Emerging Status of Multidrug-Resistant Bacteria and Fungi in the Arabian Peninsula
Abstract
Simple Summary
Abstract
1. Purpose and Methods
- Is there a common MDR organism (MDRO) reported in the Arabian Peninsula?
- Are there any MDR bacteria and fungi that have emerged in the Arabian Peninsula in the last 10 years?
- What are the logical relationships between sets of MDR bacteria and fungi in countries of the Arabian Peninsula?
- How many in-depth studies have been conducted on the molecular nature of drug-resistant micro-organisms prevalent in the Arabian Peninsula?
- What are the novel antimicrobial strategies developed in the study region?
- Is there a high mortality reported due to MDR micro-organisms in the Arabian Peninsula?
- To what extent have nanomaterials and nanoparticles been exploited against MDR micro-organisms in the Arabian Peninsula?
- Articles reporting the multidrug-resistant bacteria in the Arabian Peninsula.
- Articles reporting multidrug-resistant fungi in the Arabian Peninsula.
- Research articles published since 2010 regarding multidrug-resistant micro-organisms in the Arabian Peninsula.
- Full-text articles.
- Articles discussing the use of multidrug-resistant bacteria and fungi to screen the antimicrobials in the Arabian Peninsula.
- Articles reporting the multidrug-resistant micro-organisms from other than the Arabian Peninsula.
- Review articles reporting the multidrug-resistant micro-organisms from the Arabian Peninsula.
2. Logical Relation between Sets of Multidrug-Resistant (MDR) Microbes
2.1. Saudi Arabia
2.2. Bahrain
2.3. Kuwait
2.4. Oman
2.5. Qatar
2.6. United Arab Emirates
2.7. Jordan
2.8. Iraq
2.9. Yemen
3. Methods Used to Detect Multidrug-Resistant Organisms
4. Antimicrobial Resistance Profiles Used for Treating Multidrug-Resistant Organisms
5. Mining Natural and Novel Antimicrobials against MDR
6. Conclusions and Future Perspectives
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sonnevend, Á.; Ghazawi, A.; Alqahtani, M.; Shibl, A.; Jamal, W.; Hashmey, R.; Pal, T. Plasmid-Mediated Colistin Resistance in Escherichia coli from the Arabian Peninsula. Int. J. Infect. Dis. 2016, 50, 85–90. [Google Scholar] [CrossRef]
- Zowawi, H.M.; Sartor, A.L.; Sidjabat, H.E.; Balkhy, H.H.; Walsh, T.R.; Al Johani, S.M.; AlJindan, R.Y.; Alfaresi, M.; Ibrahim, E.; Al-Jardani, A.; et al. Molecular Epidemiology of Carbapenem-Resistant Acinetobacter Baumannii Isolates in the Gulf Cooperation Council States: Dominance of OXA-23-Type Producers. J. Clin. Microbiol. 2015, 53, 896–903. [Google Scholar] [CrossRef]
- Sonnevend, Á.; Ghazawi, A.A.; Hashmey, R.; Jamal, W.; Rotimi, V.O.; Shibl, A.M.; Al-Jardani, A.; Al-Abri, S.S.; Tariq, W.U.Z.; Weber, S.; et al. Characterization of Carbapenem-Resistant Enterobacteriaceae with High Rate of Autochthonous Transmission in the Arabian Peninsula. PLoS ONE 2015, 10, e0131372. [Google Scholar] [CrossRef]
- Rolain, J.-M.; Loucif, L.; Al-Maslamani, M.; Elmagboul, E.; Al-Ansari, N.; Taj-Aldeen, S.; Shaukat, A.; Ahmedullah, H.; Hamed, M. Emergence of Multidrug-Resistant Acinetobacter Baumannii Producing OXA-23 Carbapenemase in Qatar. New Microbes New Infect. 2016, 11, 47–51. [Google Scholar] [CrossRef] [PubMed]
- Khan, F.Y.; Abukhattab, M.; AbuKamar, M.; Anand, D. Adult K Lebsiella Pneumoniae Meningitis in Qatar: Clinical Pattern of Ten Cases. Asian Pac. J. Trop. Biomed. 2014, 4, 669–672. [Google Scholar] [CrossRef]
- Al Samawi, M.S.; Khan, F.Y.; Eldeeb, Y.; Almaslamani, M.; Alkhal, A.; Alsoub, H.; Ghadban, W.; Howady, F.; Hashim, S. Acinetobacter Infections among Adult Patients in Qatar: A 2-Year Hospital-Based Study. Can. J. Infect. Dis. Med. Microbiol. 2016, 2016, 6873689. [Google Scholar] [CrossRef]
- Khan, F.Y.; Elshafie, S.S.; Almaslamani, M.; Abu-Khattab, M.; El Hiday, A.H.; Errayes, M.; Almaslamani, E. Epidemiology of Bacteraemia in Hamad General Hospital, Qatar: A One Year Hospital-Based Study. Travel Med. Infect. Dis. 2010, 8, 377–387. [Google Scholar] [CrossRef] [PubMed]
- John Albert, M.; Bulach, D.; Alfouzan, W.; Izumiya, H.; Carter, G.; Alobaid, K.; Alatar, F.; Sheikh, A.R.; Poirel, L. Non-Typhoidal Salmonella Blood Stream Infection in Kuwait: Clinical and Microbiological Characteristics. PLoS Negl. Trop. Dis. 2019, 13, e0007293. [Google Scholar] [CrossRef]
- Alfouzan, W.; Ahmad, S.; Dhar, R.; Asadzadeh, M.; Almerdasi, N.; Abdo, N.M.; Joseph, L.; de Groot, T.; Alali, W.Q.; Khan, Z.; et al. Molecular Epidemiology of Candida Auris Outbreak in a Major Secondary-Care Hospital in Kuwait. J. Fungi 2020, 6, 307. [Google Scholar] [CrossRef] [PubMed]
- Jamal, W.; Rotimi, V.O.; Albert, M.J.; Khodakhast, F.; Nordmann, P.; Poirel, L. High Prevalence of VIM-4 and NDM-1 Metallo-β-Lactamase among Carbapenem-Resistant Enterobacteriaceae. J. Med. Microbiol. 2013, 62, 1239–1244. [Google Scholar] [CrossRef]
- Khan, Z.; Ahmad, S.; Al-Sweih, N.; Joseph, L.; Alfouzan, W.; Asadzadeh, M. Increasing Prevalence, Molecular Characterization and Antifungal Drug Susceptibility of Serial Candida Auris Isolates in Kuwait. PLoS ONE 2018, 13, e0195743. [Google Scholar] [CrossRef] [PubMed]
- Jamal, W.; Al Roomi, E.; AbdulAziz, L.R.; Rotimi, V.O. Evaluation of Curetis Unyvero, a Multiplex PCR-Based Testing System, for Rapid Detection of Bacteria and Antibiotic Resistance and Impact of the Assay on Management of Severe Nosocomial Pneumonia. J. Clin. Microbiol. 2014, 52, 2487–2492. [Google Scholar] [CrossRef]
- Jamal, W.Y.; Albert, M.J.; Khodakhast, F.; Poirel, L.; Rotimi, V.O. Emergence of New Sequence Type OXA-48 Carbapenemase-Producing Enterobacteriaceae in Kuwait. Microb. Drug Resist. 2015, 21, 329–334. [Google Scholar] [CrossRef]
- Ibrahim, M.M.; Abuelmatty, A.M.; Mohamed, G.H.; Nasr, M.A.; Hussein, A.K.; Ebaed, M.E.D.; Sarhan, H.A. Best Tigecycline Dosing for Treatment of Infections Caused by Multidrug-Resistant Pathogens in Critically Ill Patients with Different Body Weights. Drug Des. Dev. Ther. 2018, 12, 4171–4179. [Google Scholar] [CrossRef]
- Babakir-Mina, M.; Othman, N.; Najmuldeen, H.H.; Noori, C.K.; Fatah, C.F.; Perno, C.-F.; Ciotti, M. Antibiotic Susceptibility of Vancomyin and Nitrofurantoin in Staphylococcus Aureus Isolated from Burnt Patients in Sulaimaniyah, Iraqi Kurdistan. New Microbiol. 2012, 35, 439–446. [Google Scholar]
- Jaber, A.A.S.; Ibrahim, B. Evaluation of Risk Factors Associated with Drug-Resistant Tuberculosis in Yemen: Data from Centres with High Drug Resistance. BMC Infect. Dis. 2019, 19, 464. [Google Scholar] [CrossRef] [PubMed]
- Jaber, A.A.S.; Ibrahim, B. Health-Related Quality of Life of Patients with Multidrug-Resistant Tuberculosis in Yemen: Prospective Study. Health Qual. Life Outcomes 2019, 17, 142. [Google Scholar] [CrossRef]
- Abd El Hafez, M.; Khalaf, N.G.; El Ahmady, M.; Abd El Aziz, A.; Hashim, A.E.G. An Outbreak of Methicillin Resistant Staphylococcus EpidermiDis. among Neonates in a Hospital in Saudi Arabia. J. Infect. Dev. Ctries 2011, 5, 692–699. [Google Scholar] [CrossRef]
- Al Mayahi, Z.; Kamel, S.; Amer, H.; Beatty, M. Outbreak of Colistin-Resistant Organisms at a Tertiary Hospital in Riyadh, Saudi Arabia, 2016. Pan Afr. Med. J. 2019, 34, 162. [Google Scholar] [CrossRef]
- Khan, U.; Huerga, H.; Khan, A.J.; Mitnick, C.D.; Hewison, C.; Varaine, F.; Bastard, M.; Rich, M.; Franke, M.F.; Atwood, S.; et al. The EndTB Observational Study Protocol: Treatment of MDR-TB with Bedaquiline or Delamanid Containing Regimens. BMC Infect. Dis. 2019, 19, 733. [Google Scholar] [CrossRef] [PubMed]
- Al-Ghafli, H.; Kohl, T.A.; Merker, M.; Varghese, B.; Halees, A.; Niemann, S.; Al-Hajoj, S. Drug-Resistance Profiling and Transmission Dynamics of Multidrug-Resistant Mycobacterium Tuberculosis in Saudi Arabia Revealed by Whole Genome Sequencing. Infect. Drug Resist. 2018, 11, 2219–2229. [Google Scholar] [CrossRef]
- Al-Jindan, R.; Al-Eraky, D. Two Cases of the Emerging Candida Auris in a University Hospital from Saudi Arabia. Saudi J. Med. Med. Sci. 2021, 9, 71. [Google Scholar] [CrossRef]
- Alhussain, F.A.; Yenugadhati, N.; Al Eidan, F.A.; Al Johani, S.; Badri, M. Risk Factors, Antimicrobial Susceptibility Pattern and Patient Outcomes of Pseudomonas Aeruginosa Infection: A Matched Case-Control Study. J. Infect. Public Health 2021, 14, 152–157. [Google Scholar] [CrossRef]
- AlJindan, R.; AlEraky, D.M.; Mahmoud, N.; Abdalhamid, B.; Almustafa, M.; AbdulAzeez, S.; Borgio, J.F. Drug Resistance-Associated Mutations in ERG11 of Multidrug-Resistant Candida Auris in a Tertiary Care Hospital of Eastern Saudi Arabia. J. Fungi 2020, 7, 18. [Google Scholar] [CrossRef]
- Almaghrabi, R.S.; Albalawi, R.; Mutabagani, M.; Atienza, E.; Aljumaah, S.; Gade, L.; Forsberg, K.; Litvintseva, A.; Althawadi, S. Molecular Characterisation and Clinical Outcomes of Candida Auris Infection: Single-centre Experience in Saudi Arabia. Mycoses 2020, 63, 452–460. [Google Scholar] [CrossRef] [PubMed]
- Bosaeed, M.; Ahmad, A.; Alali, A.; Mahmoud, E.; Alswidan, L.; Alsaedy, A.; Aljuhani, S.; Alalwan, B.; Alshamrani, M.; Alothman, A. Experience With Ceftolozane-Tazobactam for the Treatment of Serious Pseudomonas Aeruginosa Infections in Saudi Tertiary Care Center. Infect. Dis. 2020, 13, 117863372090597. [Google Scholar] [CrossRef]
- Aljadani, R.; Ahmed, A.E.; AL-Jahdali, H. Tuberculosis Mortality and Associated Factors at King Abdulaziz Medical City Hospital. BMC Infect. Dis. 2019, 19, 427. [Google Scholar] [CrossRef] [PubMed]
- Almangour, T.A.; Alenazi, B.; Ghonem, L.; Alhifany, A.A.; Aldakheel, B.A.; Alruwaili, A. Inhaled Colistin for the Treatment of Nosocomial Pneumonia Due to Multidrug-Resistant Gram-Negative Bacteria: A Real-Life Experience in Tertiary Care Hospitals in Saudi Arabia. Saudi Pharm. J. 2020, 28, 1009–1013. [Google Scholar] [CrossRef] [PubMed]
- Abdalhamid, B.; Almaghrabi, R.; Althawadi, S.; Omrani, A. First Report of Candida Auris Infections from Saudi Arabia. J. Infect. Public Health 2018, 11, 598–599. [Google Scholar] [CrossRef]
- Alraddadi, B.M.; Saeedi, M.; Qutub, M.; Alshukairi, A.; Hassanien, A.; Wali, G. Efficacy of Ceftazidime-Avibactam in the Treatment of Infections Due to Carbapenem-Resistant Enterobacteriaceae. BMC Infect. Dis. 2019, 19, 772. [Google Scholar] [CrossRef]
- Balkhy, H.; El-Saed, A.; Maghraby, R.; Khan, R.; Rishu, A.; Al-Dorzi, H.; Arabi, Y. Drug-Resistant Ventilator Associated Pneumonia in a Tertiary Care Hospital in Saudi Arabia. Ann. Thorac. Med. 2014, 9, 104. [Google Scholar] [CrossRef]
- Älgå, A.; Wong, S.; Shoaib, M.; Lundgren, K.; Giske, C.G.; von Schreeb, J.; Malmstedt, J. Infection with High Proportion of Multidrug-Resistant Bacteria in Conflict-Related Injuries is Associated with Poor Outcomes and Excess Resource Consumption: A Cohort Study of Syrian Patients Treated in Jordan. BMC Infect. Dis. 2018, 18, 233. [Google Scholar] [CrossRef] [PubMed]
- Al-lawama, M.; Aljbour, H.; Tanash, A.; Badran, E. Intravenous Colistin in the Treatment of Multidrug-Resistant Acinetobacter in Neonates. Ann. Clin. Microbiol. Antimicrob. 2016, 15, 8. [Google Scholar] [CrossRef] [PubMed]
- Almomani, B.A.; McCullough, A.; Gharaibeh, R.; Samrah, S.; Mahasneh, F. Incidence and Predictors of 14-Day Mortality in Multidrug-Resistant Acinetobacter Baumannii in Ventilator-Associated Pneumonia. J. Infect. Dev. Ctries 2015, 9, 1323–1330. [Google Scholar] [CrossRef]
- Abd El Ghany, M.; Sharaf, H.; Al-agamy, M.H.; Shibl, A.; Hill-Cawthorne, G.A.; Hong, P.-Y. Genomic Characterization of NDM-1 and 5, and OXA-181 Carbapenemases in Uropathogenic Escherichia coli Isolates from Riyadh, Saudi Arabia. PLoS ONE 2018, 13, e0201613. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Halim, H.; Al Dajani, A.; Abdelhalim, A.; Abdelmalek, S. The Search of Potential Inhibitors of the AcrAB–TolC System of Multidrug-Resistant Escherichia coli: An in Silico Approach. Appl. Microbiol. Biotechnol. 2019, 103, 6309–6318. [Google Scholar] [CrossRef]
- Abed, A.R.; Khudhair, A.M.; Hussein, I.M. Effects of Misuse of Antibiotics on the Resistance of Escherichia coli Isolated from the Intestines of Broiler Chickens. Int. J. Drug Deliv. Technol. 2020, 10, 190–194. [Google Scholar] [CrossRef]
- Ahmad, S.; Al-Mutairi, N.M.; Mokaddas, E. Variations in the Occurrence of Specific RpoB Mutations in Rifampicin- Resistant Mycobacterium Tuberculosis Isolates from Patients of Different Ethnic Groups in Kuwait. Indian J. Med. Res. 2012, 135, 756–762. [Google Scholar]
- Ahmad Hamad, P.; Khadija, K.M. Prevalence of blaTEM, blaSHV, and blaCTX-M Genes among ESBL-Producing Klebsiella pneumoniae and Escherichia coli Isolated from Thalassemia Patients in Erbil, Iraq. Mediterr. J. Hematol. Infect. Dis. 2019, 11, e2019041. [Google Scholar] [CrossRef]
- Ahmed, M.U.; Farooq, R.; Hawashim, N.A.; Ahmed, M.; Yiannakou, N.; Sayeed, F.; Sayed, A.R.; Lutfullah, S. Sensitive, Resistant and Multi-Drug Resistant Acinetobacter Baumanii at Saudi Arabia Hospital Eastern Region. Pak. J. Pharm. Sci. 2015, 28, 825–832. [Google Scholar]
- Al Johani, S.M.; Akhter, J.; Balkhy, H.; El-Saed, A.; Younan, M.; Memish, Z. Prevalence of Antimicrobial Resistance among Gram-Negative Isolates in an Adult Intensive Care Unit at a Tertiary Care Center in Saudi Arabia. Ann. Saudi Med. 2010, 30, 364–369. [Google Scholar] [CrossRef] [PubMed]
- Al Wutayd, O.; Al Nafeesah, A.; Adam, I.; Babikir, I. The Antibiotic Susceptibility Patterns of Uropathogens Isolated in Qassim, Saudi Arabia. J. Infect. Dev. Ctries 2018, 12, 946–952. [Google Scholar] [CrossRef] [PubMed]
- Alaa, A.R.; Solhan, M.A.; Lalan, R.M.; Hiwa, L.I.; Mhamad, N.R.; Niga, K.M.; Awat, J.N.; Salar, A.I.; Brwa, H.Q.; Daryan, K.K.H.; et al. The antibiotic resistance pattern and molecular characterization of blactx and blatem genes of e. Coli isolated from different hosts based on the rate of antibiotic consumption in Sulaymaniyah/Iraq. Appl. Ecol. Environ. Res. 2020, 18, 6025–6040. [Google Scholar] [CrossRef]
- Alanazi, M.Q.; Alqahtani, F.Y.; Aleanizy, F.S. An Evaluation of E. coli in Urinary Tract Infection in Emergency Department at KAMC in Riyadh, Saudi Arabia: Retrospective Study. Ann. Clin. Microbiol. Antimicrob. 2018, 17, 3. [Google Scholar] [CrossRef] [PubMed]
- Alatoom, A.; Elsayed, H.; Lawlor, K.; AbdelWareth, L.; El-Lababidi, R.; Cardona, L.; Mooty, M.; Bonilla, M.-F.; Nusair, A.; Mirza, I. Comparison of Antimicrobial Activity between Ceftolozane–Tazobactam and Ceftazidime–Avibactam against Multidrug-Resistant Isolates of Escherichia coli, Klebsiella Pneumoniae, and Pseudomonas Aeruginosa. Int. J. Infect. Dis. 2017, 62, 39–43. [Google Scholar] [CrossRef]
- Alavudeen, S.S.; Vigneshwaran, E.; Asiri, S.A.A.; Alahmari, M.H.A.; Mohammed, M.A.; Algahtani, T.; Khan, N.A. Distribution of Multi-Resistant Bacterial Isolates from Clinical Specimens in a Hospital Environment of Kingdom of Saudi Arabia. J. Young Pharm. 2017, 9, 347–351. [Google Scholar] [CrossRef]
- Al-Ayed, M.S.Z.; Asaad, A.M.; Qureshi, M.A.; Attia, H.G.; AlMarrani, A.H. Antibacterial Activity of Salvadora Persica L. (Miswak) Extracts against Multidrug Resistant Bacterial Clinical Isolates. Evid.-Based Complementary Altern. Med. 2016, 2016, 7083964. [Google Scholar] [CrossRef]
- Albukhari, T.A.M.; Nafady-Hego, H.; Elgendy, H.; Abd Elmoneim, H.M.; Nafady, A.; Alzahrani, A.M. Analysis of Bacterial and Fungal Infections after Cytoreduction Surgery and Hyperthermic Intraperitoneal Chemotherapy: An Observational Single-Centre Study. Int. J. Microbiol. 2019, 2019, 6351874. [Google Scholar] [CrossRef]
- Al-Guranie, D.R.; Al-Mayahie, S.M. Prevalence of E. coli ST131 among Uropathogenic E. coli Isolates from Iraqi Patients in Wasit Province, Iraq. Int. J. Microbiol. 2020, 2020, 8840561. [Google Scholar] [CrossRef]
- Ali, F.A.; Hussen, B.M.; Zaki, S.M. Molecular detection of blactx-m gene among pseudomonas aeruginosa strains isolated from different clinical samples in erbil city. Ann. Trop. Med. Public Health 2020, 23, 231. [Google Scholar] [CrossRef]
- Aljanaby, A.A.J. Antibacterial Activity of an Aqueous Extracts of Alkanna Tinctoria Roots against Drug Resistant Aerobic Pathogenic Bacteria Isolated from Patients with Burns Infections. Russ. Open Med. J. 2018, 7, e0104. [Google Scholar] [CrossRef]
- Aljanaby, A.A.J.; Tuwaij, N.S.S.; Al-khilkhali, H.J.B. Antimicrobial Susceptibility Patterns of Klebsiella Pneumoniae Isolated from Older Smokers and Non-Smokers of Inpatients in Intensive Care Unit Infected with Chronic Pneumonia in AL-Najaf Hospital, Iraq. J. Pharm. Sci. 2018, 10, 6. [Google Scholar]
- Alm’amoori, K.; Hadi, Z.; Almohana, A. Molecular Investigation of Plasmid–Mediated Quinolone Resistant Genes among Aminoglycoside-Resistant Uropathogenic Escherichia coli Isolates from Babylon Hospitals, Iraq. Indian J. Forensic Med. Toxicol. 2020, 14, 493–498. [Google Scholar]
- Al-Mayahie, S.M. Phenotypic and Genotypic Comparison of ESBL Production by Vaginal Escherichia coli Isolates from Pregnant and Non-Pregnant Women. Ann. Clin. Microbiol. Antimicrob. 2013, 12, 7. [Google Scholar] [CrossRef] [PubMed]
- Al-Mulla, N.; Elshafie, S.S.; Janahi, M.; Al-Nasser, A.; Chandra, P.; Taj-Aldeen, S.J. Bacterial Bloodstream Infections and Antimicrobial Susceptibility Pattern in Pediatric Hematology/Oncology Patients after Anticancer Chemotherapy. Infect. Drug Resist. 2014, 7, 289. [Google Scholar] [CrossRef]
- AlOtair, H.A.; Hussein, M.A.; Elhoseny, M.A.; Alzeer, A.H.; Khan, M.F. Severe Pneumonia Requiring ICU Admission: Revisited. J. Taibah Univ. Med. Sci. 2015, 10, 293–299. [Google Scholar] [CrossRef]
- Alqasim, A.; Abu Jaffal, A.; Alyousef, A.A. Prevalence of Multidrug Resistance and Extended-Spectrum β -Lactamase Carriage of Clinical Uropathogenic Escherichia coli Isolates in Riyadh, Saudi Arabia. Int. J. Microbiol. 2018, 2018, 3026851. [Google Scholar] [CrossRef]
- Alshareef, H.; Alfahad, W.; Albaadani, A.; Alyazid, H.; Talib, R.B. Impact of Antibiotic De-Escalation on Hospitalized Patients with Urinary Tract Infections: A Retrospective Cohort Single Center Study. J. Infect. Public Health 2020, 13, 985–990. [Google Scholar] [CrossRef]
- Alshukairi, A.N.; Moalim, H.M.; Alsaedi, A.; Almansouri, W.Y.; Al-Zahrani, M.; Aljuaid, A.; Alraddadi, B.M.; Altorkistani, H.H.; Alrajhi, A.A.; Al-Hajoj, S.A. Family Cluster of Multi-Drug Resistant Tuberculosis in Kingdom of Saudi Arabia. J. Infect. Public Health 2020, 13, 154–157. [Google Scholar] [CrossRef]
- Altalhi, A.D.; Gherbawy, Y.A.; Hassan, S.A. Antibiotic Resistance in Escherichia coli Isolated from Retail Raw Chicken Meat in Taif, Saudi Arabia. Foodborne Pathog. Dis. 2010, 7, 281–285. [Google Scholar] [CrossRef]
- Awayid, H.S.; Sahar, B.R.; Jalil, I.S.; Nouman, K.T. Immunization against Multi Drug Resistance Uropathogenic E. coli Isolate from Urinary Tract Infection in Pregnancy. Res. J. Pharm. Technol. 2019, 12, 5444. [Google Scholar] [CrossRef]
- Azab, K.S.M.; Abdel-Rahman, M.A.; El-Sheikh, H.H.; Azab, E.; Gobouri, A.A.; Farag, M.M.S. Distribution of Extended-Spectrum β-Lactamase (ESBL)-Encoding Genes among Multidrug-Resistant Gram-Negative Pathogens Collected from Three Different Countries. Antibiotics 2021, 10, 247. [Google Scholar] [CrossRef]
- Azim, N.S.A.; Al-Harbi, M.A.; Al-Zaban, M.I.; Nofal, M.Y.; Somily, A.M. Prevalence and Antibiotic Susceptibility among Gram Negative Bacteria Isolated from Intensive Care Units at a Tertiary Care Hospital in Riyadh, Saudi Arabia. J. Pure Appl. Microbiol. 2019, 13, 201–208. [Google Scholar] [CrossRef]
- Badran, E.F.; Din, R.A.Q.; Shehabi, A.A. Low Intestinal Colonization of Escherichia coli Clone ST131 Producing CTX-M-15 in Jordanian Infants. J. Med. Microbiol. 2016, 65, 137–141. [Google Scholar] [CrossRef] [PubMed]
- Balkhy, H.H.; El-Saed, A.; Alshamrani, M.M.; Alsaedi, A.; Al Nasser, W.; El Gammal, A.; Aljohany, S.M.; Almunif, S.; Arabi, Y.; Alqahtani, S.; et al. Ten-Year Resistance Trends in Pathogens Causing Healthcare-Associated Infections; Reflection of Infection Control Interventions at a Multi-Hospital Healthcare System in Saudi Arabia, 2007–2016. Antimicrob. Resist. Infect. Control 2020, 9, 21. [Google Scholar] [CrossRef]
- Balkhy, H.H.; El-Saed, A.; Alshamrani, M.M.; Alsaedi, A.; Nasser, W.A.; Gammal, A.E.; Aljohany, S.M.; Arabi, Y.; Alqahtani, S.; Bonnie, H.B.; et al. High Burden of Resistant Gram Negative Pathogens Causing Device-Associated Healthcare Infections in a Tertiary Care Setting in Saudi Arabia, 2008–2016. J. Glob. Antimicrob. Resist. 2020, 23, 26–32. [Google Scholar] [CrossRef] [PubMed]
- Bindayna, K.M.; Khanfar, H.S.; Senok, A.C.; Botta, G.A. Predominance of CTX-M Genotype among Extended Spectrum Beta Lactamase Isolates in a Tertiary Hospital in Saudi Arabia. Saudi Med. J. 2010, 31, 859–863. [Google Scholar]
- Burjaq, S.Z.; Shehabi, A. Fresh Leafy Green Vegetables Associated with Multidrug Resistant E. coli. Int. ARABIC J. Antimicrob. Agents 2013, 3, 1–7. [Google Scholar]
- Darwish, R.M.; Aburjai, T.A. Effect of Ethnomedicinal Plants Used in Folklore Medicine in Jordan as Antibiotic Resistant Inhibitors on Escherichia coli. BMC Complement Altern. Med. 2010, 10, 9. [Google Scholar] [CrossRef]
- Dashti, A.A.; Vali, L.; El-Shazly, S.; Jadaon, M.M. The Characterization and Antibiotic Resistance Profiles of Clinical Escherichia coli O25b-B2-ST131 Isolates in Kuwait. BMC Microbiol. 2014, 14, 214. [Google Scholar] [CrossRef]
- Dawood, W.S. Molecular and Susceptibility Study of Antibiotic Resistance Genes in E. coli Isolated from Selected Iraqi Patients. Syst. Rev. Pharm. 2020, 11, 10. [Google Scholar]
- El-Ghareeb, W.R.; Abdel-Raheem, S.M.; Al-Marri, T.M.; Alaql, F.A.; Fayez, M.M. Isolation and Identification of Extended Spectrum β-Lactamases (ESBLs) Escherichia coli from Minced Camel Meat in Eastern Province, Saudi Arabia. Thai J. Vet. Med. 2020, 50, 155–161. [Google Scholar]
- El-Saed, A.; Balkhy, H.H.; Alshamrani, M.M.; Aljohani, S.; Alsaedi, A.; Al Nasser, W.; El Gammal, A.; Almohrij, S.A.; Alyousef, Z.; Almunif, S.; et al. High Contribution and Impact of Resistant Gram Negative Pathogens Causing Surgical Site Infections at a Multi-Hospital Healthcare System in Saudi Arabia, 2007–2016. BMC Infect. Dis. 2020, 20, 275. [Google Scholar] [CrossRef] [PubMed]
- Elsohaby, I.; Samy, A.; Elmoslemany, A.; Alorabi, M.; Alkafafy, M.; Aldoweriej, A.; Al-Marri, T.; Elbehiry, A.; Fayez, M. Migratory Wild Birds as a Potential Disseminator of Antimicrobial-Resistant Bacteria around Al-Asfar Lake, Eastern Saudi Arabia. Antibiotics 2021, 10, 260. [Google Scholar] [CrossRef] [PubMed]
- Eltai, N.O.; Abdfarag, E.A.; Al-Romaihi, H.; Wehedy, E.; Mahmoud, M.H.; Alawad, O.K.; Al-Hajri, M.M.; Al Thani, A.A.; Yassine, H.M. Antibiotic Resistance Profile of Commensal Escherichia coli Isolated from Broiler Chickens in Qatar. J. Food Prot. 2017, 81, 302–307. [Google Scholar] [CrossRef] [PubMed]
- Eltai, N.O.; Al Thani, A.A.; Al Hadidi, S.H.; Al Ansari, K.; Yassine, H.M. Antibiotic Resistance and Virulence Patterns of Pathogenic Escherichia coli Strains Associated with Acute Gastroenteritis among Children in Qatar. BMC Microbiol. 2020, 20, 54. [Google Scholar] [CrossRef] [PubMed]
- Fadlelmula, A.; Al-Hamam, N.A.; Al-Dughaym, A.M. A Potential Camel Reservoir for Extended-Spectrum β-Lactamase-Producing Escherichia coli Causing Human Infection in Saudi Arabia. Trop. Anim. Health Prod. 2016, 48, 427–433. [Google Scholar] [CrossRef]
- Garcell, H.G.; Arias, A.V.; Pancorbo Sandoval, C.A.; García, E.G.; Valle Gamboa, M.E.; Sado, A.B.; Alfonso Serrano, R.N. Incidence and Etiology of Surgical Site Infections in Appendectomies: A 3-Year Prospective Study. Oman Med. J. 2017, 32, 31–35. [Google Scholar] [CrossRef]
- Ghanem, B.; Haddadin, R.N. Multiple Drug Resistance and Biocide Resistance in Escherichia coli Environmental Isolates from Hospital and Household Settings. Antimicrob. Resist. Infect. Control 2018, 7, 47. [Google Scholar] [CrossRef]
- Haddadin, R.N.; Assaf, A.M.; Homsi, A.; Collier, P.J.; Shehabi, A. Investigating Possible Association between Multidrug Resistance and Isolate Origin with Some Virulence Factors of Escherichia coli Strains Isolated from Infant Faeces and Fresh Green Vegetables. J. Appl. Microbiol. 2019, 127, 88–98. [Google Scholar] [CrossRef] [PubMed]
- Hameed, T.; Al Nafeesah, A.; Chishti, S.; Al Shaalan, M.; Al Fakeeh, K. Community-Acquired Urinary Tract Infections in Children: Resistance Patterns of Uropathogens in a Tertiary Care Center in Saudi Arabia. Int. J. Pediatrics Adolesc. Med. 2019, 6, 51–54. [Google Scholar] [CrossRef] [PubMed]
- Hammoudi, A. Antibiotics Susceptibility Pattern of Some Enterobacteriaceae Isolates from Different Clinical Infectious Sources. Int. J. Res. Pharm. Sci. 2019, 10, 734–741. [Google Scholar]
- Hassan, H.; Abdalhamid, B. Molecular Characterization of Extended-Spectrum Beta-Lactamase Producing Enterobacteriaceae in a Saudi Arabian Tertiary Hospital. J. Infect. Dev. Ctries 2014, 8, 282–288. [Google Scholar] [CrossRef]
- Huang, X.-Z.; Frye, J.G.; Chahine, M.A.; Glenn, L.M.; Ake, J.A.; Su, W.; Nikolich, M.P.; Lesho, E.P. Characteristics of Plasmids in Multi-Drug-Resistant Enterobacteriaceae Isolated during Prospective Surveillance of a Newly Opened Hospital in Iraq. PLoS ONE 2012, 7, e40360. [Google Scholar] [CrossRef]
- Ibrahim, R.A.; Cryer, T.L.; Lafi, S.Q.; Basha, E.-A.; Good, L.; Tarazi, Y.H. Identification of Escherichia coli from Broiler Chickens in Jordan, Their Antimicrobial Resistance, Gene Characterization and the Associated Risk Factors. BMC Vet. Res. 2019, 15, 159. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, I.; Al- Shwaikh, R.; Ismaeil, M. Virulence and Antimicrobial Resistance of Escherichia coli Isolated from Tigris River and Children Diarrhea. Infect. Drug Resist. 2014, 317. [Google Scholar] [CrossRef][Green Version]
- Ibrahim, M.E.; Bilal, N.E.; Hamid, M.E. Comparison of Phenotypic Characteristics and Antimicrobial Resistance Patterns of Clinical Escherichia coli Collected From Two Unrelated Geographical Areas. GJHS 2014, 6, 126. [Google Scholar] [CrossRef]
- Jaloob Aljanaby, A.A.; Aljanaby, I.A.J. Antimicrobial Sensitivity Pattern of Pathogenic Bacteria Isolated from Older Women with Asymptomatic Bacteriuria. Biomed. Res. 2018, 29. [Google Scholar] [CrossRef]
- Karomi, A.S.A. Screening Study for Some Strains of E. coli Collected from Five Regions in Kurdistan-Iraq for Its Sensitivity, Resistance and MDR against Thirteen Antibiotics. Med. Leg. Update 2020, 20, 526–531. [Google Scholar] [CrossRef]
- Khairy, G.A.; Kambal, A.M.; Al-Dohayan, A.A.; Al-Shehri, M.Y.; Zubaidi, A.M.; Al-Naami, M.Y.; AlSaif, F.A.; Al-Obaid, O.A.; Al-Saif, A.A.; El-Farouk, O.Y.; et al. Surgical Site Infection in a Teaching Hospital: A Prospective Study. J. Taibah Univ. Med. Sci. 2011, 6, 114–120. [Google Scholar] [CrossRef]
- Majeed, H.T.; Aljanaby, A.A.J. Antibiotic Susceptibility Patterns and Prevalence of Some Extended Spectrum Beta- Lactamases Genes in Gram-Negative Bacteria Isolated from Patients Infected with Urinary Tract Infections in Al-Najaf City, Iraq. Avicenna J. Med. Biotechnol. 2019, 11, 192–201. [Google Scholar]
- Marie, M.A.; John, J.; Krishnappa, L.G.; Gopalkrishnan, S. Molecular Characterization of the β-Lactamases in Escherichia coli and Klebsiella Pneumoniae from a Tertiary Care Hospital in Riyadh, Saudi Arabia: β-Lactamases in Enterobacteriaceae. Microbiol. Immunol. 2013, 57, 805–810. [Google Scholar] [CrossRef]
- Mazi, W.; Begum, Z.; Abdulla, D.; Hesham, A.; Maghari, S.; Assiri, A.; Senok, A. Central Line–Associated Bloodstream Infection in a Trauma Intensive Care Unit: Impact of Implementation of Society for Healthcare Epidemiology of America/Infectious Diseases Society of America Practice Guidelines. Am. J. Infect. Control 2014, 42, 865–867. [Google Scholar] [CrossRef]
- Memish, Z.A.; Assiri, A.; Almasri, M.; Roshdy, H.; Hathout, H.; Kaase, M.; Gatermann, S.G.; Yezli, S. Molecular Characterization of Carbapenemase Production Among Gram-Negative Bacteria in Saudi Arabia. Microb. Drug Resist. 2015, 21, 307–314. [Google Scholar] [CrossRef]
- Mutti, M.; Sonnevend, Á.; Pál, T.; Junttila, S.; Ekker, H.; Galik, B.; Gyenesei, A.; Nagy, G.; Nagy, E.; Szijártó, V. Complete Genome Sequence of Escherichia coli 81009, a Representative of the Sequence Type 131 C1-M27 Clade with a Multidrug-Resistant Phenotype. Genome Announc. 2018, 6, e00056-18. [Google Scholar] [CrossRef]
- Nairoukh, Y.R.; Mahafzah, A.M.; Irshaid, A.; Shehabi, A.A. Molecular Characterization of Multidrug Resistant Uropathogenic E. coli Isolates from Jordanian Patients. Open Microbiol. J. 2018, 12, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Narchi, H.; Al-Hamdani, M. Uropathogen Resistance to Antibiotic Prophylaxis in Urinary Tract Infections. Microb. Drug Resist. 2010, 16, 151–154. [Google Scholar] [CrossRef] [PubMed]
- Nimri, L.; Abu AL- Dahab, F.; Batchoun, R. Foodborne Bacterial Pathogens Recovered from Contaminated Shawarma Meat in Northern Jordan. J. Infect. Dev. Ctries 2014, 8, 1407–1414. [Google Scholar] [CrossRef] [PubMed]
- Nimri, L.; Samara, H.; Batchoun, R. Detection of Mutations Associated with Multidrug-Resistant Mycobacterium Tuberculosis Clinical Isolates. FEMS Immunol. Med. Microbiol. 2011, 62, 321–327. [Google Scholar] [CrossRef]
- Obaidat, M.M. Prevalence and Antimicrobial Resistance of Listeria Monocytogenes, Salmonella Enterica and Escherichia coli O157:H7 in Imported Beef Cattle in Jordan. Comp. Immunol. Microbiol. Infect. Dis. 2020, 70, 101447. [Google Scholar] [CrossRef]
- Pál, T.; Ghazawi, A.; Darwish, D.; Villa, L.; Carattoli, A.; Hashmey, R.; Aldeesi, Z.; Jamal, W.; Rotimi, V.; Al-Jardani, A.; et al. Characterization of NDM-7 Carbapenemase-Producing Escherichia coli Isolates in the Arabian Peninsula. Microb. Drug Resist. 2017, 23, 871–878. [Google Scholar] [CrossRef]
- Radwan Ali, M.; Khudhair, A.M. Detection of Colony Adhesion Factors and Genetic Background of Adhesion Genes Among Multidrug-Resistant Uropathogenic Escherichia coli Isolated in Iraq. J. Pure Appl. Microbiol. 2018, 12, 2017–2026. [Google Scholar] [CrossRef]
- Rana, M.A.; Abd El Rahaman, B.; Mady, A.F.; Al Odat, M.; Al Harthy, A.; Ramadan, O.E.S.; Mumtaz, S.A.; Omrani, A.S. Intra-Pleural Colistin Methanesulfonate Therapy for Pleural Infection Caused by Carbapenem-Resistant Acinetobacter Baumannii: A Successful Case Report. Infect. Dis. Rep. 2014, 6, 38–40. [Google Scholar] [CrossRef]
- Salah, M.; Badran, E.; Shehabi, A. High Incidence of Multidrug Resistant Escherichia coli Producing CTX-M-Type ESBLs Colonizing the Intestine of Jordanian Infants. Int. Arab. J. Antimicrob. Agents 2014, 3, 1–8. [Google Scholar]
- Shobrak, M.Y.; Hassan, S.A.; Stiévenart, C.; El-Deeb, B.A.; Gherbawy, Y.A. Prevalence and Antibiotic Resistance Profile of Intestinal Bacteria Isolated from Captive Adult Houbara Bustards ( ) Exposed to Natural Weather Conditions in Saudi Arabia. Escherichia coli. Glob. Vet. 2013, 10, 276–284. [Google Scholar]
- Sonnevend, A.; Yahfoufi, N.; Ghazawi, A.; Jamal, W.; Rotimi, V.; Pal, T. Contribution of Horizontal Gene Transfer to the Emergence of VIM-4 Carbapenemase Producer Enterobacteriaceae in Kuwait. Infect. Drug Resist. 2017, 10, 469–478. [Google Scholar] [CrossRef] [PubMed]
- Taha, M.M.E.; Homeida, H.E.; Dafalla, O.M.E.; Abdelwahab, S.I. Multidrug Resistance, Prevalence and Phylogenetic Analysis of Genes Encoding Class II and III Integrons in Clinically Isolated Escherichia coli. Cell. Mol. Biol. 2018, 64, 122. [Google Scholar] [CrossRef]
- Taher, I.; Almaeen, A.; Aljourfi, H.; Bohassan, E.; Helmy, A.; El-Masry, E.; Saleh, B.; Aljaber, N. Surveillance of Antibiotic Resistance among Uropathogens in Aljouf Region Northern Saudi Arabia. Iran. J. Microbiol. 2020, 11, 468. [Google Scholar] [CrossRef]
- Tarazi, Y.H.; Abu-Basha, E.A.; Ismail, Z.B.; Tailony, R.A. In Vitro and in Vivo Efficacy Study of Cefepime, Doripenem, Tigecycline, and Tetracycline against Extended-Spectrum Beta-Lactamases Escherichia coli in Chickens. Vet. World 2020, 13, 446–451. [Google Scholar] [CrossRef]
- Thani, A.S.B. Characterization of Previously Identified Novel DNA Fragment Associated with Pathogenicity Island III536 Reveals New Bla Gene. Infect. Genet. Evol. 2019, 75, 103971. [Google Scholar] [CrossRef]
- Saeed, N.K.; Kambal, A.M.; El-Khizzi, N.A. Antimicrobial-Resistant Bacteria in a General Intensive Care Unit in Saudi Arabia. Saudi Med. J. 2010, 31, 1341–1349. [Google Scholar]
- Ahmad, S.; Mokaddas, E.; Al-Mutairi, N.; Eldeen, H.S.; Mohammadi, S. Discordance across Phenotypic and Molecular Methods for Drug Susceptibility Testing of Drug-Resistant Mycobacterium Tuberculosis Isolates in a Low TB Incidence Country. PLoS ONE 2016, 11, e0153563. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, M.M.; Mohammed, S.H.; Nasurallah, H.A.A.; Ali, M.M.; Couvin, D.; Rastogi, N. Snapshot of the Genetic Diversity of Mycobacterium Tuberculosis Isolates in Iraq. Int. J. Mycobacteriol. 2014, 3, 184–196. [Google Scholar] [CrossRef] [PubMed]
- Ahmed-Abakur, E.; Saad Alnour, T. Detection of Multidrug Resistant Mycobacterium Tuberculosis in Tabuk, Saudi Arabia, Using Genotype MTBDRplus. Int. J. Mycobacteriol. 2019, 8, 25. [Google Scholar] [CrossRef]
- Al Mahbashi, A.A.; Mukhtar, M.M.; Mahgoub, E.S. Molecular Typing of Mycobacterium spp. Isolates from Yemeni Tuberculosis Patients. East Mediterr. Health J. 2013, 19, 942–946. [Google Scholar] [CrossRef]
- AL Qurainees, G.I.; Tufenkeji, H.T. A Child with Complicated Mycobacterium Tuberculosis. Int. J. Pediatrics Adolesc. Med. 2016, 3, 28–33. [Google Scholar] [CrossRef]
- Alateah, S.M.; Othman, M.W.; Ahmed, M.; Al Amro, M.S.; Al Sherbini, N.; Ajlan, H.H. A Retrospective Study of Tuberculosis Prevalence amongst Patients Attending a Tertiary Hospital in Riyadh, Saudi Arabia. J. Clin. Tuberc. Other Mycobact. Dis. 2020, 21, 100185. [Google Scholar] [CrossRef]
- Al-Hajoj, S.; Varghese, B.; Shoukri, M.M.; Al-Omari, R.; Al-Herbwai, M.; AlRabiah, F.; Alrajhi, A.A.; Abuljadayel, N.; Al-Thawadi, S.; Zumla, A.; et al. Epidemiology of Antituberculosis Drug Resistance in Saudi Arabia: Findings of the First National Survey. Antimicrob. Agents Chemother. 2013, 57, 2161–2166. [Google Scholar] [CrossRef] [PubMed]
- Al-Hajoj, S.; Shoukri, M.; Memish, Z.; AlHakeem, R.; AlRabiah, F.; Varghese, B. Exploring the Sociodemographic and Clinical Features of Extrapulmonary Tuberculosis in Saudi Arabia. PLoS ONE 2015, 10, e0101667. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.; Howady, F.; Munir, W.; Karim, H.; Al-Suwaidi, Z.; Al-Maslamani, M.; Alkhal, A.; Elmaki, N.; Ziglam, H. Drug-Resistant Tuberculosis: An Experience from Qatar. Libyan J. Med. 2020, 15, 1744351. [Google Scholar] [CrossRef]
- Ali Chaudhry, L.; Rambhala, N.; Al-Shammri, A.S.; Al-Tawfiq, J.A. Patterns of Antituberculous Drug Resistance in Eastern Saudi Arabia: A 7-Year Surveillance Study from 1/2003 to 6/2010. J. Epidemiol. Glob. Health 2011, 2, 57. [Google Scholar] [CrossRef]
- Al-Mutairi, N.M.; Ahmad, S.; Mokaddas, E.; Eldeen, H.S.; Joseph, S. Occurrence of Disputed RpoB Mutations among Mycobacterium Tuberculosis Isolates Phenotypically Susceptible to Rifampicin in a Country with a Low Incidence of Multidrug-Resistant Tuberculosis. BMC Infect. Dis. 2019, 19, 3. [Google Scholar] [CrossRef]
- Al-Mutairi, N.M.; Ahmad, S.; Mokaddas, E. First Report of Molecular Detection of Fluoroquinolone Resistance-Associated GyrA Mutations in Multidrug-Resistant Clinical Mycobacterium Tuberculosis Isolates in Kuwait. BMC Res. Notes 2011, 4, 123. [Google Scholar] [CrossRef]
- Al-Mutairi, N.M.; Ahmad, S.; Mokaddas, E. Molecular Screening Versus Phenotypic Susceptibility Testing of Multidrug-Resistant Mycobacterium Tuberculosis Isolates for Streptomycin and Ethambutol. Microb. Drug Resist. 2018, 24, 923–931. [Google Scholar] [CrossRef] [PubMed]
- Al-Mutairi, N.M.; Ahmad, S.; Mokaddas, E.M. Correction to: Molecular Characterization of Multidrug-Resistant Mycobacterium Tuberculosis (MDR-TB) Isolates Identifies Local Transmission of Infection in Kuwait, a Country with a Low Incidence of TB and MDR-TB. Eur. J. Med. Res. 2020, 25, 14. [Google Scholar] [CrossRef]
- Al-Rubaye, D.S.; Henihan, G.; Al-Abasly, A.K.A.; Seagar, A.-L.; Al-Attraqchi, A.A.F.; Schulze, H.; Hashim, D.S.; Kamil, J.K.; Laurenson, I.F.; Bachmann, T.T. Genotypic Assessment of Drug-Resistant Tuberculosis in Baghdad and Other Iraqi Provinces Using Low-Cost and Low-Density DNA Microarrays. J. Med. Microbiol. 2016, 65, 114–122. [Google Scholar] [CrossRef] [PubMed]
- Altuwaijri, T.A.; Alhindi, G.K.; Al-Qattan, N.M.; Alkharashi, S.K.; Somily, A.M.; Altoijry, A.H. Occurrence of Venous Thromboembolism in Hospitalized Patients with Tuberculosis in Saudi Arabia: A Retrospective Cohort Study. Int. J. Mycobacteriol. 2020, 9, 4. [Google Scholar]
- Alyamani, E.J.; Marcus, S.A.; Ramirez-Busby, S.M.; Hansen, C.; Rashid, J.; El-kholy, A.; Spalink, D.; Valafar, F.; Almehdar, H.A.; Jiman-Fatani, A.A.; et al. Genomic Analysis of the Emergence of Drug-Resistant Strains of Mycobacterium Tuberculosis in the Middle East. Sci. Rep. 2019, 9, 4474. [Google Scholar] [CrossRef] [PubMed]
- Al-Zarouni, M.; Dash, N.; Al Ali, M.; Al-Shehhi, F.; Panigrahi, D. Tuberculosis and MDR-TB in the Northern Emirates of United Arab Emirates: A 5-Year Study. Southeast Asian J. Trop Med. Public Health 2010, 41, 163–168. [Google Scholar] [PubMed]
- Asaad, A.M.; Alqahtani, J.M. Primary Anti-Tuberculous Drugs Resistance of Pulmonary Tuberculosis in Southwestern Saudi Arabia. J. Infect. Public Health 2012, 5, 281–285. [Google Scholar] [CrossRef] [PubMed][Green Version]
- El Mahalli, A.A.; Al-Qahtani, M.F. Predictors of Drug Resistance in Tuberculosis Patients in the Eastern Province, Saudi Arabia. J. Egypt. Public Health Assoc. 2015, 90, 24–28. [Google Scholar] [CrossRef] [PubMed]
- Flaifel, D.K.; Al-Azawi, I.H. The Role of IL-6 Gene Polymorphism in Multidrug-Resistant Tuberculosis Patients in Iraq. Int. J. Drug Deliv. Technol. 2020, 10, 81–84. [Google Scholar] [CrossRef]
- Habous, M.; Elimam, M.; AlDabal, L.; Chidambaran, B.; AlDeesi, Z. Pattern of Primary Tuberculosis Drug Resistance and Associated Risk Factors at Dubai Health Authority in Dubai. Int. J. Mycobacteriol. 2020, 9, 6. [Google Scholar]
- Kareem, P.A.; Alsammak, E.G.H.; Abdullah, Y.J.; Bdaiwi, Q.M. Estimation of Antibacterial Activity of Zinc Oxide, Titanium Dioxide, and Silver Nanoparticles against Multidrug-Resistant Bacteria Isolated from Clinical Cases in Amara City, Iraq. Drug Invent. Today 2019, 11, 5. [Google Scholar]
- Elhassan, M.M.; Hemeg, H.A.; Elmekki, M.A.; Turkistani, K.A.; Abdul-Aziz, A.A. Burden of Multidrug Resistant Mycobacterium Tuberculosis Among New Cases in Al-Madinah Al-Monawarah, Saudi Arabia. Infect. Disord.-Drug Targets 2017, 17, 14–23. [Google Scholar] [CrossRef][Green Version]
- Merza, M.A.; Farnia, P.; Salih, A.M.; Masjedi, M.R.; Velayati, A.A. First Insight into the Drug Resistance Pattern of Mycobacterium Tuberculosis in Dohuk, Iraq: Using Spoligotyping and MIRU-VNTR to Characterize Multidrug Resistant Strains. J. Infect. Public Health 2011, 4, 41–47. [Google Scholar] [CrossRef] [PubMed]
- Mokaddas, E.; Ahmad, S.; Eldeen, H.S.; Al-Mutairi, N. Discordance between Xpert MTB/RIF Assay and Bactec MGIT 960 Culture System for Detection of Rifampin-Resistant Mycobacterium Tuberculosis Isolates in a Country with a Low Tuberculosis (TB) Incidence. J. Clin. Microbiol. 2015, 53, 1351–1354. [Google Scholar] [CrossRef]
- Sambas, M.F.M.K.; Rabbani, U.; Al-Gethamy, M.M.M.; Surbaya, S.H.; Alharbi, F.F.I.; Ahmad, R.G.A.; Qul, H.K.H.; Nassar, S.M.S.; Maddah, A.K.M.A.; Darweesh, B.A.K. Prevalence and Determinants of Multidrug-Resistant Tuberculosis in Makkah, Saudi Arabia. Infect. Drug Resist. 2020, 13, 4031–4038. [Google Scholar] [CrossRef]
- Somily, A.M.; Naeem, T.; Habib, H.A.; Sarwar, M.S.; Kunimoto, D.Y.; Kambal, A.M. Changing Epidemiology of Tuberculosis Detected by an 8-Year Retrospective Laboratory Study in a Tertiary Teaching Hospital in Central Saudi Arabia. Saudi Med. J. 2014, 35, 691–698. [Google Scholar]
- Varghese, B.; Supply, P.; Allix-Béguec, C.; Shoukri, M.; Al-Omari, R.; Herbawi, M.; Al-Hajoj, S. Admixed Phylogenetic Distribution of Drug Resistant Mycobacterium Tuberculosis in Saudi Arabia. PLoS ONE 2013, 8, e55598. [Google Scholar] [CrossRef]
- Varghese, B.; Al-Hajoj, S. First Insight Into the Fluoroquinolone and Aminoglycoside Resistance of Multidrug-Resistant Mycobacterium Tuberculosis in Saudi Arabia. Am. J. Trop. Med. Hyg. 2017, 96, 1066–1070. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Abulhasan, Y.B.; Abdullah, A.A.; Shetty, S.A.; Ramadan, M.A.; Yousef, W.; Mokaddas, E.M. Health Care-Associated Infections in a Neurocritical Care Unit of a Developing Country. Neurocrit. Care 2020, 32, 836–846. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, S.S.; Shariq, A.; Alsalloom, A.A.; Babikir, I.H.; Alhomoud, B.N. Uropathogens and Their Antimicrobial Resistance Patterns: Relationship with Urinary Tract Infections. Int. J. Health Sci. 2019, 13, 8. [Google Scholar]
- Al Bshabshe, A.; Joseph, M.R.P.; Al Hussein, A.; Haimour, W.; Hamid, M.E. Multidrug Resistance Acinetobacter Species at the Intensive Care Unit, Aseer Central Hospital, Saudi Arabia: A One Year Analysis. Asian Pac. J. Trop. Med. 2016, 9, 903–908. [Google Scholar] [CrossRef] [PubMed]
- Al-Agamy, M.H.; Jeannot, K.; El-Mahdy, T.S.; Shibl, A.M.; Kattan, W.; Plésiat, P.; Courvalin, P. First Detection of GES-5 Carbapenemase-Producing Acinetobacter Baumannii Isolate. Microb. Drug Resist. 2017, 23, 556–562. [Google Scholar] [CrossRef]
- Alamri, A.M.; Alsultan, A.A.; Ansari, M.A.; Alnimr, A.M. Biofilm-Formation in Clonally Unrelated Multidrug-Resistant Acinetobacter Baumannii Isolates. Pathogens 2020, 9, 630. [Google Scholar] [CrossRef]
- Al-Anazi, K.A.; Abdalhamid, B.; Alshibani, Z.; Awad, K.; Alzayed, A.; Hassan, H.; Alsayiegh, M. Acinetobacter Baumannii Septicemia in a Recipient of an Allogeneic Hematopoietic Stem Cell Transplantation. Case Rep. Transplant. 2012, 2012, 646195. [Google Scholar] [CrossRef] [PubMed]
- Al-Dabaibah, N.; Obeidat, N.; Shehabi, A. Epidemiology Features of Acinetobacter Baumannii Colonizing Respiratory Tracts of ICU Patients. Int. Arab. J. Antimicrob. Agents 2012, 2, 1–7. [Google Scholar]
- ALfadli, M.; EL-sehsah, E.M.; Ramadan, M.A.-M. Risk Factors and Distribution of MDROs among Patients with Healthcare Associated Burn Wound Infection. Germs 2018, 8, 199–206. [Google Scholar] [CrossRef]
- Alhaddad, M.; AlBarjas, A.; Alhammar, L.; Al Rashed, A.; Badger-Emeka, L. Molecular Characterization and Antibiotic Susceptibility Pattern of Acinetobacter Baumannii Isolated in Intensive Care Unit Patients in Al-Hassa, Kingdom of Saudi Arabia. Int. J. Appl. Basic Med. Res. 2018, 8, 19. [Google Scholar] [CrossRef]
- Al-Hamad, A.; Pal, T.; Leskafi, H.; Abbas, H.; Hejles, H.; Alsubikhy, F.; Darwish, D.; Ghazawi, A.; Sonnevend, A. Molecular Characterization of Clinical and Environmental Carbapenem Resistant Acinetobacter Baumannii Isolates in a Hospital of the Eastern Region of Saudi Arabia. J. Infect. Public Health 2020, 13, 632–636. [Google Scholar] [CrossRef]
- Alharbi, A.; Alshami, I. In Vitro Effects of Tigecycline in Combination with Other Antimicrobials against Multidrug-Resistant Acinetobacter Baumannii Isolates. J. Pure Appl. Microbiol. 2015, 8, 497–502. [Google Scholar]
- AL-Harmoosh, R.; Jarallah, E.; AL-Shamari, A. Coexistence of the BlaIMP and BlaSIM Genes in Clinical Isolates of Acinetobacter baumanniiIN Babylon Hospitals Iraq. Int. J. PharmTech Res. 2016, 9, 257–264. [Google Scholar]
- Ali, H.M.; Salem, M.Z.M.; El-Shikh, M.S.; Megeed, A.A.; Alogaibi, Y.A.; Talea, I.A. Investigation of the Virulence Factors and Molecular Characterization of the Clonal Relations of Multidrug-Resistant Acinetobacter Baumannii Isolates. J. AOAC Int. 2017, 100, 152–158. [Google Scholar] [CrossRef]
- Ali, K.M.; Al-Jaff, B.M.A. Source and Antibiotic Susceptibility of Gram-Negative Bacteria Causing Superficial Incisional Surgical Site Infections. Int. J. Surg. Open 2021, 30, 100318. [Google Scholar] [CrossRef]
- Ali, M.; Marie, M.; Gowda Krishnappa, L.; Alzahrani, A.J.; Mubaraki, M.A.; Alyousef, A.A. A Prospective Evaluation of Synergistic Effect of Sulbactam and Tazobactam Combination with Meropenem or Colistin against Multidrug Resistant Acinetobacter Baumannii. Bosn. J. Basic Med. Sci. 2015, 15, 24. [Google Scholar] [CrossRef][Green Version]
- Aljindan, R.; Bukharie, H.; Alomar, A.; Abdalhamid, B. Prevalence of Digestive Tract Colonization of Carbapenem-Resistant Acinetobacter Baumannii in Hospitals in Saudi Arabia. J. Med. Microbiol. 2015, 64, 400–406. [Google Scholar] [CrossRef] [PubMed]
- AL-Kadmy, I.; Ali, A.; Salman, I.; Khazaal, S. Molecular Characterization of Acinetobacter Baumannii Isolated from Iraqi Hospital Environment. New Microbes New Infect. 2017, 21, 51–57. [Google Scholar] [CrossRef] [PubMed]
- Almaghrabi, M.K.; Joseph, M.R.P.; Assiry, M.M.; Hamid, M.E. Multidrug-Resistant Acinetobacter Baumannii: An Emerging Health Threat in Aseer Region, Kingdom of Saudi Arabia. Can. J. Infect. Dis. Med. Microbiol. 2018, 2018, 9182747. [Google Scholar] [CrossRef]
- Almutairy, R.; Aljrarri, W.; Noor, A.; Elsamadisi, P.; Shamas, N.; Qureshi, M.; Ismail, S. Impact of Colistin Dosing on the Incidence of Nephrotoxicity in a Tertiary Care Hospital in Saudi Arabia. Antibiotics 2020, 9, 485. [Google Scholar] [CrossRef]
- Al-Obeid, S.; Jabri, L.; Al-Agamy, M.; Al-Omari, A.; Shibl, A. Epidemiology of Extensive Drug Resistant Acinetobacter Baumannii (XDRAB) at Security Forces Hospital (SFH) in Kingdom of Saudi Arabia (KSA). J. Chemother. 2015, 27, 156–162. [Google Scholar] [CrossRef]
- Al-Ouqaili, M.T.S.; Jal’oot, A.S.; Badawy, A.S. Identification of an OprD and Bla IMP Gene-Mediated Carbapenem Resistance in Acinetobacter Baumannii and Pseudomonas Aeruginosa among Patients with Wound Infections in Iraq. Asian J. Pharm. 2018, 12. [Google Scholar] [CrossRef]
- Al-Sweih, N.A.; Al-Hubail, M.A.; Rotimi, V.O. Emergence of Tigecycline and Colistin Resistance in Acinetobacter Species Isolated from Patients in Kuwait Hospitals. J. Chemother. 2011, 23, 13–16. [Google Scholar] [CrossRef]
- Al-Sweih, N.A.; Al-Hubail, M.; Rotimi, V.O. Three Distinct Clones of Carbapenem-Resistant Acinetobacter Baumannii with High Diversity of Carbapenemases Isolated from Patients in Two Hospitals in Kuwait. J. Infect. Public Health 2012, 5, 102–108. [Google Scholar] [CrossRef]
- Aly, M.; Tayeb, H.T.; Al Johani, S.M.; Alyamani, E.J.; Aldughaishem, F.; Alabdulkarim, I.; Balkhy, H.H. Genetic Diversity of OXA-51-like Genes among Multidrug-Resistant Acinetobacter Baumannii in Riyadh, Saudi Arabia. Eur. J. Clin. Microbiol. Infect. Dis. 2014, 33, 1223–1228. [Google Scholar] [CrossRef] [PubMed]
- Aly, M.M.; Abu Alsoud, N.M.; Elrobh, M.S.; Al Johani, S.M.; Balkhy, H.H. High Prevalence of the PER-1 Gene among Carbapenem-Resistant Acinetobacter Baumannii in Riyadh, Saudi Arabia. Eur. J. Clin. Microbiol. Infect. Dis. 2016, 35, 1759–1766. [Google Scholar] [CrossRef] [PubMed]
- Azeez, Z.F.; Hatite Al-Daraghi, W.A. Isolation of Lytic Acinetobacter Baumannii Phage VB_Acib_C_A10 from Iraq Pond Waters and Comparing Its Antibacterial Effect with Cefotaxime Antibiotic. Int. J. Pharm. Qual. Assur. 2019, 10, 84–89. [Google Scholar] [CrossRef]
- Bakour, S.; Alsharapy, S.A.; Touati, A.; Rolain, J.-M. Characterization of Acinetobacter Baumannii Clinical Isolates Carrying Bla OXA-23 Carbapenemase and 16S RRNA Methylase ArmA Genes in Yemen. Microb. Drug Resist. 2014, 20, 604–609. [Google Scholar] [CrossRef] [PubMed]
- Batarseh, A.; Al-Sarhan, A.; Maayteh, M.; Al-Khatirei, S.; Alarmouti, M. Antibiogram of Multidrug Resistant Acinetobacter Baumannii Isolated from Clinical Specimens at King Hussein Medical Centre, Jordan: A Retrospective Analysis. Easter Mediterr. Health J. 2015, 21, 828–834. [Google Scholar] [CrossRef]
- Conlon, J.M.; Ahmed, E.; Pal, T.; Sonnevend, A. Potent and Rapid Bactericidal Action of Alyteserin-1c and Its [E4K] Analog against Multidrug-Resistant Strains of Acinetobacter Baumannii. Peptides 2010, 31, 1806–1810. [Google Scholar] [CrossRef] [PubMed]
- Conlon, J.M.; Mechkarska, M.; Arafat, K.; Attoub, S.; Sonnevend, A. Analogues of the Frog Skin Peptide Alyteserin-2a with Enhanced Antimicrobial Activities against Gram-Negative Bacteria: ALYTESERIN-2: STRUCTURE-ACTIVITY. J. Pept. Sci. 2012, 18, 270–275. [Google Scholar] [CrossRef]
- Conlon, J.M.; Sonnevend, A.; Pál, T.; Vila-Farrés, X. Efficacy of Six Frog Skin-Derived Antimicrobial Peptides against Colistin-Resistant Strains of the Acinetobacter Baumannii Group. Int. J. Antimicrob. Agents 2012, 39, 317–320. [Google Scholar] [CrossRef] [PubMed]
- El-Ageery, S.M.; Abo-Shadi, M.A.; Elgendy, A.M.; Alghaithy, A.A.; Kandeel, A.Y. The Role of Health Care Workers and Environment on Transmission of Methicillin–Resistant Staphylococcus Aureus among Patients in a Medical Intensive Care Unit in a Saudi Hospital. Appl. Microbiol. 2011, 5, 1–8. [Google Scholar]
- Ghaima, K.; Saadedin, S.; Jassim, K. Prevalence of BlaOXA like Carbapenemase Genes in Multidrug Resistant Acinetobacter Baumannii Isolated from Burns and Wounds in Baghdad Hospitals. Res. J. Pharm. Biol. Chem. Sci. 2016, 7, 1347. [Google Scholar]
- Gowda, K.L.; Marie, M.A.M.; Al-Sheikh, Y.A.; John, J.; Gopalkrishnan, S.; Shashidhar, P.C.; Dabwan, K.H.M. A 6-Year Surveillance of Antimicrobial Resistance Patterns of Acinetobacter Baumannii Bacteremia Isolates from a Tertiary Care Hospital in Saudi Arabia during 2005–2010. Libyan J. Med. 2014, 9, 24039. [Google Scholar] [CrossRef]
- Hoang, V.-T.; Dao, T.-L.; Ly, T.D.A.; Gouriet, F.; Hadjadj, L.; Belhouchat, K.; Chaht, K.L.; Yezli, S.; Alotaibi, B.; Raoult, D.; et al. Acquisition of Multidrug-Resistant Bacteria and Encoding Genes among French Pilgrims during the 2017 and 2018 Hajj. Eur. J. Clin. Microbiol. Infect. Dis. 2021, 40, 1199–1207. [Google Scholar] [CrossRef]
- Kareem, S.M.; Al-Kadmy, I.M.S.; Al-Kaabi, M.H.; Aziz, S.N.; Ahmad, M. Acinetobacter Baumannii Virulence Is Enhanced by the Combined Presence of Virulence Factors Genes Phospholipase C (PlcN) and Elastase (LasB). Microb. Pathog. 2017, 110, 568–572. [Google Scholar] [CrossRef] [PubMed]
- Kusradze, I.; Diene, S.M.; Goderdzishvili, M.; Rolain, J.-M. Molecular Detection of OXA Carbapenemase Genes in Multidrug-Resistant Acinetobacter Baumannii Isolates from Iraq and Georgia. Int. J. Antimicrob. Agents 2011, 38, 164–168. [Google Scholar] [CrossRef]
- Lopes, B.S.; Al-Agamy, M.H.; Ismail, M.A.; Shibl, A.M.; Al-Qahtani, A.A.; Al-Ahdal, M.N.; Forbes, K.J. The Transferability of BlaOXA-23 Gene in Multidrug-Resistant Acinetobacter Baumannii Isolates from Saudi Arabia and Egypt. Int. J. Med. Microbiol. 2015, 305, 581–588. [Google Scholar] [CrossRef]
- Mahdi, L.; Mahdi, N.; Al-kakei, S.; Musafer, H.; Al-Joofy, I.; Essa, R.; Zwain, L.; Salman, I.; Mater, H.; Al-Alak, S.; et al. Treatment Strategy by Lactoperoxidase and Lactoferrin Combination: Immunomodulatory and Antibacterial Activity against Multidrug-Resistant Acinetobacter Baumannii. Microb. Pathog. 2018, 114, 147–152. [Google Scholar] [CrossRef] [PubMed]
- Mazi, W.; Alshammari, F.; Yu, J.; Saeed, A. A Descriptive Analysis of PVL-Positive Multidrug-Resistant Staphylococcus Aureus in Hospital-Associated Infections in Saudi Arabia. Bioinformation 2020, 16, 586–593. [Google Scholar] [CrossRef]
- Mechkarska, M.; Prajeep, M.; Radosavljevic, G.D.; Jovanovic, I.P.; Baloushi, A.A.; Sonnevend, A.; Lukic, M.L.; Conlon, J.M. An Analog of the Host-Defense Peptide Hymenochirin-1B with Potent Broad-Spectrum Activity against Multidrug-Resistant Bacteria and Immunomodulatory Properties. Peptides 2013, 50, 153–159. [Google Scholar] [CrossRef] [PubMed]
- Muslim, S.N.; Al-Kadmy, I.M.S.; Auda, I.G.; Ali, A.N.M.; Al-Jubori, S.S. A Novel Genetic Determination of a Lectin Gene in Iraqi Acinetobacter Baumannii Isolates and Use of Purified Lectin as an Antibiofilm Agent. J. AOAC Int. 2018, 101, 1623–1630. [Google Scholar] [CrossRef] [PubMed]
- Nasser, K.; Mustafa, A.S.; Khan, M.W.; Purohit, P.; Al-Obaid, I.; Dhar, R.; Al-Fouzan, W. Draft Genome Sequences of Six Multidrug-Resistant Clinical Strains of Acinetobacter Baumannii, Isolated at Two Major Hospitals in Kuwait. Genome Announc. 2018, 6, e00264-18. [Google Scholar] [CrossRef] [PubMed]
- Obeidat, N.; Jawdat, F.; Al-Bakri, A.G.; Shehabi, A.A. Major Biologic Characteristics of Acinetobacter Baumannii Isolates from Hospital Environmental and Patients’ Respiratory Tract Sources. Am. J. Infect. Control 2014, 42, 401–404. [Google Scholar] [CrossRef]
- Qasim, Z.J.; Kadhim, H.S.; Abdulamir, A.S. Identification of Antibiotic Resistance Genes in Multi-Drug Resistant Acinetobacter Baumannii Clinical Isolates of Iraqi Patients (Zq Strains), Using Whole-Genome Sequencing. Int. J. Pharm. Qual. Assur. 2019, 10, 670–680. [Google Scholar] [CrossRef]
- Rabaan, A.A.; Saunar, J.V.; Bazzi, A.M.; Raslan, W.F.; Taylor, D.R.; Al-Tawfiq, J.A. Epidemiology and Detection of Acinetobacter Using Conventional Culture and In-House Developed PCR Based Methods. J. Infect. Public Health 2017, 10, 124–128. [Google Scholar] [CrossRef] [PubMed]
- Radhi, S.H.; Al-Charrakh, A.H. Occurrence of MBLs and Carbapenemases among MDR and XDR Acinetobacter Baumannii Isolated from Hospitals in Iraq. Ind. J. Publ. Health Res. Dev. 2019, 10, 668. [Google Scholar] [CrossRef]
- Ridha, D.; Ali, M.; Jassim, K. Occurrence of Metallo-β-Lactamase Genes among Acinetobacter Baumannii Isolated from Different Clinical Samples. J. Pure Appl. Microbiol. 2019, 13, 1111–1119. [Google Scholar] [CrossRef]
- Salahuddin, N.; Amer, L.; Joseph, M.; El Hazmi, A.; Hawa, H.; Maghrabi, K. Determinants of Deescalation Failure in Critically Ill Patients with Sepsis: A Prospective Cohort Study. Crit. Care Res. Pract. 2016, 2016, 6794861. [Google Scholar] [CrossRef]
- Samrah, S.; Bashtawi, Y.; Hayajneh, W.; Almomani, B.; Momany, S.; Khader, Y. Impact of Colistin-Initiation Delay on Mortality of Ventilator-Associated Pneumonia Caused by A. Baumannii. J. Infect. Dev. Ctries 2016, 10, 1129–1134. [Google Scholar] [CrossRef] [PubMed]
- Senok, A.; Garaween, G.; Raji, A.; Khubnani, H.; Kim Sing, G.; Shibl, A. Genetic Relatedness of Clinical and Environmental Acinetobacter Baumanii Isolates from an Intensive Care Unit Outbreak. J. Infect. Dev. Ctries 2015, 9, 665–669. [Google Scholar] [CrossRef] [PubMed]
- Shah, M.W.; Yasir, M.; Farman, M.; Jiman-Fatani, A.A.; Almasaudi, S.B.; Alawi, M.; El-Hossary, D.; Azhar, E.I. Antimicrobial Susceptibility and Molecular Characterization of Clinical Strains of Acinetobacter Baumannii in Western Saudi Arabia. Microb. Drug Resist. 2019, 25, 1297–1305. [Google Scholar] [CrossRef]
- Somily, A.M. Comparison of E-Test and Disc Diffusion Methods for the in Vitro Evaluation of the Antimicrobial Activity of Colistin in Multi-Drug Resistant Gram-Negative Bacilli. Saudi Med. J. 2010, 5, 507–511. [Google Scholar]
- Somily, A.M.; Absar, M.M.; Arshad, M.Z.; Al Aska, A.I.; Shakoor, Z.A.; Fatani, A.J.; Siddiqui, Y.M.; Murray, T.S. Antimicrobial Susceptibility Patterns of Multidrug-Resistant Pseudomonas Aeruginosa and Acinetobacter Baumannii against Carbapenems, Colistin, and Tigecycline. Saudi Med. J. 2012, 33, 750–755. [Google Scholar]
- Vijayakumar, R.; Sandle, T.; Al-Aboody, M.S.; AlFonaisan, M.K.; Alturaiki, W.; Mickymaray, S.; Premanathan, M.; Alsagaby, S.A. Distribution of Biocide Resistant Genes and Biocides Susceptibility in Multidrug-Resistant Klebsiella Pneumoniae, Pseudomonas Aeruginosa and Acinetobacter Baumannii—A First Report from the Kingdom of Saudi Arabia. J. Infect. Public Health 2018, 11, 812–816. [Google Scholar] [CrossRef]
- Wahaab, I.T.A.; Almaroof, S.Q.M.; Yaseen, Z.T. Causative Microorganisms and Antibiotics Susceptibility in Neonatal Sepsis at Neonatal Intensive Care Unit: A Longitudinal Study from Diyala Governorate in Iraq. Indian J. Forensic Med. Toxicol. 2021, 15, 101–110. [Google Scholar] [CrossRef]
- Wibberg, D.; Salto, I.P.; Eikmeyer, F.G.; Maus, I.; Winkler, A.; Nordmann, P.; Pühler, A.; Poirel, L.; Schlüter, A. Complete Genome Sequencing of Acinetobacter Baumannii Strain K50 Discloses the Large Conjugative Plasmid PK50a Encoding Carbapenemase OXA-23 and Extended-Spectrum β-Lactamase GES-11. Antimicrob. Agents Chemother. 2018, 62, e00212-18. [Google Scholar] [CrossRef] [PubMed]
- Yasir, M.; Shah, M.W.; Jiman-Fatani, A.A.; Almasaudi, S.B.; Ahmad, H.; Alawi, M.; Azhar, E.I. Draft Genome Sequence of a Clinical Acinetobacter Baumannii Isolate of New Sequence Type ST1688 from Saudi Arabia. J. Glob. Antimicrob. Resist. 2019, 18, 151–152. [Google Scholar] [CrossRef]
- Al-Agamy, M.H.; El-Mahdy, T.S.; Radwan, H.H.; Poirel, L. Cooccurrence of NDM-1, ESBL, RmtC, AAC(6′)-Ib, and QnrB in Clonally Related Klebsiella Pneumoniae Isolates Together with Coexistence of CMY-4 and AAC(6′)-Ib in Enterobacter Cloacae Isolates from Saudi Arabia. BioMed Res. Int. 2019, 2019, 6736897. [Google Scholar] [CrossRef]
- Alatoom, A.; Sartawi, M.; Lawlor, K.; AbdelWareth, L.; Thomsen, J.; Nusair, A.; Mirza, I. Persistent Candidemia despite Appropriate Fungal Therapy: First Case of Candida Auris from the United Arab Emirates. Int. J. Infect. Dis. 2018, 70, 36–37. [Google Scholar] [CrossRef]
- Al-Baloushi, A.E.; Pál, T.; Ghazawi, A.; Sonnevend, A. Genetic Support of Carbapenemases in Double Carbapenemase Producer Klebsiella Pneumoniae Isolated in the Arabian Peninsula. Acta Microbiol. Immunol. Hung. 2018, 65, 135–150. [Google Scholar] [CrossRef]
- Alghoribi, M.F.; Binkhamis, K.; Alswaji, A.A.; Alhijji, A.; Alsharidi, A.; Balkhy, H.H.; Doumith, M.; Somily, A. Genomic Analysis of the First KPC-Producing Klebsiella Pneumoniae Isolated from a Patient in Riyadh: A New Public Health Concern in Saudi Arabia. J. Infect. Public Health 2020, 13, 647–650. [Google Scholar] [CrossRef]
- Algowaihi, R.; Ashgar, S.; Sirag, B.; Shalam, S.; Nassir, A.; Ahmed, A. Draft Genome Sequence of a Multidrug-Resistant Klebsiella Pneumoniae Strain Isolated from King Abdullah Medical City, Makkah, Saudi Arabia. Genome Announc. 2016, 4, e00375-16. [Google Scholar] [CrossRef] [PubMed]
- AL-Khikani, F.H.O.; Abadi, R.M.; Ayit, A.S. Emerging Carbapenemase Klebsiella Oxytoca with Multidrug Resistance Implicated in Urinary Tract Infection. Biomed. Biotechnol. Res. J. 2020, 2, 148–151. [Google Scholar]
- AL-Muqdadi, B.M.J.; AL-Saadi, B.Q.H. Detection Of Arma Gene, Kpc Enzyme And Molecular Typing Of K. Pneumoniae Clinical Isolate From Public Hospitals In Baghdad City, Iraq. Biochem. Cell. Arch. 2020, 20, 2101–2105. [Google Scholar]
- Al-Qahtani, A.A.; Al-Agamy, M.H.; Ali, M.S.; Al-Ahdal, M.N.; Aljohi, M.A.; Shibl, A.M. Characterization of Extended-Spectrum Beta-Lactamase-Producing Klebsiella Pneumoniae from Riyadh, Saudi Arabia. J. Chemother. 2014, 26, 139–145. [Google Scholar] [CrossRef]
- Alsanie, W.F. Molecular Diversity and Profile Analysis of Virulence-Associated Genes in Some Klebsiella Pneumoniae Isolates. Pract. Lab. Med. 2020, 19, e00152. [Google Scholar] [CrossRef] [PubMed]
- Alyousef, A.A.; Khazaal, S.S.; Ali, A.N.M.; Hussein, N.H.; Hussein, S.M.A. Detection of Prevalent Mechanism of Extended Spectrum β-Lactamases, Metallo β-Lactamases, and AmpC β Lactamases-Producing Klebsiella Pneumoniae in the Tertiary Care Hospital. Rev. Med. Microbiol. 2017, 28, 133–139. [Google Scholar] [CrossRef]
- Salman, R.A.; Ghaima, K.K. Prevalence of ESBL genes in ESBL producing Klebsiella pneumoniae isolated from patients with urinary tract infections in Baghdad, Iraq. Biosci. Res. 2018, 15, 2049–2059. [Google Scholar]
- El Nekidy, W.S.; Mooty, M.Y.; Attallah, N.; Cardona, L.; Bonilla, M.F.; Ghazi, I.M. Successful Treatment of Multidrug Resistant Klebsiella Pneumoniae Using Dual Carbapenem Regimen in Immunocompromised Patient. IDCases 2017, 9, 53–55. [Google Scholar] [CrossRef]
- Hassan, M.I.; Alkharsah, K.R.; Alzahrani, A.J.; Obeid, O.E.; Khamis, A.H.; Diab, A. Detection of Extended Spectrum Beta-Lactamases-Producing Isolates and Effect of AmpC Overlapping. J. Infect. Dev. Ctries 2013, 7, 618–629. [Google Scholar] [CrossRef] [PubMed]
- Lagha, R.; Ben Abdallah, F.; ALKhammash, A.A.H.; Amor, N.; Hassan, M.M.; Mabrouk, I.; Alhomrani, M.; Gaber, A. Molecular Characterization of Multidrug Resistant Klebsiella Pneumoniae Clinical Isolates Recovered from King Abdulaziz Specialist Hospital at Taif City, Saudi Arabia. J. Infect. Public Health 2021, 14, 143–151. [Google Scholar] [CrossRef]
- Poirel, L.; Al Maskari, Z.; Al Rashdi, F.; Bernabeu, S.; Nordmann, P. NDM-1-Producing Klebsiella Pneumoniae Isolated in the Sultanate of Oman. J. Antimicrob. Chemother. 2011, 66, 304–306. [Google Scholar] [CrossRef]
- Saaid Tuwaij, N.S.; Al-khilkhali, H.J.B.; Mohsen, H.M. Prevalence of SUL(1,2), GYR(A, B) and OXA Genes among Multidrug Resistance Klebsiella Pneumoniae Isolates Recovered from Women Suffering Urinary Tract Infection. Int. J. Res. Pharm. Sci. 2020, 11, 2424–2432. [Google Scholar] [CrossRef]
- Shibl, A.; Al-Agamy, M.; Memish, Z.; Senok, A.; Khader, S.A.; Assiri, A. The Emergence of OXA-48- and NDM-1-Positive Klebsiella Pneumoniae in Riyadh, Saudi Arabia. Int. J. Infect. Dis. 2013, 17, e1130–e1133. [Google Scholar] [CrossRef]
- Shlash, A.A.A.; Tuwaij, N.S.S. Molecular Dissemination Of Ambler Class A And C Β- Lactamase Genes Among Ceftriaxone Resistant Klebsiella Pneumoniae Infection In Najaf City, Iraq. Biochem. Cell. Arch. 2018, 18, 2511–2522. [Google Scholar]
- Vali, L.; Dashti, A.A.; Jadaon, M.M.; El-Shazly, S. The Emergence of Plasmid Mediated Quinolone Resistance QnrA2 in Extended Spectrum β-Lactamase Producing Klebsiella Pneumoniae in the Middle East. DARU J. Pharm Sci. 2015, 23, 34. [Google Scholar] [CrossRef]
- uz Zaman, T.; Alrodayyan, M.; Albladi, M.; Aldrees, M.; Siddique, M.I.; Aljohani, S.; Balkhy, H.H. Clonal Diversity and Genetic Profiling of Antibiotic Resistance among Multidrug/Carbapenem-Resistant Klebsiella Pneumoniae Isolates from a Tertiary Care Hospital in Saudi Arabia. BMC Infect. Dis. 2018, 18, 205. [Google Scholar] [CrossRef] [PubMed]
- uz Zaman, T.; Aldrees, M.; Al Johani, S.M.; Alrodayyan, M.; Aldughashem, F.A.; Balkhy, H.H. Multi-Drug Carbapenem-Resistant Klebsiella Pneumoniae Infection Carrying the OXA-48 Gene and Showing Variations in Outer Membrane Protein 36 Causing an Outbreak in a Tertiary Care Hospital in Riyadh, Saudi Arabia. Int. J. Infect. Dis. 2014, 28, 186–192. [Google Scholar] [CrossRef]
- AbdulWahab, A.; Taj-Aldeen, S.J.; Ibrahim, E.B.; Hussain, S.; Muhammed, R.; Ahmed, I.; Abdeen, Y.; Sadek, O.; Abu-Madi, M. Genetic Relatedness and Host Specificity of Pseudomonas Aeruginosa Isolates from Cystic Fibrosis and Non-Cystic Fibrosis Patients. Infect. Drug Resist. 2014, 7, 309. [Google Scholar] [CrossRef]
- AbdulWahab, A.; Zahraldin, K.; Ahmed, M.A.; Jarir, S.A.; Muneer, M.; Mohamed, S.F.; Hamid, J.M.; Hassan, A.A.; Ibrahim, E.B. The Emergence of Multidrug-Resistant Pseudomonas Aeruginosa in Cystic Fibrosis Patients on Inhaled Antibiotics. Lung India 2017, 34, 527. [Google Scholar] [CrossRef]
- Ahmad, I.; Irfan, S.; Abohashrh, M.; Wahab, S.; Abullais, S.S.; Javali, M.A.; Nisar, N.; Alam, M.M.; Srivastava, S.; Saleem, M.; et al. Inhibitory Effect of Nepeta Deflersiana on Climax Bacterial Community Isolated from the Oral Plaque of Patients with Periodontal Disease. Molecules 2021, 26, 202. [Google Scholar] [CrossRef] [PubMed]
- Alamri, A.M.; Alfifi, S.; Aljehani, Y.; Alnimr, A. Whole Genome Sequencing of Ceftolozane-Tazobactam and Ceftazidime-Avibactam Resistant Pseudomonas Aeruginosa Isolated from a Blood Stream Infection Reveals VEB and Chromosomal Metallo-Beta Lactamases as Genetic Determinants: A Case Report. Infect. Drug Resist. 2020, 13, 4215–4222. [Google Scholar] [CrossRef]
- Al-Delaimi, M.S.; Yacoob Aldosky, H.Y. Amending the Efficiency of Antimicrobials against Multidrug-Resistant Pseudomonas Aeruginosa by Low-Frequency Magnetic Fields. Bull. Exp. Biol. Med. 2020, 170, 35–39. [Google Scholar] [CrossRef]
- Alhamdani, R.J.M.; Al-Luaibi, Y.Y.Y. Detection of exoA, nan1 genes, the biofilm production with the effect of Oyster shell and two plant extracts on Pseudomonas aeroginosa isolated from burn’ patient and their surrounding environment. Syst. Rev. Pharm. 2020, 11, 11. [Google Scholar]
- Al-Zahrani, I.A.; Al-Ahmadi, B.M. Dissemination of VIM-Producing Pseudomonas Aeruginosa Associated with High-Risk Clone ST654 in a Tertiary and Quaternary Hospital in Makkah, Saudi Arabia. J. Chemother. 2021, 33, 12–20. [Google Scholar] [CrossRef]
- Asghar, A. Antimicrobial Susceptibility and Metallo-β-Lactamase Production among Pseudomonas Aeruginosa Isolated from Makkah Hospitals. Pak. J. Med. Sci. 2012, 28, 781–786. [Google Scholar]
- Behbahani, M.R.; Keshavarzi, A.; Pirbonyeh, N.; Javanmardi, F.; Khoob, F.; Emami, A. Plasmid- related β-Lactamase Genes in Pseudomonas Aeruginosa Isolates: A Molecular Study in Burn Patients. J. Med. Microbiol. 2019, 68, 1740–1746. [Google Scholar]
- Bourghli, A.; Boissiere, L.; Obeid, I. Thoracic Kyphotic Deformity Secondary to Old Pseudomonas Aeruginosa Spondylodiscitis in an Immunocompromised Patient With Persistent Infection Foci—A Case Report. Int. J. Spine Surg. 2019, 13, 392–398. [Google Scholar] [CrossRef] [PubMed]
- Hassan, S.; Najati, A.; Abass, K. Isolation and Identification of Multi-Drug Resistant “Pseudomonas Aeruginosa” from Burn Wound Infection in Kirkuk City, Iraq. Eurasian J. Biosci. 2019, 13, 1045–1050. [Google Scholar]
- Hassan, S.A.; Shobrak, M.Y. Prevalence and Antimicrobial Resistance Characteristics of Gram-Negative Bacteria Associated with Wild Animals Presenting at Live Animal Market, Taif, Western Saudi Arabia. Antimicrob. Resist. 2014, 8, 273–282. [Google Scholar]
- Jaaffar, A.I. Detection of BlaIMP Gene among Pseudomonas Aeruginosa Isolated from Different Clinical Samples. Drug Invent. Today 2019, 12, 5. [Google Scholar]
- Khan, R.; Al-Dorzi, H.M.; Tamim, H.M.; Rishu, A.H.; Balkhy, H.; El-Saed, A.; Arabi, Y.M. The Impact of Onset Time on the Isolated Pathogens and Outcomes in Ventilator Associated Pneumonia. J. Infect. Public Health 2016, 9, 161–171. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.A.; Faiz, A. Antimicrobial Resistance Patterns of Pseudomonas Aeruginosa in Tertiary Care Hospitals of Makkah and Jeddah. Ann. Saudi Med. 2016, 36, 23–28. [Google Scholar] [CrossRef]
- Mahdi, L.H.; Jabbar, H.S.; Auda, I.G. Antibacterial Immunomodulatory and Antibiofilm Triple Effect of Salivaricin LHM against Pseudomonas Aeruginosa Urinary Tract Infection Model. Int. J. Biol. Macromol. 2019, 134, 1132–1144. [Google Scholar] [CrossRef] [PubMed]
- Nasser, M.; Gayen, S.; Kharat, A.S. Prevalence of β-Lactamase and Antibiotic-Resistant Pseudomonas Aeruginosa in the Arab Region. J. Glob. Antimicrob. Resist. 2020, 22, 152–160. [Google Scholar] [CrossRef]
- Nasser, M.; Kharat, A.S. Phenotypic Demonstration of SS-Lactamase (ESßLs, MßLs, and Amp-C) among MDR Pseudomonas Aeruginosa Isolates Obtained From Burn Wound Infected in Yemen. J. Appl. Biol. Biotechnol. 2019, 7, 31–34. [Google Scholar] [CrossRef]
- Sid Ahmed, M.A.; Khan, F.A.; Sultan, A.A.; Söderquist, B.; Ibrahim, E.B.; Jass, J.; Omrani, A.S. β-Lactamase-Mediated Resistance in MDR-Pseudomonas Aeruginosa from Qatar. Antimicrob. Resist. Infect. Control 2020, 9, 170. [Google Scholar] [CrossRef] [PubMed]
- Tarazi, Y.H.; Abu-Basha, E.; Ismail, Z.B.; Al-Jawasreh, S.I. Antimicrobial Susceptibility of Multidrug-Resistant Pseudomonas Aeruginosa Isolated from Drinking Water and Hospitalized Patients in Jordan. Acta Trop. 2021, 217, 105859. [Google Scholar] [CrossRef]
- Zikri, A.; El Masri, K. Use of Ceftolozane/Tazobactam for the Treatment of Multidrug-Resistant Pseudomonas Aeruginosa Pneumonia in a Pediatric Patient with Combined Immunodeficiency (CID): A Case Report from a Tertiary Hospital in Saudi Arabia. Antibiotics 2019, 8, 67. [Google Scholar] [CrossRef] [PubMed]
- Abulreesh, H.H.; Organji, S.R.; Osman, G.E.H.; Elbanna, K.; Almalki, M.H.K.; Ahmad, I. Prevalence of Antibiotic Resistance and Virulence Factors Encoding Genes in Clinical Staphylococcus Aureus Isolates in Saudi Arabia. Clin. Epidemiol. Glob. Health 2017, 5, 196–202. [Google Scholar] [CrossRef]
- Abulreesh, H.H.; Organji, S.R. The Prevalence of Multidrug-Resistant Staphylococci in Food and the Environment of Makkah, Saudi Arabia. Res. J. Microbiol. 2011, 6, 510–523. [Google Scholar] [CrossRef]
- Al Zebary, M.K.; Yousif, S.Y.; Assafi, M.S. The Prevalence, Molecular Characterization and Antimicrobial Susceptibility of S. Aureus Isolated from Impetigo Cases in Duhok, Iraq. Open Dermatol. J. 2017, 11, 22–29. [Google Scholar] [CrossRef]
- AlFouzan, W.; Al-Haddad, A.; Udo, E.; Mathew, B.; Dhar, R. Frequency and Clinical Association of Panton-Valentine Leukocidin-Positive Staphylococcus Aureus Isolates: A Study from Kuwait. Med Princ. Pract. 2013, 22, 245–249. [Google Scholar] [CrossRef]
- Alghizzi, M.J.; Alansari, M.; Shami, A. The Prevalence of Staphylococcus Aureus and Methicillin Resistant Staphylococcus Aureus in Processed Food Samples in Riyadh, Saudi Arabia. J. Pure Appl. Microbiol. 2021, 15, 91–99. [Google Scholar] [CrossRef]
- Al-Zoubi, M.S.; Al-Tayyar, I.A.; Hussein, E.; Jabali, A.A.; Khudairat, S. Antimicrobial Susceptibility Pattern of Staphylococcus Aureus Isolated from Clinical Specimens in Northern Area of Jordan. Iran. J. Microbiol. 2015, 7, 265. [Google Scholar]
- Hamid, M.E. Prevalence of Bacterial Pathogens in Aseer Region, Kingdom of Saudi Arabia: Emphasis on Antimicrobial Susceptibility of Staphylococcus Aureus. Oman Med. J. 2011, 26, 368–369. [Google Scholar] [CrossRef]
- Ismail, Z.B. Molecular Characteristics, Antibiogram and Prevalence of Multi-Drug Resistant Staphylococcus Aureus (MDRSA) Isolated from Milk Obtained from Culled Dairy Cows and from Cows with Acute Clinical Mastitis. Asian Pac. J. Trop. Biomed. 2017, 7, 694–697. [Google Scholar] [CrossRef]
- Kanaan, M.H.G. Antibacterial Effect of Ozonated Water against Methicillin-Resistant Staphylococcus Aureus Contaminating Chicken Meat in Wasit Province, Iraq. Vet. World 2018, 11, 1445–1453. [Google Scholar] [CrossRef]
- Kanaan, M.H.G.; Al-Isawi, A.J.O. Prevalence Of Methicillin Or Multiple Drug-Resistant Staphylococcus Aureus In Cattle Meat Marketed In Wasit Province. Biochem. Cell. Arch. 2019, 19, 495–502. [Google Scholar]
- Obaidat, M.M.; Bani Salman, A.E.; Lafi, S.Q. Prevalence of Staphylococcus Aureus in Imported Fish and Correlations between Antibiotic Resistance and Enterotoxigenicity. J. Food Prot. 2015, 78, 1999–2005. [Google Scholar] [CrossRef] [PubMed]
- Vellappally, S.; Divakar, D.D.; Al Kheraif, A.A.; Ramakrishnaiah, R.; Alqahtani, A.; Dalati, M.H.N.; Anil, S.; Khan, A.A.; Harikrishna Varma, P.R. Occurrence of Vancomycin-Resistant Staphylococcus Aureus in the Oral Cavity of Patients with Dental Caries. Acta Microbiol. Et Immunol. Hung. 2017, 64, 343–351. [Google Scholar] [CrossRef]
- Abdalla, N.M.; Osman, A.A.; Haimour, W.O.; Sarhan, M.A.; Mohammed, M.N.; Zyad, E.M.; Al-ghtani, A.M. Antimicrobial Susceptibility Pattern in Nosocomial Infections Caused by Acinetobacter Species in Asir Region, Saudi Arabia. Pak. J. Biol. Sci. 2013, 16, 275–280. [Google Scholar] [CrossRef][Green Version]
- Balkhy, H.H.; Bawazeer, M.S.; Kattan, R.F.; Tamim, H.M.; Johani, S.M.; Aldughashem, F.A.; Al Alem, H.A.; Adlan, A.; Herwaldt, L.A. Epidemiology of Acinetobacter spp.-Associated Healthcare Infections and Colonization among Children at a Tertiary-Care Hospital in Saud Arabia: A 6-Year Retrospective Cohort Study. Eur. J. Clin. Microbiol. Infect. Dis. 2012, 31, 2645–2651. [Google Scholar] [CrossRef] [PubMed]
- Somily, A.M.; Al-Khattaf, A.S.; Kambal, A.M. Antimicrobial Activity of Tigecycline against Bacterial Isolates from Intensive Care Units in a Teaching Hospital in Central Saudi Arabia. Saudi Med. J. 2010, 31, 18–24. [Google Scholar] [PubMed]
- Al-Kharousi, Z.S.; Guizani, N.; Al-Sadi, A.M.; Al-Bulushi, I.M. Antibiotic Resistance of Enterobacteriaceae Isolated from Fresh Fruits and Vegetables and Characterization of Their AmpC β-Lactamases. J. Food Prot. 2019, 82, 1857–1863. [Google Scholar] [CrossRef]
- Alkofide, H.; Alhammad, A.M.; Alruwaili, A.; Aldemerdash, A.; Almangour, T.A.; Alsuwayegh, A.; Almoqbel, D.; Albati, A.; Alsaud, A.; Enani, M. Multidrug-Resistant and Extensively Drug-Resistant Enterobacteriaceae: Prevalence, Treatments, and Outcomes—A Retrospective Cohort Study. Infect. Drug Resist. 2020, 13, 4653–4662. [Google Scholar] [CrossRef] [PubMed]
- Jamal, W.Y.; Albert, M.J.; Rotimi, V.O. High Prevalence of New Delhi Metallo-β-Lactamase-1 (NDM-1) Producers among Carbapenem-Resistant Enterobacteriaceae in Kuwait. PLoS ONE 2016, 11, e0152638. [Google Scholar] [CrossRef]
- Al-Mayahi, F.S.A.; Ali, R.H. Preliminary study of emergence MDR of Providencia spp. isolates producing ESBL, AmpC and MBL among patients with RTI and in wastewater in Al-Diwaniya city, Iraq. Biochem. Cell. Arch. 2018, 18, 12. [Google Scholar]
- Ayyal Al-Gburi, N.M. Isolation and Molecular Identification and Antimicrobial Susceptibility of Providencia spp. from Raw Cow’s Milk in Baghdad, Iraq. Vet. Med. Int. 2020, 2020, 8874747. [Google Scholar] [CrossRef]
- Al-Lahham, A.; Qayyas, J.A. The Impact of the 7-Valent Pneumococcal Conjugate Vaccine on Nasopharyngeal Carriage of Streptococcus Pneumoniae in Infants of Ajlun Governorate in Jordan. Jordan J. Biol. Sci. 2018, 11, 8. [Google Scholar]
- Al-Mazrou, K.A.; Shibl, A.M.; Kandeil, W.; Pirçon, J.-Y.; Marano, C. A Prospective, Observational, Epidemiological Evaluation of the Aetiology and Antimicrobial Susceptibility of Acute Otitis Media in Saudi Children Younger than 5 Years of Age. J. Epidemiol. Glob. Health 2014, 4, 231. [Google Scholar] [CrossRef] [PubMed]
- Almazrou, Y.; Shibl, A.M.; Alkhlaif, R.; Pirçon, J.-Y.; Anis, S.; Kandeil, W.; Hausdorff, W.P. Epidemiology of Invasive Pneumococcal Disease in Saudi Arabian Children Younger than 5 Years of Age. J. Epidemiol. Glob. Health 2015, 6, 95. [Google Scholar] [CrossRef] [PubMed]
- Alnimr, A.M.; Farhat, M. Phenotypic and Molecular Study of Pneumococci Causing Respiratory Tract Infections: A 3-Year Prospective Cohort. Saudi Med. J. 2017, 38, 350–358. [Google Scholar] [CrossRef]
- Al-Sa’ady, A.T.; Hussein, F.H. Nanomedical Applications of Titanium Dioxide Nanoparticles as Antibacterial Agent against Multi-Drug Resistant Streptococcus Pneumoniae. Syst. Rev. Pharm. 2020, 11, 11. [Google Scholar]
- Elshafie, S.; Taj-Aldeen, S.J. Emerging Resistant Serotypes of Invasive Streptococcus Pneumoniae. Infect. Drug Resist. 2016, 9, 153–160. [Google Scholar] [CrossRef]
- Sallam, M. Trends in Antimicrobial Drug Resistance of Streptococcus Pneumoniae Isolates at Jordan University Hospital (2000–2018). Antibiotics 2019, 8, 41. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.; Baek, J.Y.; Kim, S.H.; Song, J.-H.; Ko, K.S. Predominance of ST320 among Streptococcus Pneumoniae Serotype 19A Isolates from 10 Asian Countries. J. Antimicrob. Chemother. 2011, 66, 1001–1004. [Google Scholar] [CrossRef] [PubMed]
- Al Baidani, A.; Elshoni, W.; Shawa, T. Antibiotic Susceptibility Pattern of Methicillin-Resistant Staphylococcus Aureus in Three Hospitals at Hodeidah City, Yemen. Glob. J. Pharmacol. 2011, 55, 105–111. [Google Scholar]
- Albarrag, A.; Shami, A.; Almutairi, A.; Alsudairi, S.; Aldakeel, S.; Al-Amodi, A. Prevalence and Molecular Genetics of Methicillin-Resistant Staphylococcus Aureus Colonization in Nursing Homes in Saudi Arabia. Can. J. Infect. Dis. Med. Microbiol. 2020, 2020, 2434350. [Google Scholar] [CrossRef] [PubMed]
- Alfouzan, W.; Udo, E.E.; Modhaffer, A.; Alosaimi, A. Molecular Characterization of Methicillin- Resistant Staphylococcus Aureus in a Tertiary Care Hospital in Kuwait. Sci. Rep. 2019, 9, 18527. [Google Scholar] [CrossRef]
- Alhussaini, M.S. Methicillin-Resistant Staphylococcus Aureus Nasal Carriage Among Patients Admitted at Shaqra General Hospital in Saudi Arabia. Pak. J. Biol. Sci. 2016, 19, 233–238. [Google Scholar] [CrossRef] [PubMed]
- Alkharsah, K.R.; Rehman, S.; Alkhamis, F.; Alnimr, A.; Diab, A.; Al-Ali, A.K. Comparative and Molecular Analysis of MRSA Isolates from Infection Sites and Carrier Colonization Sites. Ann. Clin. Microbiol. Antimicrob. 2018, 17, 7. [Google Scholar] [CrossRef]
- Aziz, Z.S.; Hassan, M.A. Phenotypic and Molecular Study of MecA Gene in MRSA Isolated from Clinical Cases in Misan Province/Iraq. Indian J. Public Health 2019, 10, 553. [Google Scholar] [CrossRef]
- Helmi, N.R.; Zaman, R.M.; Aly, M.M. Prevalence Of Gram-Positive Bacteria In Jeddah, Kingdom Of Saudi Arabia: Study Of Antimicrobial Resistance Patterns And Molecular Typing. Int. J. Pharma Biol. Sci. 2013, 4, 1231–1245. [Google Scholar]
- Obaidat, M.M.; Bani Salman, A.E.; Roess, A.A. High Prevalence and Antimicrobial Resistance of MecA Staphylococcus Aureus in Dairy Cattle, Sheep, and Goat Bulk Tank Milk in Jordan. Trop. Anim. Health Prod. 2018, 50, 405–412. [Google Scholar] [CrossRef]
- Udo, E.E.; Boswihi, S.S. Antibiotic Resistance Trends in Methicillin-Resistant Staphylococcus aureus Isolated in Kuwait Hospitals: 2011–2015. Med Princ. Pract. 2017, 26, 485–490. [Google Scholar] [CrossRef] [PubMed]
- Hala, S.; Antony, C.P.; Alshehri, M.; Althaqafi, A.O.; Alsaedi, A.; Mufti, A.; Kaaki, M.; Alhaj-Hussein, B.T.; Zowawi, H.M.; Al-Amri, A.; et al. First Report of Klebsiella Quasipneumoniae Harboring BlaKPC-2 in Saudi Arabia. Antimicrob. Resist. Infect. Control 2019, 8, 203. [Google Scholar] [CrossRef]
- Ejaz, H.; Alzahrani, B.; Hamad, M.F.S.; Abosalif, K.O.A.; Junaid, K.; Abdalla, A.E.; Elamir, M.Y.M.; Aljaber, N.J.; Hamam, S.S.M.; Younas, S. Molecular Analysis of the Antibiotic Resistant NDM-1 Gene in Clinical Isolates of Enterobacteriaceae. Clin. Lab. 2020, 66. [Google Scholar] [CrossRef]
- Ahmad, S.; Khan, Z.; Al-Sweih, N.; Alfouzan, W.; Joseph, L. Candida Auris in Various Hospitals across Kuwait and Their Susceptibility and Molecular Basis of Resistance to Antifungal Drugs. Mycoses 2020, 63, 104–112. [Google Scholar] [CrossRef] [PubMed]
- Emara, M.; Ahmad, S.; Khan, Z.; Joseph, L.; Al-Obaid, I.; Purohit, P.; Bafna, R. Candida Auris Candidemia in Kuwait, 2014. Emerg. Infect. Dis. 2015, 21, 1091–1092. [Google Scholar] [CrossRef]
- Khan, Z.; Ahmad, S.; Benwan, K.; Purohit, P.; Al-Obaid, I.; Bafna, R.; Emara, M.; Mokaddas, E.; Abdullah, A.A.; Al-Obaid, K.; et al. Invasive Candida Auris Infections in Kuwait Hospitals: Epidemiology, Antifungal Treatment and Outcome. Infection 2018, 46, 641–650. [Google Scholar] [CrossRef]
- Hassan, M.M.; Belal, E.-S.B. Antibiotic Resistance and Virulence Genes in Enterococcus Strains Isolated from Different Hospitals in Saudi Arabia. Biotechnol. Biotechnol. Equip. 2016, 30, 726–732. [Google Scholar] [CrossRef][Green Version]
- Salem-Bekhit, M.; Moussa, I.; Muharram, M.; Alanazy, F.; Hefni, H. Prevalence and Antimicrobial Resistance Pattern of Multidrug-Resistant Enterococci Isolated from Clinical Specimens. Indian J. Med. Microbiol. 2012, 30, 44–51. [Google Scholar] [CrossRef]
- Dibby, H.; Shlash, R. The Problem of Multidrug Resistance Bacterial Strains in Daily Clinical Practice in Dealing with Typhoid Fever in Mid-Euphrates Region of Iraq: A Cross Sectional Study. Indian J. Forensic Med. Toxicol. 2020, 14, 626–630. [Google Scholar]
- El-Tayeb, M.A.; Ibrahim, A.S.S.; Al-Salamah, A.A.; Almaary, K.S.; Elbadawi, Y.B. Prevalence, Serotyping and Antimicrobials Resistance Mechanism of Salmonella Enterica Isolated from Clinical and Environmental Samples in Saudi Arabia. Braz. J. Microbiol. 2017, 48, 499–508. [Google Scholar] [CrossRef] [PubMed]
- Harb, A.; O’Dea, M.; Hanan, Z.K.; Abraham, S.; Habib, I. Prevalence, Risk Factors and Antimicrobial Resistance of Salmonella Diarrhoeal Infection among Children in Thi-Qar Governorate, Iraq. Epidemiol. Infect. 2017, 145, 3486–3496. [Google Scholar] [CrossRef]
- Sahan, Z.A.; Hamed, S.L. Molecular Detection of Extended-Spectrum β-Lactamases- Producer Serratia Marcescens Causing Neonatal Sepsis in Iraq. Int. J. Res. Pharm. Sci. 2020, 11, 5803–5808. [Google Scholar] [CrossRef]
- Abdel-Haleem, A.M.; Rchiad, Z.; Khan, B.K.; Abdallah, A.M.; Naeem, R.; Nikhat Sheerin, S.; Solovyev, V.; Ahmed, A.; Pain, A. Genome Sequence of a Multidrug-Resistant Strain of Stenotrophomonas Maltophilia with Carbapenem Resistance, Isolated from King Abdullah Medical City, Makkah, Saudi Arabia. Genome Announc. 2015, 3. [Google Scholar] [CrossRef]
- El-Kersh, T.A.; Marie, M.A.; Al-Sheikh, Y.A.; Al-Agamy, M.H.; Al-Bloushy, A.A. Prevalence and Risk Factors of Early Fecal Carriage of Enterococcus Faecalis and Staphylococcus spp. and Their Antimicrobial Resistant Patterns among Healthy Neonates Born in a Hospital Setting in Central Saudi Arabia. Saudi Med. J. 2016, 37, 280–287. [Google Scholar] [CrossRef] [PubMed]
- Farman, M.; Yasir, M.; Al-Hindi, R.R.; Farraj, S.A.; Jiman-Fatani, A.A.; Alawi, M.; Azhar, E.I. Genomic Analysis of Multidrug-Resistant Clinical Enterococcus Faecalis Isolates for Antimicrobial Resistance Genes and Virulence Factors from the Western Region of Saudi Arabia. Antimicrob. Resist. Infect. Control 2019, 8, 55. [Google Scholar] [CrossRef]
- George, S.K.; Suseela, M.R.; El Safi, S.; Elnagi, E.A.; Al-Naam, Y.A.; Adam, A.A.; Jacob, A.M.; Al-Maqati, T.; Ks, H.K. Molecular Determination of van Genes among Clinical Isolates of Enterococci at a Hospital Setting. Saudi J. Biol. Sci. 2021, 28, 2895–2899. [Google Scholar] [CrossRef]
- Garaween, G.; Somily, A.; Raji, A.; Braun, S.; Al-Kattan, W.; Shibl, A.; Ehricht, R.; Senok, A. Serogenotyping and Emergence of Extended-Spectrum β-Lactamase Genes in Non-Typhoidal Salmonella: First Report from Saudi Arabia. J. Med. Microbiol. 2016, 65, 1343–1346. [Google Scholar] [CrossRef]
- Alsalem, Z.; Elhadi, N.; Aljeldah, M.; Alzahrani, F.; Nishibuchi, M. Characterization of Vibrio Vulnificus Isolated from the Coastal Areas in the Eastern Province of Saudi Arabia. J. Pure Appl. Microbiol. 2018, 12, 1355–1364. [Google Scholar] [CrossRef]
- M Kurdi Al-Dulaimi, M.; Abd Mutalib, S.; Abd Ghani, M.; Mohd Zaini, N.A.; Ariffin, A.A. Multiple Antibiotic Resistance (MAR), Plasmid Profiles, and DNA Polymorphisms among Vibrio Vulnificus Isolates. Antibiotics 2019, 8, 68. [Google Scholar] [CrossRef]
- Abro, A.H. Salmonella Typhi: Antibiotic Sensitivity Pattern in Dubai, United Arab Emirates. Pak. J. Med. Sci. 2011, 27, 6. [Google Scholar]
- AL-Fatlawy, H.N.K.; AL-Hadrawi, H.A.N. Molecular Profiling of Class I Integron Gene in MDR Salmonella Typhi Isolates. J. Pure Appl. Microbiol. 2020, 14, 1825–1833. [Google Scholar] [CrossRef]
- Yassin, M.T.; Mostafa, A.A.; Al-Askar, A.A.; Bdeer, R. In Vitro Antifungal Resistance Profile of Candida Strains Isolated from Saudi Women Suffering from Vulvovaginitis. Eur. J. Med. Res. 2020, 25, 1. [Google Scholar] [CrossRef]
- Alaboudi, A.R.; Malkawi, I.M.; Osaili, T.M.; Abu-Basha, E.A.; Guitian, J. Prevalence, Antibiotic Resistance and Genotypes of Campylobacter Jejuni and Campylobacter Coli Isolated from Chickens in Irbid Governorate, Jordan. Int. J. Food Microbiol. 2020, 327, 108656. [Google Scholar] [CrossRef] [PubMed]
- Ghunaim, H.; Behnke, J.M.; Aigha, I.; Sharma, A.; Doiphode, S.H.; Deshmukh, A.; Abu-Madi, M.M. Analysis of Resistance to Antimicrobials and Presence of Virulence/Stress Response Genes in Campylobacter Isolates from Patients with Severe Diarrhoea. PLoS ONE 2015, 10, e0119268. [Google Scholar] [CrossRef]
- Kanaan, M.H.G.; Mohammed, F.A. Antimicrobial Resistance of Campylobacter Jejuni from Poultry Meat in Local Markets of Iraq. Plant Arch. 2020, 20, 410–415. [Google Scholar]
- Jasim, S.A.; Al-abodi, H.R.; Ali, W.S. Resistance Rate and Novel Virulence Factor Determinants of Arcobacter spp., from Cattle Fresh Meat Products from Iraq. Microb. Pathog. 2021, 152, 104649. [Google Scholar] [CrossRef]
- AbdAlhussen, L.S.; Darweesh, M.F. Prevelance and Antibiotic Susceptibility Patterns of Pantoea spp. Int. J. ChemTech Res. 2016, 9, 430–437. [Google Scholar]
- Fazaa, S.A.; Darweesh, M.F. Prevalence of Pantoea spp. among Recurrent UTI Patients with Emphasis on Risk Factors and Antibiotic Resistance Patterns in Al-Qadisiyah Hospitals, Iraq. Ann. Trop. Med. Public Health 2020, 23, sp231202. [Google Scholar] [CrossRef]
- Burjaq, S.Z.; Abu-Romman, S.M. Prevalence and Antimicrobial Resistance of Salmonella spp. From Irrigation Water in Two Major Sources in Jordan. Curr. Microbiol. 2020, 77, 3760–3766. [Google Scholar] [CrossRef] [PubMed]
- Jassim, A.A.; Al-Gburi, N.M. Virulence Genes And Antimicrobial Resistance Of Salmonella Isolated From Milk In Wasit Province, Iraq. Plant Arch. 2020, 20, 2033–2039. [Google Scholar]
- Olaitan, A.O.; Dia, N.M.; Gautret, P.; Benkouiten, S.; Belhouchat, K.; Drali, T.; Parola, P.; Brouqui, P.; Memish, Z.; Raoult, D.; et al. Acquisition of Extended-Spectrum Cephalosporin- and Colistin-Resistant Salmonella Enterica Subsp. Enterica Serotype Newport by Pilgrims during Hajj. Int. J. Antimicrob. Agents 2015, 45, 600–604. [Google Scholar] [CrossRef]
- Osaili, T.M.; Al-Nabulsi, A.A.; Shaker, R.R.; Jaradat, Z.W.; Taha, M.; Al-Kherasha, M.; Meherat, M.; Holley, R. Prevalence of Salmonella Serovars, Listeria Monocytogenes, and Escherichia coli O157:H7 in Mediterranean Ready-to-Eat Meat Products in Jordan. J. Food Prot. 2014, 77, 106–111. [Google Scholar] [CrossRef]
- Abd Al-Mayahi, F.S.; Jaber, S.M. Multiple Drug Resistance of Listeria Monocytogenes Isolated from Aborted Women by Using Serological and Molecular Techniques in Diwaniyah City/Iraq. Iran. J. Microbiol. 2020, 12, 305. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, M.S.; Taha, Z.M.A.; Omer, L.T. Isolation and Molecular Identification with Resistant Profile Determination of Listeria Monocytogenes from Imported Chicken Carcasses in Duhok, Kurdistan Region, Iraq. J. Pure Appl. Microbiol. 2015, 9, 97–103. [Google Scholar]
- Alanber, M.N.; Alharbi, N.S.; Khaled, J.M. Evaluation of Multidrug-Resistant Bacillus Strains Causing Public Health Risks in Powdered Infant Milk Formulas. J. Infect. Public Health 2020, 13, 1462–1468. [Google Scholar] [CrossRef]
- Hayder, T.; Aljanaby, A.A.J.J. Antibiotics Susceptibility Patterns of Citrobacter Freundii Isolated from Pa-Tients with Urinary Tract Infection in Al-Najaf Governorate—Iraq. Int. J. Res. Pharm. Sci. 2019, 10, 1481–1488. [Google Scholar] [CrossRef]
- Talat, A.; Khalid, S.; Majeed, H.A.R.; Khan, A.U. Whole-Genome Sequence Analysis of Multidrug-Resistant Staphylococcus EpidermiDis. ST35 Strain Isolated from Human Ear Infection of an Iraqi Patient. J. Glob. Antimicrob. Resist. 2020, 21, 318–320. [Google Scholar] [CrossRef]
- Al-Muhanna, A.S.; Al-Muhanna, S.; Alzuhairi, M.A. Molecular Investigation of Extended-Spectrum Beta-Lactamase Genes and Potential Drug Resistance in Clinical Isolates of Morganella Morganii. Ann. Saudi Med. 2016, 36, 223–228. [Google Scholar] [CrossRef] [PubMed]
- Hanan, Z.K. Molecular Detection of Cholera Infection during the Outbreak in Thi-Qar Province/Iraq in 2015–2016. J. Phys. Conf. Ser. 2019, 1279, 012068. [Google Scholar] [CrossRef]
- AlJindan, R.; AlEraky, D.M.; Borgio, J.F.; AbdulAzeez, S.; Abdalhamid, B.; Mahmoud, N.; Farhat, M. Diagnostic Deficiencies of C. Difficile Infection among Patients in a Tertiary Hospital in Saudi Arabia: A Laboratory-Based Case Series. Saudi J. Biol. Sci. 2021, 28, 4472–4477. [Google Scholar] [CrossRef]
- Al-Sa’ady, A.T. Detection of Vancomycin Resistance in Multidrug-Resistant Enterococcus Faecalis Isolated from Burn Infections. Drug Invent. Today 2019, 11, 7. [Google Scholar]
- Ulger Toprak, N.; Veloo, A.C.M.; Urban, E.; Wybo, I.; Justesen, U.S.; Jean-Pierre, H.; Morris, T.; Akgul, O.; Kulekci, G.; Soyletir, G.; et al. A Multicenter Survey of Antimicrobial Susceptibility of Prevotella Species as Determined by Etest Methodology. Anaerobe 2018, 52, 9–15. [Google Scholar] [CrossRef]
- Taj-Aldeen, S.J.; Deshmukh, A.; Doiphode, S.; Abdul Wahab, A.; Allangawi, M.; AlMuzrkchi, A.; Klaassen, C.H.; Meis, J.F. Molecular Identification and Susceptibility Pattern of Clinical Nocardia Species: Emergence of Nocardia Crassostreae as an Agent of Invasive Nocardiosis. Can. J. Infect. Dis. Med. Microbiol. 2013, 24, e33–e38. [Google Scholar] [CrossRef]
- El-Mahdy, T.S.; Al-Agamy, M.H.; Al-Qahtani, A.A.; Shibl, A.M. Detection of Bla OXA-23-like and Bla NDM-1 in Acinetobacter Baumannii from the Eastern Region, Saudi Arabia. Microb. Drug Resist. 2017, 23, 115–121. [Google Scholar] [CrossRef]
- Khan, Z.; Ahmad, S.; Joseph, L.; Al-Obaid, K. Isolation of Cholesterol-Dependent, Multidrug-Resistant Candida Glabratastrains from Blood Cultures of a Candidemia Patient in Kuwait. BMC Infect. Dis. 2014, 14, 188. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Alosaimi, R.S.; Kaaki, M.M. Catheter-Related ESBL-Producing Leclercia Adecarboxylata Septicemia in Hemodialysis Patient: An Emerging Pathogen? Case Rep. Infect. Dis. 2020, 2020, 7403152. [Google Scholar] [CrossRef] [PubMed]
- Guan, Q.; Almutairi, T.S.; Alhalouli, T.; Pain, A.; Alasmari, F. Metagenomics of Imported Multidrug-Resistant Mycobacterium Leprae, Saudi Arabia, 2017. Emerg. Infect. Dis. 2020, 26, 615–617. [Google Scholar] [CrossRef] [PubMed]
- Abdalhamid, B.; Elhadi, N.; Alsamman, K.; Aljindan, R. Chryseobacterium Gleum Pneumonia in an Infant with Nephrotic Syndrome. IDCases 2016, 5, 34–36. [Google Scholar] [CrossRef] [PubMed]
- Aljindan, R.; Elhadi, N. Genetic Relationship of Multi-Resistant Acinetobacter Baumannii Isolates in Kingdom of Saudi Arabia. J. Pure Appl. Microbiol. 2018, 12, 1951–1958. [Google Scholar] [CrossRef]
- Yasir, M.; Farman, M.; Shah, M.W.; Jiman-Fatani, A.A.; Othman, N.A.; Almasaudi, S.B.; Alawi, M.; Shakil, S.; Al-Abdullah, N.; Ismaeel, N.A.; et al. Genomic and Antimicrobial Resistance Genes Diversity in Multidrug-Resistant CTX-M-Positive Isolates of Escherichia coli at a Health Care Facility in Jeddah. J. Infect. Public Health 2020, 13, 94–100. [Google Scholar] [CrossRef]
- Johani, K.; Abualsaud, D.; Costa, D.M.; Hu, H.; Whiteley, G.; Deva, A.; Vickery, K. Characterization of Microbial Community Composition, Antimicrobial Resistance and Biofilm on Intensive Care Surfaces. J. Infect. Public Health 2018, 11, 418–424. [Google Scholar] [CrossRef]
- Hamza, W.; Salama, M.; Morsi, S.; Abdo, N.; Al-Fadhli, M. Benchmarking for Surgical Site Infections among Gastrointestinal Surgeries and Related Risk Factors: Multicenter Study in Kuwait. Infect. Drug Resist. 2018, 11, 1373–1381. [Google Scholar] [CrossRef]
- Balkhair, A.; Al-Farsi, Y.M.; Al-Muharrmi, Z.; Al-Rashdi, R.; Al-Jabri, M.; Neilson, F.; Al-Adawi, S.S.; El-Beeli, M.; Al-Adawi, S. Epidemiology of Multi-Drug Resistant Organisms in a Teaching Hospital in Oman: A One-Year Hospital-Based Study. Sci. World J. 2014, 2014, 157102. [Google Scholar] [CrossRef]
- Al Awaidy, S.T. Tuberculosis Elimination in Oman: Winning the War on the Disease. ERJ Open Res. 2018, 4, 00121–02018. [Google Scholar] [CrossRef]
- Metry, A.M.; Al Salmi, I.; Al-Abri, S.; Al Ismaili, F.; Al Mahrouqi, Y.; Hola, A.; Shaheen, F.A.M. Epidemiology and Outcome of Tuberculosis in Immunocompromised Patients. Saudi J. Kidney Dis. Transpl. 2017, 28, 806–817. [Google Scholar] [PubMed]
- Hasan, M.R.; Sundaram, M.S.; Sundararaju, S.; Tsui, K.-M.; Karim, M.Y.; Roscoe, D.; Imam, O.; Janahi, M.A.; Thomas, E.; Dobson, S.; et al. Unusual Accumulation of a Wide Array of Antimicrobial Resistance Mechanisms in a Patient with Cytomegalovirus-Associated Hemophagocytic Lymphohistiocytosis: A Case Report. BMC Infect. Dis. 2020, 20, 237. [Google Scholar] [CrossRef] [PubMed]
- Alkhawaja, E.; Hammadi, S.; Abdelmalek, M.; Mahasneh, N.; Alkhawaja, B.; Abdelmalek, S.M. Antibiotic Resistant Cutibacterium Acnes among Acne Patients in Jordan: A Cross Sectional Study. BMC Dermatol. 2020, 20, 17. [Google Scholar] [CrossRef]
- Al-Shamahy, H.A.; Sabrah, A.A.; Al-Robasi, A.B.; Naser, S.M. Types of Bacteria Associated with Neonatal Sepsis in Al-Thawra University Hospital, Sana’a, Yemen, and Their Antimicrobial Profile. Sultan Qaboos Univ. Med J. 2012, 12, 48–54. [Google Scholar] [CrossRef]
- Kordalewska, M.; Guerrero, K.D.; Garcia-Rubio, R.; Jiménez-Ortigosa, C.; Mediavilla, J.R.; Cunningham, M.H.; Hollis, F.; Hong, T.; Chow, K.F.; Kreiswirth, B.N.; et al. Antifungal Drug Susceptibility and Genetic Characterization of Fungi Recovered from COVID-19 Patients. J. Fungi 2021, 7, 552. [Google Scholar] [CrossRef]
- Al-Mutairi, N.M.; Ahmad, S.; Mokaddas, E. Performance Comparison of Four Methods for Detecting Multidrug-Resistant Mycobacterium Tuberculosis Strains. Int. J. Tuberc. Lung Dis. 2011, 15, 110–115. [Google Scholar]
- Ali, R.M.; Alsudani, A.A. Discordance between GeneXpert Assay and Conventional Drug-Susceptibility Testing in Detecting Rifampicin-Resistant Tuberculosis: A Perspective of the Line Probe Assay. Int. J. Mycobacteriol. 2016, 5, S193–S194. [Google Scholar] [CrossRef] [PubMed]
- Arastehfar, A.; Fang, W.; Daneshnia, F.; Al-Hatmi, A.M.; Liao, W.; Pan, W.; Khan, Z.; Ahmad, S.; Rosam, K.; Lackner, M.; et al. Novel Multiplex Real-Time Quantitative PCR Detecting System Approach for Direct Detection of Candida Auris and Its Relatives in Spiked Serum Samples. Future Microbiol. 2019, 14, 33–45. [Google Scholar] [CrossRef] [PubMed]
- Alabdullatif, M.; Alrehaili, J. Three Years of Evaluation to Determine Reduction of Antibiotic Resistance in Gram-Negative Bacteria by the Saudi National Action Plan. Infect. Drug Resist. 2020, 13, 3657–3667. [Google Scholar] [CrossRef]
- Amoudy, H.A.; Safar, H.A.; Mustafa, A.S. Development of Escherichia coli and Mycobacterium Smegmatis Recombinants Expressing Major Mycobacterium Tuberculosis-Specific Antigenic Proteins. Int. J. Mycobacteriol. 2016, 5, S84–S85. [Google Scholar] [CrossRef] [PubMed]
- Lesho, E.; Lin, X.; Clifford, R.; Snesrud, E.; Onmus-Leone, F.; Appalla, L.; Ong, A.; Maybank, R.; Nielsen, L.; Kwak, Y.; et al. From the Battlefield to the Bedside: Supporting Warfighter and Civilian Health With the “ART” of Whole Genome Sequencing for Antibiotic Resistance and Outbreak Investigations. Mil. Med. 2016, 181, 621–624. [Google Scholar] [CrossRef] [PubMed]
- Al Tall, Y.; Abualhaijaa, A.; Alsaggar, M.; Almaaytah, A.; Masadeh, M.; Alzoubi, K.H. Design and Characterization of a New Hybrid Peptide from LL-37 and BMAP-27. Infect. Drug Resist. 2019, 12, 1035–1045. [Google Scholar] [CrossRef] [PubMed]
- Almaaytah, A.; Abualhaijaa, A.; Alqudah, O. The Evaluation of the Synergistic Antimicrobial and Antibiofilm Activity of AamAP1-Lysine with Conventional Antibiotics against Representative Resistant Strains of Both Gram-Positive and Gram-Negative Bacteria. Infect. Drug Resist. 2019, 12, 1371–1380. [Google Scholar] [CrossRef] [PubMed]
- Obeidat, M.; Shatnawi, M.; Al-alawi, M.; Al-Zu‘bi, E.; Al-Dmoor, H.; Al-Qudah, M.; El-Qudah, J.; Otri, I. Antimicrobial Activity of Crude Extracts of Some Plant Leaves. Res. J. Microbiol. 2012, 7, 59–67. [Google Scholar] [CrossRef]
- Ahmed, A.A.; Salih, F.A. Low Concentrations of Local Honey Modulate ETA Expression, and Quorum Sensing Related Virulence in Drug-Resistant Pseudomonas Aeruginosa Recovered from Infected Burn Wounds. Iran. J. Basic Med. Sci. 2019, 22, 568. [Google Scholar] [CrossRef]
- Al-Somat, M.; Al-Adhal, A.; Al-Arwali, A.; Al, K.; Al-Moyed, K.; Shopit, A.; Karbane, M.E.; AL-Kamarany, M.A. Sensitivity of Clinical Bacterial Isolates to Honey Marketed in Yemen. Int. J. Pharm. Sci. Rev. Res. 2014, 27, 267–271. [Google Scholar]
- Darwish, R.M.; Fares, R.J.A.; Zarga, M.H.A.; Nazer, I.K. Antibacterial Effect of Jordanian Propolis and Isolated Flavonoids against Human Pathogenic Bacteria. Afr. J. Biotechnol. 2010, 9, 9. [Google Scholar]
- Yang, S.-K.; Yusoff, K.; Thomas, W.; Akseer, R.; Alhosani, M.S.; Abushelaibi, A.; Lim, S.-H.-E.; Lai, K.-S. Lavender Essential Oil Induces Oxidative Stress Which Modifies the Bacterial Membrane Permeability of Carbapenemase Producing Klebsiella Pneumoniae. Sci. Rep. 2020, 10, 819. [Google Scholar] [CrossRef]
- Soliman, S.S.M.; Semreen, M.H.; El-Keblawy, A.A.; Abdullah, A.; Uppuluri, P.; Ibrahim, A.S. Assessment of Herbal Drugs for Promising Anti-Candida Activity. BMC Complement Altern Med. 2017, 17, 257. [Google Scholar] [CrossRef]
- Perveen, K.; Husain, F.M.; Qais, F.A.; Khan, A.; Razak, S.; Afsar, T.; Alam, P.; Almajwal, A.M.; Abulmeaty, M.M.A. Microwave-Assisted Rapid Green Synthesis of Gold Nanoparticles Using Seed Extract of Trachyspermum Ammi: ROS Mediated Biofilm Inhibition and Anticancer Activity. Biomolecules 2021, 11, 197. [Google Scholar] [CrossRef] [PubMed]
- Almaaytah, A.; Qaoud, M.; Khalil Mohammed, G.; Abualhaijaa, A.; Knappe, D.; Hoffmann, R.; Al-Balas, Q. Antimicrobial and Antibiofilm Activity of UP-5, an Ultrashort Antimicrobial Peptide Designed Using Only Arginine and Biphenylalanine. Pharmaceuticals 2018, 11, 3. [Google Scholar] [CrossRef] [PubMed]
- Soliman, S.S.M.; Saeed, B.Q.; Elseginy, S.A.; Al-Marzooq, F.; Ahmady, I.M.; El-Keblawy, A.A.; Hamdy, R. Critical Discovery and Synthesis of Novel Antibacterial and Resistance-Modifying Agents Inspired by Plant Phytochemical Defense Mechanisms. Chem.-Biol. Interact. 2021, 333, 109318. [Google Scholar] [CrossRef] [PubMed]
- Muthukrishnan, P.; Chithra Devi, D.; Mostafa, A.A.; Alsamhary, K.I.; Abdel-Raouf, N.; Nageh Sholkamy, E. Antimicrobial Efficacy of Nocardiopsis Sp. MK_MSt033 against Selected Multidrug Resistant Clinical Microbial Pathogens. J. Infect. Public Health 2020, 13, 1522–1532. [Google Scholar] [CrossRef]
- Almaaytah, A.; Ajingi, Y.; Abualhaijaa, A.; Tarazi, S.; Alshar’i, N.; Al-Balas, Q. Peptide Consensus Sequence Determination for the Enhancement of the Antimicrobial Activity and Selectivity of Antimicrobial Peptides. Infect. Drug Resist. 2017, 10, 1. [Google Scholar] [CrossRef]
- Almaaytah, A.; Mohammed, G.; Abualhaijaa, A.; Al-Balas, Q. Development of Novel Ultrashort Antimicrobial Peptide Nanoparticles with Potent Antimicrobial and Antibiofilm Activities against Multidrug-Resistant Bacteria. Drug Des. Dev. Ther. 2017, 11, 3159–3170. [Google Scholar] [CrossRef] [PubMed]
- Mechkarska, M.; Ahmed, E.; Coquet, L.; Leprince, J.; Jouenne, T.; Vaudry, H.; King, J.D.; Conlon, J.M. Antimicrobial Peptides with Therapeutic Potential from Skin Secretions of the Marsabit Clawed Frog Xenopus Borealis (Pipidae). Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2010, 152, 467–472. [Google Scholar] [CrossRef] [PubMed]
- Omran, Z.; Bader, A.; Porta, A.; Vandamme, T.; Anton, N.; Alehaideb, Z.; El-Said, H.; Faidah, H.; Essa, A.; Vassallo, A.; et al. Evaluation of Antimicrobial Activity of Triphala Constituents and Nanoformulation. Evid.-Based Complementary Altern. Med. 2020, 2020, 6976973. [Google Scholar] [CrossRef]
- Mahdi, L.; Musafer, H.; Zwain, L.; Salman, I.; Al-Joofy, I.; Rasool, K.; Mussa, A.; Al-kakei, S.; Al-Oqaili, R.; Al-Alak, S.; et al. Two Novel Roles of Buffalo Milk Lactoperoxidase, Antibiofilm Agent and Immunomodulator against Multidrug Resistant Salmonella Enterica Serovar Typhi and Listeria Monocytogenes. Microb. Pathog. 2017, 109, 221–227. [Google Scholar] [CrossRef]
- Rahmeh, R.; Akbar, A.; Kishk, M.; Al-Onaizi, T.; Al-Azmi, A.; Al-Shatti, A.; Shajan, A.; Al-Mutairi, S.; Akbar, B. Distribution and Antimicrobial Activity of Lactic Acid Bacteria from Raw Camel Milk. New Microbes New Infect. 2019, 30, 100560. [Google Scholar] [CrossRef]
- Abdel-Aziz, H.; Eldehna, W.; Keeton, A.; Piazza, G.; Kadi, A.; Attwa, M.; Abdelhameed, A.; Attia, M. Isatin-Benzoazine Molecular Hybrids as Potential Antiproliferative Agents: Synthesis and in Vitro Pharmacological Profiling. Drug Des. Dev. Ther. 2017, 11, 2333–2346. [Google Scholar] [CrossRef]
- Aboul-Fadl, T.; Bin-Jubair, F.A.S.; Aboul-Wafa, O. Schiff Bases of Indoline-2,3-Dione (Isatin) Derivatives and Nalidixic Acid Carbohydrazide, Synthesis, Antitubercular Activity and Pharmacophoric Model Building. Eur. J. Med. Chem. 2010, 45, 4578–4586. [Google Scholar] [CrossRef]
- Ahamed, A.; Arif, I.A.; Mateen, M.; Surendra Kumar, R.; Idhayadhulla, A. Antimicrobial, Anticoagulant, and Cytotoxic Evaluation of Multidrug Resistance of New 1,4-Dihydropyridine Derivatives. Saudi J. Biol. Sci. 2018, 25, 1227–1235. [Google Scholar] [CrossRef] [PubMed]
- Bogue, A.L.; Panmanee, W.; McDaniel, C.T.; Mortensen, J.E.; Kamau, E.; Actis, L.A.; Johannigman, J.A.; Schurr, M.J.; Satish, L.; Kotagiri, N.; et al. AB569, a Non-Toxic Combination of Acidified Nitrite and EDTA, is Effective at Killing the Notorious Iraq/Afghanistan Combat Wound Pathogens, Multi-Drug Resistant Acinetobacter Baumannii and Acinetobacter spp. PLoS ONE 2021, 16, e0247513. [Google Scholar] [CrossRef] [PubMed]
- Venugopala, K.N.; Uppar, V.; Chandrashekharappa, S.; Abdallah, H.H.; Pillay, M.; Deb, P.K.; Morsy, M.A.; Aldhubiab, B.E.; Attimarad, M.; Nair, A.B.; et al. Cytotoxicity and Antimycobacterial Properties of Pyrrolo[1,2-a]Quinoline Derivatives: Molecular Target Identification and Molecular Docking Studies. Antibiotics 2020, 9, 233. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, I.; Wahab, S.; Nisar, N.; Dera, A.A.; Alshahrani, M.Y.; Abullias, S.S.; Irfan, S.; Alam, M.M.; Srivastava, S. Evaluation of Antibacterial Properties of Matricaria Aurea on Clinical Isolates of Periodontitis Patients with Special Reference to Red Complex Bacteria. Saudi Pharm. J. 2020, 28, 1203–1209. [Google Scholar] [CrossRef]
- Al-Asmari, A.K.; Alamri, M.A.; Almasoudi, A.S.; Abbasmanthiri, R.; Mahfoud, M. Evaluation of the in Vitro Antimicrobial Activity of Selected Saudi Scorpion Venoms Tested against Multidrug-Resistant Micro-Organisms. J. Glob. Antimicrob. Resist. 2017, 10, 14–18. [Google Scholar] [CrossRef]
- Alkhedaide, A.Q.; Ismail, T.A.; Alotaibi, S.H.; Nassan, M.A.; Shehri, Z.S.A. Preventive Effect of Artemisinin Extract against Cholestasis Induced via Lithocholic Acid Exposure. Biosci. Rep. 2018, 38, BSR20181011. [Google Scholar] [CrossRef]
- El-Newehy, M.H.; Moydeen A., M.; Aldalbahi, A.K.; Thamer, B.M.; Mahmoud, Y.A.-G.; El-Hamshary, H. Biocidal Polymers: Synthesis, Characterization and Antimicrobial Activity of Bis-Quaternary Onium Salts of Poly(Aspartate-Co-Succinimide). Polymers 2020, 13, 23. [Google Scholar] [CrossRef]
- Manzoor, S.; Bilal, A.; Khan, S.; Ullah, R.; Iftikhar, S.; Emwas, A.-H.; Alazmi, M.; Gao, X.; Jawaid, A.; Saleem, R.S.Z.; et al. Identification and Characterization of SSE15206, a Microtubule Depolymerizing Agent That Overcomes Multidrug Resistance. Sci. Rep. 2018, 8, 3305. [Google Scholar] [CrossRef]
- Al-Dhabi, N.A.; Ghilan, A.-K.M.; Esmail, G.A.; Valan Arasu, M.; Duraipandiyan, V.; Ponmurugan, K. Bioactivity Assessment of the Saudi Arabian Marine Streptomyces Sp. Al-Dhabi-90, Metabolic Profiling and Its in Vitro Inhibitory Property against Multidrug Resistant and Extended-Spectrum Beta-Lactamase Clinical Bacterial Pathogens. J. Infect. Public Health 2019, 12, 549–556. [Google Scholar] [CrossRef] [PubMed]
- Alekish, M.; Ismail, Z.B.; Albiss, B.; Nawasrah, S. In Vitro Antibacterial Effects of Zinc Oxide Nanoparticles on Multiple Drug-Resistant Strains of Staphylococcus Aureus and Escherichia coli: An Alternative Approach for Antibacterial Therapy of Mastitis in Sheep. Vet. World 2018, 11, 1428–1432. [Google Scholar] [CrossRef] [PubMed]
- Alsamhary, K.I. Eco-Friendly Synthesis of Silver Nanoparticles by Bacillus Subtilis and Their Antibacterial Activity. Saudi J. Biol. Sci. 2020, 27, 2185–2191. [Google Scholar] [CrossRef]
- Al-Dhabi, N.; Ghilan, A.-K.M.; Arasu, M. Characterization of Silver Nanomaterials Derived from Marine Streptomyces Sp. Al-Dhabi-87 and Its In Vitro Application against Multidrug Resistant and Extended-Spectrum Beta-Lactamase Clinical Pathogens. Nanomaterials 2018, 8, 279. [Google Scholar] [CrossRef] [PubMed]
- Abdulmughni, J.; Mahyoub, E.M.; Alaghbari, A.T.; Al Serouri, A.A.; Khader, Y. Performance of Multidrug-Resistant Tuberculosis Surveillance in Yemen: Interview Study. JMIR Public Health Surveill 2019, 5, e14294. [Google Scholar] [CrossRef]
- Ganaie, F.; Nagaraj, G.; Govindan, V.; Basha, R.; Hussain, M.; Ashraf, N.; Ahmed, S.; Ravi Kumar, K.L. Impact of Hajj on the S. Pneumoniae Carriage among Indian Pilgrims during 2016- a Longitudinal Molecular Surveillance Study. Travel Med. Infect. Dis. 2018, 23, 64–71. [Google Scholar] [CrossRef]
- Leangapichart, T.; Gautret, P.; Griffiths, K.; Belhouchat, K.; Memish, Z.; Raoult, D.; Rolain, J.-M. Acquisition of a High Diversity of Bacteria during the Hajj Pilgrimage, Including Acinetobacter Baumannii with Bla OXA-72 and Escherichia coli with Bla NDM-5 Carbapenemase Genes. Antimicrob. Agents Chemother. 2016, 60, 5942–5948. [Google Scholar] [CrossRef]
- Zumla, A.; Saeed, A.B.; Alotaibi, B.; Yezli, S.; Dar, O.; Bieh, K.; Bates, M.; Tayeb, T.; Mwaba, P.; Shafi, S.; et al. Tuberculosis and Mass Gatherings—Opportunities for Defining Burden, Transmission Risk, and the Optimal Surveillance, Prevention, and Control Measures at the Annual Hajj Pilgrimage. Int. J. Infect. Dis. 2016, 47, 86–91. [Google Scholar] [CrossRef]
- Aurilio, C.; Sansone, P.; Paladini, A.; Barbarisi, M.; Coppolino, F.; Pota, V.; Pace, M.C. Multidrug Resistence Prevalence in COVID Area. Life 2021, 11, 601. [Google Scholar] [CrossRef]
- Vaillancourt, M.; Jorth, P. The Unrecognized Threat of Secondary Bacterial Infections with COVID-19. mBio 2020, 11, e01806-20. [Google Scholar] [CrossRef]
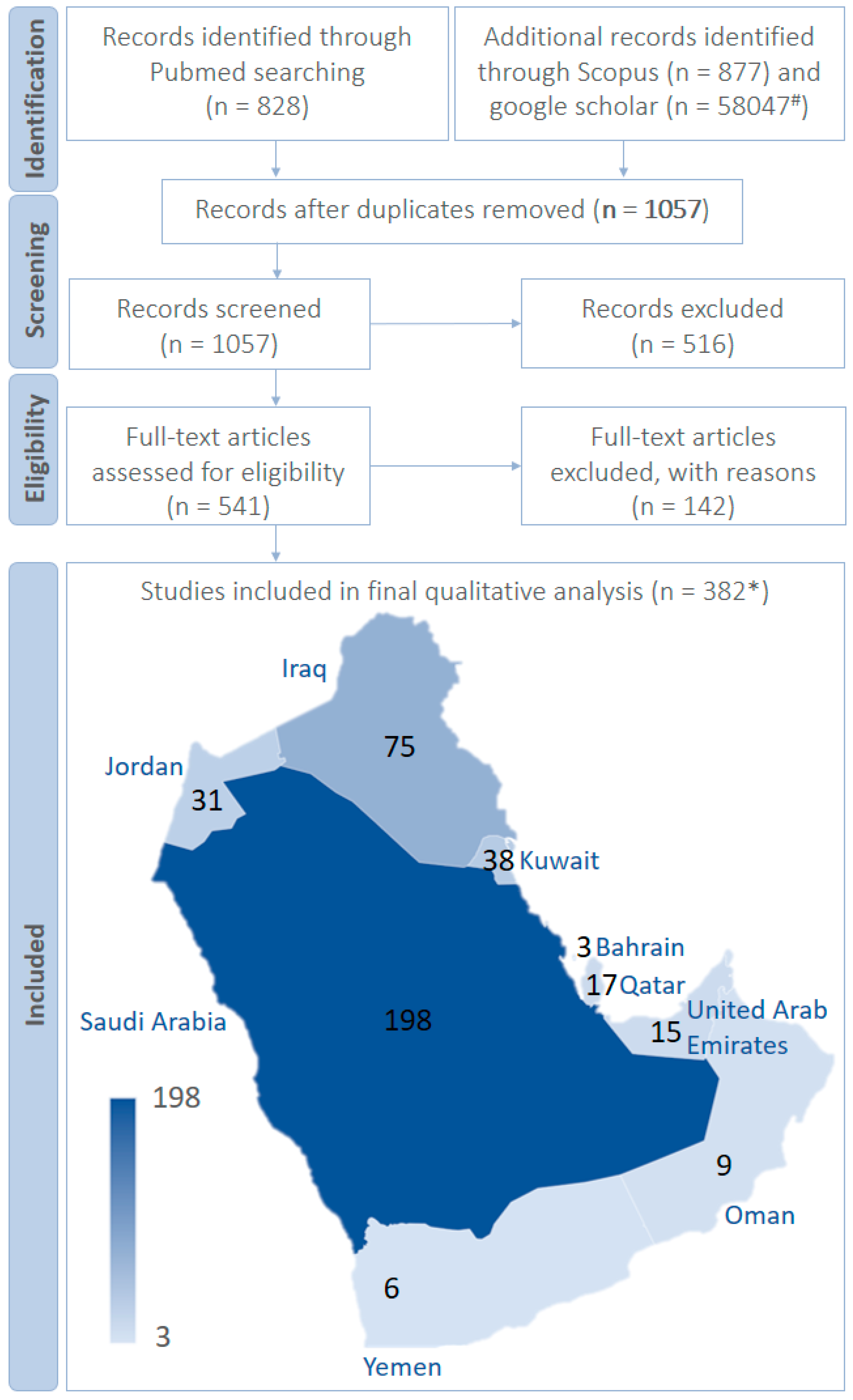
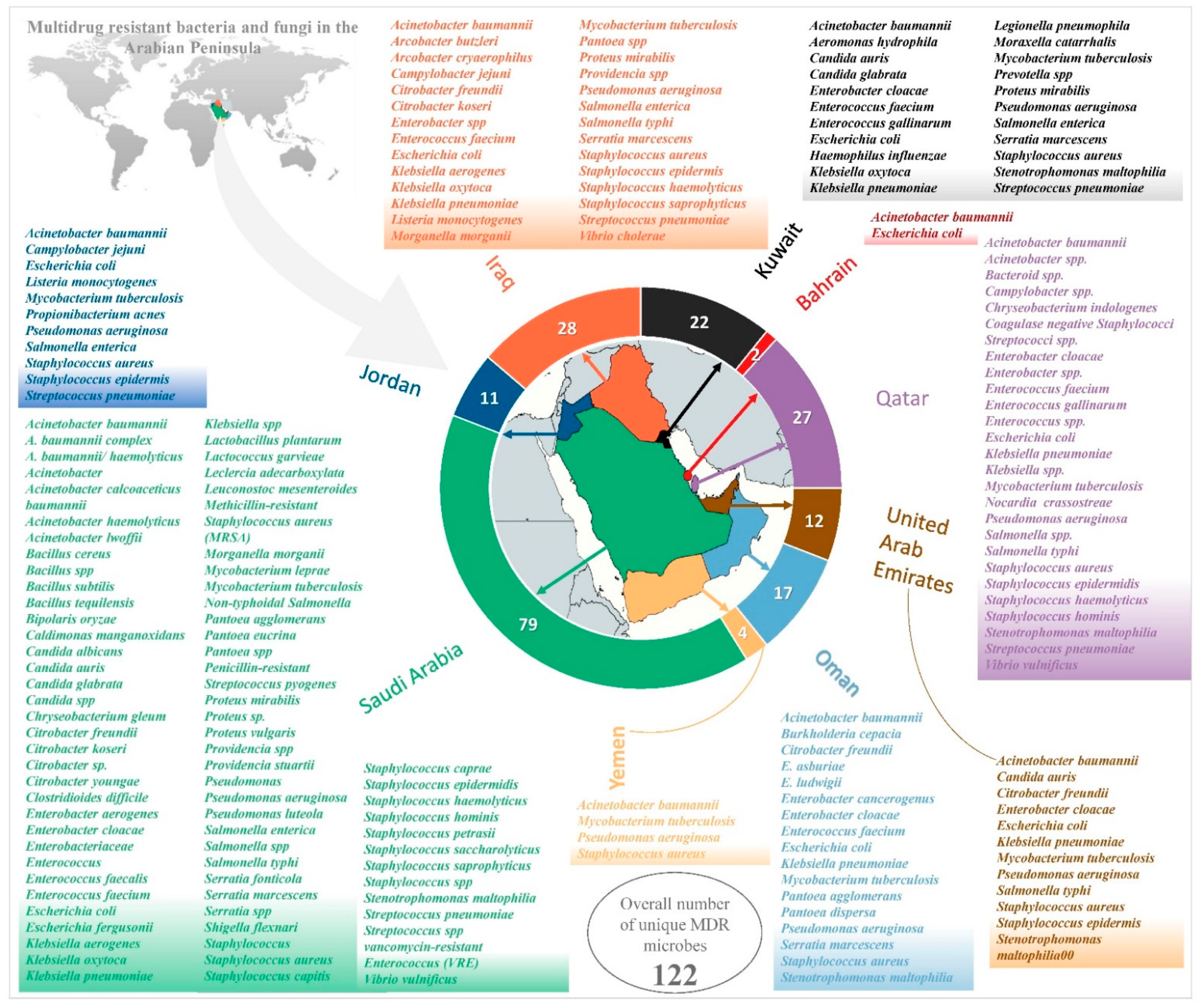
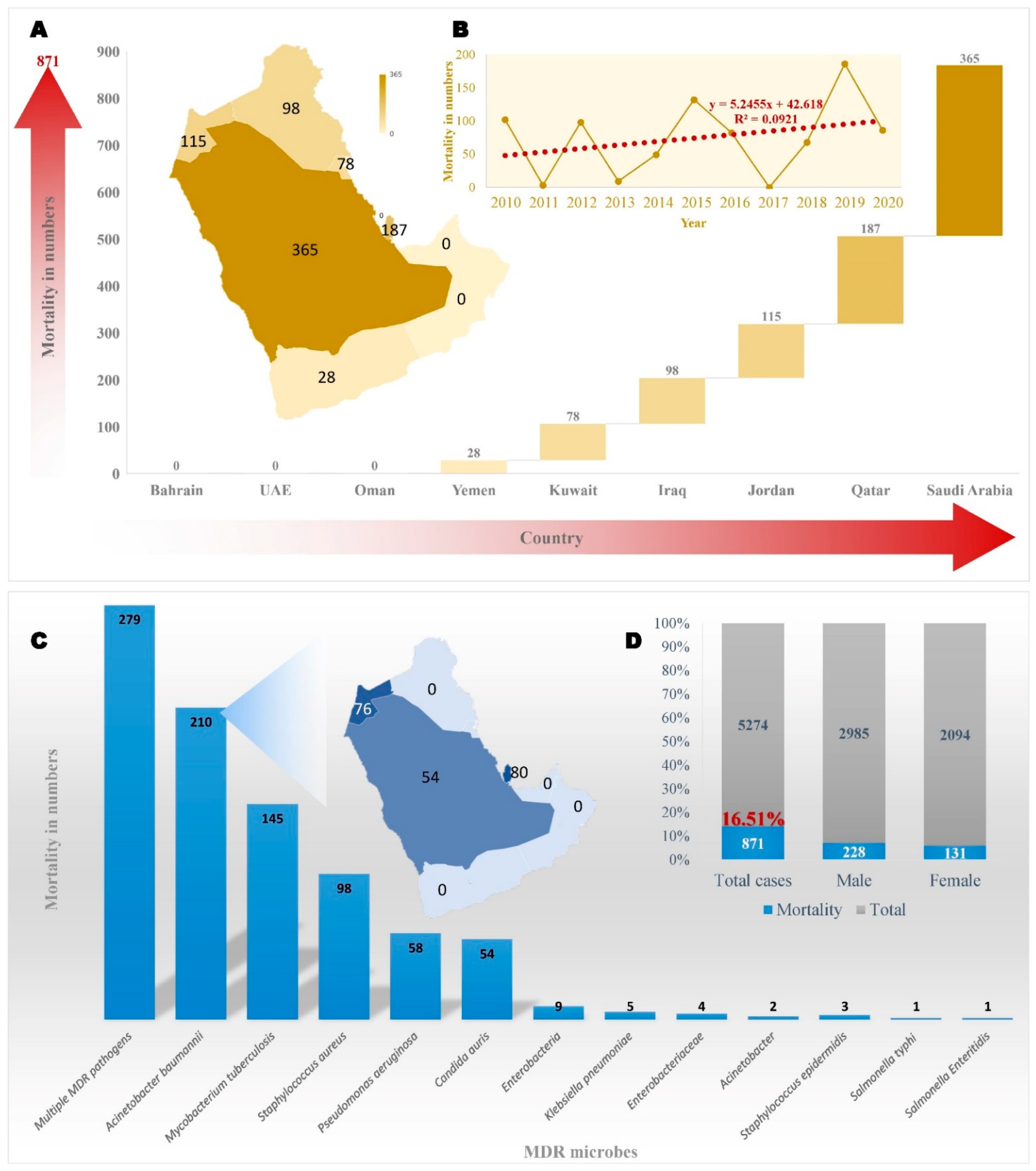
| Countries | Number of MDRO | Multidrug-Resistant Micro-Organism(s) |
|---|---|---|
| Bahrain, Iraq, Jordan, Kuwait, Oman, Qatar, Saudi Arabia, United Arab Emirates, Yemen | 1 | Acinetobacter baumannii |
| Bahrain, Iraq, Jordan, Kuwait, Oman, Qatar, Saudi Arabia, United Arab Emirates | 1 | Escherichia coli |
| Iraq, Jordan, Kuwait, Oman, Qatar, Saudi Arabia, United Arab Emirates, Yemen, | 3 | Mycobacterium tuberculosis, Staphylococcus aureus, Pseudomonas aeruginosa |
| Iraq, Kuwait, Oman, Qatar, Saudi Arabia, United Arab Emirates | 1 | Klebsiella pneumoniae |
| Kuwait, Oman, Qatar, Saudi Arabia, United Arab Emirates | 2 | Enterobacter cloacae, Stenotrophomonas maltophilia |
| Iraq, Kuwait, Oman, Qatar, Saudi Arabia | 1 | Enterococcus faecium |
| Iraq, Jordan, Kuwait, Qatar, Saudi Arabia | 1 | Streptococcus pneumoniae |
| Iraq, Jordan, Kuwait, Saudi Arabia | 1 | Salmonella enterica |
| Iraq, Qatar, Saudi Arabia, United Arab Emirates | 1 | Salmonella typhi |
| Kuwait, Saudi Arabia, United Arab Emirates | 1 | Candida auris |
| Iraq, Kuwait, Saudi Arabia, Oman | 3 | Serratia marcescens, Proteus mirabilis, Klebsiella oxytoca |
| Iraq, Saudi Arabia, United Arab Emirates, Oman | 1 | Citrobacter freundii |
| Iraq, Jordan, United Arab Emirates, Qatar | 1 | Staphylococcus epidermis |
| Kuwait, Saudi Arabia | 1 | Candida glabrata |
| Oman, Saudi Arabia | 1 | Pantoea agglomerans |
| Qatar, Saudi Arabia | 1 | Vibrio vulnificus |
| Iraq, Saudi Arabia | 6 | Klebsiella aerogenes, Morganella morganii, Citrobacter koseri, Pantoea spp., Providencia spp., Staphylococcus saprophyticus |
| Iraq, Saudi Arabia, Qatar | Staphylococcus haemolyticus | |
| Kuwait, | 1 | Enterococcus gallinarum |
| Iraq, Jordan | 2 | Listeria monocytogenes, Campylobacter jejuni |
| Saudi Arabia, Qatar | 4 | Staphylococcus hominis, Klebsiella spp., Salmonella spp., Streptococcus spp. |
| Iraq, Qatar | 1 | Enterobacter spp. |
| Saudi Arabia | 53 | Staphylococcus capitis, Staphylococcus caprae, Acinetobacter calcoaceticus baumannii, Pseudomonas, Bacillus subtilis, Leuconostoc mesenteroides, Candida albicans, Staphylococcus epidermidis, Proteus vulgaris, Proteus sp., Bacillus cereus, Lactobacillus plantarum, Bipolaris oryzae, Methicillin-resistant Staphylococcus aureus, Bacillus tequilensis, Staphylococcus, Enterococcus, Staphylococcus saccharolyticus, Leclercia adecarboxylata, Shigella flexnari, Staphylococcus spp., Pantoea eucrina, Enterobacteriaceae, Citrobacter youngae, Staphylococcus petrasii, Lactococcus garvieae, Escherichia fergusonii, Caldimonas manganoxidans, Chryseobacterium gleum, Citrobacter sp., Providencia stuartii, Vancomycin-resistant Enterococcus, Mycobacterium leprae, Clostridium spp., Stenotrophomonus maltophilia, Enterococcus faecalis, Acinetobacter, Non-typhoidal Salmonella, Enterobacter aerogenes, Acinetobacter lwoffii, A. baumannii complex, A. baumannii/haemolyticus, Acinetobacter haemolyticus, Pseudomonas luteola, Serratia fonticola, Penicillin-resistant Streptococcus pyogenes, Bacillus spp., Serratia spp., Candida spp. |
| Kuwait | 5 | Legionella pneumophila, Aeromonas hydrophila, Moraxella catarrhalis, Prevotella spp., Haemophilus influenzae |
| Oman | 5 | Pantoea dispersa, Burkholderia cepacia, Enterobacter asburiae, Enterobacter hormaechei Enterobacter ludwigii |
| Qatar | 7 | Campylobacter spp., Nocardia crassostreae, Chryseobacterium indologenes, Acinetobacter spp., Bacteroid spp., Coagulase negative Staphylococci, Enterococcus spp. |
| Jordan | 1 | Propionibacterium acnes |
| Iraq | 1 | Vibrio cholerae |
| S. No | MDR Microbe | Saudi Arabia | Bahrain | Kuwait | Oman | Qatar | United Arab Emirates | Jordan | Iraq | Yemen | Total Number of Reports * |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Acinetobacter baumannii | 59 | 1 | 8 | 2 | 2 | 5 | 6 | 12 | 1 | 96 # |
| 2 | Escherichia coli | 51 | 2 | 4 | 5 | 6 | 6 | 11 | 10 | 94 | |
| 3 | Mycobacterium tuberculosis | 31 | 11 | 2 | 1 | 2 | 1 | 6 | 4 | 58 | |
| 4 | Klebsiella pneumoniae | 36 | 3 | 4 | 4 | 4 | 7 | 58 | |||
| 5 | Pseudomonas aeruginosa | 36 | 4 | 1 | 4 | 1 | 1 | 9 | 1 | 57 | |
| 6 | Staphylococcus aureus | 16 | 4 | 1 | 1 | 2 | 5 | 5 | 1 | 35 | |
| 7 | Acinetobacter | 14 | 2 | 16 | |||||||
| 8 | Streptococcus pneumoniae | 7 | 1 | 1 | 2 | 1 | 12 | ||||
| 9 | Stenotrophomonas maltophilia | 7 | 1 | 1 | 2 | 11 | |||||
| 10 | Enterobacter cloacae | 6 | 1 | 2 | 1 | 1 | 11 | ||||
| 11 | Candida auris | 4 | 6 | 1 | 11 | ||||||
| 12 | Enterobacteriaceae | 6 | 5 | 11 | |||||||
| 13 | MRSA | 11 | 11 | ||||||||
| 14 | Stenotrophomonus maltophilia | 6 | 1 | 10 | |||||||
| 15 | Salmonella enterica | 2 | 1 | 2 | 4 | 9 | |||||
| 16 | Proteus mirabilis | 8 | 1 | 9 | |||||||
| 17 | Serratia marcescens | 3 | 3 | 1 | 2 | 8 | |||||
| 18 | Enterococcus faecalis | 8 | 8 | ||||||||
| 19 | Citrobacter freundii | 4 | 1 | 1 | 1 | 7 | |||||
| 20 | Enterococcus faecium | 6 | 6 | ||||||||
| 21 | Klebsiella oxytoca | 2 | 2 | 2 | 6 | ||||||
| 22 | Listeria monocytogenes | 2 | 3 | 5 | |||||||
| 23 | Morganella morganii | 4 | 1 | 5 | |||||||
| 24 | Staphylococcus epidermidis | 3 | 1 | 1 | 1 | 5 | |||||
| 25 | Salmonella spp. | 4 | 1 | 1 | 5 | ||||||
| 26 | Klebsiella spp. | 4 | 1 | 1 | 5 | ||||||
| 27 | Candida spp. | 4 | 1 | 5 | |||||||
| 28 | Streptococcus spp. | 2 | 3 | 1 | 5 | ||||||
| 29 | Enterobacter | 4 | 1 | 5 | |||||||
| 30 | Enterococcus | 3 | 2 | 5 | |||||||
| 31 | Staphylococcus spp. | 5 | 5 | ||||||||
| 32 | Salmonella typhi | 2 | 1 | 1 | 1 | 5 | |||||
| 33 | Enterobacter aerogenes | 5 | 5 | ||||||||
| 34 | Staphylococcus hominis | 4 | 1 | 4 | |||||||
| 35 | Candida albicans | 4 | 4 | ||||||||
| 36 | Non-typhoidal Salmonella | 2 | 2 | 4 | |||||||
| 37 | Citrobacter koseri | 2 | 1 | 4 | |||||||
| 38 | Klebsiella aerogenes | 4 | 4 | ||||||||
| 39 | Enterococcus faecalis/faecium | 1 | 1 | 1 | 3 | ||||||
| 40 | Providencia spp. | 1 | 2 | 3 | |||||||
| 41 | Pantoea spp. | 1 | 2 | 3 | |||||||
| 42 | Providencia stuartii | 3 | 3 | ||||||||
| 43 | Acinetobacter lwoffii | 3 | 3 | ||||||||
| 44 | Serratia spp. | 3 | 3 | ||||||||
| 45 | Campylobacter jejuni | 1 | 1 | 2 | |||||||
| 46 | Staphylococcus (MRSA) | 2 | 2 | ||||||||
| 47 | Proteus sp. | 1 | 1 | 2 | |||||||
| 48 | Candida glabrata | 1 | 1 | 2 | |||||||
| 49 | Pantoea agglomerans | 1 | 1 | 2 | |||||||
| 50 | Bacillus subtilis | 2 | 2 | ||||||||
| 51 | Staphylococcus capitis | 2 | 2 | ||||||||
| 52 | Staphylococcus saprophyticus | 2 | 2 |
| Country | Qatar | Qatar | Qatar | Qatar | Kuwait | Kuwait | Kuwait | Kuwait | Kuwait | Kuwait | Kuwait | Iraq | Yemen | Yemen |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MDR/Reference | [4] | [5] | [6] | [7] | [8] | [9] | [10] | [11] | [12] | [13] | [14] | [15] | [16] | [17] |
| Total cases | 48 | 10 | 239 | 452 | 61 | 71 | 14 | 15 | 49 | 11 | 1430 | 654 | 115 | 80 |
| Median age years | 48 | 43 | 49.1 | 45.98 | 58 | 59.5 | 63.9 | 56.6 | 55.6 | 49.8 | 57.5 | 18 | 45 | 45 |
| Gender F | 1 | 57 | 159 | 19 | 24 | 5 | 6 | 15 | 6 | 730 | 381 | 50 | 32 | |
| Gender M | 9 | 182 | 293 | 42 | 47 | 9 | 9 | 34 | 5 | 700 | 273 | 65 | 48 | |
| No. of Discharged patients | 5 | 174 | 350 | 34 | 6 | 46 | 9 | 1415 | 556 | 101 | 66 | |||
| Total number of non-survivors | 15 | 5 | 65 | 102 | 4 | 37 | 9 | 8 | 3 | 2 | 15 | 98 | 14 | 14 |
| Number of non-survivors F | 35 | 2 | 3 | 3 | 1 | |||||||||
| Number of non-survivors M | 5 | 67 | 2 | 6 | 5 | 1 | ||||||||
| Multiple MDR pathogens | 102 | 9 | 3 | 2 | 15 | |||||||||
| Acinetobacter baumannii | 15 | 65 | ||||||||||||
| Pseudomonas aeruginosa | ||||||||||||||
| Staphylococcus aureus | 98 | |||||||||||||
| Mycobacterium tuberculosis | 14 | 14 | ||||||||||||
| Escherichia coli | ||||||||||||||
| Klebsiella pneumoniae | 5 | |||||||||||||
| Staphylococcus epidermidis | ||||||||||||||
| Enterobacteria | ||||||||||||||
| Candida auris | 37 | 8 | ||||||||||||
| Acinetobacter | ||||||||||||||
| Enterobacteriaceae | ||||||||||||||
| Proteus sp. | ||||||||||||||
| Salmonella typhi | 1 | |||||||||||||
| Salmonella enterica | 1 |
| Country | Saudi Arabia | Saudi Arabia | Saudi Arabia | Saudi Arabia | Saudi Arabia | Saudi Arabia | Saudi Arabia | Saudi Arabia | Saudi Arabia | Saudi Arabia | Saudi Arabia | Saudi Arabia | Saudi Arabia | Saudi Arabia | Jordan | Jordan | Jordan | Jordan |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MDR/Reference | [18] | [19] | [20] | [21] | [22] | [23] | [24] | [25] | [26] | [27] | [28] | [29] | [30] | [31] | [32] | [33] | [34] | [34] |
| Total cases | 49 | 83 | 60 | 71 | 2 | 90 | 5 | 7 | 19 | 713 | 86 | 3 | 38 | 146 | 457 | 21 | 119 | 56 |
| Median age years | newborn | 50 | 50.4 | 37.5 | 73.5 | 59.9 | 63.6 | 63 | 57 | 37 | 56 | 43 | 60.5 | 47.3 | 27 | 0.6 | 55.5 | 53.4 |
| Gender F | 11 | 23 | 0 | 32 | 1 | 10 | 294 | 31 | 3 | 14 | 42 | 62 | 64 | 22 | ||||
| Gender M | 18 | 37 | 48 | 2 | 58 | 5 | 6 | 9 | 419 | 55 | 24 | 104 | 395 | 55 | 34 | |||
| No. of Discharged patients | 26 | 30 | 34 | 47 | 2 | 198 | 19 | 69 | 30 | |||||||||
| Total number of non-survivors | 3 | 31 | 54 | 7 | 2 | 54 | 3 | 3 | 4 | 110 | 39 | 1 | 13 | 41 | 37 | 2 | 50 | 26 |
| Number of non-survivors F | 0 | 34 | 14 | 1 | 26 | 12 | ||||||||||||
| Number of non-survivors M | 3 | 76 | 25 | 24 | 14 | |||||||||||||
| Multiple MDR pathogens | 31 | 39 | 41 | 37 | ||||||||||||||
| Acinetobacter baumannii | 54 | 50 | 26 | |||||||||||||||
| Pseudomonas aeruginosa | 54 | 4 | ||||||||||||||||
| Staphylococcus aureus | ||||||||||||||||||
| Mycobacterium tuberculosis | 7 | 110 | ||||||||||||||||
| Escherichia coli | ||||||||||||||||||
| K. pneumoniae | ||||||||||||||||||
| Staphylococcus epidermidis | 3 | |||||||||||||||||
| Enterobacteria | ||||||||||||||||||
| Candida auris | 2 | 3 | 3 | 1 | ||||||||||||||
| Acinetobacter | 2 | |||||||||||||||||
| Enterobacteriaceae | 13 | |||||||||||||||||
| Proteus sp. | ||||||||||||||||||
| Salmonella typhi | ||||||||||||||||||
| Salmonella Enteritidis |
| MDRO | Total Strain | Reference |
|---|---|---|
| Escherichia coli | 13254 | [1,3,7,10,11,14,31,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69,70,71,72,73,74,75,76,77,78,79,80,81,82,83,84,85,86,87,88,89,90,91,92,93,94,95,96,97,98,99,100,101,102,103,104,105,106,107,108,109,110,111] |
| Mycobacterium tuberculosis | 13087 | [16,17,21,27,38,50,51,56,99,112,113,114,115,116,117,118,119,120,121,122,123,124,125,126,127,128,129,130,131,132,133,134,135,136,137,138,139,140,141] |
| Acinetobacter baumannii | 5600 | [2,6,12,28,40,42,46,47,48,51,55,56,58,63,73,88,93,94,103,105,108,111,142,143,144,145,146,147,148,149,150,151,152,153,154,155,156,157,158,159,160,161,162,163,164,165,166,167,168,169,170,171,172,173,174,175,176,177,178,179,180,181,182,183,184,185,186,187,188,189,190,191,192,193,194,195,196,197,198,199] |
| Klebsiella pneumoniae | 5480 | [3,5,10,11,12,14,28,30,38,39,40,41,42,45,46,47,48,51,52,55,56,58,62,63,67,81,82,83,84,88,91,92,93,105,106,111,127,143,149,155,160,181,196,200,201,202,203,204,205,206,207,208,209,210,211,212,213,214,215,216,217,218,219,220] |
| Pseudomonas aeruginosa | 3445 | [11,12,14,23,26,28,31,38,41,42,45,46,47,48,50,51,52,55,56,58,62,63,81,82,83,88,90,91,93,94,109,111,143,149,160,162,180,181,184,190,194,195,196,221,222,223,224,225,226,227,228,229,230,231,232,233,234,235,236,237,238,239,240,241] |
| Staphylococcus aureus | 3133 | [7,11,12,15,42,46,56,58,62,65,73,90,108,143,149,242,243,244,245,246,247,248,249,250,251,252,253] |
| Acinetobacter | 2033 | [6,14,31,33,38,41,65,66,73,187,194,254,255,256] |
| Enterobacteriaceae | 1411 | [3,13,19,20,73,101,176,190,232,257,258,259] |
| Providencia spp. | 1216 | [19,232,260,261] |
| Streptococcus pneumoniae | 1077 | [56,262,263,264,265,266,267,268,269] |
| Methicillin-resistant Staphylococcus aureus (MRSA) | 1000 | [9,11,19,31,46,47,56,93,142,173,176,181,182,190,270,271,272,273,274,275,276,277,278] |
| Klebsiella spp. | 927 | [31,65,66,73,94,108,111,279] |
| Enterobacter | 815 | [11,31,41,48,50,65,66,94,280] |
| Proteus vulgaris | 722 | [40,42,232] |
| Candida auris | 580 | [9,11,22,24,25,29,281,282,283] |
| Enterococcus | 553 | [65,66,81,105,108,284,285] |
| Pseudomonas spp. | 481 | [46,65,66,73,83,108] |
| Klebsiella oxytoca | 434 | [12,40,42,46,50,105,143,205,232] |
| Salmonella enterica | 413 | [100,286,287,288] |
| Serratia marcescens | 289 | [12,40,41,42,46,47,51,88,111,143,149,289] |
| Stenotrophomonus maltophilia | 257 | [31,41,47,65,66,290] |
| Enterococcus faecalis | 256 | [42,47,58,93,143,181,291,292] |
| Vancomycin resistant Enterococcus (VRE) | 255 | [19,73,190,293] |
| Non-typhoidal Salmonella | 248 | [8,203,294] |
| Vibrio vulnificus | 234 | [295,296] |
| Salmonella typhi | 231 | [8,297,298] |
| Proteus sp. | 220 | [12,38,41,48,51,65,108,143] |
| Candida spp. | 180 | [65,66,299] |
| Stenotrophomonas maltophilia | 149 | [40,46,48,111,172,190,256] |
| Proteus mirabilis | 137 | [14,42,46,58,62,81,82,91,93,105,111,143,181,232] |
| Campylobacter jejuni | 125 | [250,300,301,302] |
| Arcobacter butzleri | 100 | [303] |
| Pantoea spp. | 99 | [143,304,305] |
| salmonella spp. | 85 | [46,98,105,232,306,307,308,309] |
| Listeria monocytogenes | 83 | [40,100,309,310,311] |
| Staphylococcus spp. | 78 | [46,74,108,291] |
| Bacillus spp. | 77 | [312] |
| Citrobacter freundii | 66 | [42,81,91,105,143,232,313] |
| Serratia spp. | 66 | [65,66,232] |
| Staphylococcus epidermidis | 59 | [18,46,55,314] |
| Enterobacter cloacae | 54 | [10,42,46,58,81,83,93,105,106,110,111,143] |
| Morganella morganii | 54 | [42,46,58,83,111,143,315] |
| Citrobacter spp. | 54 | [11,38,41,83,232] |
| Enterobacter aerogenes | 40 | [42,46,83,91,111,143] |
| A. baumannii/haemolyticus | 32 | [144] |
| Streptococcus spp. | 23 | [58,108] |
| Vibrio vulnificus | 23 | [296] |
| Staphylococcus saprophyticus | 20 | [42,51,88,143] |
| Arcobacter cryaerophilus | 20 | [303] |
| Vibrio cholerae | 20 | [316] |
| A. baumannii complex | 19 | [144] |
| Clostridioides difficile | 18 | [317] |
| Enterococcus faecium | 17 | [42,46,143,149,318] |
| Salmonella Enteritidies | 15 | [8] |
| Prevotella spp. | 14 | [319] |
| Nocardia crassostreae | 13 | [267,320] |
| Candida albicans | 11 | [190] |
| Staphylococcus hominis | 11 | [46,55] |
| Acinetobacter lwoffii | 11 | [42,143,144] |
| Providencia stuartii | 10 | [46,48,58] |
| Acinetobacter calcoaceticus baumannii | 10 | [321] |
| Citrobacter koseri | 9 | [46,58,111,155] |
| Pseudomonas luteola | 9 | [42] |
| Pantoea agglomerans | 9 | [42] |
| Staphylococcus capitis | 4 | [46] |
| Acinetobacter haemolyticus | 4 | [144] |
| Serratia fonticola | 4 | [42] |
| Candida glabrata | 2 | [322] |
| methicillin-resistant Staphylococcus epidermidis (MRSE) | 1 | [47] |
| penicillin-resistant Streptococcus pyogenes | 1 | [47] |
| Leclercia adecarboxylata | 1 | [323] |
| Mycobacterium leprae | 1 | [324] |
| Chryseobacterium gleum | 1 | [325] |
| Enterococcus gallinarum | 1 | [149] |
| Klebsiella oxytoca | 1 | [10] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Borgio, J.F.; Rasdan, A.S.; Sonbol, B.; Alhamid, G.; Almandil, N.B.; AbdulAzeez, S. Emerging Status of Multidrug-Resistant Bacteria and Fungi in the Arabian Peninsula. Biology 2021, 10, 1144. https://doi.org/10.3390/biology10111144
Borgio JF, Rasdan AS, Sonbol B, Alhamid G, Almandil NB, AbdulAzeez S. Emerging Status of Multidrug-Resistant Bacteria and Fungi in the Arabian Peninsula. Biology. 2021; 10(11):1144. https://doi.org/10.3390/biology10111144
Chicago/Turabian StyleBorgio, J. Francis, Alia Saeed Rasdan, Bayan Sonbol, Galyah Alhamid, Noor B. Almandil, and Sayed AbdulAzeez. 2021. "Emerging Status of Multidrug-Resistant Bacteria and Fungi in the Arabian Peninsula" Biology 10, no. 11: 1144. https://doi.org/10.3390/biology10111144
APA StyleBorgio, J. F., Rasdan, A. S., Sonbol, B., Alhamid, G., Almandil, N. B., & AbdulAzeez, S. (2021). Emerging Status of Multidrug-Resistant Bacteria and Fungi in the Arabian Peninsula. Biology, 10(11), 1144. https://doi.org/10.3390/biology10111144








