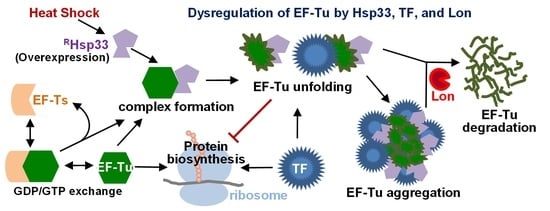
Molecular Effects of Elongation Factor Ts and Trigger Factor on the Unfolding and Aggregation of Elongation Factor Tu Induced by the Prokaryotic Molecular Chaperone Hsp33
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Protein Sample Preparation
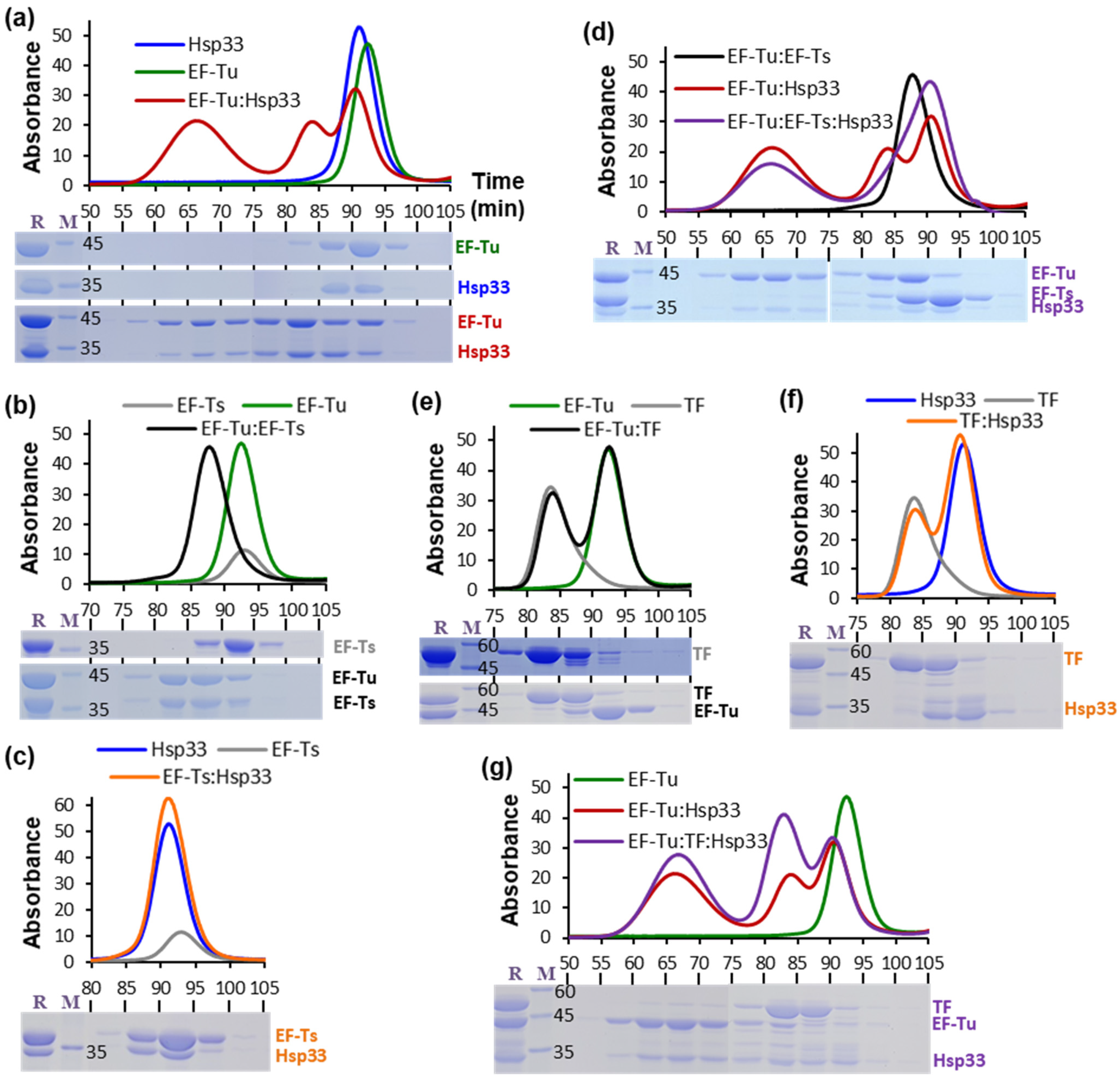
2.2. Analytical Gel Filtration
2.3. Light Scattering Measurements
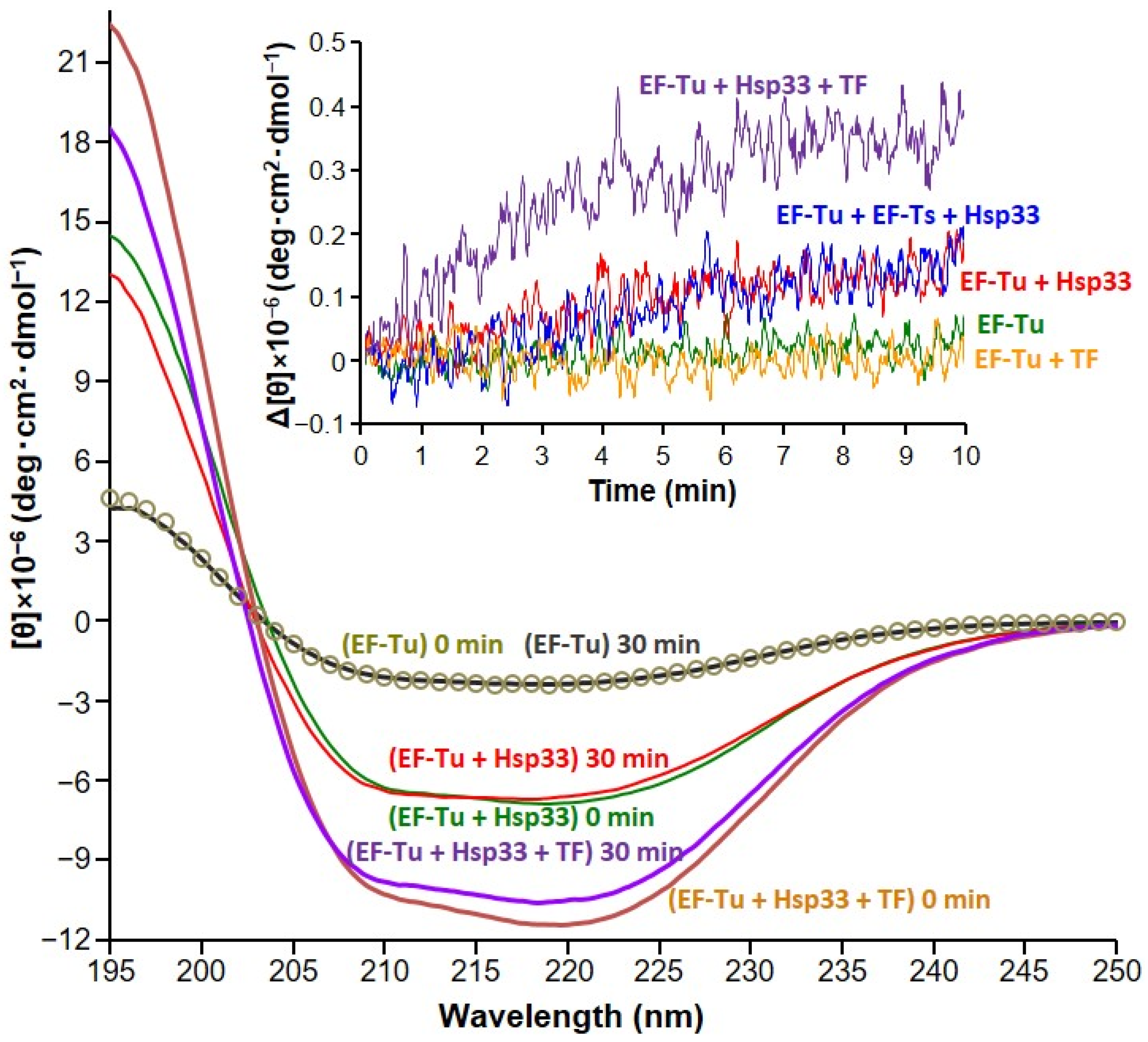
2.4. Circular Dichroism (CD) Spectroscopy
2.5. Isothermal Titration Calorimetry (ITC)
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CD | circular dichroism |
| DTT | 1,4-dithiothreitol |
| EDTA | ethylenediaminetetraacetic acid |
| EF-Ts | elongation factor thermo-stable |
| EF-Tu | elongation factor thermo-unstable |
| FPLC | fast protein liquid chromatograpy |
| HEPES | 2-[4-(2-hydroxyethyl)piperazin-1-yl]ethanesulfonic acid |
| Hsp33 | heat shock protein 33 |
| IPTG | Isopropyl β-D-1-thiogalactopyranoside |
| ITC | isothermal titration calorimetry |
| PCR | polymerase chain reaction |
| PQC | protein quality control |
| SDS-PAGE | sodium dodecyl sulfate-polyacrylamide gel electrophoresis |
| TF | trigger factor |
| Tris-HCl | tris(hydroxymethyl)aminomethane hydrochloride |
References
- Hartl, F.U.; Bracher, A.; Hayer-Hartl, M. Molecular chaperones in protein folding and proteostasis. Nature 2011, 475, 324–332. [Google Scholar] [CrossRef] [PubMed]
- Ciechanover, A.; Kwon, Y.T. Protein quality control by molecular chaperones in neurodegeneration. Front. Neurosci. 2017, 11, 185. [Google Scholar] [CrossRef] [PubMed]
- Radzinski, M.; Oppenheim, T.; Metanis, N.; Rechmann, D. The Cys sense: Thiol redox switches mediate life cycles of cellular proteins. Biomolecules 2021, 11, 469. [Google Scholar] [CrossRef] [PubMed]
- McClellan, A.J.; Tam, S.; Kaganovich, D.; Frydman, J. Protein quality control: Chaperones culling corrupt conformations. Nat. Cell Biol. 2005, 7, 736–741. [Google Scholar] [CrossRef] [PubMed]
- Bukau, B.; Weissman, J.; Horwich, A. Molecular chaperones and protein quality control. Cell 2006, 125, 443–451. [Google Scholar] [CrossRef]
- Mattoo, R.U.H.; Goloubinoff, P. Molecular chaperones are nanomachines that catalytically unfold misfolded and alternatively folded proteins. Cell. Mol. Life Sci. 2014, 71, 3311–3325. [Google Scholar] [CrossRef]
- Dahl, J.-U.; Gray, M.J.; Jakob, U. Protein quality control under oxidative stress conditions. J. Mol. Biol. 2015, 427, 1549–1563. [Google Scholar] [CrossRef] [PubMed]
- Chuang, S.E.; Blattner, F.R. Characterization of twenty-six new heat shock genes of Escherichia coli. J. Bacteriol. 1993, 175, 5242–5252. [Google Scholar] [CrossRef]
- Aramin, S.; Fassler, R.; Chikne, V.; Goldenberg, M.; Arian, T.; Eliaz, L.K.; Rimon, O.; Ram, O.; Michaeli, S.; Reichmann, D. TrypOx, a novel eukaryotic homolog of the redox-regulated chaperone Hsp33 in Trypanosoma brucei. Front. Microbiol. 2020, 11, 1844. [Google Scholar] [CrossRef]
- Segal, N.; Shapira, M. HSP33 in eukaryotes—An evolutionary tale of a chaperone adapted to photosynthetic organisms. Plant J. 2015, 82, 850–860. [Google Scholar] [CrossRef]
- Jakob, U.; Muse, W.; Eser, M.; Bardwell, J.C.A. Chaperone activity with a redox switch. Cell 1999, 96, 341–352. [Google Scholar] [CrossRef]
- Won, H.-S.; Low, L.Y.; De Guzman, R.; Martinez-Yamout, M.; Jakob, U.; Dyson, H.J. The zinc-dependent redox switch domain of the chaperone Hsp33 has a novel fold. J. Mol. Biol. 2004, 341, 893–899. [Google Scholar] [CrossRef]
- Ilbert, M.; Horst, J.; Ahrens, S.; Winter, J.; Graf, P.C.F.; Lilie, H.; Jakob, U. The redox-switch domain of Hsp33 functions as dual stress sensor. Nat. Struct. Mol. Biol. 2007, 14, 556–563. [Google Scholar] [CrossRef]
- Winter, J.; Ilbert, M.; Graf, P.C.F.; Özcelik, D.; Jakob, U. Bleach activates a redox-regulated chaperone by oxidative protein unfolding. Cell 2008, 135, 691–701. [Google Scholar] [CrossRef]
- Cremers, C.M.; Reichmann, D.; Hausmann, J.; Ilbert, M.; Jakob, U. Unfolding of metastable linker region is at the core of Hsp33 activation as a redox-regulated chaperone. J. Biol. Chem. 2010, 285, 11243–11251. [Google Scholar] [CrossRef]
- Lee, Y.-S.; Lee, J.; Ryu, K.-S.; Lee, Y.; Jung, T.-G.; Jang, J.-H.; Sim, D.-W.; Kim, E.-H.; Seo, M.-D.; Lee, K.W.; et al. Semi-empirical structure determination of Escherichia coli Hsp33 and identification of dynamic regulatory elements for the activation process. J. Mol. Biol. 2015, 427, 3850–3861. [Google Scholar] [CrossRef] [PubMed]
- Moayed, F.; Bezrukavnikov, S.; Naqvi, M.M.; Groitl, B.; Cremers, C.M.; Kramer, G.; Ghosh, K.; Jakob, U.; Tans, S.J. The anti-aggregation holdase Hsp33 promotes the formation of folded protein structures. Biophys. J. 2020, 118, 85–95. [Google Scholar] [CrossRef] [PubMed]
- Groitl, B.; Horowitz, S.; Makepeace, K.A.T.; Petrotchenko, E.V.; Borchers, C.H.; Reichmann, D.; Bardwell, J.C.A.; Jakob, U. Protein unfolding as a switch from self-recognition to high-affinity client binding. Nat. Commun. 2016, 7, 10357. [Google Scholar] [CrossRef]
- Reichmann, D.; Xu, Y.; Cremers, C.M.; Ilbert, M.; Mittelman, R.; Fitzgerald, M.C. Order out of disorder: Working cycle of an intrinsically unfolded chaperone. Cell 2012, 148, 947–957. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.-S.; Ryu, K.-S.; Kim, S.-J.; Ko, H.-S.; Sim, D.-W.; Jeon, Y.H.; Kim, E.-H.; Choi, W.-S.; Won, H.-S. Verification of the interdomain contact site in the inactive monomer, and the domain-swapped fold in the active dimer of Hsp33 in solution. FEBS Lett. 2012, 586, 411–415. [Google Scholar] [CrossRef][Green Version]
- Akhtar, M.W.; Srinivas, V.; Raman, B.; Ramakrishna, T.; Inobes, T.; Maki, K.; Arai, M.; Kuwajima, K.; Rao, C.M. Oligomeric Hsp33 with enhanced chaperone activity: Gel filtration, cross-linking, and small angle x-ray scattering (SAXS) analysis. J. Biol. Chem. 2004, 279, 55760–55769. [Google Scholar] [CrossRef]
- Voth, W.; Jakob, U. Stress-activated chaperones: A first line of defense. Trends Biochem. Sci. 2017, 42, 899–913. [Google Scholar] [CrossRef]
- Rimon, O.; Suss, O.; Goldenberg, M.; Fassler, R.; Yogev, O.; Amartely, H.; Propper, G.; Friedler, A.; Reichmann, D. A role of metastable regions and their connectivity in the inactivation of a redox-regulated chaperone and its inter-chaperone crosstalk. Antioxid. Redox Signal. 2017, 27, 1252–1267. [Google Scholar] [CrossRef] [PubMed]
- Bruel, N.; Castanié-Cornet, M.-P.; Cirinesi, A.-M.; Koningstein, G.; Georgopoulos, C.; Luirink, J.; Genevaux, P. Hsp33 controls elongation factor-Tu stability and allows Escherichia coli growth in the absence of the major DnaK and trigger factor chaperones. J. Biol. Chem. 2012, 287, 44435–44446. [Google Scholar] [CrossRef] [PubMed]
- Noble, C.G.; Song, H. Structural studies of elongation and release factors. Cell. Mol. Life Sci. 2008, 65, 1335–1346. [Google Scholar] [CrossRef]
- Maracci, C.; Rodnina, M.V. Translational GTPases. Biopolymers 2016, 105, 463–475. [Google Scholar] [CrossRef]
- Thirup, S.S.; Van, L.B.; Nielsen, T.K.; Knudsen, C.R. Structural outline of the detailed mechanism for elongation factor Ts-mediated guanine nucleotide exchange on elongation factor Tu. J. Struct. Biol. 2015, 191, 10–21. [Google Scholar] [CrossRef]
- Jo, K.-S.; Kim, J.-H.; Ryu, K.-S.; Kang, J.-S.; Wang, C.-Y.; Lee, Y.-S.; Seo, M.-D.; Lee, Y.-H.; Won, H.-S. Unique unfoldase/aggregase activity of a molecular chaperone Hsp33 in its holding-inactive state. J. Mol. Biol. 2019, 431, 1468–1480. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.Y.; Kinoshita, M.; Kume, S.; GT, H.; Sugiki, T.; Ladbury, J.E.; Kojima, C.; Ikegami, T.; Kurisu, G.; Goto, Y.; et al. Non-covalent forces tune the electron transfer complex between ferredoxin and sulfite reductase to optimize enzymatic activity. Biochem. J. 2016, 473, 3837–3854. [Google Scholar] [CrossRef]
- Hoffmann, A.; Bukau, B.; Kramer, G. Structure and function of the molecular chaperone Trigger Factor. Biochim. Biophys. Acta 2010, 1803, 650–661. [Google Scholar] [CrossRef]
- Martinez-Hackert, E.; Hendrichson, W.A. Promiscuous substrate recognition in folding and assembly activities of the trigger factor chaperone. Cell 2009, 138, 923–934. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.-P.; Perrett, S.; Zhou, J.-M. Dimeric trigger factor stabluy binds folding-competent intermediates and cooperates with the DnaK-DnaJ-GrpE chaperone system to allow refolding. J. Biol. Chem. 2005, 280, 13315–13320. [Google Scholar] [CrossRef] [PubMed]
- Van Melderen, L.; Aertsen, A. Regulation and quality control by Lon-dependent proteolysis. Res. Microbiol. 2009, 160, 645–651. [Google Scholar] [CrossRef] [PubMed]
- Shalgi, R.; Hurt, J.A.; Krykbaeva, I.; Taipale, M.; Lindquist, S.; Burge, C.B. Widespread regulation of translation by elongation pausing in heat shock. Mol. Cell 2013, 49, 439–452. [Google Scholar] [CrossRef]
- Yutthanasirikul, R.; Nagano, T.; Jimbo, H.; Hihara, Y.; Kanamori, T.; Ueda, T.; Haruyama, T.; Konno, H.; Yoshida, K.; Hisabori, T.; et al. Oxidation of a cysteine residue in elongation factor EF-Tu reversibly inhibits translation in the cyanobacterium Synechocystis sp. PCC 6803. J. Biol. Chem. 2016, 291, 5860–5870. [Google Scholar] [CrossRef]
- Wholey, W.Y.; Jakob, U. Hsp33 confers bleach resistance by protecting elongation factor Tu against oxidative degradation in Vibro cholerae. Mol. Microbiol. 2012, 83, 981–991. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Keum, M.; Ito, D.; Kim, M.-S.; Lin, Y.; Yoon, K.-H.; Kim, J.; Lee, S.-H.; Kim, J.-H.; Yu, W.; Lee, Y.-H.; et al. Molecular Effects of Elongation Factor Ts and Trigger Factor on the Unfolding and Aggregation of Elongation Factor Tu Induced by the Prokaryotic Molecular Chaperone Hsp33. Biology 2021, 10, 1171. https://doi.org/10.3390/biology10111171
Keum M, Ito D, Kim M-S, Lin Y, Yoon K-H, Kim J, Lee S-H, Kim J-H, Yu W, Lee Y-H, et al. Molecular Effects of Elongation Factor Ts and Trigger Factor on the Unfolding and Aggregation of Elongation Factor Tu Induced by the Prokaryotic Molecular Chaperone Hsp33. Biology. 2021; 10(11):1171. https://doi.org/10.3390/biology10111171
Chicago/Turabian StyleKeum, Minho, Dai Ito, Mi-Seong Kim, Yuxi Lin, Kyeong-Hyeon Yoon, Jihoon Kim, Sung-Hee Lee, Ji-Hun Kim, Wookyung Yu, Young-Ho Lee, and et al. 2021. "Molecular Effects of Elongation Factor Ts and Trigger Factor on the Unfolding and Aggregation of Elongation Factor Tu Induced by the Prokaryotic Molecular Chaperone Hsp33" Biology 10, no. 11: 1171. https://doi.org/10.3390/biology10111171
APA StyleKeum, M., Ito, D., Kim, M.-S., Lin, Y., Yoon, K.-H., Kim, J., Lee, S.-H., Kim, J.-H., Yu, W., Lee, Y.-H., & Won, H.-S. (2021). Molecular Effects of Elongation Factor Ts and Trigger Factor on the Unfolding and Aggregation of Elongation Factor Tu Induced by the Prokaryotic Molecular Chaperone Hsp33. Biology, 10(11), 1171. https://doi.org/10.3390/biology10111171