Immunostimulant Bathing Influences the Expression of Immune- and Metabolic-Related Genes in Atlantic Salmon Alevins
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals, Fish Stock, and Experimental Setup
2.2. RNA Extraction and cDNA Synthesis
2.3. qPCR
2.4. Data Analysis
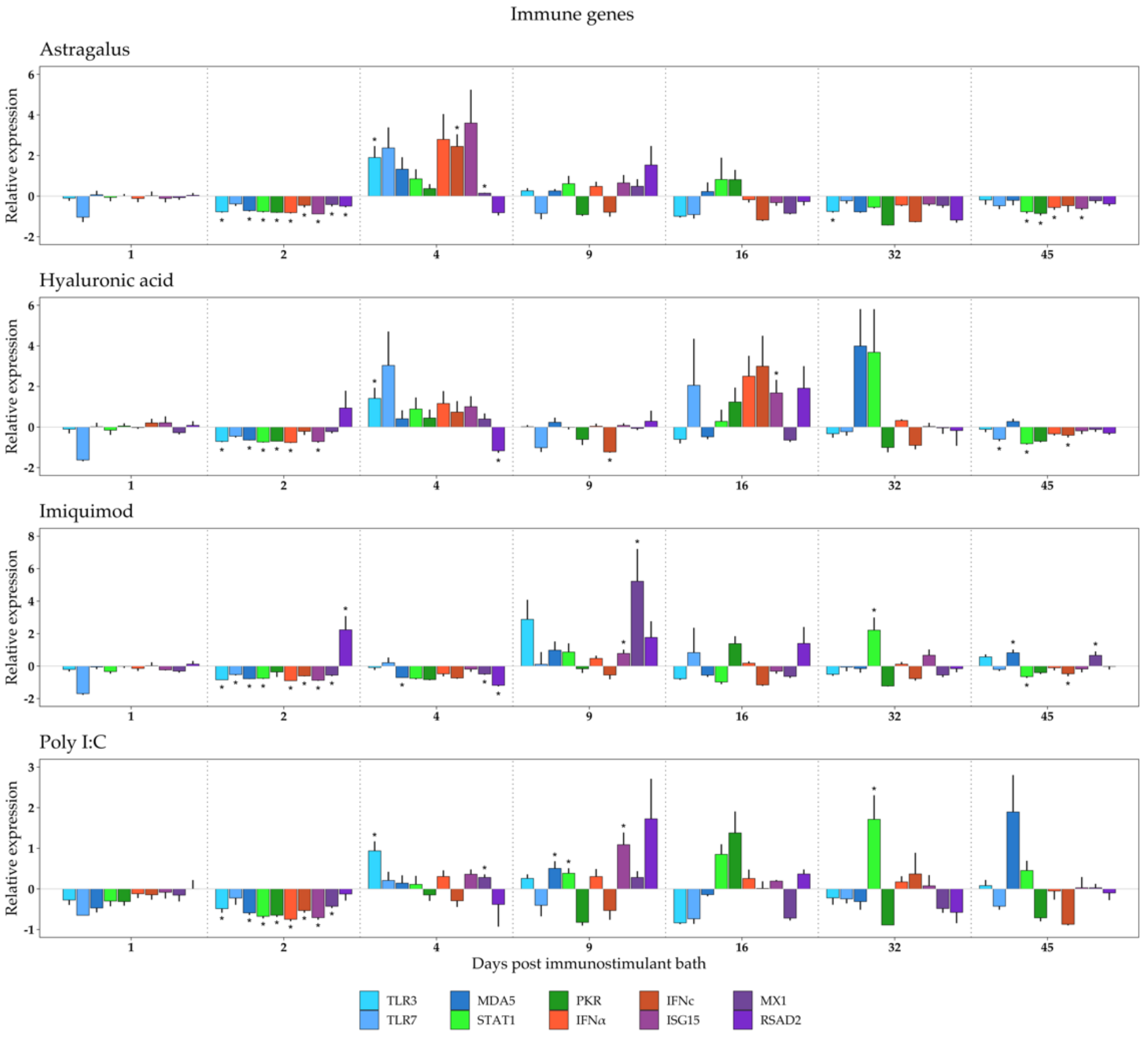
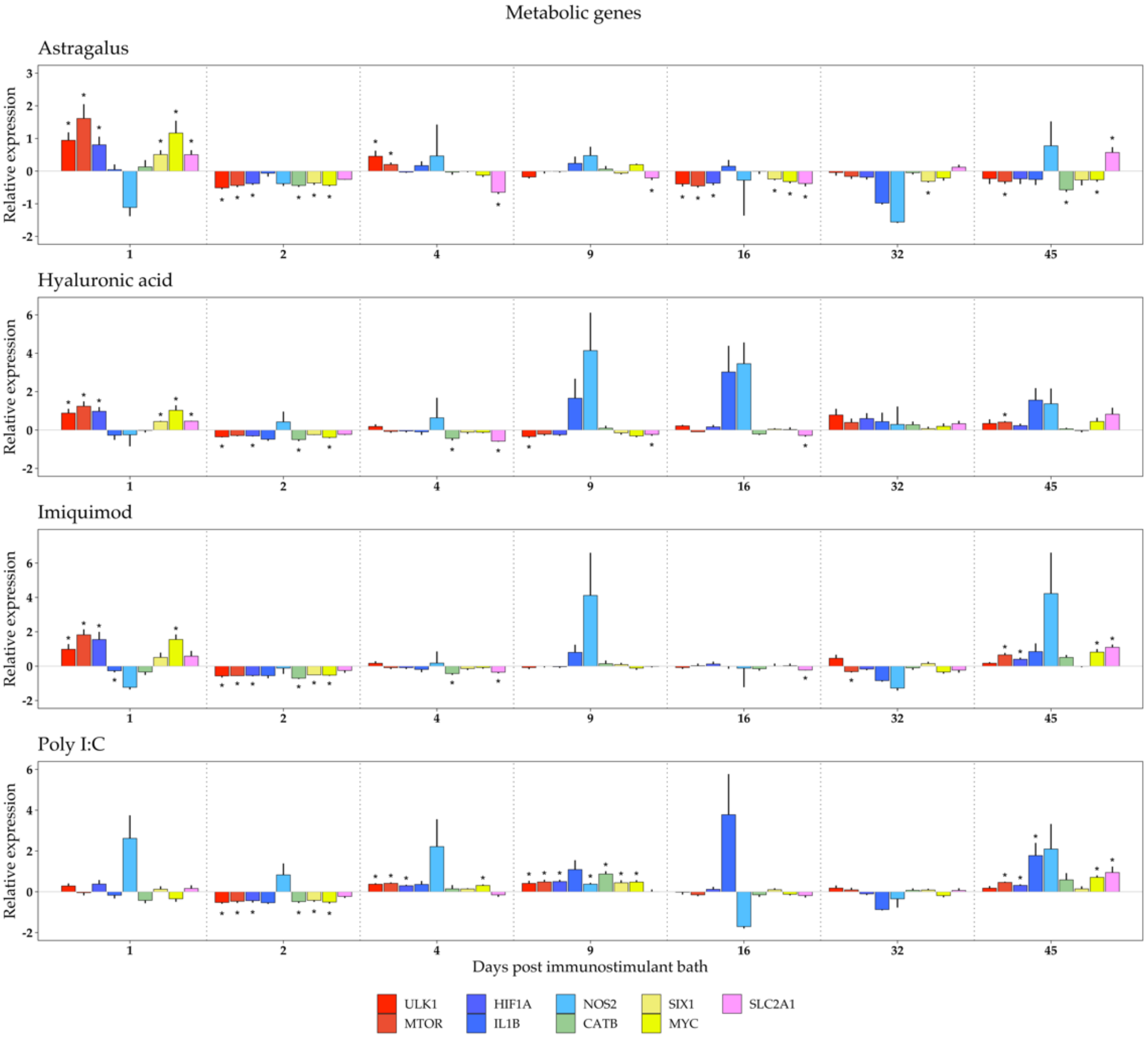
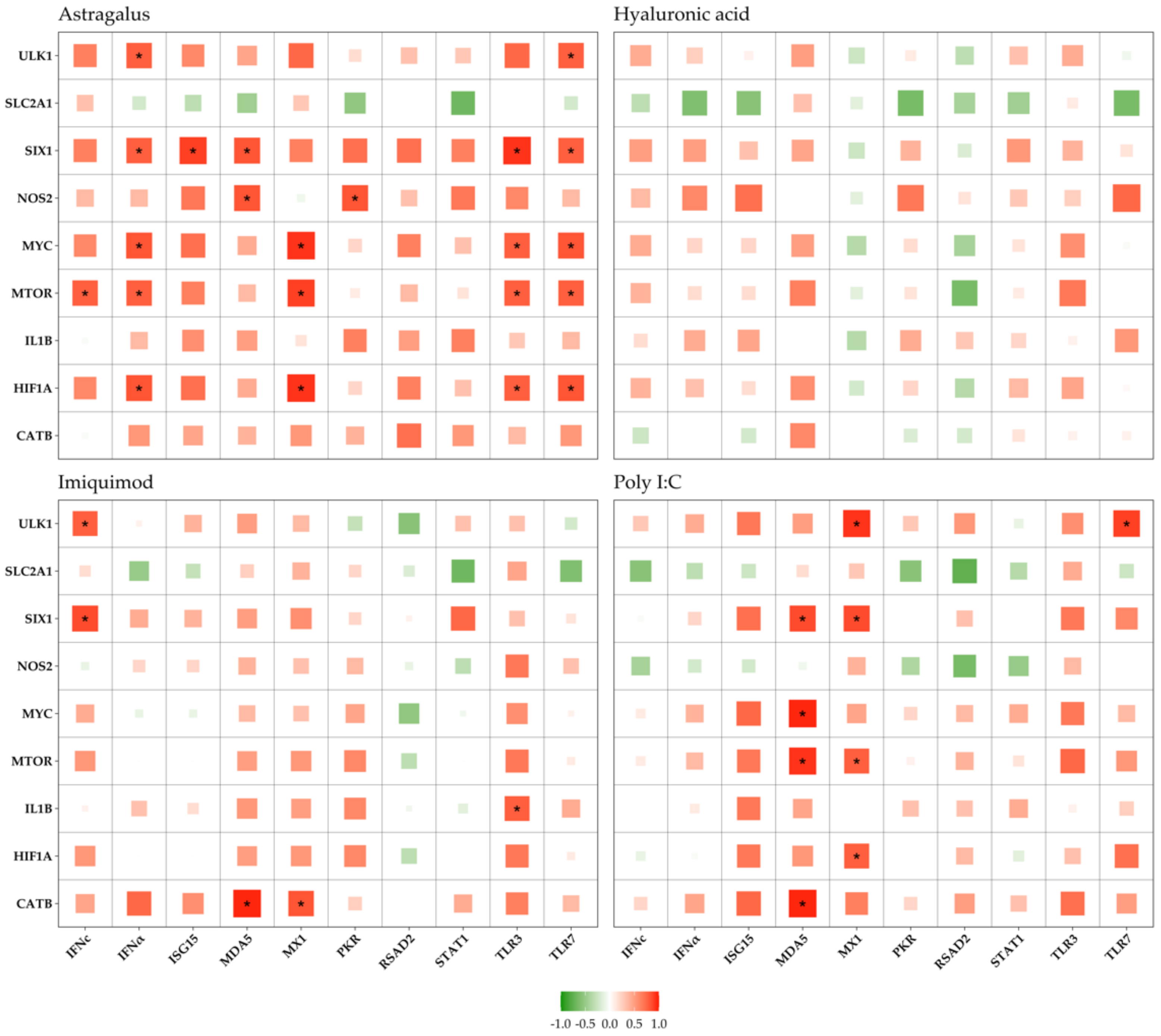
3. Results
3.1. Immune Response to Immunostimulant Bath
3.2. Metabolic Response to Immunostimulant Bath
3.3. Immune vs. Metabolic Genes
4. Discussion
4.1. Immune Genes
4.2. Metabolic Genes
4.3. Gene Correlations
4.3.1. Astragalus
4.3.2. Imiquimod
4.3.3. Poly I:C
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- FAO. The State of World Fisheries and Aquaculture 2020, Sustainability in Action; Food and Agriculture Organization: Rome, Italy, 2020. [Google Scholar] [CrossRef]
- Ingun, S.; Cecilie, S.; Walde, J.B.B.; Bornø, G.; Haukaas, A.; Brun, E. (Eds.) The Health Situation in Norwegian Aquaculture 2019, Norwegian Veterinary Institute. 2020. Available online: https://www.vetinst.no/rapporter-og-publikasjoner/rapporter/2020/fish-health-report-2019/_/attachment/download/7507c5b2-df54-4028-aee8-da5ae3010d0d:728b5711c9fb60a0ba16803064780727f5bc3b6b/Fish%20health%20report%202019.pdf (accessed on 2 July 2021).
- Vadstein, O.; Bergh, Ø.; Gatesoupe, F.-J.; Galindo-Villegas, J.; Mulero, V.; Picchietti, S.; Scapigliati, G.; Makridis, P.; Olsen, Y.; Dierckens, K.; et al. Microbiology and Immunology of Fish Larvae. Rev. Aquac. 2013, 5, S1–S25. [Google Scholar] [CrossRef] [Green Version]
- Aoki, T.; Hikima, J.; Hwang, S.D.; Jung, T.S. Innate Immunity of Finfish: Primordial Conservation and Function of Viral RNA Sensors in Teleosts. Fish Shellfish Immunol. 2013, 35, 1689–1702. [Google Scholar] [CrossRef]
- Janeway, C.A. Approaching the Asymptote? Evolution and Revolution in Immunology. Cold Spring Harb. Symp. Quant. Biol. 1989, 54, 1–13. [Google Scholar] [CrossRef]
- Murphy, K.; Weaver, C. Janeway’s Immunobiology, 9th ed.; Garland Science, Taylor & Francis Group LLC: New York, NY, USA, 2017; ISBN 978-0-8153-4505-3. [Google Scholar]
- Svingerud, T.; Holand, J.K.; Robertsen, B. Infectious Salmon Anemia Virus (ISAV) Replication Is Transiently Inhibited by Atlantic Salmon Type I Interferon in Cell Culture. Virus Res. 2013, 177, 163–170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beutler, B.; Jiang, Z.; Georgel, P.; Crozat, K.; Croker, B.; Rutschmann, S.; Du, X.; Hoebe, K. Genetic Analysis of Host Resistance: Toll-like Receptor Signaling and Immunity at Large. Annu. Rev. Immunol. 2006, 24, 353–389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, S.N.; Zou, P.F.; Nie, P. Retinoic Acid-Inducible Gene I (RIG-I)-like Receptors (RLRs) in Fish: Current Knowledge and Future Perspectives. Immunology 2017, 151, 16–25. [Google Scholar] [CrossRef]
- Zhang, L.; Gao, Z.; Yu, L.; Zhang, B.; Wang, J.; Zhou, J. Nucleotide-Binding and Oligomerization Domain (NOD)-like Receptors in Teleost Fish: Current Knowledge and Future Perspectives. J. Fish. Dis. 2018, 41, 1317–1330. [Google Scholar] [CrossRef]
- Roy, P.; Panda, S.P.; Pal, A.; Jayasankar, P.; Das, B.K. Ontogenetic Profile of Antiviral Mx Gene and Its Role in Innate Immunity in Mrigal, Cirrhinus Mrigala (Hamilton 1822). Aquac. Res. 2017, 48, 3230–3243. [Google Scholar] [CrossRef]
- Vercammen, E.; Staal, J.; Beyaert, R. Sensing of Viral Infection and Activation of Innate Immunity by Toll-like Receptor 3. Clin. Microbiol. Rev. 2008, 21, 13–25. [Google Scholar] [CrossRef] [Green Version]
- Chu, Q.; Gao, Y.; Xu, G.; Wu, C.; Xu, T. Transcriptome Comparative Analysis Revealed Poly(I:C) Activated RIG-I/MDA5-Mediated Signaling Pathway in Miiuy Croaker. Fish Shellfish Immunol. 2015, 47, 168–174. [Google Scholar] [CrossRef]
- Kitao, Y.; Kono, T.; Korenaga, H.; Iizasa, T.; Nakamura, K.; Savan, R.; Sakai, M. Characterization and Expression Analysis of Type I Interferon in Common Carp Cyprinus carpio L. Mol. Immunol. 2009, 46, 2548–2556. [Google Scholar] [CrossRef] [PubMed]
- Tanekhy, M.; Kono, T.; Sakai, M. Cloning, Characterization, and Expression Analysis of Toll-like Receptor-7 CDNA from Common Carp, Cyprinus carpio L. Comp. Biochem. Physiol. Part. D Genom. Proteom. 2010, 5, 245–255. [Google Scholar] [CrossRef] [PubMed]
- Hemmi, H.; Kaisho, T.; Takeuchi, O.; Sato, S.; Sanjo, H.; Hoshino, K.; Horiuchi, T.; Tomizawa, H.; Takeda, K.; Akira, S. Small-Antiviral Compounds Activate Immune Cells via the TLR7 MyD88-Dependent Signaling Pathway. Nat. Immunol. 2002, 3, 196–200. [Google Scholar] [CrossRef] [PubMed]
- Schön, M.P.; Schön, M. Imiquimod: Mode of Action. Br. J. Dermatol. 2007, 157, 8–13. [Google Scholar] [CrossRef] [PubMed]
- Flutter, B.; Nestle, F.O. TLRs to Cytokines: Mechanistic Insights from the Imiquimod Mouse Model of Psoriasis. Eur. J. Immunol. 2013, 43, 3138–3146. [Google Scholar] [CrossRef]
- Gong, T.; Liu, L.; Jiang, W.; Zhou, R. DAMP-Sensing Receptors in Sterile Inflammation and Inflammatory Diseases. Nat. Rev. Immunol. 2020, 20, 95–112. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, H.; Xiao, Q.; Liang, H.; Li, Z.; Jiang, C.; Wu, H.; Zheng, Q. Hyaluronic Acid Fragments Evoke Kupffer Cells via TLR4 Signaling Pathway. Sci. China C Life Sci. C 2009, 52, 147–154. [Google Scholar] [CrossRef]
- You, N.; Chu, S.; Cai, B.; Gao, Y.; Hui, M.; Zhu, J.; Wang, M. Bioactive Hyaluronic Acid Fragments Inhibit Lipopolysaccharide-Induced Inflammatory Responses via the Toll-like Receptor 4 Signaling Pathway. Front. Med. 2021, 15, 292–301. [Google Scholar] [CrossRef]
- Senbanjo, L.T.; Chellaiah, M.A. CD44: A Multifunctional Cell Surface Adhesion Receptor Is a Regulator of Progression and Metastasis of Cancer Cells. Front. Cell Dev. Biol. 2017, 5, 18. [Google Scholar] [CrossRef] [Green Version]
- Cermelli, C.; Cuoghi, A.; Scuri, M.; Bettua, C.; Neglia, R.G.; Ardizzoni, A.; Blasi, E.; Iannitti, T.; Palmieri, B. In Vitro Evaluation of Antiviral and Virucidal Activity of a High Molecular Weight Hyaluronic Acid. Virol. J. 2011, 8, 141. [Google Scholar] [CrossRef] [Green Version]
- Sun, Y.; Wang, X.; Zhou, H.; Mai, K.; He, G. Dietary Astragalus Polysaccharides Ameliorates the Growth Performance, Antioxidant Capacity and Immune Responses in Turbot (Scophthalmus maximus L.). Fish Shellfish. Immunol. 2020, 99, 603–608. [Google Scholar] [CrossRef]
- Jian, J.; Wu, Z. Effects of Traditional Chinese Medicine on Nonspecific Immunity and Disease Resistance of Large Yellow Croaker, Pseudosciaena crocea (Richardson). Aquaculture 2003, 218, 1–9. [Google Scholar] [CrossRef]
- Liu, Y.; Miao, Y.; Xu, N.; Ding, T.; Cui, K.; Chen, Q.; Zhang, J.; Fang, W.; Mai, K.; Ai, Q. Effects of Dietary Astragalus Polysaccharides (APS) on Survival, Growth Performance, Activities of Digestive Enzyme, Antioxidant Responses and Intestinal Development of Large Yellow Croaker (Larimichthys crocea) Larvae. Aquaculture 2020, 517, 734752. [Google Scholar] [CrossRef]
- Ardó, L.; Yin, G.; Xu, P.; Váradi, L.; Szigeti, G.; Jeney, Z.; Jeney, G. Chinese Herbs (Astragalus membranaceus and Lonicera japonica) and Boron Enhance the Non-Specific Immune Response of Nile Tilapia (Oreochromis niloticus) and Resistance against Aeromonas Hydrophila. Aquaculture 2008, 275, 26–33. [Google Scholar] [CrossRef]
- Elabd, H.; Wang, H.P.; Shaheen, A.; Matter, A. Astragalus membranaceus Nanoparticles Markedly Improve Immune and Anti-Oxidative Responses; and Protection against Aeromonas veronii in Nile Tilapia Oreochromis niloticus. Fish Shellfish Immunol. 2020, 97, 248–256. [Google Scholar] [CrossRef]
- Tang, J.; Cai, J.; Liu, R.; Wang, J.; Lu, Y.; Wu, Z.; Jian, J. Immunostimulatory Effects of Artificial Feed Supplemented with a Chinese Herbal Mixture on Oreochromis niloticus against Aeromonas hydrophila. Fish Shellfish Immunol. 2014, 39, 401–406. [Google Scholar] [CrossRef] [PubMed]
- Zahran, E.; Risha, E.; AbdelHamid, F.; Mahgoub, H.A.; Ibrahim, T. Effects of Dietary Astragalus Polysaccharides (APS) on Growth Performance, Immunological Parameters, Digestive Enzymes, and Intestinal Morphology of Nile Tilapia (Oreochromis niloticus). Fish Shellfish Immunol. 2014, 38, 149–157. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.M.; Jiang, Y.; Chen, Y.J.; Luo, L.; Doolgindachbaporn, S.; Yuangsoi, B. Effects of Astragalus Polysaccharides (APS) and Chitooligosaccharides (COS) on Growth, Immune Response and Disease Resistance of Juvenile Largemouth Bass, Micropterus Salmoides. Fish Shellfish Immunol. 2017, 70, 40–47. [Google Scholar] [CrossRef]
- Mo, W.Y.; Lun, C.H.I.; Choi, W.M.; Man, Y.B.; Wong, M.H. Enhancing Growth and Non-Specific Immunity of Grass Carp and Nile Tilapia by Incorporating Chinese Herbs (Astragalus membranaceus and Lycium barbarum) into Food Waste Based Pellets. Environ. Pollut. 2016, 219, 475–482. [Google Scholar] [CrossRef]
- Yuan, C.; Pan, X.; Gong, Y.; Xia, A.; Wu, G.; Tang, J.; Han, X. Effects of Astragalus Polysaccharides (APS) on the Expression of Immune Response Genes in Head Kidney, Gill and Spleen of the Common Carp, Cyprinus carpio L. Int. Immunopharmacol. 2008, 8, 51–58. [Google Scholar] [CrossRef]
- Elabd, H.; Wang, H.P.; Shaheen, A.; Yao, H.; Abbass, A. Feeding Glycyrrhiza glabra (Liquorice) and Astragalus membranaceus (AM) Alters Innate Immune and Physiological Responses in Yellow Perch (Perca flavescens). Fish Shellfish Immunol. 2016, 54, 374–384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, H.; Yu, F.; Wang, Q. Astragalus Membranaceus Polysaccharide-Enhanced Lymphocytes Proliferation of Yellow Drum Nibea albiflora In Vitro. In Proceedings of the Advances in Applied Biotechnology; Zhang, T.-C., Nakajima, M., Eds.; Springer: Berlin/Heidelberg, Germany, 2015; pp. 249–254. [Google Scholar]
- Bai, D.; Wu, X.; Zhu, G.; Guo, Y.; Yang, G.; Ning, B.; Xing, K. Astragalus Polysaccharides Enhance Cellular Immune Response and Disease Resistance in Yellow Catfish. Isr. J. Aquac. 2012, 64, 1–7. [Google Scholar]
- Jian, J.; Wu, Z. Influences of Traditional Chinese Medicine on Non-Specific Immunity of Jian Carp (Cyprinus carpio Var. Jian). Fish Shellfish Immunol. 2004, 16, 185–191. [Google Scholar] [CrossRef]
- Zhang, P.; Liu, X.; Liu, H.; Wang, W.; Liu, X.; Li, X.; Wu, X. Astragalus Polysaccharides Inhibit Avian Infectious Bronchitis Virus Infection by Regulating Viral Replication. Microb. Pathog. 2018, 114, 124–128. [Google Scholar] [CrossRef] [PubMed]
- Xue, H.; Gan, F.; Zhang, Z.; Hu, J.; Chen, X.; Huang, K. Astragalus Polysaccharides Inhibits PCV2 Replication by Inhibiting Oxidative Stress and Blocking NF-ΚB Pathway. Int. J. Biol. Macromol. 2015, 81, 22–30. [Google Scholar] [CrossRef] [PubMed]
- Shao, B.-M.; Xu, W.; Dai, H.; Tu, P.; Li, Z.; Gao, X.-M. A Study on the Immune Receptors for Polysaccharides from the Roots of Astragalus membranaceus, a Chinese Medicinal Herb. Biochem. Biophys. Res. Commun. 2004, 320, 1103–1111. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Liu, Z.; Wang, Z.; Yu, S.; Long, T.; Zhou, X.; Bao, Y. Astragalus Polysaccharides Exerts Immunomodulatory Effects via TLR4-Mediated MyD88-Dependent Signaling Pathway In Vitro and In Vivo. Sci. Rep. 2017, 7, 44822. [Google Scholar] [CrossRef] [Green Version]
- Wei, W.; Xiao, H.-T.; Bao, W.-R.; Ma, D.-L.; Leung, C.-H.; Han, X.-Q.; Ko, C.-H.; Lau, C.B.-S.; Wong, C.-K.; Fung, K.-P.; et al. TLR-4 May Mediate Signaling Pathways of Astragalus Polysaccharide RAP Induced Cytokine Expression of RAW264.7 Cells. J. Ethnopharmacol. 2016, 179, 243–252. [Google Scholar] [CrossRef] [Green Version]
- Corcoran, S.E.; O’Neill, L.A.J. HIF1α and Metabolic Reprogramming in Inflammation. J. Clin. Investig. 2016, 126, 3699–3707. [Google Scholar] [CrossRef]
- Fitzgerald, K.A.; Kagan, J.C. Toll-like Receptors and the Control of Immunity. Cell 2020, 180, 1044–1066. [Google Scholar] [CrossRef]
- Ganeshan, K.; Chawla, A. Metabolic Regulation of Immune Responses. Annu. Rev. Immunol. 2014, 32, 609–634. [Google Scholar] [CrossRef] [Green Version]
- O’Neill, L.A.J.; Kishton, R.J.; Rathmell, J. A Guide to Immunometabolism for Immunologists. Nat. Rev. Immunol. 2016, 16, 553–565. [Google Scholar] [CrossRef] [Green Version]
- Pearce, E.L.; Pearce, E.J. Metabolic Pathways in Immune Cell Activation and Quiescence. Immunity 2013, 38, 633–643. [Google Scholar] [CrossRef] [Green Version]
- Weichhart, T.; Hengstschläger, M.; Linke, M. Regulation of Innate Immune Cell Function by MTOR. Nat. Rev. Immunol. 2015, 15, 599–614. [Google Scholar] [CrossRef]
- Grodzki, A.C.G.; Giulivi, C.; Lein, P.J. Oxygen Tension Modulates Differentiation and Primary Macrophage Functions in the Human Monocytic THP-1 Cell Line. PLoS ONE 2013, 8, e54926. [Google Scholar] [CrossRef]
- Langston, P.K.; Shibata, M.; Horng, T. Metabolism Supports Macrophage Activation. Front. Immunol. 2017, 8, 61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Neill, L.A.J.; Pearce, E.J. Immunometabolism Governs Dendritic Cell and Macrophage Function. J. Exp. Med. 2015, 213, 15–23. [Google Scholar] [CrossRef]
- Liu, Y.; Xin, Z.Z.; Zhang, D.Z.; Wang, Z.F.; Zhu, X.Y.; Tang, B.P.; Jiang, S.H.; Zhang, H.B.; Zhou, C.L.; Chai, X.Y.; et al. Transcriptome Analysis of Yellow Catfish (Pelteobagrus fulvidraco) Liver Challenged with Polyriboinosinic Polyribocytidylic Acid (Poly I:C). Fish Shellfish Immunol. 2017, 68, 395–403. [Google Scholar] [CrossRef] [PubMed]
- Soleng, M.; Johansen, L.-H.; Johnsen, H.; Johansson, G.S.; Breiland, M.W.; Rørmark, L.; Pittman, K.; Pedersen, L.-F.; Lazado, C.C. Atlantic Salmon (Salmo Salar) Mounts Systemic and Mucosal Stress Responses to Peracetic Acid. Fish Shellfish Immunol. 2019, 93, 895–903. [Google Scholar] [CrossRef] [PubMed]
- Jensen, I.; Overrein, M.C.; Fredriksen, B.N.; Strandskog, G.; Seternes, T. Differences in Smolt Status Affect the Resistance of Atlantic Salmon (Salmo salar L.) against Infectious Pancreatic Necrosis, While Vaccine-Mediated Protection Is Unaffected. J. Fish Dis. 2019, 42, 1271–1282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johansen, L.-H.; Dahle, M.K.; Wessel, Ø.; Timmerhaus, G.; Løvoll, M.; Røsæg, M.; Jørgensen, S.M.; Rimstad, E.; Krasnov, A. Differences in Gene Expression in Atlantic Salmon Parr and Smolt after Challenge with Piscine orthoreovirus (PRV). Mol. Immunol. 2016, 73, 138–150. [Google Scholar] [CrossRef]
- Braden, L.M.; Barker, D.E.; Koop, B.F.; Jones, S.R.M. Comparative Defense-Associated Responses in Salmon Skin Elicited by the Ectoparasite Lepeophtheirus salmonis. Comp. Biochem. Physiol. Part D Genom. Proteom. 2012, 7, 100–109. [Google Scholar] [CrossRef]
- Ulvestad, J.S.; Kumari, J.; Seternes, T.; Chi, H.; Dalmo, R.A. Studies on the Effects of LPS, ß-Glucan and Metabolic Inhibitors on the Respiratory Burst and Gene Expression in Atlantic Salmon Macrophages. J. Fish. Dis. 2018, 41, 1117–1127. [Google Scholar] [CrossRef]
- Olsvik, P.A.; Vikeså, V.; Lie, K.K.; Hevrøy, E.M. Transcriptional Responses to Temperature and Low Oxygen Stress in Atlantic Salmon Studied with Next-Generation Sequencing Technology. BMC Genom. 2013, 14, 817. [Google Scholar] [CrossRef] [Green Version]
- Bahuaud, D.; Mørkøre, T.; Langsrud, Ø.; Sinnes, K.; Veiseth, E.; Ofstad, R.; Thomassen, M.S. Effects of −1.5 °C Super-Chilling on Quality of Atlantic Salmon (Salmo salar) Pre-Rigor Fillets: Cathepsin Activity, Muscle Histology, Texture and Liquid Leakage. Food Chem. 2008, 111, 329–339. [Google Scholar] [CrossRef]
- Svingerud, T.; Solstad, T.; Sun, B.; Nyrud, M.L.J.; Kileng, Ø.; Greiner-Tollersrud, L.; Robertsen, B. Atlantic Salmon Type I IFN Subtypes Show Differences in Antiviral Activity and Cell-Dependent Expression: Evidence for High IFNb/IFNc–Producing Cells in Fish Lymphoid Tissues. J. Immunol. 2012, 189, 5912–5923. [Google Scholar] [CrossRef] [Green Version]
- Moore, L.J.; Jarungsriapisit, J.; Nilsen, T.O.; Stefansson, S.; Taranger, G.L.; Secombes, C.J.; Morton, H.C.; Patel, S. Immune Gene Profiles in Atlantic Salmon (Salmo Salar L.) Post-Smolts Infected with SAV3 by Bath-Challenge Show a Delayed Response and Lower Levels of Gene Transcription Compared to Injected Fish. Fish Shellfish Immunol. 2017, 62, 320–331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pfaffl, M.W. A New Mathematical Model for Relative Quantification in Real-Time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef] [PubMed]
- Kassambara, A. ggpubr: “ggplot2” Based Publication Ready Plots. 2020. Available online: https://CRAN.R-project.org/package=ggpubr.
- R Core Team A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; R Core Team: Vienna, Austria, 2020; Available online: https://www.r-project.org/.
- Harrell, F.E., Jr. Hmisc: Harrell Miscellaneous. 2021. Available online: https://CRAN.R-project.org/package=Hmisc.
- Cooper, N. NCmisc: Miscellaneous Functions for Creating Adaptive Functions and Scripts. 2018. Available online: https://search.r-project.org/CRAN/refmans/NCmisc/html/00Index.html.
- Waddell, J. oaColors: OpenAnalytics Colors Package. 2015. Available online: https://CRAN.R-project.org/package=oaColors.
- Pedersen, T.L. patchwork: The Composer of Plots. 2020. Available online: https://cran.r-project.org/package=patchwork.
- Wickham, H.; Bryan, J. readxl: Read. Excel Files. 2019. Available online: https://CRAN.R-project.org/package=readxl.
- Kassambara, A. rstatix: Pipe-Friendly Framework for Basic Statistical Tests. 2021. Available online: https://cran.r-project.org/package=rstatix.
- Wickham, H.; Averick, M.; Bryan, J.; Chang, W.; McGowan, L.D.; François, R.; Grolemund, G.; Hayes, A.; Henry, L.; Hester, J.; et al. Welcome to the Tidyverse. J. Open Source Softw. 2019, 4, 1686. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B Stat. Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Pottinger, T.G.; Mosuwe, E. The Corticosteroidogenic Response of Brown and Rainbow Trout Alevins and Fry to Environmental Stress during a “Critical Period. ” Gen. Comp. Endocrinol. 1994, 95, 350–362. [Google Scholar] [CrossRef]
- Li, Y.; Ran, C.; Wei, K.; Xie, Y.; Xie, M.; Zhou, W.; Yang, Y.; Zhang, Z.; Lv, H.; Ma, X.; et al. The Effect of Astragalus Polysaccharide on Growth, Gut and Liver Health, and Anti-Viral Immunity of Zebrafish. Aquaculture 2021, 540, 736677. [Google Scholar] [CrossRef]
- Kumar, H.; Kawai, T.; Akira, S. Pathogen Recognition by the Innate Immune System. Int. Rev. Immunol. 2011, 30, 16–34. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, Y.; Cao, X.; Jin, X.; Jin, T. Pattern Recognition Receptors in Zebrafish Provide Functional and Evolutionary Insight into Innate Immune Signaling Pathways. Cell Mol. Immunol. 2017, 14, 80–89. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Renauld, J.-C. Class II Cytokine Receptors and Their Ligands: Key Antiviral and Inflammatory Modulators. Nat. Rev. Immunol. 2003, 3, 667–676. [Google Scholar] [CrossRef]
- Skjesol, A.; Hansen, T.; Shi, C.-Y.; Thim, H.L.; Jørgensen, J.B. Structural and Functional Studies of STAT1 from Atlantic Salmon (Salmo salar). BMC Immunol. 2010, 11, 17. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Xie, X.; Li, C.; Fu, P. Systematic Review of the Renal Protective Effect of Astragalus Membranaceus (Root) on Diabetic Nephropathy in Animal Models. J. Ethnopharmacol. 2009, 126, 189–196. [Google Scholar] [CrossRef]
- Abdullahi, A.Y.; Kallon, S.; Yu, X.; Zhang, Y.; Li, G. Vaccination with Astragalus and Ginseng Polysaccharides Improves Immune Response of Chickens against H5N1 Avian Influenza Virus. Biomed. Res. Int. 2016, 2016, 1510264. [Google Scholar] [CrossRef] [Green Version]
- Stenson, W.F.; Ciorba, M.A. Nonmicrobial Activation of TLRs Controls Intestinal Growth, Wound Repair, and Radioprotection. Front. Immunol. 2020, 11, 617510. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Geng, C.; Hou, S.; Fan, H.; Gong, Y. Damage-Associated Molecular Patterns and Their Signaling Pathways in Primary Blast Lung Injury: New Research Progress and Future Directions. Int. J. Mol. Sci. 2020, 21, 6303. [Google Scholar] [CrossRef]
- Eckert, R.L.; Adhikary, G.; Young, C.A.; Jans, R.; Crish, J.F.; Xu, W.; Rorke, E.A. AP1 Transcription Factors in Epidermal Differentiation and Skin Cancer. J. Skin Cancer 2013, e537028. [Google Scholar] [CrossRef] [Green Version]
- Palti, Y. Toll-like Receptors in Bony Fish: From Genomics to Function. Dev. Comp. Immunol. 2011, 35, 1263–1272. [Google Scholar] [CrossRef]
- Jensen, S.; Thomsen, A.R. Sensing of RNA Viruses: A Review of Innate Immune Receptors Involved in Recognizing RNA Virus Invasion. J. Virol. 2012, 86, 2900–2910. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lazarte, J.M.S.; Thompson, K.D.; Jung, T.S. Pattern Recognition by Melanoma Differentiation-Associated Gene 5 (MDA5) in Teleost Fish: A Review. Front. Immunol. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Avunje, S.; Jung, S.-J. Poly (I:C) and Imiquimod Induced Immune Responses and Their Effects on the Survival of Olive Flounder (Paralichthys olivaceus) from Viral Haemorrhagic Septicaemia. Fish Shellfish Immunol. 2017, 71, 338–345. [Google Scholar] [CrossRef] [PubMed]
- Bottrel, R.L.A.; Yang, Y.-L.; Levy, D.E.; Tomai, M.; Reis, L.F.L. The Immune Response Modifier Imiquimod Requires STAT-1 for Induction of Interferon, Interferon-Stimulated Genes, and Interleukin-6. Antimicrob. Agents Chemother. 1999, 43, 856–861. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, F.-F.; Zhang, Y.-B.; Liu, T.-K.; Liu, Y.; Sun, F.; Jiang, J.; Gui, J.-F. Fish Virus-Induced Interferon Exerts Antiviral Function through STAT1 Pathway. Mol. Immunol. 2010, 47, 2330–2341. [Google Scholar] [CrossRef]
- Gan, Z.; Cheng, J.; Chen, S.; Laghari, Z.A.; Hou, J.; Xia, L.; Lu, Y.; Nie, P. Functional Characterization of a Group II Interferon, IFNc in the Perciform Fish, Nile Tilapia (Oreochromis niloticus). Fish Shellfish Immunol. 2020, 105, 86–94. [Google Scholar] [CrossRef]
- Zhou, Z.; Zhang, B.; Sun, L. Poly(I:C) Induces Antiviral Immune Responses in Japanese Flounder (Paralichthys Olivaceus) That Require TLR3 and MDA5 and Is Negatively Regulated by Myd88. PLoS ONE 2014, 9, e112918. [Google Scholar] [CrossRef] [Green Version]
- Dixit, E.; Kagan, J.C. Chapter Four—Intracellular Pathogen Detection by RIG-I-Like Receptors. In Advances in Immunology; Alt, F.W., Ed.; Academic Press: Waltham, MA, USA, 2013; Volume 117, pp. 99–125. [Google Scholar]
- Jensen, I.; Albuquerque, A.; Sommer, A.-I.; Robertsen, B. Effect of Poly I:C on the Expression of Mx Proteins and Resistance against Infection by Infectious Salmon Anaemia Virus in Atlantic Salmon. Fish Shellfish Immunol. 2002, 13, 311–326. [Google Scholar] [CrossRef]
- Kileng, Ø.; Albuquerque, A.; Robertsen, B. Induction of Interferon System Genes in Atlantic Salmon by the Imidazoquinoline S-27609, a Ligand for Toll-like Receptor 7. Fish Shellfish Immunol. 2008, 24, 514–522. [Google Scholar] [CrossRef]
- Chen, A.; Diaz-Soto, M.P.; Sanmamed, M.F.; Adams, T.; Schupp, J.C.; Gupta, A.; Britto, C.; Sauler, M.; Yan, X.; Liu, Q.; et al. Single-Cell Characterization of a Model of Poly I:C-Stimulated Peripheral Blood Mononuclear Cells in Severe Asthma. Respir. Res. 2021, 22, 122. [Google Scholar] [CrossRef]
- Dempoya, J.; Matsumiya, T.; Imaizumi, T.; Hayakari, R.; Xing, F.; Yoshida, H.; Okumura, K.; Satoh, K. Double-Stranded RNA Induces Biphasic STAT1 Phosphorylation by Both Type I Interferon (IFN)-Dependent and Type I IFN-Independent Pathways. J. Virol. 2012, 86, 12760–12769. [Google Scholar] [CrossRef] [Green Version]
- Polymeropoulos, E.T.; Elliott, N.G.; Frappell, P.B. Acute but Not Chronic Hyperoxia Increases Metabolic Rate without Altering the Cardiorespiratory Response in Atlantic Salmon Alevins. Aquaculture 2019, 502, 189–195. [Google Scholar] [CrossRef]
- Zhao, J.; Benakanakere, M.R.; Hosur, K.B.; Galicia, J.C.; Martin, M.; Kinane, D.F. Mammalian Target of Rapamycin (MTOR) Regulates TLR3 Induced Cytokines in Human Oral Keratinocytes. Mol. Immunol. 2010, 48, 294–304. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, J.; Yu, Y.; Qin, X.-W.; Zeng, R.-Y.; Wang, Y.-Y.; Li, Z.-M.; Mi, S.; Weng, S.-P.; Guo, C.-J.; He, J.-G. Identification and Functional Analysis of the Mandarin Fish (Siniperca chuatsi) Hypoxia-Inducible Factor-1α Involved in the Immune Response. Fish Shellfish Immunol. 2019, 92, 141–150. [Google Scholar] [CrossRef] [PubMed]
- Scatozza, F.; D’Amore, A.; Fontanella, R.A.; DE Cesaris, P.; Marampon, F.; Padula, F.; Ziparo, E.; Riccioli, A.; Filippini, A. Toll-Iike Receptor-3 Activation Enhances Malignant Traits in Human Breast Cancer Cells Through Hypoxia-Inducible Factor-1α. Anticancer Res. 2020, 40, 5379–5391. [Google Scholar] [CrossRef] [PubMed]
- Feingold, K.R.; Shigenaga, J.K.; Kazemi, M.R.; McDonald, C.M.; Patzek, S.M.; Cross, A.S.; Moser, A.; Grunfeld, C. Mechanisms of Triglyceride Accumulation in Activated Macrophages. J. Leukoc. Biol. 2012, 92, 829–839. [Google Scholar] [CrossRef] [Green Version]
- Liang, S.; Ji, L.; Kang, L.; Hu, X. Chapter Five—Metabolic regulation of innate immunity. In Advances in Immunology in China—Part B; Dong, C., Jiang, Z., Eds.; Academic Press: Waltham, MA, USA, 2020; Volume 145, pp. 129–157. [Google Scholar]
- Saxton, R.A.; Sabatini, D.M. MTOR Signaling in Growth, Metabolism, and Disease. Cell 2017, 168, 960–976. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.; Kundu, M.; Viollet, B.; Guan, K.-L. AMPK and MTOR Regulate Autophagy through Direct Phosphorylation of Ulk1. Nat. Cell Biol. 2011, 13, 132–141. [Google Scholar] [CrossRef] [Green Version]
- Buechler, M.B.; Akilesh, H.M.; Hamerman, J.A. Cutting Edge: Direct Sensing of TLR7 Ligands and Type I IFN by the Common Myeloid Progenitor Promotes MTOR/PI3K-Dependent Emergency Myelopoiesis. J. Immunol. 2016, 197, 2577–2582. [Google Scholar] [CrossRef] [Green Version]
- Wang, B.X.; Fish, E.N. The Yin and Yang of Viruses and Interferons. Trends Immunol. 2012, 33, 190–197. [Google Scholar] [CrossRef]
- Katholnig, K.; Linke, M.; Pham, H.; Hengstschläger, M.; Weichhart, T. Immune Responses of Macrophages and Dendritic Cells Regulated by MTOR Signaling. Biochem. Soc. Trans. 2013, 41, 927–933. [Google Scholar] [CrossRef]
- Li, Y.; Sun, X.-X.; Qian, D.Z.; Dai, M.-S. Molecular Crosstalk Between MYC and HIF in Cancer. Front. Cell Dev. Biol. 2020, 8. [Google Scholar] [CrossRef] [PubMed]
- Gordan, J.D.; Bertout, J.A.; Hu, C.-J.; Diehl, J.A.; Simon, M.C. HIF-2alpha Promotes Hypoxic Cell Proliferation by Enhancing c-Myc Transcriptional Activity. Cancer Cell 2007, 11, 335–347. [Google Scholar] [CrossRef] [Green Version]
- Wei, J.; Jiang, H.; Gao, H.; Wang, G. Blocking Mammalian Target of Rapamycin (MTOR) Attenuates HIF-1α Pathways Engaged-Vascular Endothelial Growth Factor (VEGF) in Diabetic Retinopathy. Cell Physiol. Biochem. 2016, 40, 1570–1577. [Google Scholar] [CrossRef]
- Han, S.; Xu, W.; Wang, Z.; Qi, X.; Wang, Y.; Ni, Y.; Shen, H.; Hu, Q.; Han, W. Crosstalk between the HIF-1 and Toll-like Receptor/Nuclear Factor-ΚB Pathways in the Oral Squamous Cell Carcinoma Microenvironment. Oncotarget 2016, 7, 37773–37789. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Ruan, H.; Himmati, F.; Zhao, M.-T.; Chen, C.C.; Makar, M.; Chen, I.Y.; Sallam, K.; Mocarski, E.S.; Sayed, D.; et al. HIF1α Regulates Early Metabolic Changes Due to Activation of Innate Immunity in Nuclear Reprogramming. Stem Cell Rep. 2020, 14, 192–200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nicholas, S.A.; Sumbayev, V.V. The Involvement of Hypoxia-Inducible Factor 1 Alpha in Toll-like Receptor 7/8-Mediated Inflammatory Response. Cell Res. 2009, 19, 973–983. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- López, D.M.; Mesri, E.; Andreanksy, S. Activation of the Hypoxia-Inducible Factor 1 Alpha Is Necessary for Type 1 Interferon and IFN Stimulatory Gene Expression during Gammaherpesvirus Infection. J. Immunol. 2020, 204, 171.13. [Google Scholar]
- Mizushima, N. The Role of the Atg1/ULK1 Complex in Autophagy Regulation. Curr. Opin. Cell Biol. 2010, 22, 132–139. [Google Scholar] [CrossRef] [PubMed]
- Saleiro, D.; Mehrotra, S.; Kroczynska, B.; Beauchamp, E.M.; Lisowski, P.; Majchrzak-Kita, B.; Bhagat, T.D.; Stein, B.L.; McMahon, B.; Altman, J.K.; et al. Central Role of ULK1 in Type I Interferon Signaling. Cell Rep. 2015, 11, 605–617. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Delgado, M.A.; Elmaoued, R.A.; Davis, A.S.; Kyei, G.; Deretic, V. Toll-like Receptors Control Autophagy. EMBO J. 2008, 27, 1110–1121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, C.-C.; Wu, C.C.; Lin, T.L. Role of Chicken Melanoma Differentiation-Associated Gene 5 in Induction and Activation of Innate and Adaptive Immune Responses to Infectious Bursal Disease Virus in Cultured Macrophages. Arch. Virol. 2015, 160, 3021–3035. [Google Scholar] [CrossRef]
- Lemaire, P.A.; Anderson, E.; Lary, J.; Cole, J.L. Mechanism of PKR Activation by DsRNA. J. Mol. Biol. 2008, 381, 351–360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verrier, E.R.; Langevin, C.; Benmansour, A.; Boudinot, P. Early Antiviral Response and Virus-Induced Genes in Fish. Dev. Comp. Immunol. 2011, 35, 1204–1214. [Google Scholar] [CrossRef]
- Lee, H.J.; Choi, B.; Kim, Y.; Lee, S.E.; Jin, H.J.; Lee, H.-S.; Chang, E.-J.; Kim, S.W. The Upregulation of Toll-Like Receptor 3 via Autocrine IFN-β Signaling Drives the Senescence of Human Umbilical Cord Blood-Derived Mesenchymal Stem Cells Through JAK1. Front. Immunol. 2019, 10, 1659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kawasaki, T.; Kawai, T. Chapter One—Discrimination Between Self and Non-Self-Nucleic Acids by the Innate Immune System. In Nucleic Acid Sensing and Immunity, Part A; International Review of Cell and Molecular Biology; Vanpouille-Box, C., Galluzzi, L., Eds.; Academic Press: Waltham, MA, USA, 2019; Volume 344, pp. 1–30. [Google Scholar]
- Li, L.; Liang, Y.; Kang, L.; Liu, Y.; Gao, S.; Chen, S.; Li, Y.; You, W.; Dong, Q.; Hong, T.; et al. Transcriptional Regulation of the Warburg Effect in Cancer by SIX1. Cancer Cell 2018, 33, 368–385.e7. [Google Scholar] [CrossRef] [Green Version]
- Schenten, D.; Medzhitov, R. Chapter 3—The Control of Adaptive Immune Responses by the Innate Immune System. In Advances in Immunology; Alt, F.W., Ed.; Academic Press: Waltham, MA, USA, 2011; Volume 109, pp. 87–124. [Google Scholar]
- Weiss-Sadan, T.; Itzhak, G.; Kaschani, F.; Yu, Z.; Mahameed, M.; Anaki, A.; Ben-Nun, Y.; Merquiol, E.; Tirosh, B.; Kessler, B.; et al. Cathepsin L Regulates Metabolic Networks Controlling Rapid Cell Growth and Proliferation. Mol. Cell Proteomics 2019, 18, 1330–1344. [Google Scholar] [CrossRef] [Green Version]
- Gal-Ben-Ari, S.; Barrera, I.; Ehrlich, M.; Rosenblum, K. PKR: A Kinase to Remember. Front. Mol. Neurosci. 2019, 11. [Google Scholar] [CrossRef] [Green Version]
- Wang, B.; Zhang, Y.-B.; Liu, T.-K.; Shi, J.; Sun, F.; Gui, J.-F. Fish Viperin Exerts a Conserved Antiviral Function through RLR-Triggered IFN Signaling Pathway. Dev. Comp. Immunol. 2014, 47, 140–149. [Google Scholar] [CrossRef]
- Hong, S.W.; Yoo, J.W.; Kang, H.S.; Kim, S.; Lee, D.-K. HIF-1alpha-Dependent Gene Expression Program during the Nucleic Acid-Triggered Antiviral Innate Immune Responses. Mol. Cells 2009, 27, 243–250. [Google Scholar] [CrossRef] [PubMed]
| Immunostimulant | Concentration (mg L−1) | CAS No. | Producer |
|---|---|---|---|
| Astragalus root extract | 10 | 89250-26-0 | Beijing Solarbio Science & Technology Co. Ltd., Beijing, China |
| Hyaluronic acid | 10 | 9004-61-9 | Wuhan Yuancheng Gongchuang Technology Co. Ltd., Wuhan, China |
| Imiquimod | 1 | 99011-02-6 | Wuhan Yuancheng Gongchuang Technology Co. Ltd., Wuhan, China |
| Poly I:C | 10 | 42424-50-0 | Tianjin Kangtai Biotechnology Co. Ltd., Tianjin, China |
| 40 Cycles | Melting Curve Stage | ||||||
|---|---|---|---|---|---|---|---|
| Temperature (°C) | 95 | 95 | 60 | 95 | 60 | 95 | 60 |
| Time (s) | 20 | 3 | 60 | 15 | 60 | 15 | 15 |
| Gene Symbol | Direction | Primer Sequence | GenBank Accession No. | Reference | Group |
|---|---|---|---|---|---|
| 18S | Forward | TGTGCCGCTAGAGGTGAAATT | AJ427629.1 | [53] | Housekeeping |
| Reverse | GCAAATGCTTTCGCTTTCG | ||||
| β-actin | Forward | CAGCCCTCCTTCCTCGGTAT | BT059604 | [54] | Housekeeping |
| Reverse | CGTCACACTTCATGATGGAGTTG | ||||
| EF1α | Forward | CGCCAACATGGGCTGG | AN321836 | [55] | Housekeeping |
| Reverse | TCACACCATTGGCGTTACCA | ||||
| NOS2 (iNOS) | Forward | AACGAGAGCCAACAGGTGTC | AJ300555.1 | [56] | Metabolic |
| Reverse | GGTGCAGCATGTCTTTGAGA | ||||
| SLC2A1 (GLUT1) | Forward | CGCCAGCCCATCTTCATC | AF247728 | [57] | Metabolic |
| Reverse | GAAAACAGCGTTGATGCCAGA | ||||
| MYC (c-Myc) | Forward | TCTCCACCCACCAGCACAAC | DQ834862.1 | Unpublished | Metabolic |
| Reverse | CTCCAGCCTCAGCCTTTTGAC | ||||
| MTOR (mTOR) | Forward | CAGCCTGAGGCCCTGAATAA | BT072258 | [58] | Metabolic |
| Reverse | CTCCACTTGGGTTGGCACAT | ||||
| ULK1 | Forward | CTAGCGTACATTGGGGCATT | XM014128422.1 | Unpublished | Metabolic |
| Reverse | CTTTCTCCTCCGTGAAGTCG | ||||
| HIF1A (HIF-1α) | Forward | CCACCTCATGAAGACCCATCA | DY708816 | [58] | Metabolic |
| Reverse | TCTCCACCCACACAAAGCCT | ||||
| CATB (cathepsin B) | Forward | AGGGGGGAACTCCTTACTGGCT | DR696159 | [59] | Metabolic |
| Reverse | CGATGCCACAGTGGTCCTTACCT | ||||
| SIX1 | Forward | CCAGCTCGGAAGATGAGTTC | XM029676167.1 | [57] | Metabolic |
| Reverse | TAGAGGTCCCAGCAACGAGT | ||||
| IL1B (IL-1β) | Forward | GCTGGAGAGTGCTGTGGAAGA | AY617117 | [57] | Metabolic |
| Reverse | TGCTTCCCTCCTGCTCGTAG | ||||
| IFNα | Forward | TGCAGTATGCAGAGCGTGTG | DQ354152.1 | [60] | Immune |
| Reverse | TCTCCTCCCATCTGGTCCAG | ||||
| IFNc | Forward | ATGTATGATGGGCAGTGTGG | EU768890 | [60] | Immune |
| Reverse | CCAGGCGCAGTAACTGAAAT | ||||
| ISG15 | Forward | CTGAAAAACGAAAAGGGCCA | AY926456.1 | [60] | Immune |
| Reverse | GCAGGGACTCCCTCCTTGTT | ||||
| MDA5 | Forward | CTCGTGAACTACTCAAGAGAATCG | NM001195179 | [61] | Immune |
| Reverse | CCTGGCTCATCTATCAAGTTAT | ||||
| MX1 | Forward | TGCAACCACAGAGGCTTTGAA | NM001123693.1 | [60] | Immune |
| Reverse | GGCTTGGTCAGGATGCCTAAT | ||||
| PKR | Forward | TGGCATGATGGAGACGACAG | EF523422.1 | [60] | Immune |
| Reverse | GCTGGGAGATAACTGCTCGG | ||||
| STAT1 | Forward | GGTCCACACAAATCAACGTG | DW551983 | [60] | Immune |
| Reverse | CTTTGCAGGGCCTTCTTCTT | ||||
| TLR3 | Forward | TTTGATGAGTCTCCGCCAACTCCA | BK008646 | [60] | Immune |
| Reverse | AATCTGCGAGGGACACAAAGGTCT | ||||
| TLR7 | Forward | TACAGCTTGGTAACATGACTCTCC | AGKD01152847 | [60] | Immune |
| Reverse | CAACTCTCTGAGACTTGTCGGTAA | ||||
| RSAD2 (viperin) | Forward | TCCTTGATGTTGGCGTGGAA | BT047610 | [60] | Immune |
| Reverse | GCATGTCAGCTTTGCTCCACA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Figueiredo, F.; Kristoffersen, H.; Bhat, S.; Zhang, Z.; Godfroid, J.; Peruzzi, S.; Præbel, K.; Dalmo, R.A.; Xu, X. Immunostimulant Bathing Influences the Expression of Immune- and Metabolic-Related Genes in Atlantic Salmon Alevins. Biology 2021, 10, 980. https://doi.org/10.3390/biology10100980
Figueiredo F, Kristoffersen H, Bhat S, Zhang Z, Godfroid J, Peruzzi S, Præbel K, Dalmo RA, Xu X. Immunostimulant Bathing Influences the Expression of Immune- and Metabolic-Related Genes in Atlantic Salmon Alevins. Biology. 2021; 10(10):980. https://doi.org/10.3390/biology10100980
Chicago/Turabian StyleFigueiredo, Filipe, Harald Kristoffersen, Shripathi Bhat, Zuobing Zhang, Jacques Godfroid, Stefano Peruzzi, Kim Præbel, Roy Ambli Dalmo, and Xiaoli Xu. 2021. "Immunostimulant Bathing Influences the Expression of Immune- and Metabolic-Related Genes in Atlantic Salmon Alevins" Biology 10, no. 10: 980. https://doi.org/10.3390/biology10100980
APA StyleFigueiredo, F., Kristoffersen, H., Bhat, S., Zhang, Z., Godfroid, J., Peruzzi, S., Præbel, K., Dalmo, R. A., & Xu, X. (2021). Immunostimulant Bathing Influences the Expression of Immune- and Metabolic-Related Genes in Atlantic Salmon Alevins. Biology, 10(10), 980. https://doi.org/10.3390/biology10100980








