossRef]
- Melzener, L.; Verzijden, K.E.; Buijs, A.J.; Post, M.J.; Flack, J.E. Cultured beef: From small biopsy to substantial quantity. J. Sci. Food Agric. 2020, 101, 7–14. [Google Scholar] [CrossRef]
- Bryant, C.J. Culture, meat, and cultured meat. J. Anim. Sci. 2020, 98, skaa172. [Google Scholar] [CrossRef]
- Post, M.; van der Weele, C. Principles of tissue engineering for food. In Principles of Tissue Engineering; Elsevier: Amsterdam, The Netherlands, 2014; pp. 1647–1662. [Google Scholar]
- Stephens, N.; Di Silvio, L.; Dunsford, I.; Ellis, M.; Glencross, A.; Sexton, A. Bringing cultured meat to market: Technical, socio-political, and regulatory challenges in cellular agriculture. Trends Food Sci. Technol. 2018, 78, 155–166. [Google Scholar] [CrossRef] [PubMed]
- Bhat, Z.F.; Kumar, S.; Fayaz, H. In vitro meat production: Challenges and benefits over conventional meat production. J. Integr. Agric. 2015, 14, 241–248. [Google Scholar] [CrossRef]
- Warner, R.D. Review: Analysis of the process and drivers for cellular meat production. Animal 2019, 13, 3041–3058. [Google Scholar] [CrossRef]
- Stephens, N.; Sexton, A.E.; Driessen, C. Making sense of making meat: Key moments in the first 20 years of tissue engineering muscle to make food. Front. Sustain. Food Syst. 2019, 3, 45. [Google Scholar] [CrossRef]
- Dolgin, E. Will cell-based meat ever be a dinner staple? Nature 2020, 588, S64–S67. [Google Scholar] [CrossRef]
- Hopkins, P.D.; Dacey, A. Vegetarian meat: Could technology save animals and satisfy meat eaters? J. Agric. Environ. Ethics 2008, 21, 579–596. [Google Scholar] [CrossRef]
- Phillips, J.; Lewis, P.; Post, M.; Schonwald, J.; Smith, R. Taste Test: The Scientists Who Created the World’s First Test Tube Grown Burger Are Convinced It Could Be a Long-Term Solution to The Growing Demand for Beef; Informit: Melbourne, Australia, 2013. [Google Scholar]
- Morozov, E. To save everything, click here: The folly of technological solutionism. In To Save Everything, Click Here: The Folly of Technological Solutionism; Public Affairs: New York, NY, USA, 2013. [Google Scholar]
- Van Der Weele, C.; Driessen, C. How normal meat becomes stranger as cultured meat becomes more normal; Ambivalence and ambiguity below the surface of behaviour. Front. Sustain. Food Syst. 2019, 3, 69. [Google Scholar] [CrossRef]
- Stephens, N.; King, E.; Lyall, C. Blood, meat, and upscaling tissue engineering: Promises, anticipated markets, and performativity in the biomedical and agri-food sectors. Biosocieties 2018, 13, 1–21. [Google Scholar] [CrossRef]
- Arshad, M.S.; Javed, M.; Sohaib, M.; Saeed, F.; Imran, A.; Amjad, Z. Tissue engineering approaches to develop cultured meat from cells: A mini review. Cogent Food Agric. 2017, 3, 1320814. [Google Scholar] [CrossRef]
- Tomiyama, A.J.; Kawecki, N.S.; Rosenfeld, D.L.; Jay, J.A.; Rajagopal, D.; Rowat, A.C. Bridging the gap between the science of cultured meat and public perceptions. Trends Food Sci. Technol. 2020, 104, 144–152. [Google Scholar] [CrossRef]
- Godfray, H.C.J. Meat: The Future Series—Alternative Proteins|World Economic Forum; World Economic Forum: Cologny, Switzerland, 2019. [Google Scholar]
- Reynolds, M. The Clean Meat Industry is Racing to Ditch Its Reliance on Foetal Blood. Wired. 20 March 2018. Available online: https://www.wired.co.uk/article/scaling-clean-meat-serum-just-finless-foods-mosa-meat (accessed on 17 November 2018).
- Thorrez, L.; Vandenburgh, H. Challenges in the quest for “clean meat”. Nat. Biotechnol. 2019, 37, 215–216. [Google Scholar] [CrossRef]
- U.S. Food and Drug Administration (FDA); United States Department of Agriculture (USDA). Formal Agreement between the U.S. Department of Healthand Human Services Food and Drug Administration and U.S. Department of Agriculture Office of Food Safety; FDA: Silver Spring, MD, USA; USDA: Washington, DC, USA, 2019. [Google Scholar]
- European Commission. Regulation (EU) 2015/2283 of the European Parliament and of the Council of 25 November 2015 on Novel Foods. Available online: https://eur-lex.europa.eu/legal-content/en/TXT/?uri=CELEX%3A32015R2283 (accessed on 25 November 2020).
- European Commission. Regulation (EC) No. 1829/2003 of the European Parliament and of the Council of 22 September 2003 on Genetically Modified Food and Feed. Available online: https://eur-lex.europa.eu/legal-content/en/ALL/?uri=CELEX%3A32003R1829 (accessed on 25 November 2020).
- Singapore Food Agency. Requirements for the Safety Assessment of Novel Foods; Singapore Food Agency: Singapore, 2020. [Google Scholar]
- BBC News. Singapore Approves Lab-Grown “Chicken” Meat—BBC News. Available online: https://www.bbc.com/news/business-55155741 (accessed on 28 February 2021).
- Regulatory Institute Cultured Meat: How to Regulate Alternatives to Farmed Meat. Available online: https://www.howtoregulate.org/cell-cultured-meat-regulation/ (accessed on 29 November 2020).
- Nienow, A.W. Reactor engineering in large scale animal cell culture. Cytotechnology 2006, 50, 9–33. [Google Scholar] [CrossRef] [PubMed]
- Freed, L.E.; Guilak, F.; Guo, X.E.; Gray, M.L.; Tranquillo, R.; Holmes, J.W.; Radisic, M.; Sefton, M.V.; Kaplan, D.; Vunjak-Novakovic, G. Advanced tools for tissue engineering: Scaffolds, bioreactors, and signaling. Tissue Eng. 2006, 12, 3285–3305. [Google Scholar] [CrossRef]
- Verbruggen, S.; Luining, D.; van Essen, A.; Post, M.J. Bovine myoblast cell production in a microcarriers-based system. Cytotechnology 2018, 70, 503–512. [Google Scholar] [CrossRef]
- Tsai, A.-C.; Liu, Y.; Yuan, X.; Chella, R.; Ma, T. Aggregation kinetics of human mesenchymal stem cells under wave motion. Biotechnol. J. 2017, 12, 1600448. [Google Scholar] [CrossRef]
- Allan, S.J.; De Bank, P.A.; Ellis, M.J. Bioprocess design considerations for cultured meat production with a focus on the expansion bioreactor. Front. Sustain. Food Syst. 2019, 3, 44. [Google Scholar] [CrossRef]
- Eibl, R.; Werner, S.; Eibl, D. Bag bioreactor based on wave-induced motion: Characteristics and applications. Adv. Biochem. Eng. Biotechnol. 2009, 115, 55–87. [Google Scholar] [PubMed]
- De Jesus, S.S.; Moreira Neto, J.; Maciel Filho, R. Hydrodynamics and mass transfer in bubble column, conventional airlift, stirred airlift and stirred tank bioreactors, using viscous fluid: A comparative study. Biochem. Eng. J. 2017, 118, 70–81. [Google Scholar] [CrossRef]
- Oncül, A.A.; Kalmbach, A.; Genzel, Y.; Reichl, U.; Thévenin, D. Characterization of flow conditions in 2 L and 20 L wave bioreactors using computational fluid dynamics. Biotechnol. Prog. 2010, 26, 101–110. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhang, G.; Zhao, X.; Zhou, J.; Du, G.; Chen, J. A conceptual air-lift reactor design for large scale animal cell cultivation in the context of in vitro meat production. Chem. Eng. Sci. 2020, 211, 115269. [Google Scholar] [CrossRef]
- Oosterhuis, N.M.G.; van der Heiden, P. Mass Transfer in the CELL-tainer® Disposable Bioreactor. In Cells and Culture; Noll, T., Ed.; Springer: Dordrecht, The Netherlands, 2010; pp. 371–373. [Google Scholar]
- Rader, R.A.; Langer, E.S. Single-use technologies in biopharmaceutical manufacturing: A 10-year review of trends and the future. In Single-Use Technology in Biopharmaceutical Manufacture; Eibl, R., Eibl, D., Eds.; Wiley: Hoboken, NJ, USA, 2019; pp. 193–200. [Google Scholar]
- Junne, S.; Neubauer, P. How scalable and suitable are single-use bioreactors? Curr. Opin. Biotechnol. 2018, 53, 240–247. [Google Scholar] [CrossRef]
- Rawlings, B.; Pora, H. Environmental Impact of Single-Use and Reusable Bioprocess Systems. BioProcess Int. 2009, 7, 18–26. [Google Scholar]
- Hanga, M.P.; Ali, J.; Moutsatsou, P.; de la Raga, F.A.; Hewitt, C.J.; Nienow, A.; Wall, I. Bioprocess development for scalable production of cultivated meat. Biotechnol. Bioeng. 2020, 117, 3029–3039. [Google Scholar] [CrossRef]
- Gaspar, D.A.; Gomide, V.; Monteiro, F.J. The role of perfusion bioreactors in bone tissue engineering. Biomatter 2012, 2, 167–175. [Google Scholar] [CrossRef]
- Grayson, W.L.; Fröhlich, M.; Yeager, K.; Bhumiratana, S.; Chan, M.E.; Cannizzaro, C.; Wan, L.Q.; Liu, X.S.; Guo, X.E.; Vunjak-Novakovic, G. Engineering anatomically shaped human bone grafts. Proc. Natl. Acad. Sci. USA 2010, 107, 3299–3304. [Google Scholar] [CrossRef] [PubMed]
- Martin, I.; Wendt, D.; Heberer, M. The role of bioreactors in tissue engineering. Trends Biotechnol. 2004, 22, 80–86. [Google Scholar] [CrossRef]
- Chen, H.-C.; Hu, Y.-C. Bioreactors for tissue engineering. Biotechnol. Lett. 2006, 28, 1415–1423. [Google Scholar] [CrossRef]
- Yeatts, A.B.; Fisher, J.P. Bone tissue engineering bioreactors: Dynamic culture and the influence of shear stress. Bone 2011, 48, 171–181. [Google Scholar] [CrossRef]
- Avantor. Spinner Flasks. Available online: https://us.vwr.com/store/category/spinner-flasks/557302 (accessed on 27 February 2021).
- MERCK. Corning Spinner Flasks. Available online: https://www.sigmaaldrich.com/labware/labware-products.html?TablePage=17193021&gclid=Cj0KCQiA-OeBBhDiARIsADyBcE7GHpemIJBqS98mmTiXiScvvXot4dQjD2A53XgyMT 3bqGXjyVj2JmoaApJiEALw_wcB (accessed on 27 February 2021).
- Pall. Stirred Tank Bioreactors. Available online: https://www.pall.com/en/biotech/cell-culture/stirred-tank-bioreactors.html# (accessed on 27 February 2021).
- Bionet. F3 Industrial Bioreactor. Available online: https://bionet.com/technology/f3-bioreactor/ (accessed on 27 February 2021).
- Celltainer Biotech BV. Available online: https://celltainer.com/ (accessed on 27 February 2021).
- Cell Culture DISH. A New Wave for the Future. Available online: https://cellculturedish.com/a-new-wave-for-the-future/ (accessed on 27 February 2021).
- OSPIN. Modular Bioprocessing Modular Bioprocessing. Available online: https://ospin.de/ (accessed on 27 February 2021).
- SKE. Research Equipment InFlow Perfusion Bioreactor. Available online: http://www.ske.it/index.php/product/inflow-perfusion-bioreactor/ (accessed on 27 February 2021).
- Marques, M.P.; Szita, N. Bioprocess microfluidics: Applying microfluidic devices for bioprocessing. Curr. Opin. Chem. Eng. 2017, 18, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Figallo, E.; Cannizzaro, C.; Gerecht, S.; Burdick, J.A.; Langer, R.; Elvassore, N.; Vunjak-Novakovic, G. Micro-bioreactor array for controlling cellular microenvironments. Lab Chip 2007, 7, 710–719. [Google Scholar] [CrossRef]
- Chen, K.; Rong, N.; Wang, S.; Luo, C. A novel two-layer-integrated microfluidic device for high-throughput yeast proteomic dynamics analysis at the single-cell level. Integr. Biol. 2020, 12, 241–249. [Google Scholar] [CrossRef] [PubMed]
- Tavakoli, H.; Zhou, W.; Ma, L.; Perez, S.; Ibarra, A.; Xu, F.; Zhan, S.; Li, X. Recent advances in microfluidic platforms for single-cell analysis in cancer biology, diagnosis and therapy. Trends Anal. Chem. 2019, 117, 13–26. [Google Scholar] [CrossRef] [PubMed]
- Arter, W.E.; Levin, A.; Krainer, G.; Knowles, T.P.J. Microfluidic approaches for the analysis of protein-protein interactions in solution. Biophys. Rev. 2020, 12, 575–585. [Google Scholar] [CrossRef] [PubMed]
- Nasseri, B.; Soleimani, N.; Rabiee, N.; Kalbasi, A.; Karimi, M.; Hamblin, M.R. Point-of-care microfluidic devices for pathogen detection. Biosens. Bioelectron. 2018, 117, 112–128. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Li, M.; Liu, Y. Microfluidic-Based Approaches for Foodborne Pathogen Detection. Microorganisms 2019, 7, 381. [Google Scholar] [CrossRef]
- Coluccio, M.L.; Perozziello, G.; Malara, N.; Parrotta, E.; Zhang, P.; Gentile, F.; Limongi, T.; Raj, P.M.; Cuda, G.; Candeloro, P.; et al. Microfluidic platforms for cell cultures and investigations. Microelectron. Eng. 2019, 208, 14–28. [Google Scholar] [CrossRef]
- Chen, C.; Townsend, A.D.; Hayter, E.A.; Birk, H.M.; Sell, S.A.; Martin, R.S. Insert-based microfluidics for 3D cell culture with analysis. Anal. Bioanal. Chem. 2018, 410, 3025–3035. [Google Scholar] [CrossRef] [PubMed]
- Osaki, T.; Sivathanu, V.; Kamm, R.D. Vascularized microfluidic organ-chips for drug screening, disease models and tissue engineering. Curr. Opin. Biotechnol. 2018, 52, 116–123. [Google Scholar] [CrossRef]
- Young, E.W.K.; Beebe, D.J. Fundamentals of microfluidic cell culture in controlled microenvironments. Chem. Soc. Rev. 2010, 39, 1036–1048. [Google Scholar] [CrossRef] [PubMed]
- Haycock, J.W. 3D cell culture: A review of current approaches and techniques. Methods Mol. Biol. 2011, 695, 1–15. [Google Scholar]
- Huh, D.; Hamilton, G.A.; Ingber, D.E. From 3D cell culture to organs-on-chips. Trends Cell Biol. 2011, 21, 745–754. [Google Scholar] [CrossRef] [PubMed]
- Duval, K.; Grover, H.; Han, L.-H.; Mou, Y.; Pegoraro, A.F.; Fredberg, J.; Chen, Z. Modeling physiological events in 2D vs. 3D cell culture. Physiology 2017, 32, 266–277. [Google Scholar] [CrossRef] [PubMed]
- Kim, L.; Toh, Y.-C.; Voldman, J.; Yu, H. A practical guide to microfluidic perfusion culture of adherent mammalian cells. Lab Chip 2007, 7, 681–694. [Google Scholar] [CrossRef]
- Shemesh, J.; Jalilian, I.; Shi, A.; Heng Yeoh, G.; Knothe Tate, M.L.; Ebrahimi Warkiani, M. Flow-induced stress on adherent cells in microfluidic devices. Lab Chip 2015, 15, 4114–4127. [Google Scholar] [CrossRef]
- Terrell, J.A.; Jones, C.G.; Kabandana, G.K.M.; Chen, C. From cells-on-a-chip to organs-on-a-chip: Scaffolding materials for 3D cell culture in microfluidics. J. Mater. Chem. B 2020, 8, 6667–6685. [Google Scholar] [CrossRef] [PubMed]
- Pasirayi, G.; Auger, V.; Scott, S.M.; Rahman, P.K.S.M.; Islam, M.; O’Hare, L.; Ali, Z. Microfluidic bioreactors for cell culturing: A review. Micro Nanosyst. 2011, 3, 137–160. [Google Scholar] [CrossRef]
- Borenstein, J.; Tandon, V.; Tao, S.; Charest, J. (Eds.) Microfluidic Cell Culture Systems, 2nd ed.; Elsevier: Amsterdam, The Netherlands, 2019. [Google Scholar]
- Radonić, V.; Birgermajer, S.; Podunavac, I.; Djisalov, M.; Gadjanski, I.; Kitić, G. Microfluidic Sensor Based on Composite Left-Right Handed Transmission Line. Electronics 2019, 8, 1475. [Google Scholar] [CrossRef]
- Beebe, D.J.; Mensing, G.A.; Walker, G.M. Physics and applications of microfluidics in biology. Annu. Rev. Biomed. Eng. 2002, 4, 261–286. [Google Scholar] [CrossRef]
- Raj, M.K.; Chakraborty, S. PDMS microfluidics: A mini review. J. Appl. Polym. Sci. 2020, 137, 48958. [Google Scholar] [CrossRef]
- Wang, T.; Chen, J.; Zhou, T.; Song, L. Fabricating microstructures on glass for microfluidic chips by glass molding process. Micromachines 2018, 9, 269. [Google Scholar] [CrossRef] [PubMed]
- Bartsch, H.; Welker, T.; Welker, K.; Witte, H.; Müller, J. LTCC based bioreactors for cell cultivation. IOP Conf. Ser. Mater. Sci. Eng. 2016, 104, 012001. [Google Scholar] [CrossRef]
- Waheed, S.; Cabot, J.M.; Macdonald, N.P.; Lewis, T.; Guijt, R.M.; Paull, B.; Breadmore, M.C. 3D printed microfluidic devices: Enablers and barriers. Lab Chip 2016, 16, 1993–2013. [Google Scholar] [CrossRef]
- Bartholomeusz, D.A.; Boutte, R.W.; Andrade, J.D. Xurography: Rapid prototyping of microstructures using a cutting plotter. J. Microelectromech. Syst. 2005, 14, 1364–1374. [Google Scholar] [CrossRef]
- Roy, E.; Veres, T. Microfluidic Device, Composition and Method of Forming. U.S. Patent US9238346B2, 6 January 2016. [Google Scholar]
- Lachaux, J.; Alcaine, C.; Gómez-Escoda, B.; Perrault, C.M.; Duplan, D.O.; Wu, P.-Y.J.; Ochoa, I.; Fernandez, L.; Mercier, O.; Coudreuse, D.; et al. Thermoplastic elastomer with advanced hydrophilization and bonding performances for rapid (30s) and easy molding of microfluidic devices. Lab Chip 2017, 17, 2581–2594. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.-C.; Pan, C.-W.; Lee, W.-C.; Li, K.-M. An experimental study of micromilling parameters to manufacture microchannels on a PMMA substrate. Int. J. Adv. Manuf. Technol. 2014, 71, 1623–1630. [Google Scholar] [CrossRef]
- Chen, X.; Shen, J.; Zhou, M. Rapid fabrication of a four-layer PMMA-based microfluidic chip using CO2-laser micromachining and thermal bonding. J. Micromech. Microeng. 2016, 26, 107001. [Google Scholar] [CrossRef]
- Matellan, C.; Del Río Hernández, A.E. Cost-effective rapid prototyping and assembly of poly(methyl methacrylate) microfluidic devices. Sci. Rep. 2018, 8, 6971. [Google Scholar] [CrossRef]
- Sanjay, S.T.; Li, M.; Zhou, W.; Li, X.; Li, X. A reusable PMMA/paper hybrid plug-and-play microfluidic device for an ultrasensitive immunoassay with a wide dynamic range. Microsyst. Nanoeng. 2020, 6, 28. [Google Scholar] [CrossRef]
- Fazlikeshteli, S. Fabrication of PMMA/PANI/Fe3O4 as a Novel Conducting Hybrid Coating Fabrication of PMMA/PANI/Fe3O4 as a Novel Conducting Hybrid Coating. Polym. Plast. Technol. Eng. 2018, 57, 591–599. [Google Scholar]
- Bodiou, V.; Moutsatsou, P.; Post, M.J. Microcarriers for upscaling cultured meat production. Front. Nutr. 2020, 7, 10. [Google Scholar] [CrossRef] [PubMed]
- Kubis, H.-P.; Scheibe, R.J.; Decker, B.; Hufendiek, K.; Hanke, N.; Gros, G.; Meissner, J.D. Primary skeletal muscle cells cultured on gelatin bead microcarriers develop structural and biochemical features characteristic of adult skeletal muscle. Cell Biol. Int. 2016, 40, 364–374. [Google Scholar] [CrossRef] [PubMed]
- Tavassoli, H.; Alhosseini, S.N.; Tay, A.; Chan, P.P.Y.; Oh, S.K.W.; Warkiani, M.E. Large-scale production of stem cells utilizing microcarriers: A biomaterials engineering perspective from academic research to commercialized products. Biomaterials 2018, 181, 333–346. [Google Scholar] [CrossRef] [PubMed]
- Shit, S.C.; Shah, P.M. Edible polymers: Challenges and opportunities. J. Polym. 2014, 2014, 1–13. [Google Scholar] [CrossRef]
- Li, B.; Wang, X.; Wang, Y.; Gou, W.; Yuan, X.; Peng, J.; Guo, Q.; Lu, S. Past, present, and future of microcarrier-based tissue engineering. J. Orthop. Transl. 2015, 3, 51–57. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Shi, G.; Bei, J.; Wang, S.; Cao, Y.; Shang, Q.; Yang, G.; Wang, W. Fabrication and surface modification of macroporous poly(L-lactic acid) and poly(L-lactic-co-glycolic acid) (70/30) cell scaffolds for human skin fibroblast cell culture. J. Biomed. Mater. Res. 2002, 62, 438–446. [Google Scholar] [CrossRef]
- Wu, C.-Y.; Stoecklein, D.; Kommajosula, A.; Lin, J.; Owsley, K.; Ganapathysubramanian, B.; Di Carlo, D. Shaped 3D microcarriers for adherent cell culture and analysis. Microsyst. Nanoeng. 2018, 4, 21. [Google Scholar] [CrossRef]
- Chang, C.; Zhang, L. Cellulose-based hydrogels: Present status and application prospects. Carbohydr. Polym. 2011, 84, 40–53. [Google Scholar] [CrossRef]
- Ahmadi, F.; Oveisi, Z.; Samani, S.M.; Amoozgar, Z. Chitosan based hydrogels: Characteristics and pharmaceutical applications. Res. Pharm. Sci. 2015, 10, 1–16. [Google Scholar]
- Rafiq, Q.A.; Brosnan, K.M.; Coopman, K.; Nienow, A.W.; Hewitt, C.J. Culture of human mesenchymal stem cells on microcarriers in a 5 l stirred-tank bioreactor. Biotechnol. Lett. 2013, 35, 1233–1245. [Google Scholar] [CrossRef]
- Merten, O.-W. Advances in cell culture: Anchorage dependence. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2015, 370, 20140040. [Google Scholar] [CrossRef] [PubMed]
- Post, M.J. Proteins in cultured beef. In Proteins in Food Processing; Elsevier: Amsterdam, The Netherlands, 2018; pp. 289–298. [Google Scholar]
- Datar, I.; Betti, M. Possibilities for an in vitro meat production system. Innov. Food Sci. Emerg. Technol. 2010, 11, 13–22. [Google Scholar] [CrossRef]
- Ben-Arye, T.; Shandalov, Y.; Ben-Shaul, S.; Landau, S.; Zagury, Y.; Ianovici, I.; Lavon, N.; Levenberg, S. Textured soy protein scaffolds enable the generation of three-dimensional bovine skeletal muscle tissue for cell-based meat. Nat. Food 2020, 1, 210–220. [Google Scholar] [CrossRef]
- Cunha, A.G.; Gandini, A. Turning polysaccharides into hydrophobic materials: A critical review. Part 2. Hemicelluloses, chitin/chitosan, starch, pectin and alginates. Cellulose 2010, 17, 1045–1065. [Google Scholar]
- Bilirgen, A.C.; Toker, M.; Odabas, S.; Yetisen, A.K.; Garipcan, B.; Tasoglu, S. Plant-Based Scaffolds in Tissue Engineering. ACS Biomater. Sci. Eng. 2021. [Google Scholar] [CrossRef] [PubMed]
- Aleph Farms. The Science behind the World’s First Cultivated Steak—Grown Directly from Cells. Available online: https://alephfarms.medium.com/the-science-behind-the-worlds-first-cultivated-steak-grown-directly-from-cells-47f210d88560 (accessed on 29 November 2020).
- Bugnicourt, E. Polyhydroxyalkanoate (PHA): Review of synthesis, characteristics, processing and potential applications in packaging. Express Polym. Lett. 2014, 8, 791–808. [Google Scholar] [CrossRef]
- Woodard, L.N.; Grunlan, M.A. Hydrolytic degradation and erosion of polyester biomaterials. Acs Macro Lett. 2018, 7, 976–982. [Google Scholar] [CrossRef]
- Modulevsky, D.J.; Lefebvre, C.; Haase, K.; Al-Rekabi, Z.; Pelling, A.E. Apple derived cellulose scaffolds for 3D mammalian cell culture. PLoS ONE 2014, 9, e97835. [Google Scholar]
- Gershlak, J.R.; Hernandez, S.; Fontana, G.; Perreault, L.R.; Hansen, K.J.; Larson, S.A.; Binder, B.Y.K.; Dolivo, D.M.; Yang, T.; Dominko, T.; et al. Crossing kingdoms: Using decellularized plants as perfusable tissue engineering scaffolds. Biomaterials 2017, 125, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Holmes, J.T.; Jaberansari, Z.; Collins, W.; Leblanc Latour, M.; Modulevsky, D.J.; Pelling, A.E. Homemade Bread: Repurposing an Ancient Technology for Low Cost in vitro Tissue Engineering. bioRxiv 2020. [Google Scholar] [CrossRef]
- Levy-Mishali, M.; Zoldan, J.; Levenberg, S. Effect of scaffold stiffness on myoblast differentiation. Tissue Eng. Part A 2009, 15, 935–944. [Google Scholar] [CrossRef] [PubMed]
- Comley, K.; Fleck, N.A. The toughness of adipose tissue: Measurements and physical basis. J. Biomech. 2010, 43, 1823–1826. [Google Scholar] [CrossRef]
- Derby, B. Printing and prototyping of tissues and scaffolds. Science 2012, 338, 921–926. [Google Scholar] [CrossRef]
- Murphy, S.V.; Atala, A. 3D bioprinting of tissues and organs. Nat. Biotechnol. 2014, 32, 773–785. [Google Scholar] [CrossRef]
- Mironov, V.; Kasyanov, V.; Drake, C.; Markwald, R.R. Organ printing: Promises and challenges. Regen Med. 2008, 3, 93–103. [Google Scholar] [CrossRef]
- Dey, M.; Ozbolat, I.T. 3D bioprinting of cells, tissues and organs. Sci. Rep. 2020, 10, 14023. [Google Scholar] [CrossRef]
- Beauchamp, M.J.; Nordin, G.P.; Woolley, A.T. Moving from millifluidic to truly microfluidic sub-100-μm cross-section 3D printed devices. Anal. Bioanal. Chem. 2017, 409, 4311–4319. [Google Scholar] [CrossRef] [PubMed]
- Distler, T.; Solisito, A.A.; Schneidereit, D.; Friedrich, O.; Detsch, R.; Boccaccini, A.R. 3D printed oxidized alginate-gelatin bioink provides guidance for C2C12 muscle precursor cell orientation and differentiation via shear stress during bioprinting. Biofabrication 2020, 12, 045005. [Google Scholar] [CrossRef] [PubMed]
- Handral, H.K.; Hua Tay, S.; Wan Chan, W.; Choudhury, D. 3D Printing of cultured meat products. Crit. Rev. Food Sci. Nutr. 2020, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Zhang, M.; Bhandari, B.; Wang, Y. 3D printing: Printing precision and application in food sector. Trends Food Sci. Technol. 2017, 69, 83–94. [Google Scholar] [CrossRef]
- Bryant, C.; Barnett, J. Consumer acceptance of cultured meat: An updated review (2018–2020). Appl. Sci. 2020, 10, 5201. [Google Scholar] [CrossRef]
- Food Navigator. Aleph Farms Prints Lab-Meat in Space. Available online: https://www.foodnavigator.com/Article/2019/10/08/Aleph-Farms-prints-lab-meat-in-space (accessed on 29 November 2020).
- PR Newswire. Aleph Farms and The Technion Reveal World’s First Cultivated Ribeye Steak. Available online: https://www.prnewswire.com/il/news-releases/aleph-farms-and-the-technion-reveal-worlds-first-cultivated-ribeye-steak-301224800.html (accessed on 28 February 2021).
- Kahan, S.; Camphuijsen, J.; Cannistra, C.; Potter, G.; Consenza, Z.; Shmulevich, I. Cultivated Meat Modeling Consortium: Inaugural Meeting Whitepaper. Authorea 2020. [Google Scholar] [CrossRef]
- Li, X.; van der Steen, G.; van Dedem GW, K.; van der Wielen LA, M.; van Leeuwen, M.; van Gulik, W.M.; Heijnen, J.J.; Ottens, M.; Krommenhoek, E.E.; Gardeniers, J.G.E.; et al. Application of direct fluid flow oscillations to improve mixing in microbioreactors. AIChE J. 2009, 55, 2725–2736. [Google Scholar] [CrossRef]
- Yu, P.; Lee, T.S.; Zeng, Y.; Low, H.T. A 3D analysis of oxygen transfer in a low-cost micro-bioreactor for animal cell suspension culture. Comput. Methods Programs Biomed. 2007, 85, 59–68. [Google Scholar] [CrossRef]
- Ungerböck, B.; Pohar, A.; Mayr, T.; Plazl, I. Online oxygen measurements inside a microreactor with modeling of transport phenomena. Microfluid. Nanofluidics 2013, 14, 565–574. [Google Scholar] [CrossRef]
- Mendonça da Silva, J.; Erro, E.; Awan, M.; Chalmers, S.-A.; Fuller, B.; Selden, C. Small-Scale Fluidized Bed Bioreactor for Long-Term Dynamic Culture of 3D Cell Constructs and in vitro Testing. Front. Bioeng. Biotechnol. 2020, 8, 895. [Google Scholar] [CrossRef]
- Semenova, D.; Fernandes, A.C.; Bolivar, J.M.; Rosinha Grundtvig, I.P.; Vadot, B.; Galvanin, S.; Mayr, T.; Nidetzky, B.; Zubov, A.; Gernaey, K.V. Model-based analysis of biocatalytic processes and performance of microbioreactors with integrated optical sensors. New Biotechnol. 2020, 56, 27–37. [Google Scholar] [CrossRef] [PubMed]
- Ebrahimi, M.; Tamer, M.; Villegas, R.M.; Chiappetta, A.; Ein-Mozaffari, F. Application of CFD to Analyze the Hydrodynamic Behaviour of a Bioreactor with a Double Impeller. Processes 2019, 7, 694. [Google Scholar] [CrossRef]
- Azargoshasb, H.; Mousavi, S.M.; Jamialahmadi, O.; Shojaosadati, S.A.; Mousavi, S.B. Experiments and a three-phase computational fluid dynamics (CFD) simulation coupled with population balance equations of a stirred tank bioreactor for high cell density cultivation. Can. J. Chem. Eng. 2016, 94, 20–32. [Google Scholar] [CrossRef]
- Bustamante, M.C.C.; Cerri, M.O.; Badino, A.C. Comparison between average shear rates in conventional bioreactor with Rushton and Elephant ear impellers. Chem. Eng. Sci. 2013, 90, 92–100. [Google Scholar] [CrossRef]
- Aubin, J.; Fletcher, D.F.; Xuereb, C. Modeling turbulent flow in stirred tanks with CFD: The influence of the modeling approach, turbulence model and numerical scheme. Exp. Therm. Fluid Sci. 2004, 28, 431–445. [Google Scholar] [CrossRef]
- Vrábel, P.; van der Lans, R.G.J.; Luyben, K.C.A.; Boon, L.; Nienow, A.W. Mixing in large-scale vessels stirred with multiple radial or radial and axial up-pumping impellers: Modelling and measurements. Chem. Eng. Sci. 2000, 55, 5881–5896. [Google Scholar] [CrossRef]
- Monod, J. The Growth of Bacterial Cultures. Annu. Rev. Microbiol. 1949, 3, 371–394. [Google Scholar] [CrossRef]
- Elqotbi, M.; Vlaev, S.D.; Montastruc, L.; Nikov, I. CFD modelling of two-phase stirred bioreaction systems by segregated solution of the Euler–Euler model. Comput. Chem. Eng. 2013, 48, 113–120. [Google Scholar] [CrossRef]
- Rudniak, L.; Machniewski, P.M.; Milewska, A.; Molga, E. CFD modelling of stirred tank chemical reactors: Homogeneous and heterogeneous reaction systems. Chem. Eng. Sci. 2004, 59, 5233–5239. [Google Scholar] [CrossRef]
- Zhan, C.; Hagrot, E.; Brandt, L.; Chotteau, V. Study of hydrodynamics in wave bioreactors by computational fluid dynamics reveals a resonance phenomenon. Chem. Eng. Sci. 2019, 193, 53–65. [Google Scholar] [CrossRef]
- Singh, V. Disposable bioreactor for cell culture using wave-induced agitation. Cytotechnology 1999, 30, 149–158. [Google Scholar] [CrossRef] [PubMed]
- Whitman, W.G. The two film theory of gas absorption. Int. J. Heat Mass Transf. 1962, 5, 429–433. [Google Scholar] [CrossRef]
- Danckwerts, P.V. Significance of Liquid-Film Coefficients in Gas Absorption. Ind. Eng. Chem. 1951, 43, 1460–1467. [Google Scholar] [CrossRef]
- Fortescue, G.E.; Pearson, J.R.A. On gas absorption into a turbulent liquid. Chem. Eng. Sci. 1967, 22, 1163–1176. [Google Scholar] [CrossRef]
- Lamont, J.C.; Scott, D.S. An eddy cell model of mass transfer into the surface of a turbulent liquid. Aiche J. 1970, 16, 513–519. [Google Scholar] [CrossRef]
- McCready, M.J.; Vassiliadou, E.; Hanratty, T.J. Computer simulation of turbulent mass transfer at a mobile interface. AIChE J. 1986, 32, 1108–1115. [Google Scholar] [CrossRef]
- Turney, D.E.; Banerjee, S. Air–water gas transfer and near-surface motions. J. Fluid Mech. 2013, 733, 588–624. [Google Scholar] [CrossRef]
- Bai, Y.; Moo-Young, M.; Anderson, W.A. A mechanistic model for gas-liquid mass transfer prediction in a rocking disposable bioreactor. Biotechnol. Bioeng. 2019, 116, 1986–1998. [Google Scholar] [CrossRef]
- Starly, B.; Choubey, A. Enabling sensor technologies for the quantitative evaluation of engineered tissue. Ann. Biomed. Eng. 2008, 36, 30–40. [Google Scholar] [CrossRef] [PubMed]
- Wendt, D.; Riboldi, S.A.; Cioffi, M.; Martin, I. Bioreactors in tissue engineering: Scientific challenges and clinical perspectives. In Advances in Biochemical Engineering/Biotechnology; Springer: Berlin/Heidelberg, Germany, 2008. [Google Scholar]
- Biechele, P.; Busse, C.; Solle, D.; Scheper, T.; Reardon, K. Sensor systems for bioprocess monitoring. Eng. Life Sci. 2015, 15, 469–488. [Google Scholar] [CrossRef]
- Zhao, L.; Fu, H.-Y.; Zhou, W.; Hu, W.-S. Advances in process monitoring tools for cell culture bioprocesses. Eng. Life Sci. 2015, 15, 459–468. [Google Scholar] [CrossRef]
- Zhang, A.; Tsang, V.L.; Moore, B.; Shen, V.; Huang, Y.-M.; Kshirsagar, R.; Ryll, T. Advanced process monitoring and feedback control to enhance cell culture process production and robustness. Biotechnol. Bioeng. 2015, 112, 2495–2504. [Google Scholar] [CrossRef]
- De León, S.E.; Pupovac, A.; McArthur, S.L. Three-Dimensional (3D) cell culture monitoring: Opportunities and challenges for impedance spectroscopy. Biotechnol. Bioeng. 2020, 117, 1230–1240. [Google Scholar] [CrossRef] [PubMed]
- Hassan, Q.; Ahmadi, S.; Kerman, K. Recent Advances in Monitoring Cell Behavior Using Cell-Based Impedance Spectroscopy. Micromachines 2020, 11, 590. [Google Scholar] [CrossRef]
- Kieninger, J.; Weltin, A.; Flamm, H.; Urban, G.A. Microsensor Systems for Cell Metabolism—From 2D Culture to Organ-on-Chip. Lab Chip 2018, 18, 1274–1291. [Google Scholar] [CrossRef]
- Rubio, N.R.; Fish, K.D.; Trimmer, B.A.; Kaplan, D.L. Possibilities for engineered insect tissue as a food source. Front. Sustain. Food Syst. 2019, 3, 24. [Google Scholar] [CrossRef]
- Rubio, N.; Datar, I.; Stachura, D.; Kaplan, D.; Krueger, K. Cell-Based Fish: A Novel Approach to Seafood Production and an Opportunity for Cellular Agriculture. Front. Sustain. Food Syst. 2019, 3, 43. [Google Scholar] [CrossRef]
- O’Mara, P.; Farrell, A.; Bones, J.; Twomey, K. Staying alive! Sensors used for monitoring cell health in bioreactors. Talanta 2018, 176, 130–139. [Google Scholar] [CrossRef]
- IST. Temperature Sensors. Available online: https://www.ist-ag.com/en/products-services/temperature-sensors (accessed on 25 November 2020).
- Direct Industry. Reactor Temperature Sensor. Available online: https://www.directindustry.com/industrial-manufacturer/reactor-temperature-sensor-251712.html (accessed on 25 November 2020).
- Rosemount RosemountTM Multipoint Thermocoupleand RTD Profiling Sensors. Available online: https://www.emerson.com/documents/automation/product-data-sheet-rosemount-multipoint-thermocouple-rtd-profiling-sensors-type-tx-mtx-wx-mwx-en-88382.pdf (accessed on 25 November 2020).
- PyroScience. PyroScience Sensors. Available online: https://www.pyroscience.com/en/products/all-sensors/attributes/pH (accessed on 26 November 2020).
- Burns Engineering. Bioreactor Temperature Sensors. Available online: http://www.burnsengineering.com/local/uploads/files/bioreactords.pdf (accessed on 25 November 2020).
- Busse, C.; Biechele, P.; de Vries, I.; Reardon, K.F.; Solle, D.; Scheper, T. Sensors for disposable bioreactors. Eng. Life Sci. 2017, 17, 940–952. [Google Scholar] [CrossRef]
- Steinwedel, T.; Dahlmann, K.; Solle, D.; Scheper, T.; Reardon, K.F.; Lammers, F. Sensors for disposable bioreactor systems. In Single-Use Technology in Biopharmaceutical Manufacture; Eibl, R., Eibl, D., Eds.; Wiley: Hoboken, NJ, USA, 2019; pp. 69–82. [Google Scholar]
- Beutel, S.; Henkel, S. In situ sensor techniques in modern bioprocess monitoring. Appl. Microbiol. Biotechnol. 2011, 91, 1493–1505. [Google Scholar] [CrossRef]
- Werner, J.; Belz, M.; Klein, K.-F.; Sun, T.; Grattan, K.T.V. Fast response time fiber optical pH and oxygen sensors. In Optical Fibers and Sensors for Medical Diagnostics and Treatment Applications XX; Gannot, I., Ed.; SPIE: Bellingham, WA, USA, 2020; p. 56. [Google Scholar]
- Mettler Toledo. In-Line Measurements for Batch-to-Batch Consistency and Quality—Bio Pharmaceutical—Fermentation. Available online: https://www.mt.com/es/en/home/microsites/Pharmaceutical/Bio_Pharma/Fermentation_P.html (accessed on 26 November 2020).
- PreSens. pH Optical pH Sensors for Contactless or Minimally Invasive Measurements. Available online: https://www.presens.de/products/ph/sensors (accessed on 26 November 2020).
- Instrument Shanghai Boqu Instrument Co., Ltd. Available online: https://shboqu.en.made-in-china.com/product/jKBxtdCycZWV/China-Bioreactor-pH-Probe.html (accessed on 26 November 2020).
- Mettler Toledo. pH Sensors, Probes & Electrodes. Available online: https://www.mt.com/int/en/home/perm-lp/product-organizations/pro/ph-sensors-probes-electrodes-2.html (accessed on 28 February 2021).
- Wei, Y.; Jiao, Y.; An, D.; Li, D.; Li, W.; Wei, Q. Review of dissolved oxygen detection technology: From laboratory analysis to online intelligent detection. Sensors 2019, 19, 3995. [Google Scholar] [CrossRef]
- Quaranta, M.; Borisov, S.M.; Klimant, I. Indicators for optical oxygen sensors. Bioanal. Rev. 2012, 4, 115–157. [Google Scholar] [CrossRef]
- Systech Instruments Ltd. Paramagnetic O2 Analyzer. Available online: https://www.systechillinois.com/en/support/technologies/paramagnetic-cells (accessed on 29 November 2020).
- Mettler Toledo. Dissolved Oxygen Sensors for Process and Pure Water. Available online: https://www.mt.com/int/en/home/products/Process-Analytics/DO-CO2-ozone-sensor/dissolved-oxygen-meter.html (accessed on 26 November 2020).
- PreSens. Biopharma Single-Use Sensors for Biopharmaceutical Production. Available online: https://www.presens.de/products/oem-components/single-use-sensors-for-biopharmaceutical-production (accessed on 26 November 2020).
- Verma, A.; Verma, M.; Singh, A. Animal tissue culture principles and applications. In Animal Biotechnology; Elsevier: Amsterdam, The Netherlands, 2020; pp. 269–293. [Google Scholar]
- Ge, X.; Kostov, Y.; Rao, G. Low-cost noninvasive optical CO2 sensing system for fermentation and cell culture. Biotechnol. Bioeng. 2005, 89, 329–334. [Google Scholar] [CrossRef]
- Mills, A. Optical sensors for carbon dioxide and their applications. In Sensors for Environment, Health and Security; Springer: Dordrecht, The Netherlands, 2009; pp. 347–370. [Google Scholar]
- Zosel, J.; Oelßner, W.; Decker, M.; Gerlach, G.; Guth, U. The measurement of dissolved and gaseous carbon dioxide concentration. Meas. Sci. Technol. 2011, 22, 072001. [Google Scholar] [CrossRef]
- Li, X.; Wang, Q. Recent progress of dissolved carbon dioxide measurement technologies based on optical methods. Trans. Inst. Meas. Control 2018, 41, 014233121879122. [Google Scholar] [CrossRef]
- Mettler Toledo. CO2 Sensor|Carbon Dioxide ProbeReliable Dissolved CO2 Probe for In Situ CO2 Monitoring. Available online: https://www.mt.com/ca/en/home/products/Process-Analytics/DO-CO2-ozone-sensor/dissolved-carbon-dioxide.html (accessed on 26 February 2021).
- Borisov, S.M.; Seifner, R.; Klimant, I. A novel planar optical sensor for simultaneous monitoring of oxygen, carbon dioxide, pH and temperature. Anal. Bioanal. Chem. 2011, 400, 2463–2474. [Google Scholar] [CrossRef] [PubMed]
- Tamburini, E.; Marchetti, M.G.; Pedrini, P. Monitoring key parameters in bioprocesses using near-infrared technology. Sensors 2014, 14, 18941–18959. [Google Scholar] [CrossRef]
- Yardley, J.E.; Kell, D.B.; Barrett, J.; Davey, C.L. On-line, real-time measurements of cellular biomass using dielectric spectroscopy. Biotechnol. Genet. Eng. Rev. 2000, 17, 3–35. [Google Scholar] [CrossRef]
- Alpfmedical. Standard Bioreactor Sensors 13221 Introduction—Cell Culture. Available online: https://www.alpfmedical.info/cell-culture/standard-bioreactor-sensors-13221-introduction.html (accessed on 23 November 2020).
- Ozturk, S.S.; Thrift, J.C.; Blackie, J.D.; Naveh, D. Real-time monitoring and control of glucose and lactate concentrations in a mammalian cell perfusion reactor. Biotechnol. Bioeng. 1997, 53, 372–378. [Google Scholar] [CrossRef]
- Eyer, K.; Heinzle, E. On-line estimation of viable cells in a hybridoma culture at various DO levels using ATP balancing and redox potential measurement. Biotechnol. Bioeng. 2000, 49, 277–283. [Google Scholar] [CrossRef]
- Lourenço, N.D.; Lopes, J.A.; Almeida, C.F.; Sarraguça, M.C.; Pinheiro, H.M. Bioreactor monitoring with spectroscopy and chemometrics: A review. Anal. Bioanal. Chem. 2012, 404, 1211–1237. [Google Scholar] [CrossRef]
- Savage, S. Redorbit. Available online: https://www.redorbit.com/news/science/281073/microbial_biomass_estimation/ (accessed on 21 November 2020).
- Afguard. Faudi Aviatio—Quality Guarantees Safety. Available online: https://www.faudi-aviation.com/en/products/sensor-technology/afguard/ (accessed on 21 November 2020).
- Fan, R.; Ebrahimi, M.; Quitmann, H.; Aden, M.; Czermak, P. An innovative optical sensor for the online monitoring and control of biomass concentration in a membrane bioreactor system for lactic acid production. Sensors 2016, 16, 411. [Google Scholar] [CrossRef]
- Ude, C.; Schmidt-Hager, J.; Findeis, M.; John, G.T.; Scheper, T.; Beutel, S. Application of an online-biomass sensor in an optical multisensory platform prototype for growth monitoring of biotechnical relevant microorganism and cell lines in single-use shake flasks. Sensors 2014, 14, 17390–17405. [Google Scholar] [CrossRef]
- Carvell, J.P.; Dowd, J.E. On-line Measurements and Control of Viable Cell Density in Cell Culture Manufacturing Processes using Radio-frequency Impedance. Cytotechnology 2006, 50, 35–48. [Google Scholar] [CrossRef]
- Markx, G.H.; Davey, C.L. The dielectric properties of biological cells at radiofrequencies: Applications in biotechnology. Enzym. Microb. Technol. 1999, 25, 161–171. [Google Scholar] [CrossRef]
- Cannizzaro, C.; Gügerli, R.; Marison, I.; von Stockar, U. On-line biomass monitoring of CHO perfusion culture with scanning dielectric spectroscopy. Biotechnol. Bioeng. 2003, 84, 597–610. [Google Scholar] [CrossRef]
- Kiviharju, K.; Salonen, K.; Moilanen, U.; Meskanen, E.; Leisola, M.; Eerikäinen, T. On-line biomass measurements in bioreactor cultivations: Comparison study of two on-line probes. J. Ind. Microbiol. Biotechnol. 2007, 34, 561–566. [Google Scholar] [CrossRef]
- Metze, S.; Ruhl, S.; Greller, G.; Grimm, C.; Scholz, J. Monitoring online biomass with a capacitance sensor during scale-up of industrially relevant CHO cell culture fed-batch processes in single-use bioreactors. Bioprocess. Biosyst Eng. 2020, 43, 193–205. [Google Scholar] [CrossRef]
- Carvell, J.; Bhat, A.; Tindal, S.; Scholz, J.; van Santen, P.; Das, R.; Roosloot, R. Monitoring Live Biomass in Disposable Bioreactors—BioProcess. Available online: https://bioprocessintl.com/upstream-processing/upstream-single-use-technologies/monitoring-live-biomass-in-disposable-bioreactors/ (accessed on 21 November 2020).
- Fogale Biotech. Available online: http://www.fogalebiotech.com/PHP/products-ibiomass.php (accessed on 21 November 2020).
- Reinecke, T.; Biechele, P.; Schulte, V.; Scheper, T.; Zimmermann, S. Low-cost Sensor System for Non-invasive Monitoring of Cell Growth in Disposable Bioreactors. Proc. Eng. 2015, 120, 548–551. [Google Scholar] [CrossRef][Green Version]
- Sartorius BIOPAT VIAMASS | Sartorius. Available online: https://www.sartorius.com/shop/ww/en/brl/products-bioprocess-process-analysers/biopatviamass/p/BIOPATVIAMASS (accessed on 4 March 2021).
- Aber Instruments Biomass Sensor. Standard Remote Futura. Available online: https://aberinstruments.com/product/standard-remote-futura/ (accessed on 4 March 2021).
- Aber Instruments Biomass Sensor. Standard Futura. Available online: https://aberinstruments.com/product/standard-futura-standard-remote-futura/ (accessed on 4 March 2021).
- Fogale Nanotech iBiomass 465. Available online: http://www.fogalebiotech.com/images/PDF/PDF/IB465.pdf (accessed on 24 November 2020).
- Schneider, M.; Marison, I.W.; von Stockar, U. The importance of ammonia in mammalian cell culture. J. Biotechnol. 1996, 46, 161–185. [Google Scholar] [CrossRef]
- Mirabet, M.; Navarro, A.; Lopez, A.; Canela, E.I.; Mallol, J.; Lluis, C.; Franco, R. Ammonium toxicity in different cell lines. Biotechnol. Bioeng. 1997, 56, 530–537. [Google Scholar] [CrossRef]
- Thévenot, D.R.; Toth, K.; Durst, R.A.; Wilson, G.S. Electrochemical biosensors: Recommended definitions and classification1International Union of Pure and Applied Chemistry: Physical Chemistry Division, Commission I.7 (Biophysical Chemistry); Analytical Chemistry Division, Commission V.5 (Electroanalytical Chemistry). Biosens. Bioelectron. 2001, 16, 121–131. [Google Scholar]
- Ronkainen, N.J.; Halsall, H.B.; Heineman, W.R. Electrochemical biosensors. Chem. Soc. Rev. 2010, 39, 1747–1763. [Google Scholar] [CrossRef]
- ScienceDirect Topics. Glucose Sensor—An Overview. Available online: https://www.sciencedirect.com/topics/engineering/glucose-sensor (accessed on 19 November 2020).
- Oliver, N.S.; Toumazou, C.; Cass, A.E.G.; Johnston, D.G. Glucose sensors: A review of current and emerging technology. Diabet. Med. 2009, 26, 197–210. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, C.R. Labiotech. Available online: https://www.labiotech.eu/diabetes/needle-free-glucose-monitoring-for-diabetes-medtech/ (accessed on 25 November 2020).
- Bruen, D.; Delaney, C.; Florea, L.; Diamond, D. Glucose sensing for diabetes monitoring: Recent developments. Sensors 2017, 17, 1866. [Google Scholar] [CrossRef]
- Berry, B.N.; Dobrowsky, T.M.; Timson, R.C.; Kshirsagar, R.; Ryll, T.; Wiltberger, K. Quick generation of Raman spectroscopy based in-process glucose control to influence biopharmaceutical protein product quality during mammalian cell culture. Biotechnol. Prog. 2016, 32, 224–234. [Google Scholar] [CrossRef]
- Matthews, T.E.; Smelko, J.P.; Berry, B.; Romero-Torres, S.; Hill, D.; Kshirsagar, R.; Wiltberger, K. Glucose monitoring and adaptive feeding of mammalian cell culture in the presence of strong autofluorescence by near infrared Raman spectroscopy. Biotechnol. Prog. 2018, 34, 1574–1580. [Google Scholar] [CrossRef]
- Duarte-Delgado, D.; Narváez-Cuenca, C.-E.; Restrepo-Sánchez, L.-P.; Kushalappa, A.; Mosquera-Vásquez, T. Development and validation of a liquid chromatographic method to quantify sucrose, glucose, and fructose in tubers of Solanum tuberosum Group Phureja. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2015, 975, 18–23. [Google Scholar] [CrossRef] [PubMed]
- Lewis, C.B.; McNichols, R.J.; Gowda, A.; Coté, G.L. Investigation of Near-Infrared Spectroscopy for Periodic Determination of Glucose in Cell Culture Media in situ. Appl. Spectrosc. 2000, 54, 1453–1457. [Google Scholar] [CrossRef]
- Adams, A.G.; Bulusu RK, M.; Mukhitov, N.; Mendoza-Cortes, J.L.; Roper, M.G. Online Measurement of Glucose Consumption from HepG2 Cells Using an Integrated Bioreactor and Enzymatic Assay. Anal. Chem. 2019, 91, 5184–5190. [Google Scholar] [CrossRef]
- Tric, M.; Lederle, M.; Neuner, L.; Dolgowjasow, I.; Wiedemann, P.; Wölfl, S.; Werner, T. Optical biosensor optimized for continuous in-line glucose monitoring in animal cell culture. Anal. Bioanal. Chem. 2017, 409, 5711–5721. [Google Scholar] [CrossRef]
- Talaei, S.; Frey, O.; Psoma, S.; van der Wal, P.D.; de Rooij, N.F. Smart SU-8 pillars implemented in a microfluidic bioreactor for continuous measurement of glucose. Procedia Eng. 2010, 5, 448–451. [Google Scholar] [CrossRef][Green Version]
- Bauer, I.; John, G.T.; Spichiger, S.; Spichiger Keller, U.E. Online Monitoring of Glucose, pH, and DO in Shake Flask Culture. Available online: https://www.presens.de/company/press/article/novel-single-use-sensors-for-online-measurement-of-glucose-999 (accessed on 19 November 2020).
- Bauer, I.; Poggendorf, I.; Spichiger, S.; Spichiger-Keller, U.; John, G. Novel Single-Use Sensors for Online Measurement of Glucose. Available online: https://www.presens.de/fileadmin/user_upload/press/2012_09_BPI_Novel-Single-Use-Sensors-for-Online-Measurement-of-Glucose_IBauer.pdf (accessed on 29 November 2020).
- Tang, Y.; Petropoulos, K.; Kurth, F.; Gao, H.; Migliorelli, D.; Guenat, O.; Generelli, S. Screen-Printed Glucose Sensors Modified with Cellulose Nanocrystals (CNCs) for Cell Culture Monitoring. Biosensors 2020, 10, 125. [Google Scholar] [CrossRef] [PubMed]
- Esmaeili, C.; Abdi, M.M.; Mathew, A.P.; Jonoobi, M.; Oksman, K.; Rezayi, M. Synergy effect of nanocrystalline cellulose for the biosensing detection of glucose. Sensors 2015, 15, 24681–24697. [Google Scholar] [CrossRef]
- Hwa, K.-Y.; Subramani, B. Synthesis of zinc oxide nanoparticles on graphene-carbon nanotube hybrid for glucose biosensor applications. Biosens. Bioelectron. 2014, 62, 127–133. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Chen, Z.; Tian, R.; Yan, W.; Yao, C. Glucose biosensor based on three dimensional ordered macroporous self-doped polyaniline/Prussian blue bicomponent film. Anal. Chim. Acta 2012, 723, 94–100. [Google Scholar] [CrossRef] [PubMed]
- Bihar, E.; Wustoni, S.; Pappa, A.M.; Salama, K.N.; Baran, D.; Inal, S. A fully inkjet-printed disposable glucose sensor on paper. NPJ Flex. Electron. 2018, 2, 30. [Google Scholar] [CrossRef]
- Boero, C.; Carrara, S.; Del Vecchio, G.; Calzà, L.; De Micheli, G. Highly sensitive carbon nanotube-based sensing for lactate and glucose monitoring in cell culture. IEEE Trans. Nanobiosci. 2011, 10, 59–67. [Google Scholar] [CrossRef]
- Tseng, T.T.-C.; Chang, C.-F.; Chan, W.-C. Fabrication of implantable, enzyme-immobilized glutamate sensors for the monitoring of glutamate concentration changes in vitro and in vivo. Molecules 2014, 19, 7341–7355. [Google Scholar] [CrossRef]
- Qin, S.; Van der Zeyden, M.; Oldenziel, W.H.; Cremers, T.I.; Westerink, B.H. Microsensors for in vivo Measurement of Glutamate in Brain Tissue. Sensors 2008, 8, 6860–6884. [Google Scholar] [CrossRef]
- Schultz, J.; Uddin, Z.; Singh, G.; Howlader, M.M.R. Glutamate sensing in biofluids: Recent advances and research challenges of electrochemical sensors. Analyst 2020, 145, 321–347. [Google Scholar] [CrossRef]
- Bollella, P.; Gorton, L.; Ludwig, R.; Antiochia, R. A Third Generation Glucose Biosensor Based on Cellobiose Dehydrogenase Immobilized on a Glassy Carbon Electrode Decorated with Electrodeposited Gold Nanoparticles: Characterization and Application in Human Saliva. Sensors 2017, 17, 1912. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Yang, H.; Yang, W.; Li, Y.; Sun, C. Gold nanoparticles-mesoporous silica composite used as an enzyme immobilization matrix for amperometric glucose biosensor construction. Sens. Actuators B Chem. 2007, 124, 179–186. [Google Scholar] [CrossRef]
- Li, H.; He, J.; Zhao, Y.; Wu, D.; Cai, Y.; Wei, Q.; Yang, M. Immobilization of glucose oxidase and platinum on mesoporous silica nanoparticles for the fabrication of glucose biosensor. Electrochim. Acta 2011, 56, 2960–2965. [Google Scholar] [CrossRef]
- Ges, I.A.; Baudenbacher, F. Enzyme electrodes to monitor glucose consumption of single cardiac myocytes in sub-nanoliter volumes. Biosens. Bioelectron. 2010, 25, 1019–1024. [Google Scholar] [CrossRef]
- Batra, B.; Kumari, S.; Pundir, C.S. Construction of glutamate biosensor based on covalent immobilization of glutamate oxidase on polypyrrole nanoparticles/polyaniline modified gold electrode. Enzym. Microb. Technol. 2014, 57, 69–77. [Google Scholar] [CrossRef]
- Batra, B.; Yadav, M.; Pundir, C.S. l-Glutamate biosensor based on l-glutamate oxidase immobilized onto ZnO nanorods/polypyrrole modified pencil graphite electrode. Biochem. Eng. J. 2016, 105, 428–436. [Google Scholar] [CrossRef]
- Özel, R.E.; Ispas, C.; Ganesana, M.; Leiter, J.C.; Andreescu, S. Glutamate oxidase biosensor based on mixed ceria and titania nanoparticles for the detection of glutamate in hypoxic environments. Biosens. Bioelectron. 2014, 52, 397–402. [Google Scholar] [CrossRef]
- Scoggin, J.L.; Tan, C.; Nguyen, N.H.; Kansakar, U.; Madadi, M.; Siddiqui, S.; Arumugam, P.U.; DeCoster, M.A.; Murray, T.A. An enzyme-based electrochemical biosensor probe with sensitivity to detect astrocytic versus glioma uptake of glutamate in real time in vitro. Biosens. Bioelectron. 2019, 126, 751–757. [Google Scholar] [CrossRef]
- Hernández-Ibáñez, N.; García-Cruz, L.; Montiel, V.; Foster, C.W.; Banks, C.E.; Iniesta, J. Electrochemical lactate biosensor based upon chitosan/carbon nanotubes modified screen-printed graphite electrodes for the determination of lactate in embryonic cell cultures. Biosens. Bioelectron. 2016, 77, 1168–1174. [Google Scholar] [CrossRef]
- Shimomura, T.; Sumiya, T.; Ono, M.; Ito, T.; Hanaoka, T. Amperometric L-lactate biosensor based on screen-printed carbon electrode containing cobalt phthalocyanine, coated with lactate oxidase-mesoporous silica conjugate layer. Anal. Chim. Acta 2012, 714, 114–120. [Google Scholar] [CrossRef]
- Timmer, B.; Olthuis, W.; Berg, A.v.d. Ammonia sensors and their applications—A review. Sens. Actuators B Chem. 2005, 107, 666–677. [Google Scholar] [CrossRef]
- Ryll, T.; Valley, U.; Wagner, R. Biochemistry of growth inhibition by ammonium ions in mammalian cells. Biotechnol. Bioeng. 1994, 44, 184–193. [Google Scholar] [CrossRef] [PubMed]
- Hansen, H.A.; Emborg, C. Influence of ammonium on growth, metabolism, and productivity of a continuous suspension Chinese hamster ovary cell culture. Biotechnol. Prog. 1994, 10, 121–124. [Google Scholar] [CrossRef] [PubMed]
- Gawlitzek, M.; Ryll, T.; Lofgren, J.; Sliwkowski, M.B. Ammonium alters N-glycan structures of recombinant TNFR-IgG: Degradative versus biosynthetic mechanisms. Biotechnol. Bioeng. 2000, 68, 637–646. [Google Scholar] [CrossRef]
- Hassell, T.; Gleave, S.; Butler, M. Growth inhibition in animal cell culture. The effect of lactate and ammonia. Appl. Biochem. Biotechnol. 1991, 30, 29–41. [Google Scholar] [CrossRef]
- Derfus, G.E.; Abramzon, D.; Tung, M.; Chang, D.; Kiss, R.; Amanullah, A. Cell culture monitoring via an auto-sampler and an integrated multi-functional off-line analyzer. Biotechnol. Prog. 2010, 26, 284–292. [Google Scholar] [CrossRef]
- Abu-Absi, N.R.; Kenty, B.M.; Cuellar, M.E.; Borys, M.C.; Sakhamuri, S.; Strachan, D.J.; Hausladen, M.C.; Li, Z.J. Real time monitoring of multiple parameters in mammalian cell culture bioreactors using an in-line Raman spectroscopy probe. Biotechnol. Bioeng. 2011, 108, 1215–1221. [Google Scholar] [CrossRef] [PubMed]
- Popiel, S.; Sankowska, M. Determination of chemical warfare agents and related compounds in environmental samples by solid-phase microextraction with gas chromatography. J. Chromatogr. A 2011, 1218, 8457–8479. [Google Scholar] [CrossRef]
- Borsdorf, H.; Mayer, T.; Zarejousheghani, M.; Eiceman, G.A. Recent developments in ion mobility spectrometry. Appl. Spectrosc. Rev. 2011, 46, 472–521. [Google Scholar] [CrossRef]
- Kientz, C.E. Chromatography and mass spectrometry of chemical warfare agents, toxins and related compounds: State of the art and future prospects. J. Chromatogr. A 1998, 814, 1–23. [Google Scholar] [CrossRef]
- Fortner, E.C.; Zheng, J.; Zhang, R.; Berk Knighton, W.; Volkamer, R.M.; Sheehy, P.; Molina, L.; André, M. Measurements of Volatile Organic Compounds Using Proton Transfer Reaction—Mass Spectrometry during the MILAGRO 2006 Campaign. Atmos. Chem. Phys. 2009, 9, 467–481. [Google Scholar] [CrossRef]
- Tanguy, N.R.; Thompson, M.; Yan, N. A review on advances in application of polyaniline for ammonia detection. Sens. Actuators B Chem. 2018, 257, 1044–1064. [Google Scholar] [CrossRef]
- Bielecki, Z.; Stacewicz, T.; Smulko, J.; Wojtas, J. Ammonia Gas Sensors: Comparison of Solid-State and Optical Methods. Appl. Sci. 2020, 10, 5111. [Google Scholar] [CrossRef]
- Orion OrionTM Ammonia Gas Sensing ISE Electrodes. Available online: https://www.thermofisher.com/order/catalog/product/9512BNWP#/9512BNWP (accessed on 27 November 2020).
- Kwak, D.; Lei, Y.; Maric, R. Ammonia gas sensors: A comprehensive review. Talanta 2019, 204, 713–730. [Google Scholar] [CrossRef]
- Dervisevic, E.; Voelcker, N.H.; Risbridger, G.; Tuck, K.L.; Cadarso, V.J. High-Aspect-Ratio SU-8-Based Optofluidic Device for Ammonia Detection in Cell Culture Media. ACS Sens. 2020, 5, 2523–2529. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Song, Y.; Yang, Z. A reversible spectrophotometric method based on a coupled microfluidic chip for highly selective ammonium detection. J. Chem. 2019, 2019, 1–8. [Google Scholar] [CrossRef]
- Timmer, B.H.; van Delft, M.; Koelmans, W.W.; Olthuis, W.; van den Berg, A. Selective low concentration ammonia sensing in a microfluidic lab-on-a-chip. IEEE Sens. J. 2006, 6, 829–835. [Google Scholar] [CrossRef]
- Zhu, Z.; Lu, J.J.; Almeida MI, G.S.; Pu, Q.; Kolev, S.D.; Liu, S. A microfabricated electroosmotic pump coupled to a gas-diffusion microchip for flow injection analysis of ammonia. Microchim. Acta 2015, 182, 1063–1070. [Google Scholar] [CrossRef]
- Oliveira, S.M.; Lopes, T.I.; Toth, I.V.; Rangel, A.O. A multi-commuted flow injection system with a multi-channel propulsion unit placed before detection: Spectrophotometric determination of ammonium. Anal. Chim. Acta 2007, 600, 29–34. [Google Scholar] [CrossRef] [PubMed]
- Bäcker, M.; Rakowski, D.; Poghossian, A.; Biselli, M.; Wagner, P.; Schöning, M.J. Chip-based amperometric enzyme sensor system for monitoring of bioprocesses by flow-injection analysis. J. Biotechnol. 2013, 163, 371–376. [Google Scholar] [CrossRef] [PubMed]
- Arif, M.F.H.; Ahmed, K.; Asaduzzaman, S.; Azad, M.A.K. Design and optimization of photonic crystal fiber for liquid sensing applications. Photonic Sens. 2016, 6, 279–288. [Google Scholar] [CrossRef]
- Hartings, M.R.; Castro, N.J.; Gill, K.; Ahmed, Z. A photonic pH sensor based on photothermal spectroscopy. Sens. Actuators B Chem. 2019, 301, 127076. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Sciarrino, F.; Laing, A.; Thompson, M.G. Integrated photonic quantum technologies. Nat. Photonics 2020, 14, 273–284. [Google Scholar] [CrossRef]
- Kozma, P.; Kehl, F.; Ehrentreich-Förster, E.; Stamm, C.; Bier, F.F. Integrated planar optical waveguide interferometer biosensors: A comparative review. Biosens. Bioelectron. 2014, 58, 287–307. [Google Scholar] [CrossRef]
- Estevez, M.C.; Alvarez, M.; Lechuga, L.M. Integrated optical devices for lab-on-a-chip biosensing applications. Laser Photonics Rev. 2012, 6, 463–487. [Google Scholar] [CrossRef]
- Bar-On, O.; Brenner, P.; Siegle, T.; Gvishi, R.; Kalt, H.; Lemmer, U.; Scheuer, J. High Quality 3D Photonics using Nano Imprint Lithography of Fast Sol-gel Materials. Sci. Rep. 2018, 8, 7833. [Google Scholar] [CrossRef] [PubMed]
- Paniccia, M.; Morse, M.; Salib, M. Integrated Photonics. In Silicon Photonics. Topics in Applied Physics; Springer: Berlin, Germany, 2004; Volume 94. [Google Scholar]
- Misiakos, K.; Makarona, E.; Hoekman, M.; Fyrogenis, R.; Tukkiniemi, K.; Jobst, G.; Petrou, P.S.; Kakabakos, S.E.; Salapatas, A.; Goustouridis, D.; et al. All-Silicon Spectrally Resolved Interferometric Circuit for Multiplexed Diagnostics: A Monolithic Lab-on-a-Chip Integrating All Active and Passive Components. ACS Photonics 2019, 6, 1694–1705. [Google Scholar] [CrossRef]
- Fernández-Gavela, A.; Herranz, S.; Chocarro, B.; Falke, F.; Schreuder, E.; Leeuwis, H.; Heideman, R.G.; Lechuga, L.M. Full integration of photonic nanoimmunosensors in portable platforms for on-line monitoring of ocean pollutants. Sens. Actuators B Chem. 2019, 297, 126758. [Google Scholar] [CrossRef]
- Bai, W.; Yang, H.; Ma, Y.; Chen, H.; Shin, J.; Liu, Y.; Yang, Q.; Kandela, I.; Liu, Z.; Kang, S.-K.; et al. Flexible Transient Optical Waveguides and Surface-Wave Biosensors Constructed from Monocrystalline Silicon. Adv. Mater. 2018, 30, e1801584. [Google Scholar] [CrossRef] [PubMed]
- Stein, E.W.; Grant, P.S.; Zhu, H.; McShane, M.J. Microscale enzymatic optical biosensors using mass transport limiting nanofilms. Fabrication and characterization using glucose as a model analyte. Anal. Chem. 2007, 79, 1339–1348. [Google Scholar] [CrossRef] [PubMed]
- Xia, F.; Mueller, T.; Lin, Y.-M.; Valdes-Garcia, A.; Avouris, P. Ultrafast graphene photodetector. Nat. Nanotechnol. 2009, 4, 839–843. [Google Scholar] [CrossRef] [PubMed]
- Qu, Z.; Nedeljkovic, M.; Soler-Penadés, J.; Khokhar, A.Z.; Cao, W.; Wu, Y.; Osman, A.; Qi, Y.; Aspiotis, N.; Morgan, K.; et al. Waveguide integrated graphene mid-infrared photodetector. In Silicon Photonics XIII; Reed, G.T., Knights, A.P., Eds.; SPIE: Bellingham, WA, USA, 2018; p. 59. [Google Scholar]
- Schuler, S.; Schall, D.; Neumaier, D.; Schwarz, B.; Watanabe, K.; Taniguchi, T.; Mueller, T. Graphene photodetector integrated on a photonic crystal defect waveguide. ACS Photonics 2018, 5, 4758–4763. [Google Scholar] [CrossRef]
- Kim, J.; Kasture, M.; Hwang, T.; Kulkarni, A.; Amin, R.; Park, S.; Kim, T.; Gosavi, S. Graphene-based waveguides: Novel method for detecting biological activity. Appl. Biochem. Biotechnol. 2012, 167, 1069–1075. [Google Scholar] [CrossRef] [PubMed]
- Hwang, T.; Kim, J.A.; Kulkarni, A.; Kim, T. Graphene photo detector with integrated waveguide biochemical sensors. Sens. Actuators B Chem. 2013, 187, 319–322. [Google Scholar] [CrossRef]
- Wang, J.; Cheng, Z.; Li, X. Progress on Waveguide-Integrated Graphene Optoelectronics. Adv. Condens. Matter Phys. 2018, 2018, 1–9. [Google Scholar] [CrossRef]
- Drago, L.; Toscano, M.; Bottagisio, M. Recent Evidence on Bioactive Glass Antimicrobial and Antibiofilm Activity: A Mini-Review. Materials 2018, 11, 326. [Google Scholar] [CrossRef] [PubMed]
- Azeredo, J.; Azevedo, N.F.; Briandet, R.; Cerca, N.; Coenye, T.; Costa, A.R.; Desvaux, M.; Di Bonaventura, G.; Hébraud, M.; Jaglic, Z.; et al. Critical review on biofilm methods. Crit. Rev. Microbiol. 2017, 43, 313–351. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Z.; Sun, D.-W.; Pu, H.; Gao, W.; Dai, Q. Applications of emerging imaging techniques for meat quality and safety detection and evaluation: A review. Crit. Rev. Food Sci. Nutr. 2017, 57, 755–768. [Google Scholar] [CrossRef] [PubMed]
- Jensen, E.C. Overview of live-cell imaging: Requirements and methods used. Anat. Rec. 2013, 296, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Cole, R. Live-cell imaging: The cell’s perspective. Cell Adhes. Migr. 2014, 8, 452–459. [Google Scholar] [CrossRef] [PubMed]
- Shen, N.; Riedl, J.A.; Carvajal Berrio, D.A.; Davis, Z.; Monaghan, M.G.; Layland, S.L.; Hinderer, S.; Schenke-Layland, K. A flow bioreactor system compatible with real-time two-photon fluorescence lifetime imaging microscopy. Biomed. Mater. 2018, 13, 024101. [Google Scholar] [CrossRef]
- PerkinElmer. MuviCyte Live-Cell Imaging Kit. Available online: https://www.perkinelmer.com/Product/muvicyte-live-cell-imaging-kit-hh40000000 (accessed on 29 November 2020).
- Mulas, C.; Hodgson, A.C.; Kohler, T.N.; Agley, C.C.; Humphreys, P.; Kleine-Brüggeney, H.; Hollfelder, F.; Smith, A.; Chalut, K.J. Microfluidic platform for 3D cell culture with live imaging and clone retrieval. Lab Chip 2020, 20, 2580–2591. [Google Scholar] [CrossRef] [PubMed]
- Ovizio-iLine F. Available online: https://ovizio.com/iline-f/ (accessed on 29 November 2020).
- Ovizio. Mesenchymal Stem Cells Using Cytodex 1 Microcarriers. Available online: https://ovizio.com/portfolio/mesenchymal-stem-cells/ (accessed on 30 November 2020).
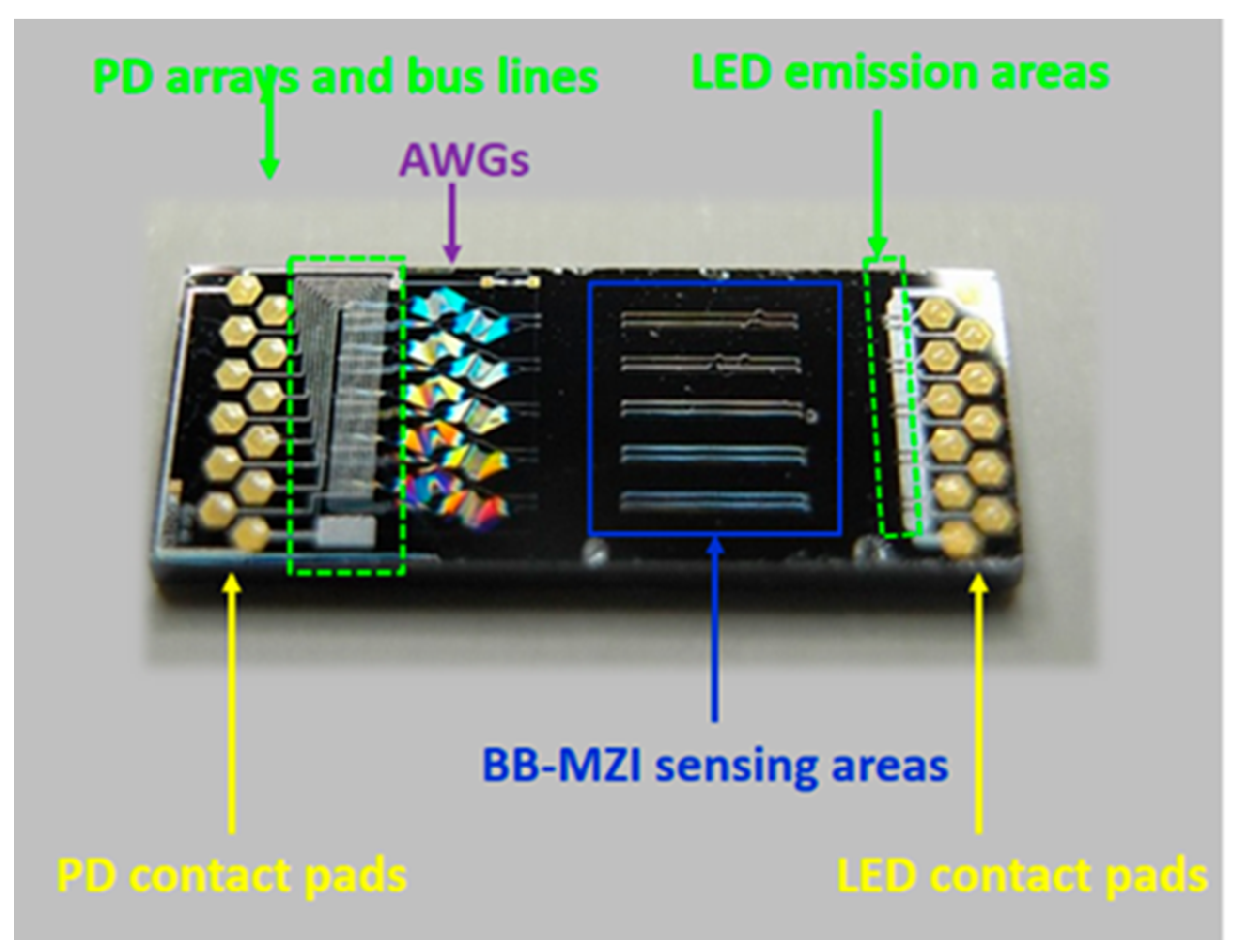
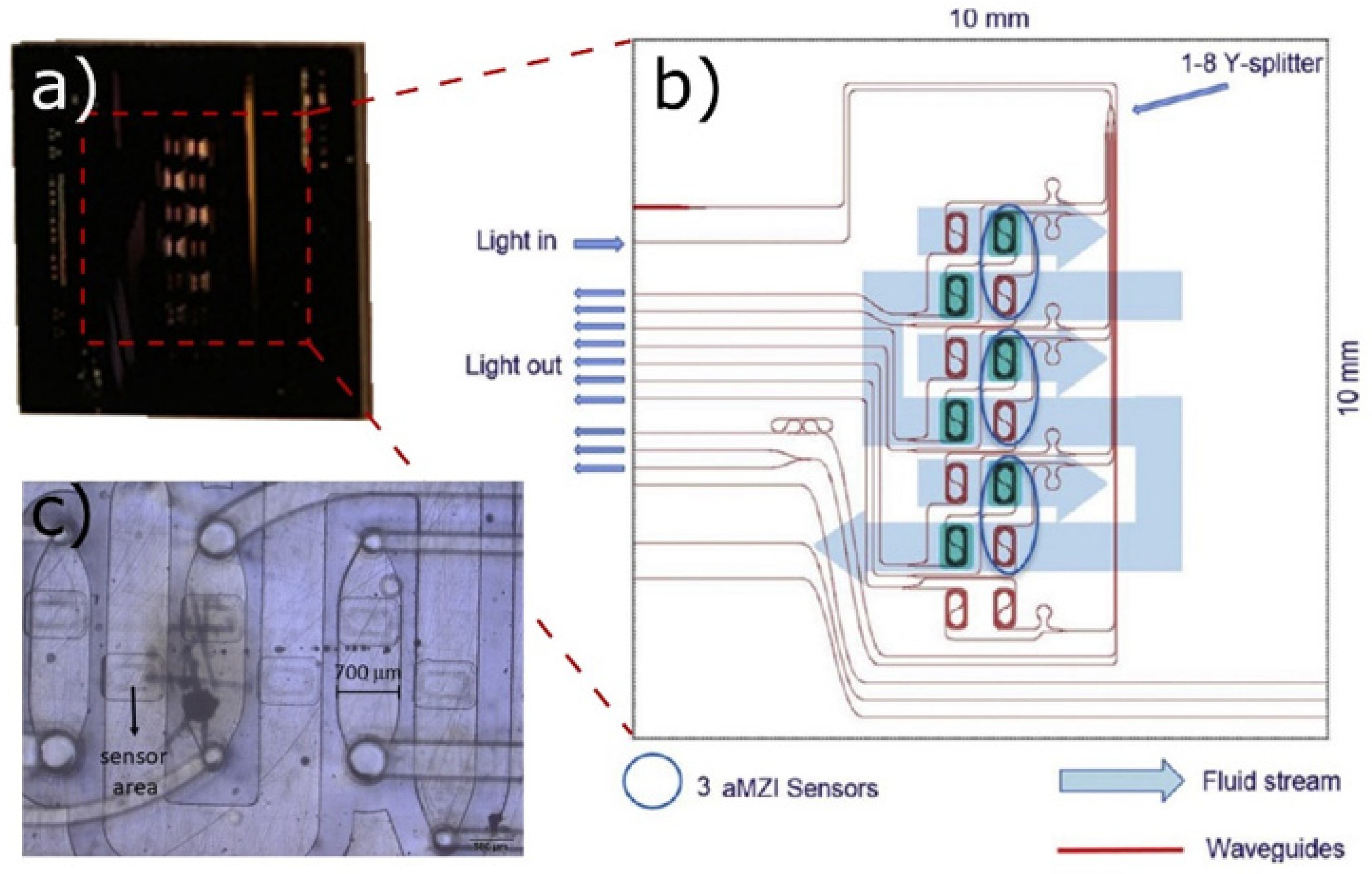
| Type of Bioreactor | Volume | Phase of CM Cultivation | Type of Agitation/Medium Flow | Integrated Sensors | Ref. |
|---|---|---|---|---|---|
| Spinner flask | 60–500 mL | Proliferation | Impeller-driven | n/a | [78,79] |
| Stirred tank | 1–5 L–2 × 104 L * | Proliferation | Impeller-driven | pH, dissolved oxygen and temperature control | [80] |
| Air, N2, CO2 and O2, pressure, optical density, viable cells, exhaust gas composition, redox, weight for reactor | [81] | ||||
| Rocking/wave | 1–100 L * | Proliferation | Rocking motion-driven | pH and dissolved oxygen control | [82] |
| internal floating filter (retains the cells in the bioreactor-filters only the media) | [83] | ||||
| Perfusion bioreactors | up to 6000 L * | Differentiation/maturation | Perfusing medium through the scaffold | pH, temperature, automatic medium exchange, glucose measurement, mechanical and electrical cell stimulation | [84] |
| bidirectional and interstitial perfusion, flow rate control | [85] |
| Principle | Sensor | Temperature Range | Accuracy/Class | Ref. |
|---|---|---|---|---|
| Resistance sensors | Platinum | −200 to 1000 °C | offered in class F0.3 (0.12%), class F0.15 (0.06%) and F0.1 (0.04%) | [188] |
| Nickel | −60 to 300 °C | 6180 ppm/K (Nickel ND), 5000 ppm/K (Nickel NL), 6370 ppm/K (Nickel NJ), 6720 ppm/K (Nickel NA) | ||
| TSic | +10 to +90 °C | ±0.5 K to ±0.1 K | ||
| United Electric Controls | −196 to 482 °C | RTP1 (std.) ± 0.12% RTP1A ± 0.06% RTP1AA ± 0.01% | [189] | |
| Thermocouple | IST, Rosemount™ | −40 to 750 °C | 1.5 °C or 0.004 |t| t is in degrees Celsius. | [188,190] |
| Krohne | −40 to 600 °C | ±0.1% or ±0.15% | [189] | |
| Pyroscience, Burns | 0 to 50 °C | ±0.10 °C | [191,192] |
| Principle | Sensor | Range | Accuracy | Ref. | |
|---|---|---|---|---|---|
| Optical | Pyroscience | pH Sensor Spots | Different ranges available (4–6; 5–7; 6–8; 7–9; total scale) | ±0.05 after 2-point calibration | [191] |
| pH Flow Through Cell | Different ranges available (4–6; 5–7; 6–8; 7–9; total scale) | ±0.05 after 2-point calibration | |||
| pH Sensor Cap for Under water devices | Different ranges available (4–6; 5–7; 6–8; 7–9; total scale) | ±0.05 after 2-point calibration | |||
| PreSens Sensors | pH-1 SMA LG1 | 4.5–7 | resolution: ±0.1 °C accuracy: ±1.0 °C | [198] | |
| Self-adhesive pH Sensor Spots SP-LG1-SA | 4.5–7 | resolution at pH = 7 ± 0.01 accuracy ±0.05/±0.10 | |||
| Single-Use pH Flow-Through Cell FTC-SU-HP5-S | 5.5–8.5 | resolution: ±0.02 accuracy: ±0.05 | |||
| Profiling pH Microsensor PM-HP5 | 5.5–8.5 | resolution: ±0.01 accuracy at pH = 7 ± 0.1 | |||
| Electrochemical | pH Probes | Total scale | n/a | [197] | |
| Hygienic pH Probe for Sterile Applications | Total scale | n/a | [200] | ||
| Bioreactor pH Probe | Total scale | Accuracy: ±0.1 | [199] | ||
| Principle | Sensor | Range/Accuracy | Ref. | |
|---|---|---|---|---|
| Paramagnetic Cells Technology | Paramagnetic O2 Analyser | Different ranges available: 0–2%, 0–10%, 0–30%, 0–100%, 98–100% and 20–22%. | [203] | |
| Optical | Mettler Toledo | Optical Dissolved Oxygen Sensors | 8 ppb to 25 ppm with accuracy ±1% | [204] |
| PreSens Oxygen Sensors | OXY-4 SMA (G3) | 0–100% O2detection limit 15 ppb dissolved oxygen | [205] | |
| Self-adhesive Oxygen Sensor Spot SP-PSt3-SA | 0–100% O2 Dissolved O2: 0–45 mg/L Accuracy ±0.4% O2 at 20.9% O2 | |||
| O2 Flow-Through Cell FTC-PSt3 | Dissolved O2: 0–45 mg/L ± 0.4% O2 at 20.9% O2 | |||
| Electrochemical | Polarographic Dissolved Oxygen Sensors | 0–10.000 ppb Accuracy ± 1% | [204] | |
| Principle | Sensor | Range/Accuracy | Ref. | |
|---|---|---|---|---|
| Optical | PreSens CO2 Sensors | CO2-1 SMA | range: 1–25% accuracy: ±0.06% at 2% CO2, ±0.15% at 6% CO2 | [205] |
| CO2 Sensor Spot SP-CD1 | range: 1–25% accuracy: ±0.06% at 2% CO2, ±0.15 % at 6% CO2 | |||
| CO2 Microsensor IMP-CDM1 | range: 0.04%–5% CO2 accuracy: ±0.01% at 0.1% CO2, ±0.1% at 1% CO2 | |||
| Potentiometric | CO2 Sensor InPro5000i/12/120 | range: 0.145–14.5 psig pCO2 accuracy: ±10 | [211] | |
| Sensor | Frequency Range | Capacity | Conductivity Range | Resolution | Type of BR | Ref. |
|---|---|---|---|---|---|---|
| Standard Remote Futura | 50 kHz–20 MHz | 0–400 pF/cm | 1–40 mS/cm | Bacteria 2 × 109 cells/mL for Escherichia coli Yeast or Animal cells 105 cells/ml | Small bioreactors (up to 100 mL working volume) | [232] |
| Standard Futura | 50 kHz–20 MHz | 0–400 pF/cm | 1–40 mS/cm | Bacteria 2 × 109 cells/mL for E. coli Yeast or Animal cells 105 cells/mL | Suitable for most BRs | [233] |
| BioPAT® ViaMass | 50 kHz–20 MHz | 0–400 pF/cm | 1–40 mS/cm | Yeast Bacteria Plant Cell | Suitable for single-use fermentation bags such as the Flexsafe® RM | [231] |
| i-Biomass | n/a | 0–700 pF/cm | 0.5–100 mS/cm | 105 cell/mL for animal cells | Single-use BR | [234] |
| Glucose Sensors | ||||
|---|---|---|---|---|
| Principle | Structure | Glucose Concentration | Limit of Detection | Ref. |
| Optical | commercially available oxygen sensor that is coated with cross-linked glucose oxidase | 0–20 mM | 0.45 mM | [248] |
| Amperometric | SU-8 pillars with immobilized enzymes | 0–12 mM | n/a | [249] |
| Amperometric | screen-printed sensor modified with cellulose nanocrystals | 0.1–2 mM | 0.004 mM | [252] |
| Electrochemical | nanocrystalline cellulose | 1.0 to 20 mM | 50 ± 10 µM | [253] |
| Electrochemical | zinc oxide nanoparticles on graphene–carbon nanotube | 10 μM to 6.5 mM | 4.5 (±0.08) μM | [254] |
| Electrochemical | three dimensional ordered macroporous self-doped polyaniline/Prussian blue bicomponent film | 2 to 1600 μM | 0.4 μM | [255] |
| Electrochemical | carbon nanotubes | 0.073 to 4 mM | 73 μM | [257] |
| Electrochemical | gold nanoparticles-mesoporous silica composite | 0.02–14 mM | n/a | [262] |
| Electrochemical | glucose oxidase and platinum on mesoporous silica nanoparticles | 0.001–26 mM | 0.2 µM | [263] |
| Amperometric | enzyme electrodes | 0–20 mM | n/a | [264] |
| Amperometric | enzyme-based sensors | 0–20 mM | 0.05 mM | [291] |
| Glutamate Sensors | ||||
| Principle | Structure | Glutamate concentration | Limit of Detection | Ref. |
| Amperometric | glutamate oxidase adsorpted on electrodeposited chitosan | 20–352 μM | 2.5 ± 1.1 μM | [258] |
| Amperometric | crosslinking of glutaraldehyde on platinum microelectrode | 20–217 μM | 6.5 ± 1.7 μM | |
| Amperometric | covalent immobilization of glutamate oxidase on polypyrrole nanoparticles/polyaniline modified gold electrode | 0.02 to 400 μM | 0.1 nM | [265] |
| Electrochemical | l-glutamate oxidase immobilized onto ZnO nanorods/polypyrrole modified pencil graphite electrode | 0.02–500 μM | 0.18 nM | [266] |
| Electrochemical | neurochemical probe | 10–570 µM | 6.3 ± 0.95 µM | [268] |
| Amperometric | enzyme-based sensors | 0–10 mM | 0.05 mM | [291] |
| Lactate Sensors | ||||
| Principle | Structure | Glutamate Concentration | Limit of Detection | Ref. |
| Electrochemical | chitosan/carbon nanotubes modified screen-printed graphite electrodes | 30.4–243.9 µM | 22.6 µM | [269] |
| Amperometric | carbon nanotube | 0.028–2 mM | 28 µM | [257] |
| Amperometric | screen-printed carbon electrode | 18.3 μM–1.5 mM | n/a | [270] |
| Ammonia sensors | ||||
| Principle | Structure | Ammonia Concentration | Limit of Detection | Ref. |
| Optical | SU-8 microfluidic device | 3–70 μM | 2.5 μM | [286] |
| Conductivity | Lab-on-Chip with channel system | 0–234 ppb | 1.1 ppb | [288] |
| Electroosmotic | microfabricated electroosmotic pump coupled to a gas-diffusion microchip | 0.25–5 mg/L | 0.10 mg/L | [289] |
| Optical | microfluidic chip coupled with spectrophotometric method | Ammonium 0.2–50 mg/L | n/a | [287] |
| Optical | flow injection system coupled with spectrophotometric method | Ammonium 50–1000 μg/L | 42 μg/L | [290] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Djisalov, M.; Knežić, T.; Podunavac, I.; Živojević, K.; Radonic, V.; Knežević, N.Ž.; Bobrinetskiy, I.; Gadjanski, I. Cultivating Multidisciplinarity: Manufacturing and Sensing Challenges in Cultured Meat Production. Biology 2021, 10, 204. https://doi.org/10.3390/biology10030204
Djisalov M, Knežić T, Podunavac I, Živojević K, Radonic V, Knežević NŽ, Bobrinetskiy I, Gadjanski I. Cultivating Multidisciplinarity: Manufacturing and Sensing Challenges in Cultured Meat Production. Biology. 2021; 10(3):204. https://doi.org/10.3390/biology10030204
Chicago/Turabian StyleDjisalov, Mila, Teodora Knežić, Ivana Podunavac, Kristina Živojević, Vasa Radonic, Nikola Ž. Knežević, Ivan Bobrinetskiy, and Ivana Gadjanski. 2021. "Cultivating Multidisciplinarity: Manufacturing and Sensing Challenges in Cultured Meat Production" Biology 10, no. 3: 204. https://doi.org/10.3390/biology10030204
APA StyleDjisalov, M., Knežić, T., Podunavac, I., Živojević, K., Radonic, V., Knežević, N. Ž., Bobrinetskiy, I., & Gadjanski, I. (2021). Cultivating Multidisciplinarity: Manufacturing and Sensing Challenges in Cultured Meat Production. Biology, 10(3), 204. https://doi.org/10.3390/biology10030204