Genetic, Epigenetic, Genomic and Microbial Approaches to Enhance Salt Tolerance of Plants: A Comprehensive Review
Abstract
Simple Summary
Abstract
1. Introduction
2. Physiological and Biochemical Basis of Salt Tolerance
2.1. Modulation of Ion Uptake and Transport
2.2. Ion Homeostasis and Compartmentalization
2.3. Synthesis of Osmoprotectants and Antioxidant Compounds
2.4. Regulation of Hormones during Salt Stress
2.5. Activation of Stress-Signaling Pathways
3. The Genetic Basis of Tolerance to Salinity in Plants
Identification and Introgression of QTLs Controlling Salt Tolerance
4. Genomic Approaches for Enhancing Salinity Tolerance
5. Genetic Engineering for Salinity Tolerance in Plants
Genetic Manipulation of Ion Transporters and Other Genes Associated with Salinity Tolerance
6. Genome Editing to Enhance Salt Tolerance in Plants
| Crop Plant Species | Target Genes | Gene Function | References |
|---|---|---|---|
| Arabidopsis (Arabidopsis thaliana) | AITR | ABA-induced transcriptional repressor | [194] |
| CBF | C-repeat binding factor | [195] | |
| SIZ1 | C2H2 type zinc finger protein | [196] | |
| Tomato (Solanum lycopersicum) | SP5G, SP | Day length sensitivity regulators | [197,198] |
| WUS | Act as both transcriptional activator and repressor of genes in the shoot apical meristem | [197] | |
| GGP1 | Vitamin C synthesis | [197] | |
| HKT1;2 | High affinity potassium transporter | [199,200,201] | |
| ARF4 | Auxin signaling | [191] | |
| HyPRP1 | Multistress tolerance | [192,193] | |
| CLV3 | Regulates shoot and floral meristem development | [197,202] | |
| Maize (Zea mays) | HKT1 | High affinity potassium transporter | [203] |
| Rice (Oryza sativa) | DOF15 | Transcription factor | [204] |
| NCA1a, NCA1b | Catalase activity-regulating chaperone | [205] | |
| PQT3 | Ubiquitin ligase | [206] | |
| FLN2 | Involved in sucrose metabolism | [207] | |
| BBS1 | Chaperone-mediated signaling | [208] | |
| NAC041 | Transcription factor | [209] | |
| BG3 | Cytokinin transporter | [210] | |
| MIR528 | Salt stress response regulator | [211] | |
| DST | Zinc finger transcription factor | [212] | |
| SPL10 | Transcription factor | [188] | |
| RR9, RR10 | Cytokinin signaling | [213] | |
| RR22 | Transcription factor | [189,190] | |
| OTS1 | Salt stress response regulator | [189,214] | |
| SAPK1, SAPK2 | ABA signaling regulator | [215] | |
| PIL14 | Transcription factor | [216] | |
| Soybean (Glycine max) | MYB118 | Transcription factor | [217] |
| NAC06 | Transcription factor | [218] |
6.1. Current Challenges and Opportunities with CRISPR-Based Approaches
7. Epigenetic/Epigenomic Approaches to Enhance Salinity Tolerance
7.1. Development of Epigenetic Recombinant Inbred Lines (EpiRILs)
| Recombinant Inbred Lines (RILs) | Epigenetic Recombinant Inbred Lines (epiRILs) | Related References Pertaining to epiRILs |
|---|---|---|
| 1. Mainly vary genetically; each RIL has a different combination of alleles. | 1. Mainly vary for epialleles (variation with respect to epigenetic marks like methylation, acetylation, and others. Each epiRIL has a different combination of epialleles | [250] |
| 2. QTLs governing a trait can be identified and introgressed into a genotype of choice | 2. epiQTLs governing a trait can be identified and introgressed into a genotype of choice | [251] |
| 3. Typically, the parents involved in the generation of RILs are genetically diverse | 3. The parents involved in the generation of epiRILs can be isogenic or near-isogenic, or genetically diverse, but they differ significantly for the epigenome | [250] |
| 4. No need to create/induce specific mutations in parents to create RILs | 4. To create epiRILs, one of the parents should be an epigenetic mutant | [250] |
| 5. In RILs, genetic variation can also bring in some epigenetic variation, particularly when the variation is related to an epigenetic modifier. However, such a variation has not been systematically documented in RILs. | 5. In epiRILs, epigenetic variation can also cause genetic variation by enhancing meiotic crossing over and activation of transposons | [250,252] |
| 6. Most of the genetic variation of RILs is heritable | 6. In epiRILs, some epigenetic variation is heritable (not all) | [250,253,254,255,256] |
7.2. Generation of Epigenetic Variants Using Inhibitors of Epigenetic Modifiers
8. Role of Plant Growth-Promoting Rhizobacteria in Enhancing Salt Tolerance of Plants
8.1. Expression of Key Stress-Inducible Genes
8.2. Modulation of Stress-Induced Compatible Solutes, Phytohormone Homeostasis, and Redox Status of Plants
8.3. Release of Volatile Compounds
9. Conclusions
Supplementary Materials
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Akram, S.; Siddiqui, M.N.; Hussain, B.M.N.; Al Bari, M.A.; Mostofa, M.G.; Hossain, M.A.; Tran, L.-S.P. Exogenous Glutathione Modulates Salinity Tolerance of Soybean [Glycine max (L.) Merrill] at Reproductive Stage. J. Plant Growth Regul. 2017, 36, 877–888. [Google Scholar] [CrossRef]
- Wang, H.; Zhang, M.; Guo, R.; Shi, D.; Liu, B.; Lin, X.; Yang, C. Effects of salt stress on ion balance and nitrogen metabolism of old and young leaves in rice (Oryza sativa L.). BMC Plant Biol. 2012, 12, 194. [Google Scholar] [CrossRef] [PubMed]
- Cheeseman, J.M. The evolution of halophytes, glycophytes and crops, and its implications for food security under saline conditions. New Phytol. 2015, 206, 557–570. [Google Scholar] [CrossRef]
- He, Q.; Altieri, A.H.; Cui, B. Herbivory drives zonation of stress-tolerant marsh plants. Ecology 2015, 96, 1318–1328. [Google Scholar] [CrossRef]
- Khan, W.U.; Ahmad, S.R.; Yasin, N.A.; Ali, A.; Ahmad, A.; Akram, W. Application of Bacillus megaterium MCR-8 improved phytoextraction and stress alleviation of nickel in Vinca rosea. Int. J. Phytoremediat. 2017, 19, 813–824. [Google Scholar] [CrossRef]
- Patil, G.; Valliyodan, B.; Deshmukh, R.; Prince, S.; Nicander, B.; Zhao, M.; Sonah, H.; Song, L.; Lin, L.; Chaudhary, J.; et al. Soybean (Glycine max) SWEET gene family: Insights through comparative genomics, transcriptome profiling and whole genome re-sequence analysis. BMC Genom. 2015, 16, 520. [Google Scholar] [CrossRef]
- van Hoorn, J.W.; Katerji, N.; Hamdy, A.; Mastrorilli, M. Effect of salinity on yield and nitrogen uptake of four grain legumes and on biological nitrogen contribution from the soil. Agric. Water Manag. 2001, 51, 87–98. [Google Scholar] [CrossRef]
- Kumar, A.; Kumar, A.; Kumar, P.; Lata, C.; Kumar, S. Effect of individual and interactive alkalinity and salinity on physiological, biochemical and nutritional traits of Marvel grass. Indian J. Exp. Biol. 2018, 56, 573–581. [Google Scholar]
- Wani, S.H.; Kumar, V.; Shriram, V.; Sah, S.K. Phytohormones and their metabolic engineering for abiotic stress tolerance in crop plants. Crop J. 2016, 4, 162–176. [Google Scholar] [CrossRef]
- Bohra, A.; Chand Jha, U.; Godwin, I.D.; Kumar Varshney, R. Genomic interventions for sustainable agriculture. Plant Biotechnol. J. 2020, 18, 2388–2405. [Google Scholar] [CrossRef] [PubMed]
- Varshney, R.K.; Sinha, P.; Singh, V.K.; Kumar, A.; Zhang, Q.; Bennetzen, J.L. 5Gs for crop genetic improvement. Curr. Opin. Plant Biol. 2020, 56, 190–196. [Google Scholar] [CrossRef] [PubMed]
- Varshney, R.K.; Godwin, I.D.; Mohapatra, T.; Jones, J.D.G.; McCouch, S.R. A SWEET solution to rice blight. Nat. Biotechnol. 2019, 37, 1280–1282. [Google Scholar] [CrossRef]
- Lehmann, J.; Bossio, D.A.; Kögel-Knabner, I.; Rillig, M.C. The concept and future prospects of soil health. Nat. Rev. Earth Environ. 2020, 1, 544–553. [Google Scholar] [CrossRef]
- Kibblewhite, M.G.; Ritz, K.; Swift, M.J. Soil health in agricultural systems. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2008, 363, 685–701. [Google Scholar] [CrossRef]
- Shrivastava, P.; Kumar, R. Soil salinity: A serious environmental issue and plant growth promoting bacteria as one of the tools for its alleviation. Saudi J. Biol. Sci. 2015, 22, 123–131. [Google Scholar] [CrossRef] [PubMed]
- James, R.A.; Blake, C.; Byrt, C.S.; Munns, R. Major genes for Na+ exclusion, Nax1 and Nax2 (wheat HKT1; 4 and HKT1; 5), decrease Na+ accumulation in bread wheat leaves under saline and waterlogged conditions. J. Exp. Bot. 2011, 62, 2939–2947. [Google Scholar] [CrossRef]
- Rahnama, A.; James, R.A.; Poustini, K.; Munns, R. Stomatal conductance as a screen for osmotic stress tolerance in durum wheat growing in saline soil. Funct. Plant Biol. 2010, 37, 255–263. [Google Scholar] [CrossRef]
- Munns, R. Genes and salt tolerance: Bringing them together. New Phytol. 2005, 167, 645–663. [Google Scholar] [CrossRef]
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef]
- Colmer, T.D.; Flowers, T.J.; Munns, R. Use of wild relatives to improve salt tolerance in wheat. J. Exp. Bot. 2006, 57, 1059–1078. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, R.; Morales, P.; Tronchoni, J.; Cordero-Bueso, G.; Vaudano, E.; Quirós, M.; Novo, M.; Torres-Pérez, R.; Valero, E. New Genes Involved in Osmotic Stress Tolerance in Saccharomyces cerevisiae. Front. Microbiol. 2016, 7, 1545. [Google Scholar] [CrossRef]
- Munns, R.; James, R.A.; Läuchli, A. Approaches to increasing the salt tolerance of wheat and other cereals. J. Exp. Bot. 2006, 57, 1025–1043. [Google Scholar] [CrossRef] [PubMed]
- Yeo, A.; Flowers, T. Accumulation and localisation of sodium ions within the shoots of rice (Oryza sativa) varieties differing in salinity resistance. Physiol. Plant 1982, 56, 343–348. [Google Scholar] [CrossRef]
- Yeo, A.; Flowers, T. Salinity resistance in rice (Oryza sativa L.) and a pyramiding approach to breeding varieties for saline soils. Funct. Plant Biol. 1986, 13, 161–173. [Google Scholar] [CrossRef]
- Carden, D.E.; Walker, D.J.; Flowers, T.J.; Miller, A.J. Single-cell measurements of the contributions of cytosolic Na+ and K+ to salt tolerance. Plant Physiol. 2003, 131, 676–683. [Google Scholar] [CrossRef]
- Flowers, T.J.; Colmer, T.D. Salinity tolerance in halophytes. New Phytol. 2008, 179, 945–963. [Google Scholar] [CrossRef]
- Bassil, E.; Ohto, M.-a.; Esumi, T.; Tajima, H.; Zhu, Z.; Cagnac, O.; Belmonte, M.; Peleg, Z.; Yamaguchi, T.; Blumwald, E. The Arabidopsis intracellular Na+/H+ antiporters NHX5 and NHX6 are endosome associated and necessary for plant growth and development. Plant Cell 2011, 23, 224–239. [Google Scholar] [CrossRef]
- Krebs, M.; Beyhl, D.; Görlich, E.; Al-Rasheid, K.A.; Marten, I.; Stierhof, Y.-D.; Hedrich, R.; Schumacher, K. Arabidopsis V-ATPase activity at the tonoplast is required for efficient nutrient storage but not for sodium accumulation. Proc. Natl. Acad. Sci. USA 2010, 107, 3251–3256. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, T.; Hamamoto, S.; Uozumi, N. Sodium transport system in plant cells. Front. Plant Sci. 2013, 4, 410. [Google Scholar] [CrossRef] [PubMed]
- Horie, T.; Hauser, F.; Schroeder, J.I. HKT transporter-mediated salinity resistance mechanisms in Arabidopsis and monocot crop plants. Trends Plant Sci. 2009, 14, 660–668. [Google Scholar] [CrossRef]
- Mäser, P.; Eckelman, B.; Vaidyanathan, R.; Horie, T.; Fairbairn, D.J.; Kubo, M.; Yamagami, M.; Yamaguchi, K.; Nishimura, M.; Uozumi, N. Altered shoot/root Na+ distribution and bifurcating salt sensitivity in Arabidopsis by genetic disruption of the Na+ transporter AtHKT1. FEBS Lett. 2002, 531, 157–161. [Google Scholar] [CrossRef]
- Uozumi, N.; Kim, E.J.; Rubio, F.; Yamaguchi, T.; Muto, S.; Tsuboi, A.; Bakker, E.P.; Nakamura, T.; Schroeder, J.I. The Arabidopsis HKT1 gene homolog mediates inward Na+ currents in Xenopus laevis oocytes and Na+ uptake in Saccharomyces cerevisiae. Plant Physiol. 2000, 122, 1249–1260. [Google Scholar] [CrossRef] [PubMed]
- Rubio, F.; Gassmann, W.; Schroeder, J.I. Sodium-driven potassium uptake by the plant potassium transporter HKT1 and mutations conferring salt tolerance. Science 1995, 270, 1660–1663. [Google Scholar] [CrossRef]
- Dietz, K.-J.; Tavakoli, N.; Kluge, C.; Mimura, T.; Sharma, S.; Harris, G.; Chardonnens, A.; Golldack, D. Significance of the V-type ATPase for the adaptation to stressful growth conditions and its regulation on the molecular and biochemical level. J. Exp. Bot. 2001, 52, 1969–1980. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, J.I.; Ward, J.M.; Gassmann, W. Perspectives on the physiology and structure of inward-rectifying K+ channels in higher plants: Biophysical implications for K+ uptake. Annu. Rev. Biophys. Biomol. Struct. 1994, 23, 441–471. [Google Scholar] [CrossRef] [PubMed]
- Barragán, V.; Leidi, E.O.; Andrés, Z.; Rubio, L.; De Luca, A.; Fernández, J.A.; Cubero, B.; Pardo, J.M. Ion exchangers NHX1 and NHX2 mediate active potassium uptake into vacuoles to regulate cell turgor and stomatal function in Arabidopsis. Plant Cell 2012, 24, 1127–1142. [Google Scholar] [CrossRef] [PubMed]
- Verslues, P.E.; Agarwal, M.; Katiyar-Agarwal, S.; Zhu, J.; Zhu, J.K. Methods and concepts in quantifying resistance to drought, salt and freezing, abiotic stresses that affect plant water status. Plant J. 2006, 45, 523–539. [Google Scholar] [CrossRef]
- Ryu, H.; Cho, Y.-G. Plant hormones in salt stress tolerance. J. Plant Biol. 2015, 58, 147–155. [Google Scholar] [CrossRef]
- Yu, Z.; Duan, X.; Luo, L.; Dai, S.; Ding, Z.; Xia, G. How Plant Hormones Mediate Salt Stress Responses. Trends Plant Sci. 2020, 25, 1117–1130. [Google Scholar] [CrossRef]
- Ahmad, P.; Rasool, S.; Gul, A.; Sheikh, S.A.; Akram, N.A.; Ashraf, M.; Kazi, A.; Gucel, S. Jasmonates: Multifunctional roles in stress tolerance. Front. Plant Sci. 2016, 7, 813. [Google Scholar] [CrossRef]
- Formentin, E.; Barizza, E.; Stevanato, P.; Falda, M.; Massa, F.; Tarkowskà, D.; Novák, O.; Lo Schiavo, F. Fast regulation of hormone metabolism contributes to salt tolerance in rice (Oryza sativa spp. Japonica, L.) by inducing specific morpho-physiological responses. Plants 2018, 7, 75. [Google Scholar] [CrossRef]
- Talla, S.K.; Panigrahy, M.; Kappara, S.; Nirosha, P.; Neelamraju, S.; Ramanan, R. Cytokinin delays dark-induced senescence in rice by maintaining the chlorophyll cycle and photosynthetic complexes. J. Exp. Bot. 2016, 67, 1839–1851. [Google Scholar] [CrossRef]
- Zwack, P.J.; Rashotte, A.M. Interactions between cytokinin signalling and abiotic stress responses. J. Exp. Bot. 2015, 66, 4863–4871. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.K. Abiotic Stress Signaling and Responses in Plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.K. Genetic analysis of plant salt tolerance using Arabidopsis. Plant Physiol. 2000, 124, 941–948. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Qiu, Q.-S.; Quintero, F.J.; Pardo, J.M.; Ohta, M.; Zhang, C.; Schumaker, K.S.; Zhu, J.-K. Transgenic evaluation of activated mutant alleles of SOS2 reveals a critical requirement for its kinase activity and C-terminal regulatory domain for salt tolerance in Arabidopsis thaliana. Plant Cell 2004, 16, 435–449. [Google Scholar] [CrossRef]
- Ji, H.; Pardo, J.M.; Batelli, G.; Van Oosten, M.J.; Bressan, R.A.; Li, X. The Salt Overly Sensitive (SOS) pathway: Established and emerging roles. Mol. Plant 2013, 6, 275–286. [Google Scholar] [CrossRef]
- Shi, H.; Quintero, F.J.; Pardo, J.M.; Zhu, J.-K. The putative plasma membrane Na+/H+ antiporter SOS1 controls long-distance Na+ transport in plants. Plant Cell 2002, 14, 465–477. [Google Scholar] [CrossRef]
- Oh, D.-H.; Lee, S.Y.; Bressan, R.A.; Yun, D.-J.; Bohnert, H.J. Intracellular consequences of SOS1 deficiency during salt stress. J. Exp. Bot. 2010, 61, 1205–1213. [Google Scholar] [CrossRef]
- Rolly, N.K.; Imran, Q.M.; Lee, I.-J.; Yun, B.-W. Salinity stress-mediated suppression of expression of salt overly sensitive signaling pathway genes suggests negative regulation by AtbZIP62 transcription factor in Arabidopsis thaliana. Int. J. Mol. Sci. 2020, 21, 1726. [Google Scholar] [CrossRef]
- Teige, M.; Scheikl, E.; Eulgem, T.; Dóczi, R.; Ichimura, K.; Shinozaki, K.; Dangl, J.L.; Hirt, H. The MKK2 pathway mediates cold and salt stress signaling in Arabidopsis. Mol. Cell 2004, 15, 141–152. [Google Scholar] [CrossRef] [PubMed]
- Thalmann, M.; Pazmino, D.; Seung, D.; Horrer, D.; Nigro, A.; Meier, T.; Kölling, K.; Pfeifhofer, H.W.; Zeeman, S.C.; Santelia, D. Regulation of leaf starch degradation by abscisic acid is important for osmotic stress tolerance in plants. Plant Cell 2016, 28, 1860–1878. [Google Scholar] [CrossRef] [PubMed]
- Kiegerl, S.; Cardinale, F.; Siligan, C.; Gross, A.; Baudouin, E.; Liwosz, A.; Eklöf, S.; Till, S.; Bögre, L.; Hirt, H. SIMKK, a mitogen-activated protein kinase (MAPK) kinase, is a specific activator of the salt stress–induced MAPK, SIMK. Plant Cell 2000, 12, 2247–2258. [Google Scholar] [PubMed]
- Wang, F.; Jing, W.; Zhang, W. The mitogen-activated protein kinase cascade MKK1-MPK4 mediates salt signaling in rice. Plant Sci. 2014, 227, 181–189. [Google Scholar] [CrossRef]
- Solis, C.A.; Yong, M.T.; Vinarao, R.; Jena, K.; Holford, P.; Shabala, L.; Zhou, M.; Shabala, S.; Chen, Z.-H. Back to the wild: On a quest for donors toward salinity tolerant rice. Front. Plant Sci. 2020, 11, 323. [Google Scholar] [CrossRef]
- Krishnamurthy, S.; Pundir, P.; Warraich, A.S.; Rathor, S.; Lokeshkumar, B.; Singh, N.K.; Sharma, P.C. Introgressed saltol QTL lines improves the salinity tolerance in rice at seedling stage. Front. Plant Sci. 2020, 11, 833. [Google Scholar] [CrossRef]
- Thomson, M.J.; de Ocampo, M.; Egdane, J.; Rahman, M.A.; Sajise, A.G.; Adorada, D.L.; Tumimbang-Raiz, E.; Blumwald, E.; Seraj, Z.I.; Singh, R.K. Characterizing the Saltol quantitative trait locus for salinity tolerance in rice. Rice 2010, 3, 148–160. [Google Scholar] [CrossRef]
- Qin, H.; Li, Y.; Huang, R. Advances and challenges in the breeding of salt-tolerant rice. Int. J. Mol. Sci. 2020, 21, 8385. [Google Scholar] [CrossRef] [PubMed]
- Akbar, M.; Yabuno, T.; Nakao, S. Breeding for Saline-resistant Varieties of Rice: I. Variability for Salt Tolerance among Some Rice Varietles. Jpn. J. Breed. 1972, 22, 277–284. [Google Scholar] [CrossRef]
- Flowers, T.; Yeo, A. Variability in the resistance of sodium chloride salinity within rice (Oryza sativa L.) varieties. New Phytol. 1981, 88, 363–373. [Google Scholar] [CrossRef]
- Razzaque, S.; Elias, S.M.; Haque, T.; Biswas, S.; Jewel, G.N.A.; Rahman, S.; Weng, X.; Ismail, A.M.; Walia, H.; Juenger, T.E. Gene expression analysis associated with salt stress in a reciprocally crossed rice population. Sci. Rep. 2019, 9, 1–17. [Google Scholar] [CrossRef]
- Arraouadi, S.; Badri, M.; Abdelly, C.; Huguet, T.; Aouani, M.E. QTL mapping of physiological traits associated with salt tolerance in Medicago truncatula Recombinant Inbred Lines. Genomics 2012, 99, 118–125. [Google Scholar] [CrossRef]
- Genc, Y.; Oldach, K.; Gogel, B.; Wallwork, H.; McDonald, G.K.; Smith, A.B. Quantitative trait loci for agronomic and physiological traits for a bread wheat population grown in environments with a range of salinity levels. Mol. Breed. 2013, 32, 39–59. [Google Scholar] [CrossRef]
- Hamwieh, A.; Xu, D. Conserved salt tolerance quantitative trait locus (QTL) in wild and cultivated soybeans. Breed. Sci. 2008, 58, 355–359. [Google Scholar] [CrossRef]
- Nguyen, V.L.; Ribot, S.A.; Dolstra, O.; Niks, R.E.; Visser, R.G.; van der Linden, C.G. Identification of quantitative trait loci for ion homeostasis and salt tolerance in barley (Hordeum vulgare L.). Mol. Breed. 2013, 31, 137–152. [Google Scholar] [CrossRef]
- Soren, K.R.; Madugula, P.; Kumar, N.; Barmukh, R.; Sengar, M.S.; Bharadwaj, C.; Sharma, P.C.; Singh, S.; Bhandari, A.; Singh, J.; et al. Genetic Dissection and Identification of Candidate Genes for Salinity Tolerance Using Axiom®CicerSNP Array in Chickpea. Int. J. Mol. Sci. 2020, 21, 5058. [Google Scholar] [CrossRef]
- Puram, V.R.R.; Ontoy, J.; Subudhi, P.K. Identification of QTLs for salt tolerance traits and prebreeding lines with enhanced salt tolerance in an introgression line population of rice. Plant Mol. Biol. Report. 2018, 36, 695–709. [Google Scholar] [CrossRef]
- Bimpong, I.K.; Manneh, B.; El-Namaky, R.; Diaw, F.; Amoah, N.K.A.; Sanneh, B.; Ghislain, K.; Sow, A.; Singh, R.; Gregorio, G. Mapping QTLs related to salt tolerance in rice at the young seedling stage using 384-plex single nucleotide polymorphism SNP, marker sets. Mol. Plant Breed. 2014, 5, 47–63. [Google Scholar] [CrossRef]
- Mohammadi, R.; Mendioro, M.S.; Diaz, G.Q.; Gregorio, G.B.; Singh, R.K. Genetic analysis of salt tolerance at seedling and reproductive stages in rice (O ryza sativa). Plant Breed. 2014, 133, 548–559. [Google Scholar] [CrossRef]
- Raghavendra, P.; Kumar, B.D.; Kumar, H.S.; Madhuri, R.; Gangaprasad, S.; Murthy, S.K.; Dhananjaya, B.; Halingali, B.; Hittalmani, S. Exploration of Genetic Diversity in Traditional Landraces of Rice for Yield and Its Attributing Traits under Saline Stress Condition. Int. J. Curr. Microbiol. Appl. Sci. 2018, 7, 3359–3366. [Google Scholar] [CrossRef]
- Rahman, M.A.; Bimpong, I.K.; Bizimana, J.; Pascual, E.D.; Arceta, M.; Swamy, B.M.; Diaw, F.; Rahman, M.S.; Singh, R. Mapping QTLs using a novel source of salinity tolerance from Hasawi and their interaction with environments in rice. Rice 2017, 10, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.K.; Kota, S.; Flowers, T.J. Salt Tolerance in Rice: Seedling and Reproductive Stage QTL Mapping Come of Age. Theor. Appl. Genet. 2021, 134, 3495–3533. [Google Scholar] [CrossRef]
- Bhatt, T.; Sharma, A.; Puri, S.; Minhas, A.P. Salt Tolerance Mechanisms and Approaches: Future Scope of Halotolerant Genes and Rice Landraces. Rice Sci. 2020, 27, 368–383. [Google Scholar] [CrossRef]
- Mansuri, R.M.; Shobbar, Z.-S.; Jelodar, N.B.; Ghaffari, M.; Mohammadi, S.M.; Daryani, P. Salt tolerance involved candidate genes in rice: An integrative meta-analysis approach. BMC Plant Biol. 2020, 20, 1–14. [Google Scholar]
- Bohar, R.; Chitkineni, A.; Varshney, R.K. Genetic molecular markers to accelerate genetic gains in crops. Biotechniques 2020, 69, 158–160. [Google Scholar] [CrossRef] [PubMed]
- Quan, R.; Wang, J.; Hui, J.; Bai, H.; Lyu, X.; Zhu, Y.; Zhang, H.; Zhang, Z.; Li, S.; Huang, R. Improvement of salt tolerance using wild rice genes. Front. Plant Sci. 2018, 8, 2269. [Google Scholar] [CrossRef]
- Asif, M.A.; Schilling, R.K.; Tilbrook, J.; Brien, C.; Dowling, K.; Rabie, H.; Short, L.; Trittermann, C.; Garcia, A.; Barrett-Lennard, E.G.; et al. Mapping of novel salt tolerance QTL in an Excalibur × Kukri doubled haploid wheat population. Theor. Appl. Genet. 2018, 131, 2179–2196. [Google Scholar] [CrossRef]
- Xue, D.; Huang, Y.; Zhang, X.; Wei, K.; Westcott, S.; Li, C.; Chen, M.; Zhang, G.; Lance, R. Identification of QTLs associated with salinity tolerance at late growth stage in barley. Euphytica 2009, 169, 187–196. [Google Scholar] [CrossRef]
- Li, J.; Liu, L.; Bai, Y.; Zhang, P.; Finkers, R.; Du, Y.; Visser, R.G.; van Heusden, A.W. Seedling salt tolerance in tomato. Euphytica 2011, 178, 403–414. [Google Scholar] [CrossRef]
- Vadez, V.; Krishnamurthy, L.; Thudi, M.; Anuradha, C.; Colmer, T.D.; Turner, N.C.; Siddique, K.H.; Gaur, P.M.; Varshney, R.K. Assessment of ICCV 2× JG 62 chickpea progenies shows sensitivity of reproduction to salt stress and reveals QTL for seed yield and yield components. Mol. Breed. 2012, 30, 9–21. [Google Scholar] [CrossRef]
- Lee, G.; Boerma, H.; Villagarcia, M.; Zhou, X.; Carter, T.; Li, Z.; Gibbs, M. A major QTL conditioning salt tolerance in S-100 soybean and descendent cultivars. Theor. Appl. Genet. 2004, 109, 1610–1619. [Google Scholar] [CrossRef]
- Hossain, H.; Rahman, M.; Alam, M.; Singh, R. Mapping of quantitative trait loci associated with reproductive-stage salt tolerance in rice. J. Agron. Crop Sci. 2015, 201, 17–31. [Google Scholar] [CrossRef]
- Kumar, V.; Singh, A.; Mithra, S.A.; Krishnamurthy, S.; Parida, S.K.; Jain, S.; Tiwari, K.K.; Kumar, P.; Rao, A.R.; Sharma, S. Genome-wide association mapping of salinity tolerance in rice (Oryza sativa). DNA Res. 2015, 22, 133–145. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, S.; Sl, K.; Kumar, V.; Singh, B.; Rao, A.; Mithra SV, A.; Rai, V.; Singh, A.K.; Singh, N.K. Mapping QTLs for salt tolerance in rice (Oryza sativa L.) by bulked segregant analysis of recombinant inbred lines using 50K SNP chip. PLoS ONE 2016, 11, e0153610. [Google Scholar] [CrossRef]
- Calapit-Palao, C.D.; Vina, C.B.; Thomson, M.J.; Singh, R.K. QTL identification for reproductive-stage salinity tolerance in rice (Oryza sativa L.). In Proceedings of the SABRAO 13th Congress and International Conference, Bogor, Indonesia, 14–16 September 2015. [Google Scholar]
- De León, J.L.D.; Escoppinichi, R.; Geraldo, N.; Castellanos, T.; Mujeeb-Kazi, A.; Röder, M.S. Quantitative trait loci associated with salinity tolerance in field grown bread wheat. Euphytica 2011, 181, 371–383. [Google Scholar] [CrossRef]
- Asif, M.A.; Garcia, M.; Tilbrook, J.; Brien, C.; Dowling, K.; Berger, B.; Schilling, R.K.; Short, L.; Trittermann, C.; Gilliham, M. Identification of salt tolerance QTL in a wheat RIL mapping population using destructive and non-destructive phenotyping. Funct. Plant Biol. 2020, 48, 131–140. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.; Xu, Y.; Teng, W.; Li, B.; Lin, T. QTLs for seedling traits under salinity stress in hexaploid wheat. Cienc. Rural 2018, 48, 1–9. [Google Scholar] [CrossRef]
- Xu, R.; Wang, J.; Li, C.; Johnson, P.; Lu, C.; Zhou, M. A single locus is responsible for salinity tolerance in a Chinese landrace barley (Hordeum vulgare L.). PLoS ONE 2012, 7, e43079. [Google Scholar] [CrossRef]
- Ma, Y.; Shabala, S.; Li, C.; Liu, C.; Zhang, W.; Zhou, M. Quantitative trait loci for salinity tolerance identified under drained and waterlogged conditions and their association with flowering time in barley (Hordeum vulgare. L). PLoS ONE 2015, 10, e0134822. [Google Scholar] [CrossRef] [PubMed]
- Mwando, E.; Angessa, T.T.; Han, Y.; Zhou, G.; Li, C. Quantitative trait loci mapping for vigour and survival traits of barley seedlings after germinating under salinity stress. Agronomy 2021, 11, 103. [Google Scholar] [CrossRef]
- Singh, A.; Gopalakrishnan, S.; Singh, V.; Prabhu, K.; Mohapatra, T.; Singh, N.; Sharma, T.; Nagarajan, M.; Vinod, K.; Singh, D. Marker assisted selection: A paradigm shift in Basmati breeding. Indian J. Genet. Plant Breed. 2011, 71, 120. [Google Scholar]
- Singh, R.; Singh, Y.; Xalaxo, S.; Verulkar, S.; Yadav, N.; Singh, S.; Singh, N.; Prasad, K.; Kondayya, K.; Rao, P.R. From QTL to variety-harnessing the benefits of QTLs for drought, flood and salt tolerance in mega rice varieties of India through a multi-institutional network. Plant Sci. 2016, 242, 278–287. [Google Scholar] [CrossRef] [PubMed]
- Huyen, L.T.N.; Cuc, L.M.; Ham, L.; Khanh, T. Introgression the SALTOL QTL into Q5DB, the elite variety of Vietnam using marker-assisted-selection (MAS). Am. J. BioSci. 2013, 1, 80–84. [Google Scholar] [CrossRef]
- Huyen, L.T.; Cuc, L.M.; Ismail, A.M.; Ham, L.H. Introgression the salinity tolerance QTLs Saltol into AS996, the elite rice variety of Vietnam. Am. J. Plant Sci. 2012, 3, 981–987. [Google Scholar] [CrossRef]
- Linh, L.H.; Linh, T.H.; Xuan, T.D.; Ham, L.H.; Ismail, A.M.; Khanh, T.D. Molecular breeding to improve salt tolerance of rice (Oryza sativa L.) in the Red River Delta of Vietnam. Int. J. Plant Genom. 2012, 2012, 949038. [Google Scholar] [CrossRef] [PubMed]
- Vu, H.T.T.; Le, D.D.; Ismail, A.M.; Le, H.H. Marker-assisted backcrossing (MABC) for improved salinity tolerance in rice (Oryza sativa L.) to cope with climate change in Vietnam. Aust. J. Crop Sci. 2012, 6, 1649–1654. [Google Scholar]
- Hoque, A.; Haque, M.A.; Sarker, M.R.A.; Rahman, M.A. Marker-assisted introgression of saltol locus into genetic background of BRRI Dhan-49. Int. J. Biosci. 2015, 6, 71–80. [Google Scholar]
- Usatov, A.; Alabushev, A.; Kostylev, P.; Azarin, K.; Makarenko, M.; Usatova, O. Introgression the saltol QTL into the elite rice variety of Russia by marker-assisted selection. Am. J. Agric. Biol. Sci. 2015, 10, 165–169. [Google Scholar] [CrossRef]
- Gorham, J.; Hardy, C.; Jones, R.W.; Joppa, L.; Law, C. Chromosomal location of a K/Na discrimination character in the D genome of wheat. Theor. Appl. Genet. 1987, 74, 584–588. [Google Scholar] [CrossRef]
- Masoudi, B.; Mardi, M.; Hervan, E.M.; Bihamta, M.R.; Naghavi, M.R.; Nakhoda, B.; Amini, A. QTL mapping of salt tolerance traits with different effects at the seedling stage of bread wheat. Plant Mol. Biol. Rep. 2015, 33, 1790–1803. [Google Scholar] [CrossRef]
- Huang, S.; Spielmeyer, W.; Lagudah, E.S.; James, R.A.; Platten, J.D.; Dennis, E.S.; Munns, R. A sodium transporter (HKT7) is a candidate for Nax1, a gene for salt tolerance in durum wheat. Plant Physiol. 2006, 142, 1718–1727. [Google Scholar] [CrossRef] [PubMed]
- Lindsay, M.P.; Lagudah, E.S.; Hare, R.A.; Munns, R. A locus for sodium exclusion (Nax1), a trait for salt tolerance, mapped in durum wheat. Funct. Plant Biol. 2004, 31, 1105–1114. [Google Scholar] [CrossRef]
- Byrt, C.S.; Platten, J.D.; Spielmeyer, W.; James, R.A.; Lagudah, E.S.; Dennis, E.S.; Tester, M.; Munns, R. HKT1; 5-like cation transporters linked to Na+ exclusion loci in wheat, Nax2 and Kna1. Plant Physiol. 2007, 143, 1918–1928. [Google Scholar] [CrossRef]
- James, R.A.; Davenport, R.J.; Munns, R. Physiological characterization of two genes for Na+ exclusion in durum wheat, Nax1 and Nax2. Plant Physiol. 2006, 142, 1537–1547. [Google Scholar] [CrossRef] [PubMed]
- Smýkal, P.; Nelson, M.; Berger, J.; Von Wettberg, E. The Impact of Genetic Changes during Crop Domestication. Agronomy 2018, 8, 119. [Google Scholar] [CrossRef]
- McCouch, S.; Navabi, Z.K.; Abberton, M.; Anglin, N.L.; Barbieri, R.L.; Baum, M.; Bett, K.; Booker, H.; Brown, G.L.; Bryan, G.J.; et al. Mobilizing Crop Biodiversity. Mol. Plant 2020, 13, 1341–1344. [Google Scholar] [CrossRef] [PubMed]
- Varshney, R.K.; Nayak, S.N.; May, G.D.; Jackson, S.A. Next-generation sequencing technologies and their implications for crop genetics and breeding. Trends Biotechnol. 2009, 27, 522–530. [Google Scholar] [CrossRef]
- Rhee, M.; Burns, M.A. Nanopore sequencing technology: Nanopore preparations. Trends Biotechnol. 2007, 25, 174–181. [Google Scholar] [CrossRef]
- Eid, J.; Fehr, A.; Gray, J.; Luong, K.; Lyle, J.; Otto, G.; Peluso, P.; Rank, D.; Baybayan, P.; Bettman, B.; et al. Real-time DNA sequencing from single polymerase molecules. Science 2009, 323, 133–138. [Google Scholar] [CrossRef]
- Varshney, R.K.; Thudi, M.; Roorkiwal, M.; He, W.; Upadhyaya, H.D.; Yang, W.; Bajaj, P.; Cubry, P.; Rathore, A.; Jian, J.; et al. Resequencing of 429 chickpea accessions from 45 countries provides insights into genome diversity, domestication and agronomic traits. Nat. Genet. 2019, 51, 857–864. [Google Scholar] [CrossRef]
- Thudi, M.; Palakurthi, R.; Schnable, J.C.; Chitikineni, A.; Dreisigacker, S.; Mace, E.; Srivastava, R.K.; Satyavathi, C.T.; Odeny, D.; Tiwari, V.K.; et al. Genomic resources in plant breeding for sustainable agriculture. J. Plant Physiol. 2021, 257, 153351. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Mauleon, R.; Hu, Z.; Chebotarov, D.; Tai, S.; Wu, Z.; Li, M.; Zheng, T.; Fuentes, R.R.; Zhang, F.; et al. Genomic variation in 3,010 diverse accessions of Asian cultivated rice. Nature 2018, 557, 43–49. [Google Scholar] [CrossRef]
- Milner, S.G.; Jost, M.; Taketa, S.; Mazón, E.R.; Himmelbach, A.; Oppermann, M.; Weise, S.; Knüpffer, H.; Basterrechea, M.; König, P.; et al. Genebank genomics highlights the diversity of a global barley collection. Nat. Genet. 2019, 51, 319–326. [Google Scholar] [CrossRef] [PubMed]
- Varshney, R.K.; Saxena, R.K.; Upadhyaya, H.D.; Khan, A.W.; Yu, Y.; Kim, C.; Rathore, A.; Kim, D.; Kim, J.; An, S.; et al. Whole-genome resequencing of 292 pigeonpea accessions identifies genomic regions associated with domestication and agronomic traits. Nat. Genet. 2017, 49, 1082–1088. [Google Scholar] [CrossRef]
- Bhandari, A.; Sandhu, N.; Bartholome, J.; Cao-Hamadoun, T.-V.; Ahmadi, N.; Kumari, N.; Kumar, A. Genome-Wide Association Study for Yield and Yield Related Traits under Reproductive Stage Drought in a Diverse indica-aus Rice Panel. Rice 2020, 13, 53. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Guo, Z.; Lv, Y.; Cen, X.; Ding, X.; Wu, H.; Li, X.; Huang, J.; Xiong, L. Genetic control of the root system in rice under normal and drought stress conditions by genome-wide association study. PLoS Genet. 2017, 13, e1006889. [Google Scholar] [CrossRef]
- Li, L.; Mao, X.; Wang, J.; Chang, X.; Reynolds, M.; Jing, R. Genetic dissection of drought and heat-responsive agronomic traits in wheat. Plant Cell Environ. 2019, 42, 2540–2553. [Google Scholar] [CrossRef] [PubMed]
- Mao, H.; Wang, H.; Liu, S.; Li, Z.; Yang, X.; Yan, J.; Li, J.; Tran, L.-S.P.; Qin, F. A transposable element in a NAC gene is associated with drought tolerance in maize seedlings. Nat. Commun. 2015, 6, 8326. [Google Scholar] [CrossRef]
- Xiao, Y.; Liu, H.; Wu, L.; Warburton, M.; Yan, J. Genome-wide Association Studies in Maize: Praise and Stargaze. Mol. Plant 2017, 10, 359–374. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, H.; Li, L.; Lan, H.; Ren, Z.; Liu, D.; Wu, L.; Liu, H.; Jaqueth, J.; Li, B.; et al. Characterizing the population structure and genetic diversity of maize breeding germplasm in Southwest China using genome-wide SNP markers. BMC Genom. 2016, 17, 697. [Google Scholar] [CrossRef]
- Hoyos-Villegas, V.; Song, Q.; Kelly, J.D. Genome-wide Association Analysis for Drought Tolerance and Associated Traits in Common Bean. Plant Genome 2017, 10, 1–17. [Google Scholar] [CrossRef]
- Bevan, M.W.; Uauy, C.; Wulff, B.B.H.; Zhou, J.; Krasileva, K.; Clark, M.D. Genomic innovation for crop improvement. Nature 2017, 543, 346–354. [Google Scholar] [CrossRef]
- Shi, Y.; Gao, L.; Wu, Z.; Zhang, X.; Wang, M.; Zhang, C.; Zhang, F.; Zhou, Y.; Li, Z. Genome-wide association study of salt tolerance at the seed germination stage in rice. BMC Plant Biol. 2017, 17, 92. [Google Scholar] [CrossRef]
- Yu, J.; Zhao, W.; Tong, W.; He, Q.; Yoon, M.Y.; Li, F.P.; Choi, B.; Heo, E.B.; Kim, K.W.; Park, Y.J. A Genome-Wide Association Study Reveals Candidate Genes Related to Salt Tolerance in Rice (Oryza sativa) at the Germination Stage. Int. J. Mol. Sci. 2018, 19, 3145. [Google Scholar] [CrossRef]
- Dilnur, T.; Peng, Z.; Pan, Z.; Palanga, K.K.; Jia, Y.; Gong, W.; Du, X. Association analysis of salt tolerance in Asiatic cotton (Gossypium arboretum) with SNP markers. Int. J. Mol. Sci. 2019, 20, 2168. [Google Scholar] [CrossRef]
- Hazzouri, K.M.; Khraiwesh, B.; Amiri, K.M.A.; Pauli, D.; Blake, T.; Shahid, M.; Mullath, S.K.; Nelson, D.; Mansour, A.L.; Salehi-Ashtiani, K.; et al. Mapping of HKT1;5 Gene in Barley Using GWAS Approach and Its Implication in Salt Tolerance Mechanism. Front. Plant Sci. 2018, 9, 156. [Google Scholar] [CrossRef] [PubMed]
- Wan, H.; Chen, L.; Guo, J.; Li, Q.; Wen, J.; Yi, B.; Ma, C.; Tu, J.; Fu, T.; Shen, J. Genome-wide association study reveals the genetic architecture underlying salt tolerance-related traits in rapeseed (Brassica napus L.). Front. Plant Sci. 2017, 8, 593. [Google Scholar] [CrossRef] [PubMed]
- Hussain, B.; Lucas, S.J.; Ozturk, L.; Budak, H. Mapping QTLs conferring salt tolerance and micronutrient concentrations at seedling stage in wheat. Sci. Rep. 2017, 7, 15662. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.; Zhao, Y.; Zhang, R.; Xing, J.; Duan, M.; Li, J.; Wang, N.; Wang, W.; Zhang, S.; Chen, Z.; et al. Mapping of a major QTL for salt tolerance of mature field-grown maize plants based on SNP markers. BMC Plant Biol. 2017, 17, 140. [Google Scholar] [CrossRef]
- Pushpavalli, R.; Krishnamurthy, L.; Thudi, M.; Gaur, P.M.; Rao, M.V.; Siddique, K.H.; Colmer, T.D.; Turner, N.C.; Varshney, R.K.; Vadez, V. Two key genomic regions harbour QTLs for salinity tolerance in ICCV 2× JG 11 derived chickpea (Cicer arietinum L.) recombinant inbred lines. BMC Plant Biol. 2015, 15, 124. [Google Scholar] [CrossRef]
- Lang, L.; Xu, A.; Ding, J.; Zhang, Y.; Zhao, N.; Tian, Z.; Liu, Y.; Wang, Y.; Liu, X.; Liang, F.; et al. Quantitative Trait Locus Mapping of Salt Tolerance and Identification of Salt-Tolerant Genes in Brassica napus L. Front. Plant Sci. 2017, 8, 1000. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.E.; Verbyla, K.L.; Verbyla, A.P.; Raghavan, C.; Singh, V.K.; Gaur, P.; Leung, H.; Varshney, R.K.; Cavanagh, C.R. MAGIC populations in crops: Current status and future prospects. Theor. Appl. Genet. 2015, 128, 999–1017. [Google Scholar] [CrossRef]
- Kover, P.X.; Valdar, W.; Trakalo, J.; Scarcelli, N.; Ehrenreich, I.M.; Purugganan, M.D.; Durrant, C.; Mott, R. A Multiparent Advanced Generation Inter-Cross to Fine-Map Quantitative Traits in Arabidopsis thaliana. PLoS Genet. 2009, 5, e1000551. [Google Scholar] [CrossRef] [PubMed]
- Garg, R.; Shankar, R.; Thakkar, B.; Kudapa, H.; Krishnamurthy, L.; Mantri, N.; Varshney, R.K.; Bhatia, S.; Jain, M. Transcriptome analyses reveal genotype-and developmental stage-specific molecular responses to drought and salinity stresses in chickpea. Sci. Rep. 2016, 6, 19228. [Google Scholar] [CrossRef]
- Mantri, N.L.; Ford, R.; Coram, T.E.; Pang, E.C. Transcriptional profiling of chickpea genes differentially regulated in response to high-salinity, cold and drought. BMC Genom. 2007, 8, 303. [Google Scholar] [CrossRef]
- Singh, D.; Singh, C.K.; Taunk, J.; Sharma, S.; Gaikwad, K.; Singh, V.; Sanwal, S.K.; Singh, D.; Sharma, P.C.; Pal, M. Transcriptome skimming of lentil (Lens culinaris Medikus) cultivars with contrast reaction to salt stress. Funct. Amp. Integr. Genom. 2021, 21, 139–156. [Google Scholar] [CrossRef] [PubMed]
- Subudhi, P.K.; Shankar, R.; Jain, M. Whole genome sequence analysis of rice genotypes with contrasting response to salinity stress. Sci. Rep. 2020, 10, 21259. [Google Scholar] [CrossRef]
- Luo, M.; Zhao, Y.; Wang, Y.; Shi, Z.; Zhang, P.; Zhang, Y.; Song, W.; Zhao, J. Comparative Proteomics of Contrasting Maize Genotypes Provides Insights into Salt-Stress Tolerance Mechanisms. J. Proteome Res. 2018, 17, 141–153. [Google Scholar] [CrossRef]
- Yu, H.; Du, Q.; Campbell, M.; Yu, B.; Walia, H.; Zhang, C. Genome-wide discovery of natural variation in pre-mRNA splicing and prioritising causal alternative splicing to salt stress response in rice. New Phytol. 2021, 230, 1273–1287. [Google Scholar] [CrossRef]
- Meuwissen, T.H.; Hayes, B.J.; Goddard, M.E. Prediction of total genetic value using genome-wide dense marker maps. Genetics 2001, 157, 1819–1829. [Google Scholar] [CrossRef]
- Crossa, J.; Pérez-Rodríguez, P.; Cuevas, J.; Montesinos-López, O.; Jarquín, D.; de Los Campos, G.; Burgueño, J.; González-Camacho, J.M.; Pérez-Elizalde, S.; Beyene, Y.; et al. Genomic Selection in Plant Breeding: Methods, Models, and Perspectives. Trends Plant Sci. 2017, 22, 961–975. [Google Scholar] [CrossRef]
- Atanda, S.A.; Olsen, M.; Burgueño, J.; Crossa, J.; Dzidzienyo, D.; Beyene, Y.; Gowda, M.; Dreher, K.; Zhang, X.; Prasanna, B.M.; et al. Maximizing efficiency of genomic selection in CIMMYT’s tropical maize breeding program. Theor. Appl. Genet. 2021, 134, 279–294. [Google Scholar] [CrossRef] [PubMed]
- Juliana, P.; Singh, R.P.; Braun, H.-J.; Huerta-Espino, J.; Crespo-Herrera, L.; Govindan, V.; Mondal, S.; Poland, J.; Shrestha, S. Genomic Selection for Grain Yield in the CIMMYT Wheat Breeding Program—Status and Perspectives. Front. Plant Sci. 2020, 11, 1418. [Google Scholar] [CrossRef]
- Veenstra, L.D.; Poland, J.; Jannink, J.L.; Sorrells, M.E. Recurrent genomic selection for wheat grain fructans. Crop Sci. 2020, 60, 1499–1512. [Google Scholar] [CrossRef]
- Cui, Y.; Li, R.; Li, G.; Zhang, F.; Zhu, T.; Zhang, Q.; Ali, J.; Li, Z.; Xu, S. Hybrid breeding of rice via genomic selection. Plant Biotechnol. J. 2020, 18, 57–67. [Google Scholar] [CrossRef] [PubMed]
- Spindel, J.; Iwata, H. Genomic Selection in Rice Breeding; Springer: Singapore, 2018. [Google Scholar] [CrossRef]
- Ankamah-Yeboah, T.; Janss, L.L.; Jensen, J.D.; Hjortshøj, R.L.; Rasmussen, S.K. Genomic Selection Using Pedigree and Marker-by-Environment Interaction for Barley Seed Quality Traits From Two Commercial Breeding Programs. Front. Plant Sci. 2020, 11, 539. [Google Scholar] [CrossRef] [PubMed]
- Thorwarth, P.; Ahlemeyer, J.; Bochard, A.M.; Krumnacker, K.; Blümel, H.; Laubach, E.; Knöchel, N.; Cselényi, L.; Ordon, F.; Schmid, K.J. Genomic prediction ability for yield-related traits in German winter barley elite material. Appl. Genet. 2017, 130, 1669–1683. [Google Scholar] [CrossRef] [PubMed]
- Tiede, T.; Smith, K.P. Evaluation and retrospective optimization of genomic selection for yield and disease resistance in spring barley. Mol. Breed. 2018, 38, 55. [Google Scholar] [CrossRef]
- Roorkiwal, M.; Jarquin, D.; Singh, M.K.; Gaur, P.M.; Bharadwaj, C.; Rathore, A.; Howard, R.; Srinivasan, S.; Jain, A.; Garg, V.; et al. Genomic-enabled prediction models using multi-environment trials to estimate the effect of genotype × environment interaction on prediction accuracy in chickpea. Sci. Rep. 2018, 8, 11701. [Google Scholar] [CrossRef]
- Roorkiwal, M.; Rathore, A.; Das, R.R.; Singh, M.K.; Jain, A.; Srinivasan, S.; Gaur, P.M.; Chellapilla, B.; Tripathi, S.; Li, Y.; et al. Genome-Enabled Prediction Models for Yield Related Traits in Chickpea. Front. Plant Sci. 2016, 7, 1666. [Google Scholar] [CrossRef]
- Santantonio, N.; Atanda, S.A.; Beyene, Y.; Varshney, R.K.; Olsen, M.; Jones, E.; Roorkiwal, M.; Gowda, M.; Bharadwaj, C.; Gaur, P.M.; et al. Strategies for Effective Use of Genomic Information in Crop Breeding Programs Serving Africa and South Asia. Front. Plant Sci. 2020, 11, 353. [Google Scholar] [CrossRef]
- Pandey, M.K.; Chaudhari, S.; Jarquin, D.; Janila, P.; Crossa, J.; Patil, S.C.; Sundravadana, S.; Khare, D.; Bhat, R.S.; Radhakrishnan, T.; et al. Genome-based trait prediction in multi- environment breeding trials in groundnut. Theor. Appl. Genet. 2020, 133, 3101–3117. [Google Scholar] [CrossRef]
- Chiurugwi, T.; Kemp, S.; Powell, W.; Hickey, L.T. Speed breeding orphan crops. Theor. Appl. Genet. 2019, 132, 607–616. [Google Scholar] [CrossRef] [PubMed]
- Hickey, L.T.; Hafeez, A.N.; Robinson, H.; Jackson, S.A.; Leal-Bertioli, S.C.M.; Tester, M.; Gao, C.; Godwin, I.D.; Hayes, B.J.; Wulff, B.B.H. Breeding crops to feed 10 billion. Nat. Biotechnol. 2019, 37, 744–754. [Google Scholar] [CrossRef]
- Watson, A.; Ghosh, S.; Williams, M.J.; Cuddy, W.S.; Simmonds, J.; Rey, M.-D.; Hatta, M.A.M.; Hinchliffe, A.; Steed, A.; Reynolds, D. Speed breeding is a powerful tool to accelerate crop research and breeding. Nat. Plants 2018, 4, 23–29. [Google Scholar] [CrossRef]
- Mann, A.; Kumar, N.; Lata, C.; Kumar, A.; Kumar, A.; Meena, B. Functional annotation of differentially expressed genes under salt stress in Dichanthium annulatum. Plant Physiol. Rep. 2019, 24, 104–111. [Google Scholar] [CrossRef]
- Joshi, R.; Wani, S.H.; Singh, B.; Bohra, A.; Dar, Z.A.; Lone, A.A.; Pareek, A.; Singla-Pareek, S.L. Transcription Factors and Plants Response to Drought Stress: Current Understanding and Future Directions. Front. Plant Sci. 2016, 7, 1029. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Xu, L.; Wang, Y.; Luo, X.; Zhu, X.; Kinuthia, K.B.; Nie, S.; Feng, H.; Li, C.; Liu, L. Transcriptome-based gene expression profiling identifies differentially expressed genes critical for salt stress response in radish (Raphanus sativus L.). Plant Cell Rep. 2016, 35, 329–346. [Google Scholar] [CrossRef] [PubMed]
- Kotula, L.; Garcia Caparros, P.; Zörb, C.; Colmer, T.D.; Flowers, T.J. Improving crop salt tolerance using transgenic approaches: An update and physiological analysis. Plant Cell Environ. 2020, 43, 2932–2956. [Google Scholar] [CrossRef] [PubMed]
- Peleg, Z.; Apse, M.P.; Blumwald, E. Engineering salinity and water-stress tolerance in crop plants: Getting closer to the field. Adv. Bot. Res. 2011, 57, 405–443. [Google Scholar]
- Wani, S.H.; Kumar, V.; Khare, T.; Guddimalli, R.; Parveda, M.; Solymosi, K.; Suprasanna, P.; Kavi Kishor, P.B. Engineering salinity tolerance in plants: Progress and prospects. Planta 2020, 251, 76. [Google Scholar] [CrossRef]
- Varshney, R.K.; Bansal, K.C.; Aggarwal, P.K.; Datta, S.K.; Craufurd, P.Q. Agricultural biotechnology for crop improvement in a variable climate: Hope or hype? Trends Plant Sci. 2011, 16, 363–371. [Google Scholar] [CrossRef] [PubMed]
- Hima Kumari, P.; Anil Kumar, S.; Ramesh, K.; Sudhakar Reddy, P.; Nagaraju, M.; Bhanu Prakash, A.; Shah, T.; Henderson, A.; Srivastava, R.K.; Rajasheker, G. Genome-wide identification and analysis of Arabidopsis sodium proton antiporter (NHX) and human sodium proton exchanger (NHE) homologs in sorghum bicolor. Genes 2018, 9, 236. [Google Scholar] [CrossRef]
- Cao, Y.; Jin, X.; Huang, H.; Derebe, M.G.; Levin, E.J.; Kabaleeswaran, V.; Pan, Y.; Punta, M.; Love, J.; Weng, J.; et al. Crystal structure of a potassium ion transporter, TrkH. Nature 2011, 471, 336–340. [Google Scholar] [CrossRef]
- Gouiaa, S.; Khoudi, H. Co-expression of vacuolar Na+/H+ antiporter and H+-pyrophosphatase with an IRES-mediated dicistronic vector improves salinity tolerance and enhances potassium biofortification of tomato. Phytochemistry 2015, 117, 537–546. [Google Scholar] [CrossRef]
- He, X.; Huang, X.; Shen, Y.; Huang, Z. Wheat V-H+-ATPase subunit genes significantly affect salt tolerance in Arabidopsis thaliana. PLoS ONE 2014, 9, e86982. [Google Scholar] [CrossRef][Green Version]
- Liu, J.; Hua, W.; Zhan, G.; Wei, F.; Wang, X.; Liu, G.; Wang, H. Increasing seed mass and oil content in transgenic Arabidopsis by the overexpression of wri1-like gene from Brassica napus. Plant Physiol. Biochem. 2010, 48, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Shen, G.; Wei, J.; Qiu, X.; Hu, R.; Kuppu, S.; Auld, D.; Blumwald, E.; Gaxiola, R.; Payton, P.; Zhang, H. Co-overexpression of AVP1 and AtNHX1 in cotton further improves drought and salt tolerance in transgenic cotton plants. Plant Mol. Biol. Rep. 2015, 33, 167–177. [Google Scholar] [CrossRef]
- Yang, Q.; Chen, Z.Z.; Zhou, X.F.; Yin, H.B.; Li, X.; Xin, X.F.; Hong, X.H.; Zhu, J.K.; Gong, Z. Overexpression of SOS (Salt Overly Sensitive) genes increases salt tolerance in transgenic Arabidopsis. Mol. Plant 2009, 2, 22–31. [Google Scholar] [CrossRef]
- Do, T.D.; Chen, H.; Hien, V.T.T.; Hamwieh, A.; Yamada, T.; Sato, T.; Yan, Y.; Cong, H.; Shono, M.; Suenaga, K. Ncl Synchronously Regulates Na+, K+ and Cl− in Soybean and Greatly Increases the Grain Yield in Saline Field Conditions. Sci. Rep. 2016, 6, 19147. [Google Scholar] [CrossRef]
- Baisakh, N.; RamanaRao, M.V.; Rajasekaran, K.; Subudhi, P.; Janda, J.; Galbraith, D.; Vanier, C.; Pereira, A. Enhanced salt stress tolerance of rice plants expressing a vacuolar H+ -ATPase subunit c1 (SaVHAc1) gene from the halophyte grass Spartina alterniflora Löisel. Plant Biotechnol. J. 2012, 10, 453–464. [Google Scholar] [CrossRef]
- Mansour, M.M.F. The plasma membrane transport systems and adaptation to salinity. J. Plant Physiol. 2014, 171, 1787–1800. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Wan, S.; Jiang, Y.; Xia, Y.; Chen, X.; Gao, M.; Cao, Y.; Luo, Y.; Zhou, Y.; Jiang, X. Over-Expression of a plasma membrane H+-ATPase SpAHA1 conferred salt tolerance to transgenic Arabidopsis. Protoplasma 2018, 255, 1827–1837. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Yin, X.; Xie, Q.; Xia, Y.; Wang, Z.; Song, J.; Zhou, Y.; Jiang, X. Co-expression of SpSOS1 and SpAHA1 in transgenic Arabidopsis plants improves salinity tolerance. BMC Plant Biol. 2019, 19, 74. [Google Scholar] [CrossRef]
- Yang, H.; Deng, L.; Liu, H.; Fan, S.; Hua, W.; Liu, J. Overexpression of BnaAOX1b Confers Tolerance to Osmotic and Salt Stress in Rapeseed. G3 Genes Genomes Genet. 2019, 9, 3501–3511. [Google Scholar] [CrossRef] [PubMed]
- Yoon, Y.; Seo, D.H.; Shin, H.; Kim, H.J.; Kim, C.M.; Jang, G. The Role of Stress-Responsive Transcription Factors in Modulating Abiotic Stress Tolerance in Plants. Agronomy 2020, 10, 788. [Google Scholar] [CrossRef]
- Schmidt, R.; Mieulet, D.; Hubberten, H.M.; Obata, T.; Hoefgen, R.; Fernie, A.R.; Fisahn, J.; San Segundo, B.; Guiderdoni, E.; Schippers, J.H.; et al. Salt-responsive ERF1 regulates reactive oxygen species-dependent signaling during the initial response to salt stress in rice. Plant Cell 2013, 25, 2115–2131. [Google Scholar] [CrossRef]
- Jin, X.; Xue, Y.; Wang, R.; Xu, R.; Bian, L.; Zhu, B.; Han, H.; Peng, R.; Yao, Q. Transcription factor OsAP21 gene increases salt/drought tolerance in transgenic Arabidopsis thaliana. Mol. Biol. Rep. 2013, 40, 1743–1752. [Google Scholar] [CrossRef]
- Maheshwari, P.; Kiran, B.; Punita, D.L.; Kavi Kishor, P.B. Overexpression of SbAP37 in rice alleviates concurrent imposition of combination stresses and modulates different sets of leaf protein profiles. Plant Cell Rep. 2017, 36, 773–786. [Google Scholar] [CrossRef]
- Nguyen, Q.H.; Vu, L.T.K.; Nguyen, L.T.N.; Pham, N.T.T.; Nguyen, Y.T.H.; Le, S.V.; Chu, M.H. Overexpression of the GmDREB6 gene enhances proline accumulation and salt tolerance in genetically modified soybean plants. Sci. Rep. 2019, 9, 19663. [Google Scholar] [CrossRef]
- Tang, Y.; Bao, X.; Zhi, Y.; Wu, Q.; Guo, Y.; Yin, X.; Zeng, L.; Li, J.; Zhang, J.; He, W.; et al. Overexpression of a MYB Family Gene, OsMYB6, Increases Drought and Salinity Stress Tolerance in Transgenic Rice. Front. Plant Sci. 2019, 10, 168. [Google Scholar] [CrossRef]
- Kamburova, V.S.; Nikitina, E.V.; Shermatov, S.E.; Buriev, Z.T.; Kumpatla, S.P.; Emani, C.; Abdurakhmonov, I.Y. Genome Editing in Plants: An Overview of Tools and Applications. Int. J. Agron. 2017, 2017, 7315351. [Google Scholar] [CrossRef]
- Zhu, C.; Bortesi, L.; Baysal, C.; Twyman, R.M.; Fischer, R.; Capell, T.; Schillberg, S.; Christou, P. Characteristics of Genome Editing Mutations in Cereal Crops. Trends Plant Sci. 2017, 22, 38–52. [Google Scholar] [CrossRef]
- Debbarma, J.; Sarki, Y.N.; Saikia, B.; Boruah, H.P.D.; Singha, D.L.; Chikkaputtaiah, C. Ethylene Response Factor (ERF) Family Proteins in Abiotic Stresses and CRISPR-Cas9 Genome Editing of ERFs for Multiple Abiotic Stress Tolerance in Crop Plants: A Review. Mol. Biotechnol. 2019, 61, 153–172. [Google Scholar] [CrossRef]
- Hussain, B.; Lucas, S.J.; Budak, H. CRISPR/Cas9 in plants: At play in the genome and at work for crop improvement. Brief. Funct. Genom. 2018, 17, 319–328. [Google Scholar] [CrossRef]
- Lan, T.; Zheng, Y.; Su, Z.; Yu, S.; Song, H.; Zheng, X.; Lin, G.; Wu, W. OsSPL10, a SBP-Box Gene, Plays a Dual Role in Salt Tolerance and Trichome Formation in Rice (Oryza sativa L.). G3 Genes Genomes Genet 2019, 9, 4107–4114. [Google Scholar] [CrossRef]
- Zhang, A.; Liu, Y.; Wang, F.; Li, T.; Chen, Z.; Kong, D.; Bi, J.; Zhang, F.; Luo, X.; Wang, J.; et al. Enhanced rice salinity tolerance via CRISPR/Cas9-targeted mutagenesis of the OsRR22 gene. Mol. Breed. 2019, 39, 47. [Google Scholar] [CrossRef] [PubMed]
- Takagi, H.; Tamiru, M.; Abe, A.; Yoshida, K.; Uemura, A.; Yaegashi, H.; Obara, T.; Oikawa, K.; Utsushi, H.; Kanzaki, E.; et al. MutMap accelerates breeding of a salt-tolerant rice cultivar. Nat. Biotechnol. 2015, 33, 445–449. [Google Scholar] [CrossRef] [PubMed]
- Bouzroud, S.; Gasparini, K.; Hu, G.; Barbosa, M.A.M.; Rosa, B.L.; Fahr, M.; Bendaou, N.; Bouzayen, M.; Zsögön, A.; Smouni, A.; et al. Down Regulation and Loss of Auxin Response Factor 4 Function Using CRISPR/Cas9 Alters Plant Growth, Stomatal Function and Improves Tomato Tolerance to Salinity and Osmotic Stress. Genes 2020, 11, 272. [Google Scholar] [CrossRef] [PubMed]
- Saikia, B.; Debbarma, J.; Maharana, J.; Singha, D.L.; Velmuruagan, N.; Dekaboruah, H.; Arunkumar, K.P.; Chikkaputtaiah, C. SlHyPRP1 and DEA1, the multiple stress responsive eight-cysteine motif family genes of tomato (Solanum lycopersicum L.) are expressed tissue specifically, localize and interact at cytoplasm and plasma membrane in vivo. Physiol. Mol. Biol. Plants 2020, 26, 2553–2568. [Google Scholar] [CrossRef] [PubMed]
- Tran, M.T.; Doan, D.T.H.; Kim, J.; Song, Y.J.; Sung, Y.W.; Das, S.; Kim, E.J.; Son, G.H.; Kim, S.H.; Van Vu, T.; et al. CRISPR/Cas9-based precise excision of SlHyPRP1 domain(s) to obtain salt stress-tolerant tomato. Plant Cell Rep. 2020, 40, 999–1011. [Google Scholar] [CrossRef]
- Chen, S.; Zhang, N.; Zhou, G.; Hussain, S.; Ahmed, S.; Tian, H.; Wang, S. Knockout of the entire family of AITR genes in Arabidopsis leads to enhanced drought and salinity tolerance without fitness costs. BMC Plant Biol. 2021, 21, 137. [Google Scholar] [CrossRef]
- Zhao, C.; Zhang, Z.; Xie, S.; Si, T.; Li, Y.; Zhu, J.K. Mutational Evidence for the Critical Role of CBF Transcription Factors in Cold Acclimation in Arabidopsis. Plant Physiol. 2016, 171, 2744–2759. [Google Scholar] [CrossRef]
- Han, G.; Yuan, F.; Guo, J.; Zhang, Y.; Sui, N.; Wang, B. AtSIZ1 improves salt tolerance by maintaining ionic homeostasis and osmotic balance in Arabidopsis. Plant Sci. 2019, 285, 55–67. [Google Scholar] [CrossRef]
- Li, T.; Yang, X.; Yu, Y.; Si, X.; Zhai, X.; Zhang, H.; Dong, W.; Gao, C.; Xu, C. Domestication of wild tomato is accelerated by genome editing. Nat. Biotechnol. 2018, 36, 1160–1163. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Jiao, Z.; Liu, L.; Wang, K.; Zhong, D.; Li, S.; Zhao, T.; Xu, X.; Cui, X. Enhancer-Promoter Interaction of SELF PRUNING 5G Shapes Photoperiod Adaptation. Plant Physiol. 2018, 178, 1631–1642. [Google Scholar] [CrossRef] [PubMed]
- Vu, T.V.; Sivankalyani, V.; Kim, E.J.; Doan, D.T.H.; Tran, M.T.; Kim, J.; Sung, Y.W.; Park, M.; Kang, Y.J.; Kim, J.Y. Highly efficient homology-directed repair using CRISPR/Cpf1-geminiviral replicon in tomato. Plant Biotechnol. J. 2020, 18, 2133–2143. [Google Scholar] [CrossRef]
- Ali, Z.; Park, H.C.; Ali, A.; Oh, D.-H.; Aman, R.; Kropornicka, A.; Hong, H.; Choi, W.; Chung, W.S.; Kim, W.-Y.; et al. TsHKT1;2, a HKT1 homolog from the extremophile Arabidopsis relative Thellungiella salsuginea, shows K+ specificity in the presence of NaCl. Plant Physiol. 2012, 158, 1463–1474. [Google Scholar] [CrossRef] [PubMed]
- Ali, A.; Raddatz, N.; Aman, R.; Kim, S.; Park, H.C.; Jan, M.; Baek, D.; Khan, I.U.; Oh, D.-H.; Lee, S.Y.; et al. A Single Amino-Acid Substitution in the Sodium Transporter HKT1 Associated with Plant Salt Tolerance. Plant Physiol. 2016, 171, 2112–2126. [Google Scholar] [CrossRef] [PubMed]
- Eck, V.; Keen, P.; Tjahjadi, M. Agrobacterium tumefaciens-Mediated Transformation of Tomato. Methods Mol. Biol. 2019, 1864, 225–234. [Google Scholar] [PubMed]
- Zhang, M.; Cao, Y.; Wang, Z.; Wang, Z.Q.; Shi, J.; Liang, X.; Song, W.; Chen, Q.; Lai, J.; Jiang, C. A retrotransposon in an HKT1 family sodium transporter causes variation of leaf Na+ exclusion and salt tolerance in maize. New Phytol. 2018, 217, 1161–1176. [Google Scholar] [CrossRef] [PubMed]
- Qin, H.; Wang, J.; Chen, X.; Wang, F.; Peng, P.; Zhou, Y.; Miao, Y.; Zhang, Y.; Gao, Y.; Qi, Y.; et al. Rice OsDOF15 contributes to ethylene-inhibited primary root elongation under salt stress. New Phytol. 2019, 223, 798–813. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Cui, L.; Xie, Z.; Zhang, Z.; Liu, E.; Peng, X. Two NCA1 isoforms interact with catalase in a mutually exclusive manner to redundantly regulate its activity in rice. BMC Plant Biol. 2019, 19, 105. [Google Scholar] [CrossRef] [PubMed]
- Alfatih, A.; Wu, J.; Jan, S.U.; Zhang, Z.S.; Xia, J.Q.; Xiang, C.B. Loss of rice PARAQUAT TOLERANCE 3 confers enhanced resistance to abiotic stresses and increases grain yield in field. Plant Cell Environ. 2020, 43, 2743–2754. [Google Scholar] [CrossRef]
- Chen, G.; Hu, J.; Dong, L.; Zeng, D.; Guo, L.; Zhang, G.; Zhu, L.; Qian, Q. The Tolerance of Salinity in Rice Requires the Presence of a Functional Copy of FLN2. Biomolecules 2019, 10, 17. [Google Scholar] [CrossRef]
- Zeng, D.D.; Yang, C.C.; Qin, R.; Alamin, M.; Yue, E.K.; Jin, X.L.; Shi, C.H. A guanine insert in OsBBS1 leads to early leaf senescence and salt stress sensitivity in rice (Oryza sativa L.). Plant Cell Rep. 2018, 37, 933–946. [Google Scholar] [CrossRef]
- Wang, B.; Zhong, Z.; Zhang, H.; Wang, X.; Liu, B.; Yang, L.; Han, X.; Yu, D.; Zheng, X.; Wang, C.; et al. Targeted Mutagenesis of NAC Transcription Factor Gene, OsNAC041, Leading to Salt Sensitivity in Rice. Rice Sci. 2019, 26, 98–108. [Google Scholar]
- Yin, W.; Xiao, Y.; Niu, M.; Meng, W.; Li, L.; Zhang, X.; Liu, D.; Zhang, G.; Qian, Y.; Sun, Z.; et al. ARGONAUTE2 Enhances Grain Length and Salt Tolerance by Activating BIG GRAIN3 to Modulate Cytokinin Distribution in Rice. Plant Cell 2020, 32, 2292–2306. [Google Scholar] [CrossRef]
- Zhou, J.; Deng, K.; Cheng, Y.; Zhong, Z.; Tian, L.; Tang, X.; Tang, A.; Zheng, X.; Zhang, T.; Qi, Y.; et al. CRISPR-Cas9 Based Genome Editing Reveals New Insights into MicroRNA Function and Regulation in Rice. Front. Plant Sci. 2017, 8, 1598. [Google Scholar] [CrossRef]
- Santosh Kumar, V.V.; Verma, R.K.; Yadav, S.K.; Yadav, P.; Watts, A.; Rao, M.V.; Chinnusamy, V. (2020) CRISPR-Cas9 mediated genome editing of drought and salt tolerance (OsDST) gene in indica mega rice cultivar MTU1010. Physiol. Mol. Biol. Plants 2020, 26, 1099–1110. [Google Scholar] [CrossRef]
- Wang, W.C.; Lin, T.C.; Kieber, J.; Tsai, Y.C. Response Regulators 9 and 10 Negatively Regulate Salinity Tolerance in Rice. Plant Cell Physiol. 2019, 60, 2549–2563. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, A.K.; Zhang, C.; Sadanandom, A. Rice OVERLY TOLERANT TO SALT 1 (OTS1) SUMO protease is a positive regulator of seed germination and root development. Plant Signal. Behav. 2016, 11, e1173301. [Google Scholar] [CrossRef]
- Lou, D.; Wang, H.; Yu, D. The sucrose non-fermenting-1-related protein kinases SAPK1 and SAPK2 function collaboratively as positive regulators of salt stress tolerance in rice. BMC Plant Biol. 2018, 18, 203. [Google Scholar] [CrossRef] [PubMed]
- Mo, W.; Tang, W.; Du, Y.; Jing, Y.; Bu, Q.; Lin, R. PHYTOCHROME-INTERACTING FACTOR-LIKE14 and SLENDER RICE1 Interaction Controls Seedling Growth under Salt Stress. Plant Physiol. 2020, 184, 506–517. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.T.; Zhao, M.J.; Wang, C.T.; Gao, Y.; Wang, Y.X.; Liu, Y.W.; Chen, M.; Chen, J.; Zhou, Y.B.; Xu, Z.S.; et al. Identification and characterization of GmMYB118 responses to drought and salt stress. BMC Plant Biol. 2018, 18, 320. [Google Scholar] [CrossRef]
- Li, M.; Chen, R.; Jiang, Q.; Sun, X.; Zhang, H.; Hu, Z. GmNAC06, a NAC domain transcription factor enhances salt stress tolerance in soybean. Plant Mol. Biol. 2021, 105, 333–345. [Google Scholar] [CrossRef] [PubMed]
- Yin, W.; Hu, Z.; Cui, B.; Guo, X.; Hu, J.; Zhu, Z.; Chen, G. Suppression of the MADS-box gene SlMBP8 accelerates fruit ripening of tomato (Solanum lycopersicum). Plant Physiol. Biochem. 2017, 118, 235–244. [Google Scholar] [CrossRef] [PubMed]
- Heigwer, F.; Kerr, G.; Boutros, M. E-CRISP: Fast CRISPR target site identification. Nat. Methods 2014, 11, 122–123. [Google Scholar] [CrossRef]
- Lei, Y.; Lu, L.; Liu, H.Y.; Li, S.; Xing, F.; Chen, L.L. CRISPR-P: A web tool for synthetic single-guide RNA design of CRISPR-system in plants. Mol. Plant 2014, 7, 1494–1496. [Google Scholar] [CrossRef]
- Brazelton, V.A., Jr.; Zarecor, S.; Wright, D.A.; Wang, Y.; Liu, J.; Chen, K.; Yang, B.; Lawrence-Dill, C.J. A quick guide to CRISPR sgRNA design tools. GM Crop Food 2015, 6, 266–276. [Google Scholar] [CrossRef]
- Mali, P.; Aach, J.; Stranges, P.B.; Esvelt, K.M.; Moosburner, M.; Kosuri, S.; Yang, L.; Church, G.M. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat. Biotechnol. 2013, 31, 833–838. [Google Scholar] [CrossRef]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A.; et al. Multiplex genome engineering using CRISPR/Cas systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef]
- Guilinger, J.P.; Thompson, D.B.; Liu, D.R. Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification. Nat. Biotechnol. 2014, 32, 577–582. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Zhang, Q.; Zhu, Q.; Liu, W.; Chen, Y.; Qiu, R.; Wang, B.; Yang, Z.; Li, H.; Lin, Y.; et al. A Robust CRISPR/Cas9 System for Convenient, High-Efficiency Multiplex Genome Editing in Monocot and Dicot Plants. Mol. Plant 2015, 8, 1274–1284. [Google Scholar] [CrossRef]
- Xu, R.; Qin, R.; Li, H.; Li, D.; Li, L.; Wei, P.; Yang, J. Generation of targeted mutant rice using a CRISPR-Cpf1 system. Plant Biotechnol. J. 2017, 15, 713–717. [Google Scholar] [CrossRef]
- Bin Moon, S.; Lee, J.M.; Kang, J.G.; Lee, N.E.; Ha, D.I.; Kim, D.Y.; Kim, S.H.; Yoo, K.; Kim, D.; Ko, J.H.; et al. Highly efficient genome editing by CRISPR-Cpf1 using CRISPR RNA with a uridinylate-rich 3’-overhang. Nat. Commun. 2018, 9, 3651. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Han, J.; Chen, Z.; Wu, H.; Dong, H.; Nie, G. Engineering cell signaling using tunable CRISPR–Cpf1-based transcription factors. Nat. Commun. 2017, 8, 2095. [Google Scholar] [CrossRef] [PubMed]
- Zetsche, B.; Gootenberg, J.S.; Abudayyeh, O.O.; Slaymaker, I.M.; Makarova, K.S.; Essletzbichler, P.; Volz, S.E.; Joung, J.; van der Oost, J.; Regev, A.; et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 2015, 163, 759–771. [Google Scholar] [CrossRef] [PubMed]
- Pausch, P.; Al-Shayeb, B.; Bisom-Rapp, E.; Tsuchida, C.A.; Li, Z.; Cress, B.F.; Knott, G.J.; Jacobsen, S.E.; Banfield, J.F.; Doudna, J.A. CRISPR-CasΦ from huge phages is a hypercompact genome editor. Science 2020, 369, 333–337. [Google Scholar] [CrossRef]
- Liu, H.; Ding, Y.; Zhou, Y.; Jin, W.; Xie, K.; Chen, L.L. CRISPR-P 2.0: An Improved CRISPR-Cas9 Tool for Genome Editing in Plants. Mol. Plant 2017, 10, 530–532. [Google Scholar] [CrossRef] [PubMed]
- Butler, N.M.; Baltes, N.J.; Voytas, D.F.; Douches, D.S. Geminivirus-Mediated Genome Editing in Potato (Solanum tuberosum L.) Using Sequence-Specific Nucleases. Front. Plant Sci. 2016, 7, 1045. [Google Scholar] [CrossRef]
- Gil-Humanes, J.; Wang, Y.; Liang, Z.; Shan, Q.; Ozuna, C.V.; Sánchez-León, S.; Baltes, N.J.; Starker, C.; Barro, F.; Gao, C.; et al. High-efficiency gene targeting in hexaploid wheat using DNA replicons and CRISPR/Cas9. Plant J. 2017, 89, 1251–1262. [Google Scholar] [CrossRef]
- Hummel, A.W.; Chauhan, R.D.; Cermak, T.; Mutka, A.M.; Vijayaraghavan, A.; Boyher, A.; Starker, C.G.; Bart, R.; Voytas, D.F.; Taylor, N.J. Allele exchange at the EPSPS locus confers glyphosate tolerance in cassava. Plant Biotechnol. J. 2018, 16, 1275–1282. [Google Scholar] [CrossRef]
- Suárez-López, P.; Gutiérrez, C. DNA replication of wheat dwarf geminivirus vectors: Effects of origin structure and size. Virology 1997, 227, 389–399. [Google Scholar] [CrossRef][Green Version]
- Baltes, N.J.; Gil-Humanes, J.; Cermak, T.; Atkins, P.A.; Voytas, D.F. DNA replicons for plant genome engineering. Plant Cell 2014, 26, 151–163. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Zhu, M.; Wang, L.; Wu, J.; Wang, Q.; Wang, R.; Zhao, Y. Programmed Self-Elimination of the CRISPR/Cas9 Construct Greatly Accelerates the Isolation of Edited and Transgene-Free Rice Plants. Mol. Plant 2018, 11, 1210–1213. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liang, Z.; Zong, Y.; Wang, Y.; Liu, J.; Chen, K.; Qiu, J.L.; Gao, C. Efficient and transgene-free genome editing in wheat through transient expression of CRISPR/Cas9 DNA or RNA. Nat. Commun. 2016, 7, 12617. [Google Scholar] [CrossRef]
- Gao, X.; Chen, J.; Dai, X.; Zhang, D.; Zhao, Y. An Effective Strategy for Reliably Isolating Heritable and Cas9-Free Arabidopsis Mutants Generated by CRISPR/Cas9-Mediated Genome Editing. Plant Physiol. 2016, 171, 1794–1800. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Ye, X.; Guo, R.; Huang, J.; Wang, W.; Tang, J.; Tan, L.; Zhu, J.-k.; Chu, C.; Qian, Y. Genome-wide targeted mutagenesis in rice using the CRISPR/Cas9 system. Mol. Plant 2017, 10, 1242–1245. [Google Scholar] [CrossRef] [PubMed]
- Veillet, F.; Perrot, L.; Chauvin, L.; Kermarrec, M.P.; Guyon-Debast, A.; Chauvin, J.E.; Nogué, F.; Mazier, M. Transgene-Free Genome Editing in Tomato and Potato Plants Using Agrobacterium-Mediated Delivery of a CRISPR/Cas9 Cytidine Base Editor. Int. J. Mol. Sci. 2019, 20, 402. [Google Scholar] [CrossRef]
- Lindbo, J.A. TRBO: A High-Efficiency Tobacco Mosaic Virus RNA-Based Overexpression Vector. Plant Physiol. 2007, 145, 1232–1240. [Google Scholar] [CrossRef]
- Cody, W.B.; Scholthof, H.B.; Mirkov, T.E. Multiplexed Gene Editing and Protein Overexpression Using a Tobacco mosaic virus Viral Vector. Plant Physiol. 2017, 175, 23–35. [Google Scholar] [CrossRef]
- Flint-Garcia, S.A. Genetics and consequences of crop domestication. J. Agric. Food Chem. 2013, 61, 8267–8276. [Google Scholar] [CrossRef]
- Govindaraj, M.; Vetriventhan, M.; Srinivasan, M. Importance of genetic diversity assessment in crop plants and its recent advances: An overview of its analytical perspectives. Genet. Res. Int. 2015, 2015, 431487. [Google Scholar] [CrossRef]
- Agarwal, G.; Kudapa, H.; Ramalingam, A.; Choudhary, D.; Sinha, P.; Garg, V.; Singh, V.K.; Patil, G.B.; Pandey, M.K.; Nguyen, H.T.; et al. Epigenetics and epigenomics: Underlying mechanisms, relevance, and implications in crop improvement. Funct. Integr. Genom. 2020, 20, 739–761. [Google Scholar] [CrossRef]
- Kumar, S. Epigenetic Regulation of Abiotic Stress Tolerance in Plants. Adv. Plants Agric. Res. 2016, 5, 517–521. [Google Scholar] [CrossRef]
- Sahu, P.P.; Pandey, G.; Sharma, N.; Puranik, S.; Muthamilarasan, M.; Prasad, M. Epigenetic mechanisms of plant stress responses and adaptation. Plant Cell Rep. 2013, 32, 1151–1159. [Google Scholar] [CrossRef]
- Johannes, F.; Porcher, E.; Teixeira, F.K.; Saliba-Colombani, V.; Simon, M.; Agier, N.; Bulski, A.; Albuisson, J.; Heredia, F.; Audigier, P.; et al. Assessing the impact of transgenerational epigenetic variation on complex traits. PLoS Genet. 2009, 5, e1000530. [Google Scholar] [CrossRef]
- Furci, L.; Jain, R.; Stassen, J.; Berkowitz, O.; Whelan, J.; Roquis, D.; Baillet, V.; Colot, V.; Johannes, F.; Ton, J. Identification and characterisation of hypomethylated DNA loci controlling quantitative resistance in Arabidopsis. eLife 2019, 8, e40655. [Google Scholar] [CrossRef] [PubMed]
- Colome-Tatche, M.; Cortijo, S.; Wardenaar, R.; Morgado, L.; Lahouze, B.; Sarazin, A.; Etcheverry, M.; Martin, A.; Feng, S.; Duvernois-Berthet, E.; et al. Features of the Arabidopsis recombination landscape resulting from the combined loss of sequence variation and DNA methylation. Proc. Natl. Acad. Sci. USA 2012, 109, 16240–16245. [Google Scholar] [CrossRef] [PubMed]
- Roux, F.; Colomé-Tatché, M.; Edelist, C.; Wardenaar, R.; Guerche, P.; Hospital, F.; Colot, V.; Jansen, R.C.; Johannes, F. Genome-wide epigenetic perturbation jump-starts patterns of heritable variation found in nature. Genetics 2011, 188, 1015–1017. [Google Scholar] [CrossRef]
- Latzel, V.; Zhang, Y.; Karlsson Moritz, K.; Fischer, M.; Bossdorf, O. Epigenetic variation in plant responses to defence hormones. Ann. Bot. 2012, 110, 1423–1428. [Google Scholar] [CrossRef]
- Zhang, Y.Y.; Fischer, M.; Colot, V.; Bossdorf, O. Epigenetic variation creates potential for evolution of plant phenotypic plasticity. New Phytol. 2013, 197, 314–322. [Google Scholar] [CrossRef]
- Latzel, V.; Allan, E.; Bortolini Silveira, A.; Colot, V.; Fischer, M.; Bossdorf, O. Epigenetic diversity increases the productivity and stability of plant populations. Nat. Commun. 2013, 4, 2875. [Google Scholar] [CrossRef]
- Vongs, A.; Kakutani, T.; Martienssen, R.A.; Richards, E.J. Arabidopsis thaliana DNA methylation mutants. Science 1993, 260, 1926–1928. [Google Scholar] [CrossRef]
- Kakutani, T.; Jeddeloh, J.A.; Richards, E.J. Characterization of an Arabidopsis thaliana DNA hypomethylation mutant. Nucleic Acids Res. 1995, 23, 130–137. [Google Scholar] [CrossRef] [PubMed]
- Jeddeloh, J.A.; Stokes, T.L.; Richards, E.J. Maintenance of genomic methylation requires a SWI2/SNF2-like protein. Nat. Genet. 1999, 22, 94–97. [Google Scholar] [CrossRef] [PubMed]
- Lippman, Z.; Gendrel, A.V.; Black, M.; Vaughn, M.W.; Dedhia, N.; McCombie, W.R.; Lavine, K.; Mittal, V.; May, B.; Kasschau, K.D.; et al. Role of transposable elements in heterochromatin and epigenetic control. Nature 2004, 430, 471–476. [Google Scholar] [CrossRef] [PubMed]
- Lauss, K.; Wardenaar, R.; Oka, R.; van Hulten, M.H.A.; Guryev, V.; Keurentjes, J.J.B.; Stam, M.; Johannes, F. Parental DNA Methylation States Are Associated with Heterosis in Epigenetic Hybrids. Plant Physiol. 2018, 176, 1627–1645. [Google Scholar] [CrossRef]
- Kawakatsu, T.; Huang, S.-S.C.; Jupe, F.; Sasaki, E.; Schmitz, R.J.; Urich, M.A.; Castanon, R.; Nery, J.R.; Barragan, C.; He, Y.; et al. Epigenomic Diversity in a Global Collection of Arabidopsis thaliana Accessions. Cell 2016, 166, 492–505. [Google Scholar] [CrossRef] [PubMed]
- Melamed-Bessudo, C.; Levy, A.A. Deficiency in DNA methylation increases meiotic crossover rates in euchromatic but not in heterochromatic regions in Arabidopsis. Proc. Natl. Acad. Sci. USA 2012, 109, E981–E988. [Google Scholar] [CrossRef] [PubMed]
- Mirouze, M.; Lieberman-Lazarovich, M.; Aversano, R.; Bucher, E.; Nicolet, J.; Reinders, J.; Paszkowski, J. Loss of DNA methylation affects the recombination landscape in Arabidopsis. Proc. Natl. Acad. Sci. USA 2012, 109, 5880–5885. [Google Scholar] [CrossRef] [PubMed]
- Yelina, N.E.; Choi, K.; Chelysheva, L.; Macaulay, M.; de Snoo, B.; Wijnker, E.; Miller, N.; Drouaud, J.; Grelon, M.; Copenhaver, G.P.; et al. Epigenetic remodeling of meiotic crossover frequency in Arabidopsis thaliana DNA methyltransferase mutants. PLoS Genet. 2012, 8, e1002844. [Google Scholar] [CrossRef]
- Yelina, N.E.; Ziolkowski, P.A.; Miller, N.; Zhao, X.; Kelly, K.A.; Muñoz, D.F.; Mann, D.J.; Copenhaver, G.P.; Henderson, I.R. High-throughput analysis of meiotic crossover frequency and interference via flow cytometry of fluorescent pollen in Arabidopsis thaliana. Nat. Protoc. 2013, 8, 2119–2134. [Google Scholar] [CrossRef]
- Taagen, E.; Bogdanove, A.J.; Sorrells, M.E. Counting on Crossovers: Controlled Recombination for Plant Breeding. Trends Plant Sci. 2020, 25, 455–465. [Google Scholar] [CrossRef] [PubMed]
- Jackson, J.P.; Lindroth, A.M.; Cao, X.; Jacobsen, S.E. Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature 2002, 416, 556–560. [Google Scholar] [CrossRef]
- Blevins, T.; Pontvianne, F.; Cocklin, R.; Podicheti, R.; Chandrasekhara, C.; Yerneni, S.; Braun, C.; Lee, B.; Rusch, D.; Mockaitis, K.; et al. A two-step process for epigenetic inheritance in Arabidopsis. Mol. Cell 2014, 54, 30–42. [Google Scholar] [CrossRef] [PubMed]
- Ono, T.; Kaya, H.; Takeda, S.; Abe, M.; Ogawa, Y.; Kato, M.; Kakutani, T.; Mittelsten Scheid, O.; Araki, T.; Shibahara, K. Chromatin assembly factor 1 ensures the stable maintenance of silent chromatin states in Arabidopsis. Genes Cells 2006, 11, 153–162. [Google Scholar] [CrossRef]
- Mohannath, G.; Pontvianne, F.; Pikaard, C.S. Selective nucleolus organizer inactivation in Arabidopsis is a chromosome position-effect phenomenon. Proc. Natl. Acad. Sci. USA 2016, 113, 13426–13431. [Google Scholar] [CrossRef]
- Luo, M.; Cheng, K.; Xu, Y.; Yang, S.; Wu, K. Plant Responses to Abiotic Stress Regulated by Histone Deacetylases. Front. Plant Sci. 2017, 8, 2147. [Google Scholar] [CrossRef]
- Huang, L.; Sun, Q.; Qin, F.; Li, C.; Zhao, Y.; Zhou, D.X. Down-regulation of a SILENT INFORMATION REGULATOR2-related histone deacetylase gene, OsSRT1, induces DNA fragmentation and cell death in rice. Plant Physiol. 2007, 144, 1508–1519. [Google Scholar] [CrossRef]
- To, T.K.; Nakaminami, K.; Kim, J.M.; Morosawa, T.; Ishida, J.; Tanaka, M.; Yokoyama, S.; Shinozaki, K.; Seki, M. Arabidopsis HDA6 is required for freezing tolerance. Biochem. Biophys. Res. Commun. 2011, 406, 414–419. [Google Scholar] [CrossRef]
- Tanaka, M.; Kikuchi, A.; Kamada, H. The Arabidopsis histone deacetylases HDA6 and HDA19 contribute to the repression of embryonic properties after germination. Plant Physiol. 2008, 146, 149–161. [Google Scholar] [CrossRef] [PubMed]
- Tiricz, H.; Nagy, B.; Ferenc, G.; Török, K.; Nagy, I.; Dudits, D.; Ayaydin, F. Relaxed chromatin induced by histone deacetylase inhibitors improves the oligonucleotide-directed gene editing in plant cells. J. Plant Res. 2018, 131, 179–189. [Google Scholar] [CrossRef]
- Compant, S.; Samad, A.; Faist, H.; Sessitsch, A. A review on the plant microbiome: Ecology, functions, and emerging trends in microbial application. J. Adv. Res. 2019, 19, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Dodd, I.C.; Pérez-Alfocea, F. Microbial amelioration of crop salinity stress. J. Exp. Bot. 2012, 63, 3415–3428. [Google Scholar] [CrossRef]
- Fahad, S.; Hussain, S.; Matloob, A.; Khan, F.A.; Khaliq, A.; Saud, S.; Hassan, S.; Shan, D.; Khan, F.; Ullah, N.; et al. Phytohormones and plant responses to salinity stress: A review. Plant Growth Regul. 2015, 75, 391–404. [Google Scholar] [CrossRef]
- Numan, M.; Bashir, S.; Khan, Y.; Mumtaz, R.; Shinwari, Z.K.; Khan, A.L.; Khan, A.; Al-Harrasi, A. Plant growth promoting bacteria as an alternative strategy for salt tolerance in plants: A review. Microbiol. Res. 2018, 209, 21–32. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.N.; Zhu, C.; Jiang, J.; Zhang, H.; Zhu, J.K.; Duan, C.G. Epigenetic regulation in plant abiotic stress responses. J. Integr. Plant Biol. 2020, 62, 563–580. [Google Scholar] [CrossRef]
- Yasmeen, T.; Ahmad, A.; Arif, M.S.; Mubin, M.; Rehman, K.; Shahzad, S.M.; Iqbal, S.; Rizwan, M.; Ali, S.; Alyemeni, M.N.; et al. Biofilm forming rhizobacteria enhance growth and salt tolerance in sunflower plants by stimulating antioxidant enzymes activity. Plant Physiol. Biochem. 2020, 156, 242–256. [Google Scholar] [CrossRef] [PubMed]
- Habib, S.H.; Kausar, H.; Saud, H.M. Plant Growth-Promoting Rhizobacteria Enhance Salinity Stress Tolerance in Okra through ROS-Scavenging Enzymes. Biomed. Res. Int. 2016, 2016, 6284547. [Google Scholar] [CrossRef] [PubMed]
- Sapre, S.; Gontia-Mishra, I.; Tiwari, S. Klebsiella sp. confers enhanced tolerance to salinity and plant growth promotion in oat seedlings (Avena sativa). Microbiol. Res. 2018, 206, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Yasin, N.A.; Akram, W.; Khan, W.U.; Ahmad, S.R.; Ahmad, A.; Ali, A. Halotolerant plant-growth promoting rhizobacteria modulate gene expression and osmolyte production to improve salinity tolerance and growth in Capsicum annum L. Environ. Sci. Pollut. Res. Int. 2018, 25, 23236–23250. [Google Scholar] [CrossRef]
- Chu, T.N.; Tran, B.T.H.; Van Bui, L.; Hoang, M.T.T. Plant growth-promoting rhizobacterium Pseudomonas PS01 induces salt tolerance in Arabidopsis thaliana. BMC Res. Notes 2019, 12, 11. [Google Scholar] [CrossRef] [PubMed]
- Ha-Tran, D.M.; Nguyen, T.T.M.; Hung, S.H.; Huang, E.; Huang, C.C. Roles of Plant Growth-Promoting Rhizobacteria (PGPR) in Stimulating Salinity Stress Defense in Plants: A Review. Int. J. Mol. Sci. 2021, 22, 3154. [Google Scholar] [CrossRef]
- Srivastava, S.; Chaudhry, V.; Mishra, A.; Chauhan, P.S.; Rehman, A.; Yadav, A.; Tuteja, N.; Nautiyal, C.S. Gene expression profiling through microarray analysis in Arabidopsis thaliana colonized by Pseudomonas putida MTCC5279, a plant growth promoting rhizobacterium. Plant Signal. Behav. 2012, 7, 235–245. [Google Scholar] [CrossRef]
- Safdarian, M.; Askari, H.; Shariati, J.V.; Nematzadeh, G. Transcriptional responses of wheat roots inoculated with Arthrobacter nitroguajacolicus to salt stress. Sci. Rep. 2019, 9, 1792. [Google Scholar] [CrossRef] [PubMed]
- Jatan, R.; Chauhan, P.S.; Lata, C. Pseudomonas putida modulates the expression of miRNAs and their target genes in response to drought and salt stresses in chickpea (Cicer arietinum L.). Genomics 2019, 111, 509–519. [Google Scholar] [CrossRef]
- Ghosh, D.; Sen, S.; Mohapatra, S. Modulation of proline metabolic gene expression in Arabidopsis thaliana under water-stressed conditions by a drought-mitigating Pseudomonas putida strain. Ann. Microbiol. 2017, 67, 655–668. [Google Scholar] [CrossRef]
- Ghosh, D.; Sen, S.; Mohapatra, S. Drought-mitigating Pseudomonas putida GAP-P45 modulates proline turnover and oxidative status in Arabidopsis thaliana under water stress. Ann. Microbiol. 2018, 68, 579–594. [Google Scholar] [CrossRef]
- Hahm, M.S.; Son, J.S.; Hwang, Y.J.; Kwon, D.K.; Ghim, S.Y. Alleviation of Salt Stress in Pepper (Capsicum annum L.) Plants by Plant Growth-Promoting Rhizobacteria. J. Microbiol. Biotechnol. 2017, 27, 1790–1797. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.-M.; Khan, A.L.; Waqas, M.; You, Y.-H.; Kim, J.-H.; Kim, J.-G.; Hamayun, M.; Lee, I.-J. Plant growth-promoting rhizobacteria reduce adverse effects of salinity and osmotic stress by regulating phytohormones and antioxidants in Cucumis sativus. J. Plant Interact. 2014, 9, 673–682. [Google Scholar] [CrossRef]
- Jalili, F.; Khavazi, K.; Pazira, E.; Nejati, A.; Rahmani, H.A.; Sadaghiani, H.R.; Miransari, M. Isolation and characterization of ACC deaminase-producing fluorescent pseudomonads, to alleviate salinity stress on canola (Brassica napus L.) growth. J. Plant Physiol. 2009, 166, 667–674. [Google Scholar] [CrossRef] [PubMed]
- Bal, H.B.; Nayak, L.; Das, S.; Adhya, T.K. Isolation of ACC deaminase producing PGPR from rice rhizosphere and evaluating their plant growth promoting activity under salt stress. Plant Soil 2013, 366, 93–105. [Google Scholar] [CrossRef]
- Kohler, J.; Hernández, J.A.; Caravaca, F.; Roldán, A. Plant-growth-promoting rhizobacteria and arbuscular mycorrhizal fungi modify alleviation biochemical mechanisms in water-stressed plants. Funct. Plant Biol. 2008, 35, 141–151. [Google Scholar] [CrossRef]
- Han, Q.-Q.; Wu, Y.-N.; Gao, H.-J.; Xu, R.; Paré, P.W.; Shi, H.; Zhao, Q.; Li, H.-R.; Khan, S.A.; Wang, Y.-Q.; et al. Improved salt tolerance of medicinal plant Codonopsis pilosula by Bacillus amyloliquefaciens GB03. Acta Physiol. Plant. 2017, 39. [Google Scholar] [CrossRef]
- Aziz, M.; Nadipalli, R.K.; Xie, X.; Sun, Y.; Surowiec, K.; Zhang, J.L.; Paré, P.W. Augmenting Sulfur Metabolism and Herbivore Defense in Arabidopsis by Bacterial Volatile Signaling. Front. Plant Sci. 2016, 7, 458. [Google Scholar] [CrossRef]
- Vaishnav, A.; Kumari, S.; Jain, S.; Varma, A.; Choudhary, D.K. Putative bacterial volatile-mediated growth in soybean (Glycine max L. Merrill) and expression of induced proteins under salt stress. J. Appl. Microbiol. 2015, 119, 539–551. [Google Scholar] [CrossRef]
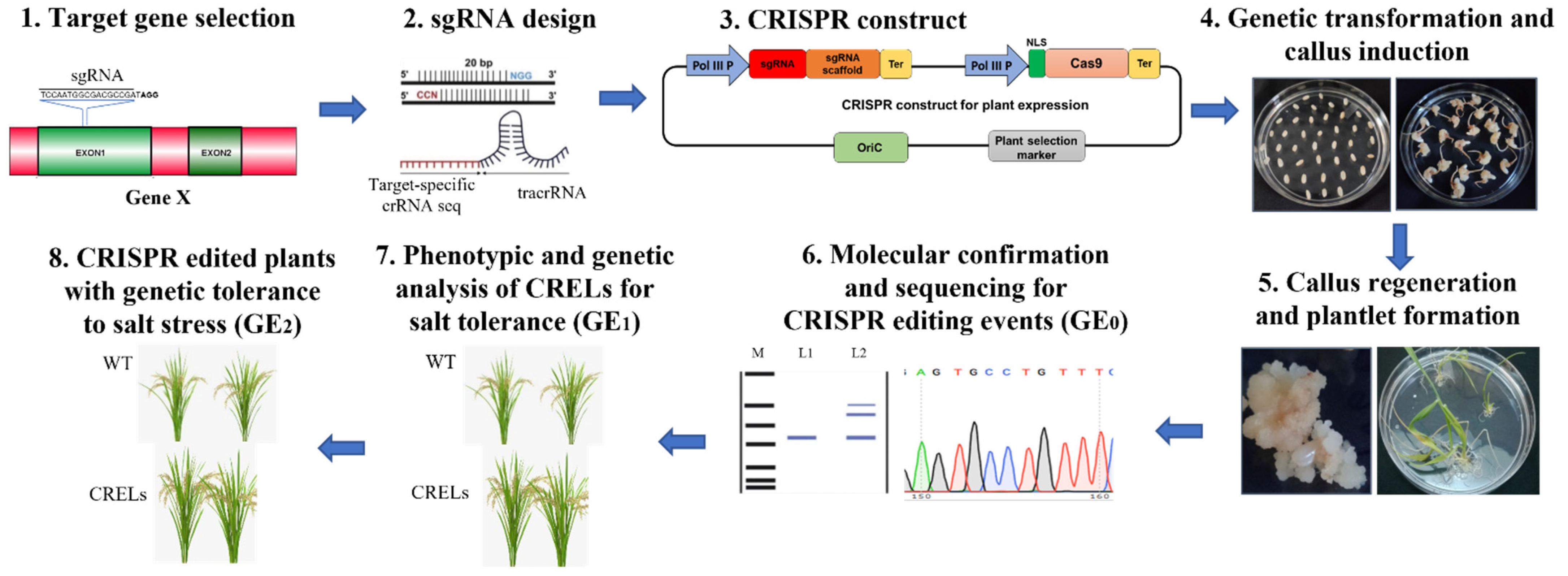
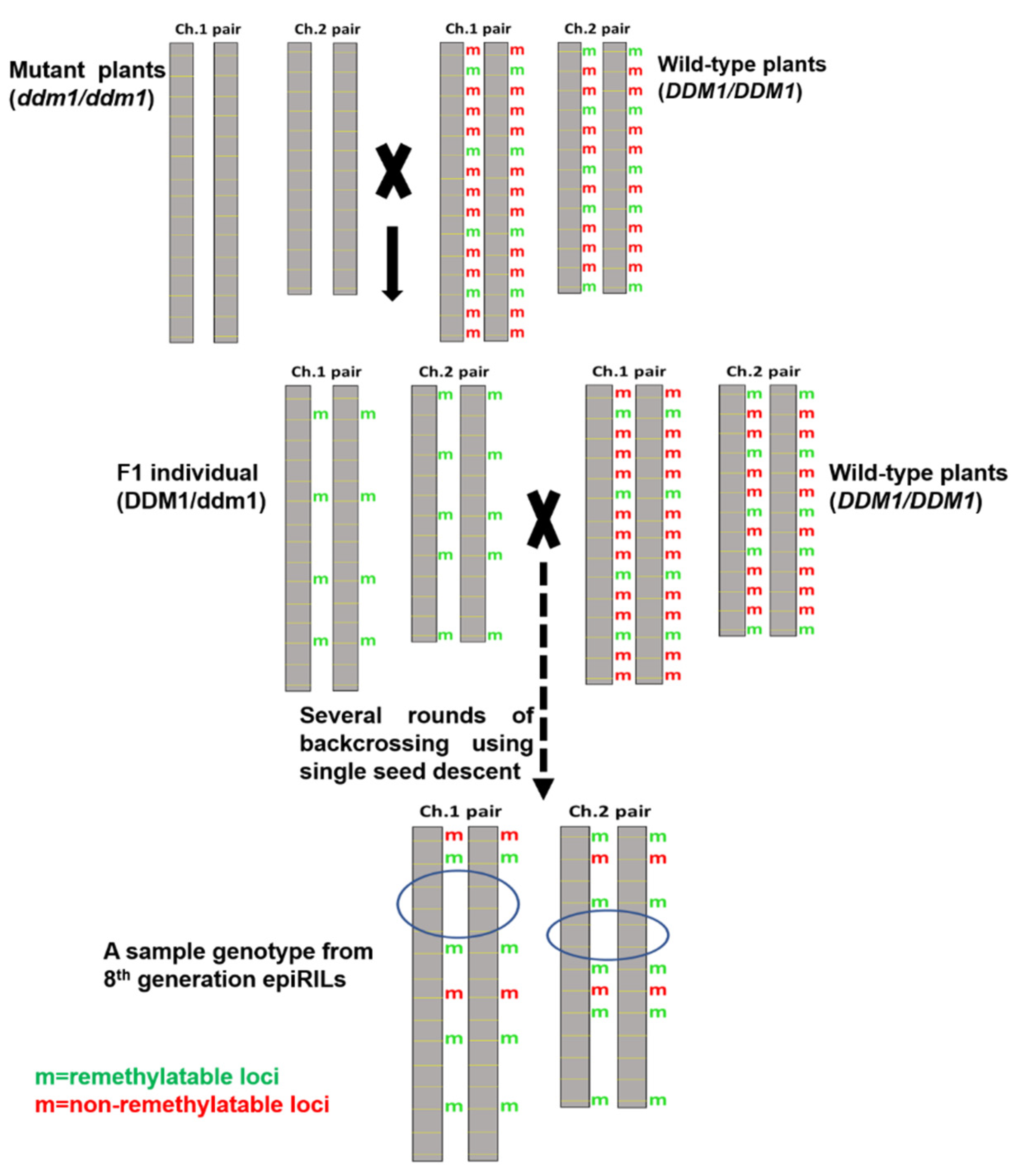
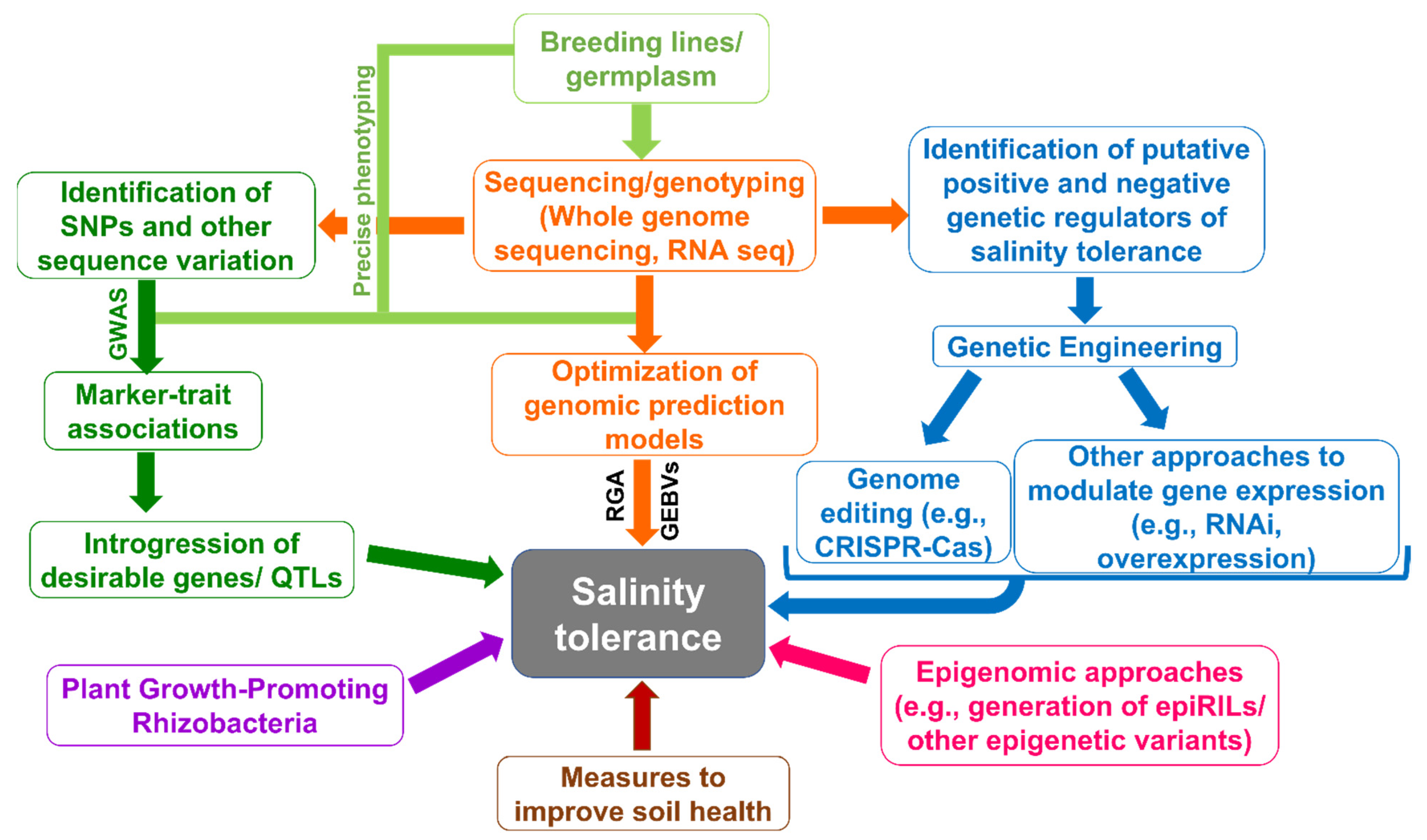
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saradadevi, G.P.; Das, D.; Mangrauthia, S.K.; Mohapatra, S.; Chikkaputtaiah, C.; Roorkiwal, M.; Solanki, M.; Sundaram, R.M.; Chirravuri, N.N.; Sakhare, A.S.; et al. Genetic, Epigenetic, Genomic and Microbial Approaches to Enhance Salt Tolerance of Plants: A Comprehensive Review. Biology 2021, 10, 1255. https://doi.org/10.3390/biology10121255
Saradadevi GP, Das D, Mangrauthia SK, Mohapatra S, Chikkaputtaiah C, Roorkiwal M, Solanki M, Sundaram RM, Chirravuri NN, Sakhare AS, et al. Genetic, Epigenetic, Genomic and Microbial Approaches to Enhance Salt Tolerance of Plants: A Comprehensive Review. Biology. 2021; 10(12):1255. https://doi.org/10.3390/biology10121255
Chicago/Turabian StyleSaradadevi, Gargi Prasad, Debajit Das, Satendra K. Mangrauthia, Sridev Mohapatra, Channakeshavaiah Chikkaputtaiah, Manish Roorkiwal, Manish Solanki, Raman Meenakshi Sundaram, Neeraja N. Chirravuri, Akshay S. Sakhare, and et al. 2021. "Genetic, Epigenetic, Genomic and Microbial Approaches to Enhance Salt Tolerance of Plants: A Comprehensive Review" Biology 10, no. 12: 1255. https://doi.org/10.3390/biology10121255
APA StyleSaradadevi, G. P., Das, D., Mangrauthia, S. K., Mohapatra, S., Chikkaputtaiah, C., Roorkiwal, M., Solanki, M., Sundaram, R. M., Chirravuri, N. N., Sakhare, A. S., Kota, S., Varshney, R. K., & Mohannath, G. (2021). Genetic, Epigenetic, Genomic and Microbial Approaches to Enhance Salt Tolerance of Plants: A Comprehensive Review. Biology, 10(12), 1255. https://doi.org/10.3390/biology10121255







