A Study of the Coevolution of Digital Organisms with an Evolutionary Cellular Automaton
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. General Description of the Cellular Automaton
2.2. Notation of ECA
- 1
- Habitat related parameters defining selective pressure:
- (a)
- NumberOfCells: number of cells that form the habitat.
- (b)
- NumberOfRsrcsInEachCell: discrete number of resources for each cell.
- (c)
- Distribution: redistribution of the populations or permeability among cells after every generation. It defines the redistribution strategy and the distance from the average after every generation.
- 2
- Species parameters that define each digital organism:
- (d)
- id: name of the species.
- (e)
- NumberOfItems: the size of the initial population.
- (f)
- DirectOffspring: number of direct offspring for each generation.
- (g)
- IndirectOffspring: number of indirect offspring or offspring given to their associates in each generation.
- (h)
- Distribution: akin to the above and defined with the habitat parameters, but each species is specified here. If it is not, the global value of Distribution is taken by default. This way, whereas habitat structural dispersion would be its permeability, species functional dispersion would be its vagility (i.e., the ability of a species to move about freely).
- (i)
- GroupPartner: list of identifiers (id) of the species with which a given species can group. Organisms in a cell are grouped with the organisms listed in their GroupPartner in the proportion indicated by their PhenotypicFlexibility parameter. To have partners requires defining not only the grouped species but also each group as a new species. Such groups are identified syntactically by joining the identifiers of each component with a vertical bar (|), e.g., A|B. Each group can be composed of two or more partners, e.g., A|B|C.
- (j)
- PhenotypicFlexibility: as explained in the definition of GroupPartner, it is the proportion of organisms of the species that must be grouped. When applied to the group, PhenotypicFlexibility defines the proportion of that group that remains grouped into the next generation—the greater the PhenotypicFlexibility, the more significant the number of groupings in the habitat.
- (k)
- AssociatedSpecies: id list of identifiers with which a given species is associated. As we will see below, one of the differences between grouping and association-based interactions is that all the organisms that meet their associate are associated with the latter. In contrast, grouping is only possible according to the proportion indicated in their PhenotypicFlexibility, as previously indicated. It is also remarkable that, whereas groups are new organisms with characteristics that are different from those of their components, associations are dynamic forms that do not generate new organisms but only indicate cooperation between species.
- (l)
- FitnessVariationLimit: maximum variation of DirectOffspring in the event that random variations of DirectFitness and IndirectFitness parameters are allowed. In any case, the sum of both is always constant.
2.3. Fitness of the Model
2.4. Characteristics of ECA
- (a)
- Immutable biological efficiency: a consumed resource unit always provides a descendant.
- (b)
- Resource uniformity: every cell has the same resources. Every cell representing the dynamic patches that divide the habitat [53] has the same amount of resources at the beginning of every generation, and the extra resources of each generation disappear for the next.
- (c)
- One-to-one interactions: in every cell, each simple or grouped organism relates to each other by pairs, 1 to 1. If an organism interacts with a list of species, the number of couples with each species is set up proportionally to their populations, as we assume that the greater the population density, the higher the probability of interaction.
- (d)
- Randomness accessing resources: there are no hierarchies to access resources; every organism has the same opportunities to obtain them by queuing randomly.
- (e)
- Simultaneous generational changes: generational changes are simultaneous for the whole habitat so that all the organisms consume their resources and have their offspring simultaneously.
- (f)
- Parents do not survive replication: only descendants survive the next generation.
- (g)
- There are no mutations: mutations are not considered in studying the effects of the parameters that are the objective of our study. Nevertheless, ECA can introduce mutations in species by (i) pausing the execution of the simulation; (ii) modifying the values to mutate in the intermediate configuration automatically saved in the folder cont.json; and (iii) by restarting the execution.
- (h)
- Habitat changes are not considered: for the same reasons as above, habitat changes can be introduced manually following the above-mentioned instructions.
- (i)
- Limited variability: we have implemented the argument varia to study the influence of selective pressure on species association capacity through variability and phenotypic accommodation of the associations [54]. The varia parameter sets random variability in the DirectOffspring of the descendants. The range of such variability is set with the parameter FitnessVariationLimit, which is also configured initially. Nevertheless, as nature limits evolutionary adaptations, the sum DirectOffspring + IndirectOffspring = InclusiveFitness of each organism will always remain unchanged, avoiding the coevolutionary “arms race” in biological fitness [55].
2.5. Other Features Not Considered in ECA
2.6. ECA Is Multi-Hierarchical
2.7. Biological Interactions in ECA
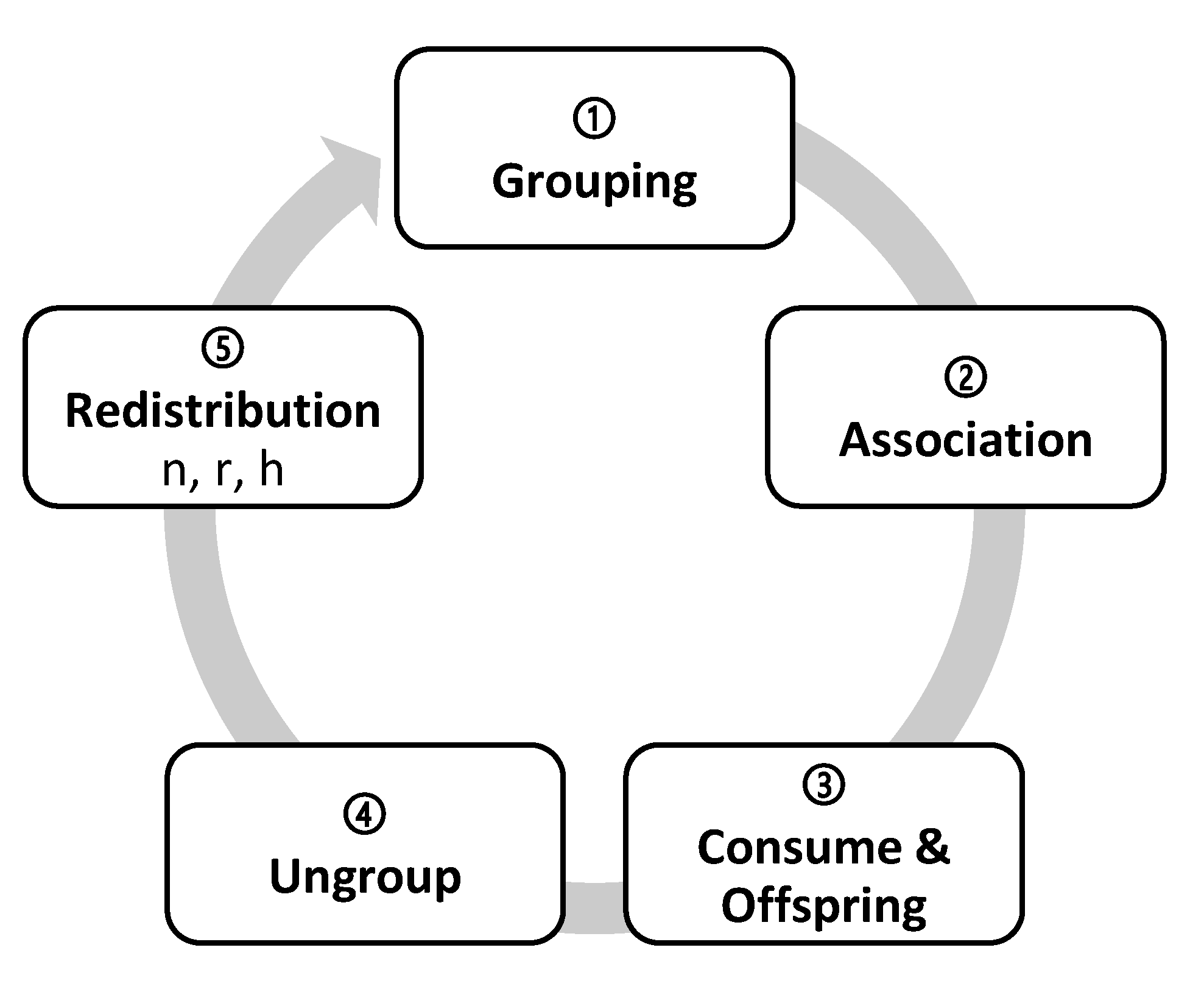
2.8. Transitional function in ECA
- (a)
- Grouping (doGrouping): the organisms that have one GroupPartner or more are grouped randomly, in proportion with the PhenotypicFlexibility of the first grouped organism.
- (b)
- Association (doAssociation): organisms with associates associate with them and can take up to five possible association roles: solitary, actors, recipients, intra-specific reciprocal, and inter-specific reciprocal (Figure 1).
- (c)
- Consumption and replication (doEnqueuingConsumeAndOffspring): organisms randomly queue to eat and reproduce. As long as there are resources they simultaneously eat and have their offspring, either directly (DirectOffspring), that they give to their species, or indirectly (IndirectOffspring), that they give to their associate. With this, randomness is implemented in access to resources, as the likelihood to occupy a specific spot in the queue is the same for all the organisms. We have studied three ways for the associates to access resources: (i) to place the associates together in a single spot of the queue; (ii) to place them in two different spots so that when is the turn of the first, it calls upon the second to consume together; and (iii) as in the second option, to place them independently but when is the turn of the first, it does not call upon the second to consume together. In the first option, associations are penalized, as they have half the chance that solitary organisms do to access resources; in the second option, associations double their chances, as they take two spots in the queue and either of the organisms ensures that both consume. We finally chose the third option, in which its recipients who do not consume will not have direct offspring, but they could receive indirect offspring as a result of the actor recipient interaction.
- (d)
- Ungrouping (doUngroup): recently formed groups ungroup proportionally to their phenotypic flexibility. The most complex groups must be ungrouped first, followed by the simplest ones to reach the maximum number of ungrouping. The more PhenotypicFlexibility the group has, the more groups remain.
- (e)
- Redistribution (doDistribute): preparing the next generation of descendants, they distribute between the cells, more or less uniformly. Depending on the cells involved, the distribution can be (i) local (combining the neighbors only) or (ii) global (ignoring the place of origin). There are, therefore, two strategies: (i) The n strategy of neigbours_distribution generates a local distribution: descendants randomly distribute from one generation to the next, one by one but within a range of neighboring cells. The user can set the range of adjacent cells or neighborhood range in the initial configuration (Distribution) from the value 0n, in which organisms remain in their cell of origin, to the value 100n, in which organisms distribute among any of the cells of the reticular network. According to the law of large numbers, the distribution becomes more uniform for larger populations and broader ranges, decreasing the variance. (ii) The r strategy of random_global_avg generates a global redistribution: all the organisms globally disperse, regardless of their origin. Groups of random size are constructed and assigned to target cells randomly. Although the neighborhood range is maxed out, the whole reticular network can distribute from 0r with zero variance (as all the values are the same) up to 100r, where each cell receives a random number of organisms, thus obtaining a high variance. The h strategy of random_global_by_cell is another alternative: the number of target cells is reduced while the distribution r takes place, increasing the variance with respect to the mean. This produces a higher or lower number of empty cells depending on the chosen value of h. Thus, 0h means no empty cells and variance 0, where all the values are the same (equivalent to 0r), whereas 50h means a distribution with 50% of empty cells and 50% of randomly occupied cells also with a distribution 50r. The extreme value 100h means that all organisms end in only one cell.
2.9. Software
3. Results and Discussion
3.1. ECA as a Virtual Lab
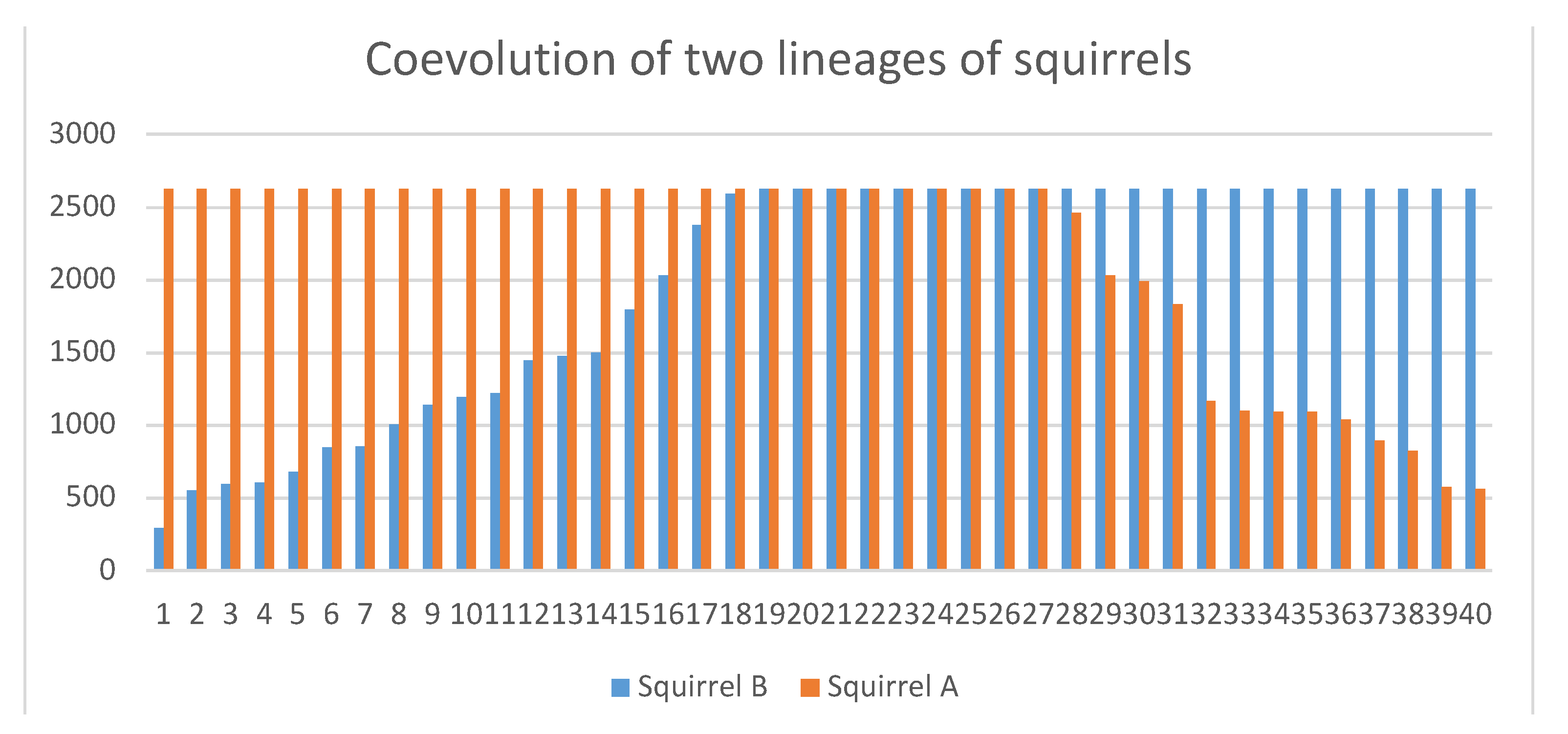
3.2. Specific Scenarios Do Not Favor Collaboration
3.3. The Emergence of Obligate Mutualism
3.4. The Well-Known Random Genetic Drift as a Selective Phenomenon in ACE
3.5. Kin Selection and Relevance of Benefit in Fitness
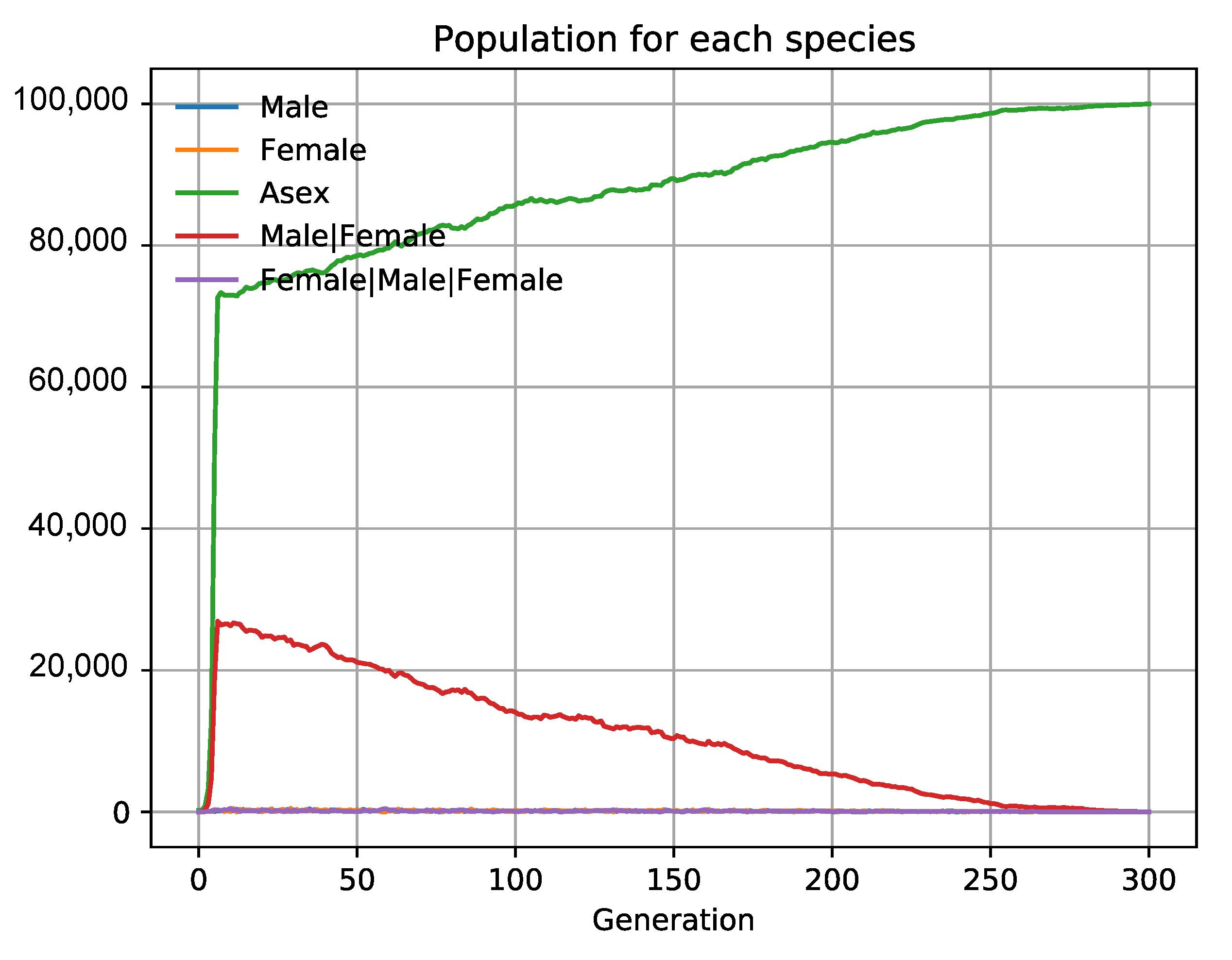
3.6. The Evolution of Sex
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Thompson, J.N. The Geographic Mosaic of Coevolution; University of Chicago Press: Chicago, IL, USA, 2005; ISBN 022611869X. [Google Scholar]
- Medeiros, L.P.; Garcia, G.; Thompson, J.N.; Guimarães, P.R. The geographic mosaic of coevolution in mutualistic networks. Proc. Natl. Acad. Sci. USA 2018, 115, 12017–12022. [Google Scholar] [CrossRef]
- Janzen, D.H. When is it Coevolution? Evolution 1980, 34, 611. [Google Scholar] [CrossRef]
- Hall, A.R.; Ashby, B.; Bascompte, J.; King, K.C. Measuring Coevolutionary Dynamics in Species-Rich Communities. Trends Ecol. Evol. 2020, 35, 539–550. [Google Scholar] [CrossRef]
- Thompson, J.N. Interaction and Coevolution; University of Chicago Press: Chicago, IL, USA, 2014; ISBN 022612732X. [Google Scholar]
- Gaba, S.; Ebert, D. Time-shift experiments as a tool to study antagonistic coevolution. Trends Ecol. Evol. 2009, 24, 226–232. [Google Scholar] [CrossRef]
- Lenski, R.E. Two-step resistance by Escherichia coli B to bacteriophage T2. Genetics 1984, 107, 1–7. [Google Scholar] [CrossRef]
- Frank, S.A. Coevolutionary genetics of hosts and parasites with quantitative inheritance. Evol. Ecol. 1994, 8, 74–94. [Google Scholar] [CrossRef][Green Version]
- Sasaki, A. Host-parasite coevolution in a multilocus gene-for-gene system. Proc. R. Soc. London B Biol. Sci. 2000, 267, 2183–2188. [Google Scholar] [CrossRef] [PubMed]
- Woodford, P. Evaluating inclusive fitness. R. Soc. Open Sci. 2019, 6, 190644. [Google Scholar] [CrossRef] [PubMed]
- Tack, A.J.M.; Thrall, P.H.; Barrett, L.G.; Burdon, J.J.; Laine, A.-L. Variation in infectivity and aggressiveness in space and time in wild host-pathogen systems: Causes and consequences. J. Evol. Biol. 2012, 25, 1918–1936. [Google Scholar] [CrossRef]
- Dawkins, R.; Krebs, J.R. Arms races between and within species. Proc. R. Soc. London. Ser. B Biol. Sci. 1979, 205, 489–511. [Google Scholar]
- Ostrowski, E.A.; Shen, Y.; Tian, X.; Sucgang, R.; Jiang, H.; Qu, J.; Katoh-Kurasawa, M.; Brock, D.A.; Dinh, C.; Lara-Garduno, F.; et al. Genomic Signatures of Cooperation and Conflict in the Social Amoeba. Curr. Biol. 2015, 25, 1661–1665. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.; Liu, H. Coexistence in a two species chemostat model with Markov switchings. Appl. Math. Lett. 2019, 94, 266–271. [Google Scholar] [CrossRef]
- Herbert, D.; Elsworth, R.; Telling, R.C. The Continuous Culture of Bacteria; a Theoretical and Experimental Study. J. Gen. Microbiol. 1956, 14, 601–622. [Google Scholar] [CrossRef]
- Abruzzi, K.C.; Zadina, A.; Luo, W.; Wiyanto, E.; Rahman, R.; Guo, F.; Shafer, O.; Rosbash, M. RNA-seq analysis of Drosophila clock and non-clock neurons reveals neuron-specific cycling and novel candidate neuropeptides. PLoS Genet. 2017, 13, e1006613. [Google Scholar] [CrossRef] [PubMed]
- Hosokawa, T.; Ishii, Y.; Nikoh, N.; Fujie, M.; Satoh, N.; Fukatsu, T. Obligate bacterial mutualists evolving from environmental bacteria in natural insect populations. Nat. Microbiol. 2016, 1, 15011. [Google Scholar] [CrossRef] [PubMed]
- Alba, D.M.; Moyà-Solà, S.; Köhler, M. Morphological affinities of the Australopithecus afarensis hand on the basis of manual proportions and relative thumb length. J. Hum. Evol. 2003, 44, 225–254. [Google Scholar] [CrossRef]
- Chapman, D.S.; Dytham, C.; Oxford, G.S. Modelling population redistribution in a leaf beetle: An evaluation of alternative dispersal functions. J. Anim. Ecol. 2007, 76, 36–44. [Google Scholar] [CrossRef]
- Miller, T.E.X.; Shaw, A.K.; Inouye, B.D.; Neubert, M.G. Sex-biased dispersal and the speed of two-sex invasions. Am. Nat. 2011, 177, 549–561. [Google Scholar] [CrossRef]
- Radinger, J.; Wolter, C. Patterns and predictors of fish dispersal in rivers. Fish Fish. 2014, 15, 456–473. [Google Scholar] [CrossRef]
- Zhou, Y.; Kot, M. Discrete-time growth-dispersal models with shifting species ranges. Theor. Ecol. 2011, 4, 13–25. [Google Scholar] [CrossRef]
- Von Neumann, J.; Burks, A.W. Theory of Self-Reproducing Automata; University of Illinois Press: London, UK, 1966. [Google Scholar]
- Adami, C. Digital genetics: Unravelling the genetic basis of evolution. Nat. Rev. Genet. 2006, 7, 109–118. [Google Scholar] [CrossRef] [PubMed]
- Lehman, J.; Clune, J.; Misevic, D. The surprising creativity of digital evolution: A collection of anecdotes from the evolutionary computation and artificial life research communities. Artif. Life 2020, 26, 274–306. [Google Scholar] [CrossRef] [PubMed]
- Dalquen, D.A.; Anisimova, M.; Gonnet, G.H.; Dessimoz, C. ALF-A simulation framework for genome evolution. Mol. Biol. Evol. 2012, 29, 1115–1123. [Google Scholar] [CrossRef]
- Pigliucci, M. Phenotypic Plasticity: Beyond Nature and Nurture; JHU Press: Baltimore, MD, USA, 2001; ISBN 0801867886. [Google Scholar]
- Gianoli, E. Plasticidad fenotípica adaptativa en plantas. In Fisiología Ecológica en Plantas. Mecanismos y Respuestas a Estrés en los Ecosistemas; EUV: Valparaíso, Chile, 2004; pp. 13–25. [Google Scholar]
- Piersma, T.; Drent, J. Phenotypic flexibility and the evolution of organismal design. Trends Ecol. Evol. 2003, 18, 228–233. [Google Scholar] [CrossRef]
- Sober, E. What Is Evolutionary Altruism? Can. J. Philos. 1988, 18, 75–99. [Google Scholar] [CrossRef]
- Cracraft, J. Species concepts and speciation analysis. Curr. Ornithol. 1983, 1, 159–187. [Google Scholar] [CrossRef]
- Falgueras-Cano, J.; Carretero-Díaz, J.M.; Moya, A. Weighted fitness theory: An approach to symbiotic communities. Environ. Microbiol. Rep. 2017, 9, 44–46. [Google Scholar] [CrossRef][Green Version]
- Heisler, I.L.; Damuth, J. A method for analyzing selection in hierarchically structured populations. Am. Nat. 1987, 130, 582–602. [Google Scholar] [CrossRef]
- Hamilton, W.D. The Evolution of Altruistic Behavior. Am. Nat. 1963, 97, 354–356. [Google Scholar] [CrossRef]
- Jørgensen, S.E.; Fath, B.D. Individual-Based Models. In Developments in Environmental Modelling; Elsevier: Amsterdam, The Netherlands, 2011; Volume 23, pp. 291–308. [Google Scholar]
- Golestani, A.; Gras, R.; Cristescu, M. Speciation with gene flow in a heterogeneous virtual world: Can physical obstacles accelerate speciation? Proc. R. Soc. B Biol. Sci. 2012, 279, 3055–3064. [Google Scholar] [CrossRef]
- Scott, R.; MacPherson, B.; Gras, R. Ecosim, an enhanced artificial ecosystem: Addressing deeper behavioral, ecological, and evolutionary questions. In Intelligent Systems, Control and Automation: Science and Engineering; Springer: Cham, Switzerland, 2019; Volume 94, pp. 223–278. [Google Scholar]
- Ofria, C.; Wilke, C.O. Avida: A software platform for research in computational evolutionary biology. Artif. Life 2004, 10, 191–229. [Google Scholar] [CrossRef]
- Zaman, L.; Meyer, J.R.; Devangam, S.; Bryson, D.M.; Lenski, R.E.; Ofria, C. Coevolution Drives the Emergence of Complex Traits and Promotes Evolvability. PLoS Biol. 2014, 12, e1002023. [Google Scholar] [CrossRef] [PubMed]
- Chow, S.S.; Wilke, C.O.; Ofria, C.; Lenski, R.E.; Adami, C. Adaptive radiation from resource competition in digital organisms. Science 2004, 305, 84–86. [Google Scholar] [CrossRef] [PubMed]
- Bohm, C.; Ackles, A.L.; Ofria, C.; Hintze, A. On Sexual Selection in the Presence of Multiple Costly Displays. In Proceedings of the 2019 Conference on Artificial Life: How Can Artificial Life Help Solve Societal Challenges, ALIFE 2019, Newcastle upon Tyne, UK, 29 July 2019; MIT Press: Cambridge, MA, USA, 2020; pp. 247–254. [Google Scholar]
- Meurant, G. The Ecology of Natural Disturbance and Patch Dynamics; Academic Press: Cambridge, MA, USA, 2012; ISBN 0323138934. [Google Scholar]
- Weiner, J.; Freckleton, R.P. Constant Final Yield. Annu. Rev. Ecol. Evol. Syst. 2010, 41, 173–192. [Google Scholar] [CrossRef]
- Hui, C. Carrying capacity, population equilibrium, and environment’s maximal load. Ecol. Modell. 2006, 192, 317–320. [Google Scholar] [CrossRef]
- West, S.A.; Griffin, A.S.; Gardner, A. Social semantics: Altruism, cooperation, mutualism, strong reciprocity and group selection. J. Evol. Biol. 2007, 20, 415–432. [Google Scholar] [CrossRef]
- Holling, C.S. The Components of Predation as Revealed by a Study of Small-Mammal Predation of the European Pine Sawfly. Can. Entomol. 1959, 91, 293–320. [Google Scholar] [CrossRef]
- Moskát, C.; Honza, M. Effect of nest and nest site characteristics on the risk of cuckoo Cuculus canorus parasitism in the great reed warbler Acrocephalus arundinaceus. Ecography 2000, 23, 335–341. [Google Scholar] [CrossRef]
- Takasu, F.; Kawasaki, K.; Nakamura, H.; Cohen, J.E.; Shigesada, N. Modeling the population dynamics of a cuckoo-host association and the evolution of host defenses. Am. Nat. 1993, 142, 819–839. [Google Scholar] [CrossRef]
- Gause, G.F. Experimental studies on the struggle for existence: I. Mixed population of two species of yeast. J. Exp. Biol. 1932, 9, 389–402. [Google Scholar] [CrossRef]
- Odum, E.P.; Odum, H.T. Fundamentals of Ecology; B. Saunders Co.: Philadelphia, PA, USA, 1963. [Google Scholar]
- Sazima, I.; Moura, R.L.; Rodrigues, M.C.M. A juvenile sharksucker, Echeneis naucrates (Echeneidae), acting as a station-based cleaner fish. Cybium 1999, 23, 377–380. [Google Scholar]
- Herbert-Read, J.E.; Romanczuk, P.; Krause, S.; Strömbom, D.; Couillaud, P.; Domenici, P.; Kurvers, R.H.J.M.; Marras, S.; Steffensen, J.F.; Wilson, A.D.M.; et al. Proto-Cooperation: Group hunting sailfish improve hunting success by alternating attacks on grouping prey. Proc. R. Soc. B Biol. Sci. 2016, 283, 20161671. [Google Scholar] [CrossRef] [PubMed]
- Pickett, S.T.A.; Rogers, K.H. Patch Dynamics: The Transformation of Landscape Structure and Function. In Wildlife and Landscape Ecology; Springer: New York, NY, USA, 1997; pp. 101–127. [Google Scholar]
- English, S.; Browning, L.E.; Raihani, N.J. Developmental plasticity and social specialization in cooperative societies. Anim. Behav. 2015, 106, 37–42. [Google Scholar] [CrossRef]
- Scheinberg, L. The Red Queen. Arch. Neurol. 1983, 40, 189. [Google Scholar] [CrossRef]
- Moyer, J.T.; Nakazono, A. Protandrous hermaphroditism in six species of the anemonefish genus Amphiprion in Japan. Jpn. J. Ichthyol. 1978, 25, 101–106. [Google Scholar] [CrossRef]
- Hull, D.L. Individuality and selection. Annu. Rev. Ecol. Syst. 1980, 11, 311–332. [Google Scholar] [CrossRef]
- Brandon, R. Adaptation and Environment; Princeton University Press: Princeton, NJ, USA, 2014. [Google Scholar]
- Waters, C.M.; Bassler, B.L. Quorum sensing: Cell-to-cell communication in bacteria. Annu. Rev. Cell Dev. Biol. 2005, 21, 319–346. [Google Scholar] [CrossRef]
- Spiers, A.J.; Bohannon, J.; Gehrig, S.M.; Rainey, P.B. Biofilm formation at the air-liquid interface by the Pseudomonas fluorescens SBW25 wrinkly spreader requires an acetylated form of cellulose. Mol. Microbiol. 2003, 50, 15–27. [Google Scholar] [CrossRef]
- Rainey, P.B.; Rainey, K. Evolution of cooperation and conflict in experimental bacterial populations. Nature 2003, 425, 72–74. [Google Scholar] [CrossRef]
- Travisano, M.; Velicer, G. Strategies of microbial cheater control.pdf. Trends Microbiol. 2004, 12, 77–78. [Google Scholar] [CrossRef]
- Mitri, S.; Xavier, J.B.; Foster, K.R. Social evolution in multispecies biofilms. Proc. Natl. Acad. Sci. USA 2011, 108, 10839–10846. [Google Scholar] [CrossRef]
- Sakagami, S.; Maeta, Y. Sociality, Induced and/or Natural, in the Basically Solitary Small Carpenter Bees (Ceratina); Japan Scientific Society Press: Tokyo, Japan, 1987. [Google Scholar]
- Cornforth, D.M.; Foster, K.R. Competition sensing: The social side of bacterial stress responses. Nat. Rev. Microbiol. 2013, 11, 285–293. [Google Scholar] [CrossRef]
- Cohen, D. Optimizing reproduction in a randomly varying environment. J. Theor. Biol. 1966, 12, 119–129. [Google Scholar] [CrossRef]
- Lindenmayer, D.B.; Fischer, J. Habitat Fragmentation and Landscape Change; Island Press: Washington, DC, USA, 2006. [Google Scholar]
- Beier, P.; Spencer, W.; Baldwin, R.F.; Mcrae, B.H. Toward Best Practices for Developing Regional Connectivity Maps. Conserv. Biol. 2011, 25, 879–892. [Google Scholar] [CrossRef] [PubMed]
- Pommier, S.; Strehaiano, P.; Délia, M.L. Modelling the growth dynamics of interacting mixed cultures: A case of amensalism. Int. J. Food Microbiol. 2005, 100, 131–139. [Google Scholar] [CrossRef]
- Guo, S.W.; Thompson, E.A. Performing the Exact Test of Hardy-Weinberg Proportion for Multiple Alleles. Biometrics 1992, 48, 361. [Google Scholar] [CrossRef] [PubMed]
- Hepburn, H.R. The Bees of the World. African Zool. 2001, 36, 117. [Google Scholar] [CrossRef]
- Cane, J.H. Habitat Fragmentation and Native Bees. Conserv. Ecol. 2001. [Google Scholar]
- Nowak, M.A.; Tarnita, C.E.; Wilson, E.O. The evolution of eusociality. Nature 2010, 466, 1057–1062. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, L.; Keller, L.; West, S.; Roze, D. Group selection and kin selection: Two concepts but one process. Proc. Natl. Acad. Sci. USA 2007, 104, 6736–6739. [Google Scholar] [CrossRef]
- Marshall, J.A.R. Group selection and kin selection: Formally equivalent approaches. Trends Ecol. Evol. 2011, 26, 325–332. [Google Scholar] [CrossRef]
- Masel, J. Genetic drift. Curr. Biol. 2011, 21, R837–R838. [Google Scholar] [CrossRef] [PubMed]
- Cormack, R.M.; Hartl, D.L.; Clark, A.G. Principles of Population Genetics. Biometrics 1990, 46, 546. [Google Scholar] [CrossRef]
- Gilbert, B.; Levine, J.M. Ecological drift and the distribution of species diversity. Proc. R. Soc. B Biol. Sci. 2017, 284, 20170507. [Google Scholar] [CrossRef]
- Hamilton, W.D. Selfish and spiteful behaviour in an evolutionary model. Nature 1970, 228, 1218–1220. [Google Scholar] [CrossRef] [PubMed]
- Krakauer, A.H. Kin selection and cooperative courtship in wild turkeys. Nature 2005, 434, 69–72. [Google Scholar] [CrossRef]
- Gorrell, J.C.; McAdam, A.G.; Coltman, D.W.; Humphries, M.M.; Boutin, S. Adopting kin enhances inclusive fitness in asocial red squirrels. Nat. Commun. 2010, 1, 22. [Google Scholar] [CrossRef] [PubMed]
- Goodenough, U.; Heitman, J. Origins of eukaryotic sexual reproduction. Cold Spring Harb. Perspect. Biol. 2014, 6, a016154. [Google Scholar] [CrossRef] [PubMed]
- Williams, G.C. Sex and Evolution; Princeton University Press: Princeton, NJ, USA, 1975; ISBN 0691081522. [Google Scholar]
- Smith, J.M.; Maynard-Smith, J. The Evolution of Sex; Cambridge University Press: Cambridge, MA, USA, 1978; Volume 4. [Google Scholar]
- Cabrero, J.; Camacho, J. Evolucion, la base de la Biología; Proyecto Sur de Ediciones; UADER: Granada, Spain, 2002; ISBN 8482541390. [Google Scholar]
- Nunney, L. The maintenance of sex by group selection. Evolution 1989, 43, 245–257. [Google Scholar] [CrossRef]
- Hamilton, W.D. Gamblers Since Life Began: Barnacles, Aphids, Elms. Q. Rev. Biol. 1975, 50, 175–180. [Google Scholar] [CrossRef]
- Hamilton, W.D. Sex versus Non-Sex versus Parasite. Oikos 1980, 35, 282. [Google Scholar] [CrossRef]
- Wilkinson, D.M. Running with the Red Queen: Reflections on “Sex versus non-sex versus parasite”. Oikos 2000, 91, 589–596. [Google Scholar] [CrossRef]
- Orians, G.H. On the Evolution of Mating Systems in Birds and Mammals. Am. Nat. 1969, 103, 589–603. [Google Scholar] [CrossRef]
- Fisher, R.A. The Genetical Theory of Natural Selection.; Clarendon Press: Oxford, UK, 2011. [Google Scholar]
- Harris, C.R.; Millman, K.J.; van der Walt, S.J.; Gommers, R.; Virtanen, P.; Cournapeau, D.; Wieser, E.; Taylor, J.; Berg, S.; Smith, N.J.; et al. Array Programming with NumPy. Nature 2020, 585, 357. [Google Scholar] [CrossRef] [PubMed]






| Relations | File | Results in ECA |
|---|---|---|
| Predation | 1Pred.json | Resources are prey |
| Amensalism | 1Amen2.json | Eucalyptus kills other plants |
| Parasitism | 1Para.json | The cuckoo survives |
| Exclusion | 1Exclu.json | The most efficient excludes the other |
| Intra-specific competition | 1Intra.json | Only the fittest live |
| Neutralism | 1Neu.json | They keep in balance |
| Commensalism | 1Comm.json | Only one benefits |
| Proto-Cooperation | 1Proto.json | Proto-cooperation prevails |
| Intra-specific social collaboration | 1Colab.json | Social bees prevail |
| Subsociality | Symb.json (simulation 4) | Sometimes solitary and other mutualists |
| Symbiosis | Symb.json (simulation 5) | Symbiosis prevails |
| Role | Direct Offspring | Indirect Offspring | Personal Offspring | Inclusive Offspring |
|---|---|---|---|---|
| Actor | ||||
| Recipient | + | |||
| Reciprocal | + |
| Partnerships | Individual | Recipient | Actor | Reciprocal |
|---|---|---|---|---|
| Consumption | ||||
| Offspring |
| Theories | File | Results in ECA |
|---|---|---|
| Law of Constant Final Yield | 1LCFY.json | Resources control population |
| Numerical and functional answer | 1Pred.json | Resources grow, the population grows |
| Competitive exclusion principle | 1Exclu.json | The most efficient excludes the other |
| Random Genetic drift | 1Deri.json | It tends towards homozygosity |
| Ecological drift | 1DeriSp.json | Populations are extinct due to sampling errors |
| Hardy–Weinberg principle | 1Hardy.json | Allele balance without natural selection |
| Fisher’s principle | 1Fish.json | The sex ratio 1:1 prevails |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Falgueras-Cano, J.; Falgueras-Cano, J.-A.; Moya, A. A Study of the Coevolution of Digital Organisms with an Evolutionary Cellular Automaton. Biology 2021, 10, 1147. https://doi.org/10.3390/biology10111147
Falgueras-Cano J, Falgueras-Cano J-A, Moya A. A Study of the Coevolution of Digital Organisms with an Evolutionary Cellular Automaton. Biology. 2021; 10(11):1147. https://doi.org/10.3390/biology10111147
Chicago/Turabian StyleFalgueras-Cano, Javier, Juan-Antonio Falgueras-Cano, and Andrés Moya. 2021. "A Study of the Coevolution of Digital Organisms with an Evolutionary Cellular Automaton" Biology 10, no. 11: 1147. https://doi.org/10.3390/biology10111147
APA StyleFalgueras-Cano, J., Falgueras-Cano, J.-A., & Moya, A. (2021). A Study of the Coevolution of Digital Organisms with an Evolutionary Cellular Automaton. Biology, 10(11), 1147. https://doi.org/10.3390/biology10111147







