Multi-Faceted Post-Transcriptional Functions of HIV-1 Rev
Abstract
:1. Introduction
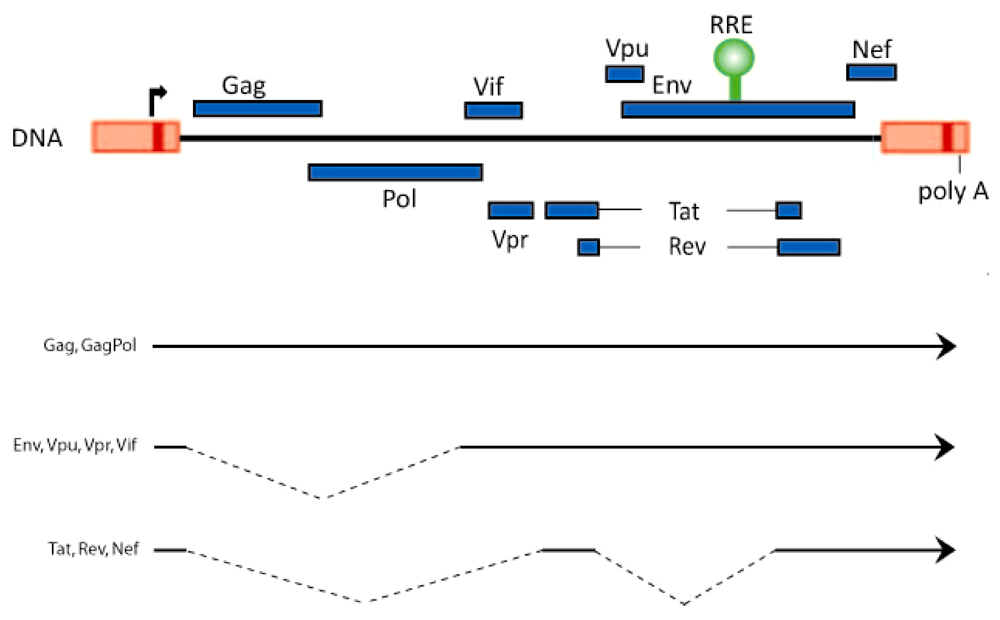
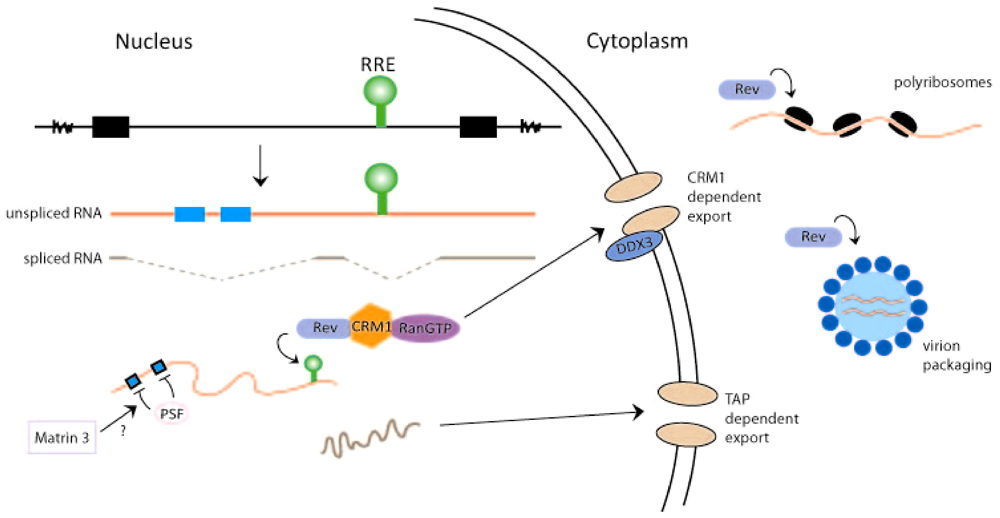
2. Rev-Dependent Export of Partially Spliced and Unspliced HIV-1 RNAs from the Nucleus
3. Rev and the Translation of HIV-1 RNA
4. Rev and HIV-1 RNA Packaging
5. Concluding Remarks
Acknowledgements
References
- Purcell, D.F.; Martin, M.A. Alternative splicing of human immunodeficiency virus type 1 mRNA modulates viral protein expression, replication, and infectivity. J. Virol. 1993, 67, 6365–6378. [Google Scholar]
- McLaren, M.; Marsh, K.; Cochrane, A. Modulating HIV-1 RNA processing and utilization. Front. Biosci. 2008, 13, 5693–5707. [Google Scholar]
- Exline, C.M.; Feng, Z.; Stoltzfus, C.M. Negative and positive mRNA splicing elements act competitively to regulate human immunodeficiency virus type 1 vif gene expression. J. Virol. 2008, 82, 3921–3931. [Google Scholar] [CrossRef]
- Stoltzfus, C.M.; Madsen, J.M. Role of viral splicing elements and cellular RNA binding proteins in regulation of HIV-1 alternative RNA splicing. Curr. HIV. Res. 2006, 4, 43–55. [Google Scholar] [CrossRef]
- Kammler, S.; Otte, M.; Hauber, I.; Kjems, J.; Hauber, J.; Schaal, H. The strength of the HIV-1 3' splice sites affects Rev function. Retrovirology 2006, 3, 89. [Google Scholar] [CrossRef]
- Cochrane, A.W.; McNally, M.T.; Mouland, A.J. The retrovirus RNA trafficking granule: From birth to maturity. Retrovirology 2006, 3, 18. [Google Scholar] [CrossRef]
- Bolinger, C.; Boris-Lawrie, K. Mechanisms employed by retroviruses to exploit host factors for translational control of a complicated proteome. Retrovirology 2009, 6, 8. [Google Scholar] [CrossRef]
- Saliou, J.M.; Bourgeois, C.F.; yadi-Ben, M.L.; Ropers, D.; Jacquenet, S.; Marchand, V.; Stevenin, J.; Branlant, C. Role of RNA structure and protein factors in the control of HIV-1 splicing. Front. Biosci. 2009, 14, 2714–2729. [Google Scholar]
- Berkhout, B.; Silverman, R.H.; Jeang, K.T. Tat trans-activates the human immunodeficiency virus through a nascent RNA target. Cell 1989, 59, 273–282. [Google Scholar] [CrossRef]
- Lever, A.M.; Jeang, K.T. Insights into cellular factors that regulate HIV-1 replication in human cells. Biochemistry 2011, 50, 920–931. [Google Scholar]
- Nekhai, S.; Jeang, K.T. Transcriptional and post-transcriptional regulation of HIV-1 gene expression: Role of cellular factors for Tat and Rev. Future Microbiol. 2006, 1, 417–426. [Google Scholar] [CrossRef]
- Lever, A.M.; Jeang, K.T. Replication of human immunodeficiency virus type 1 from entry to exit. Int. J. Hematol. 2006, 84, 23–30. [Google Scholar] [CrossRef]
- Shida, H. Role of Nucleocytoplasmic RNA transport during the life cycle of retroviruses. Front. Microbiol. 2012, 3, 179. [Google Scholar]
- Okada, H.; Zhang, X.; Ben, F.I.; Nagai, M.; Suzuki, H.; Ohashi, T.; Shida, H. Synergistic effect of human CycT1 and CRM1 on HIV-1 propagation in rat T cells and macrophages. Retrovirology 2009, 6, 43. [Google Scholar] [CrossRef]
- Daugherty, M.D.; D'Orso, I.; Frankel, A.D. A solution to limited genomic capacity: Using adaptable binding surfaces to assemble the functional HIV Rev oligomer on RNA. Mol. Cell 2008, 31, 824–834. [Google Scholar] [CrossRef]
- Daugherty, M.D.; Booth, D.S.; Jayaraman, B.; Cheng, Y.; Frankel, A.D. HIV Rev response element (RRE) directs assembly of the Rev homooligomer into discrete asymmetric complexes. Proc. Natl. Acad. Sci. USA 2010, 107, 12481–12486. [Google Scholar]
- Daugherty, M.D.; Liu, B.; Frankel, A.D. Structural basis for cooperative RNA binding and export complex assembly by HIV Rev. Nat. Struct. Mol. Biol. 2010, 17, 1337–1342. [Google Scholar]
- DiMattia, M.A.; Watts, N.R.; Stahl, S.J.; Rader, C.; Wingfield, P.T.; Stuart, D.I.; Steven, A.C.; Grimes, J.M. Implications of the HIV-1 Rev dimer structure at 3.2 A resolution for multimeric binding to the Rev response element. Proc. Natl. Acad. Sci. USA 2010, 107, 5810–5814. [Google Scholar]
- Pond, S.J.; Ridgeway, W.K.; Robertson, R.; Wang, J.; Millar, D.P. HIV-1 Rev protein assembles on viral RNA one molecule at a time. Proc. Natl. Acad. Sci. USA 2009, 106, 1404–1408. [Google Scholar]
- Bray, M.; Prasad, S.; Dubay, J.W.; Hunter, E.; Jeang, K.T.; Rekosh, D.; Hammarskjold, M.L. A small element from the Mason-Pfizer monkey virus genome makes human immunodeficiency virus type 1 expression and replication Rev-independent. Proc. Natl. Acad. Sci. USA 1994, 91, 1256–1260. [Google Scholar]
- Braun, I.C.; Rohrbach, E.; Schmitt, C.; Izaurralde, E. TAP binds to the constitutive transport element (CTE) through a novel RNA-binding motif that is sufficient to promote CTE-dependent RNA export from the nucleus. EMBO J. 1999, 18, 1953–1965. [Google Scholar] [CrossRef]
- Zolotukhin, A.S.; Michalowski, D.; Smulevitch, S.; Felber, B.K. Retroviral constitutive transport element evolved from cellular TAP(NXF1)-binding sequences. J. Virol. 2001, 75, 5567–5575. [Google Scholar] [CrossRef]
- Gruter, P.; Tabernero, C.; von, K.C.; Schmitt, C.; Saavedra, C.; Bachi, A.; Wilm, M.; Felber, B.K.; Izaurralde, E. TAP, the human homolog of Mex67p, mediates CTE-dependent RNA export from the nucleus. Mol. Cell 1998, 1, 649–659. [Google Scholar] [CrossRef]
- Fornerod, M.; Ohno, M.; Yoshida, M.; Mattaj, I.W. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 1997, 90, 1051–1060. [Google Scholar] [CrossRef]
- Hakata, Y.; Yamada, M.; Mabuchi, N.; Shida, H. The carboxy-terminal region of the human immunodeficiency virus type 1 protein Rev has multiple roles in mediating CRM1-related Rev functions. J. Virol. 2002, 76, 8079–8089. [Google Scholar]
- Schwartz, S.; Campbell, M.; Nasioulas, G.; Harrison, J.; Felber, B.K.; Pavlakis, G.N. Mutational inactivation of an inhibitory sequence in human immunodeficiency virus type 1 results in Rev-independent gag expression. J. Virol. 1992, 66, 7176–7182. [Google Scholar]
- Schneider, R.; Campbell, M.; Nasioulas, G.; Felber, B.K.; Pavlakis, G.N. Inactivation of the human immunodeficiency virus type 1 inhibitory elements allows Rev-independent expression of Gag and Gag/protease and particle formation. J. Virol. 1997, 71, 4892–4903. [Google Scholar]
- Black, A.C.; Luo, J.; Chun, S.; Bakker, A.; Fraser, J.K.; Rosenblatt, J.D. Specific binding of polypyrimidine tract binding protein and hnRNP A1 to HIV-1 CRS elements. Virus Genes 1996, 12, 275–285. [Google Scholar]
- Zolotukhin, A.S.; Michalowski, D.; Bear, J.; Smulevitch, S.V.; Traish, A.M.; Peng, R.; Patton, J.; Shatsky, I.N.; Felber, B.K. PSF acts through the human immunodeficiency virus type 1 mRNA instability elements to regulate virus expression. Mol. Cell. Biol. 2003, 23, 6618–6630. [Google Scholar]
- Afonina, E.; Neumann, M.; Pavlakis, G.N. Preferential binding of poly(A)-binding protein 1 to an inhibitory RNA element in the human immunodeficiency virus type 1 gag mRNA. J. Biol. Chem. 1997, 272, 2307–2311. [Google Scholar]
- Kula, A.; Guerra, J.; Knezevich, A.; Kleva, D.; Myers, M.P.; Marcello, A. Characterization of the HIV-1 RNA associated proteome identifies Matrin 3 as a nuclear cofactor of Rev function. Retrovirology 2011, 8, 60. [Google Scholar] [CrossRef]
- Yedavalli, V.S.; Jeang, K.T. Matrin 3 is a co-factor for HIV-1 Rev in regulating post-transcriptional viral gene expression. Retrovirology 2011, 8, 61. [Google Scholar] [CrossRef]
- Neville, M.; Stutz, F.; Lee, L.; Davis, L.I.; Rosbash, M. The importin-beta family member Crm1p bridges the interaction between Rev and the nuclear pore complex during nuclear export. Curr. Biol. 1997, 7, 767–775. [Google Scholar] [CrossRef]
- Jones, T.; Sheer, D.; Bevec, D.; Kappel, B.; Hauber, J.; Steinkasserer, A. The human HIV-1 Rev binding-protein hRIP/Rab (HRB) maps to chromosome 2q36. Genomics 1997, 40, 198–199. [Google Scholar]
- Yedavalli, V.S.; Neuveut, C.; Chi, Y.H.; Kleiman, L.; Jeang, K.T. Requirement of DDX3 DEAD box RNA helicase for HIV-1 Rev-RRE export function. Cell 2004, 119, 381–392. [Google Scholar] [CrossRef]
- Fang, J.; Kubota, S.; Yang, B.; Zhou, N.; Zhang, H.; Godbout, R.; Pomerantz, R.J. A DEAD box protein facilitates HIV-1 replication as a cellular co-factor of Rev. Virology 2004, 330, 471–480. [Google Scholar] [CrossRef]
- Edgcomb, S.P.; Carmel, A.B.; Naji, S.; mbrus-Aikelin, G.; Reyes, J.R.; Saphire, A.C.; Gerace, L.; Williamson, J.R. DDX1 is an RNA-dependent ATPase involved in HIV-1 Rev function and virus replication. J. Mol. Biol. 2012, 415, 61–74. [Google Scholar] [CrossRef]
- Li, J.; Tang, H.; Mullen, T.M.; Westberg, C.; Reddy, T.R.; Rose, D.W.; Wong-Staal, F. A role for RNA helicase A in post-transcriptional regulation of HIV type 1. Proc. Natl. Acad. Sci. USA 1999, 96, 709–714. [Google Scholar]
- Krishnan, V.; Zeichner, S.L. Alterations in the expression of DEAD-box and other RNA binding proteins during HIV-1 replication. Retrovirology 2004, 1, 42. [Google Scholar] [CrossRef]
- Naji, S.; Ambrus, G.; Cimermancic, P.; Reyes, J.R.; Johnson, J.R.; Filbrandt, R.; Huber, M.D.; Vesely, P.; Krogan, N.J.; Yates, J.R., III; et al. Host cell interactome of HIV-1 Rev includes RNA helicases involved in multiple facets of virus production. Mol. Cell. Proteomics 2012, 11, M111. [Google Scholar]
- Yedavalli, V.S.; Jeang, K.T. Trimethylguanosine capping selectively promotes expression of Rev-dependent HIV-1 RNAs. Proc. Natl. Acad. Sci. USA 2010, 107, 14787–14792. [Google Scholar] [CrossRef]
- Plessel, G.; Fischer, U.; Luhrmann, R. m3G cap hypermethylation of U1 small nuclear ribonucleoprotein (snRNP) in vitro: Evidence that the U1 small nuclear RNA-(guanosine-N2)-methyltransferase is a non-snRNP cytoplasmic protein that requires a binding site on the Sm core domain. Mol. Cell. Biol. 1994, 14, 4160–4172. [Google Scholar]
- Terns, M.P.; Grimm, C.; Lund, E.; Dahlberg, J.E. A common maturation pathway for small nucleolar RNAs. EMBO J. 1995, 14, 4860–4871. [Google Scholar]
- Arrigo, S.J.; Chen, I.S. Rev is necessary for translation but not cytoplasmic accumulation of HIV-1 vif, vpr, and env/vpu 2 RNAs. Genes Dev. 1991, 5, 808–819. [Google Scholar] [CrossRef]
- D'Agostino, D.M.; Felber, B.K.; Harrison, J.E.; Pavlakis, G.N. The Rev protein of human immunodeficiency virus type 1 promotes polysomal association and translation of gag/pol and vpu/env mRNAs. Mol. Cell. Biol. 1992, 12, 1375–1386. [Google Scholar]
- Suhasini, M.; Reddy, T.R. Cellular proteins and HIV-1 Rev function. Curr. HIV Res. 2009, 7, 91–100. [Google Scholar] [CrossRef]
- He, J.J.; Henao-Mejia, J.; Liu, Y. Sam68 functions in nuclear export and translation of HIV-1 RNA. RNA Biol. 2009, 6, 384–386. [Google Scholar] [CrossRef]
- Hartman, T.R.; Qian, S.; Bolinger, C.; Fernandez, S.; Schoenberg, D.R.; Boris-Lawrie, K. RNA helicase A is necessary for translation of selected messenger RNAs. Nat. Struct. Mol. Biol. 2006, 13, 509–516. [Google Scholar] [CrossRef]
- Groom, H.C.; Anderson, E.C.; Dangerfield, J.A.; Lever, A.M. Rev regulates translation of human immunodeficiency virus type 1 RNAs. J. Gen. Virol. 2009, 90, 1141–1147. [Google Scholar] [CrossRef]
- Boeras, I.; Sakalian, M.; West, J.T. Translation of MMTV Gag requires nuclear events involving splicing motifs in addition to the viral Rem protein and RmRE. Retrovirology 2012, 9, 8. [Google Scholar] [CrossRef]
- Berkowitz, R.D.; Hammarskjold, M.L.; Helga-Maria, C.; Rekosh, D.; Goff, S.P. 5' regions of HIV-1 RNAs are not sufficient for encapsidation: Implications for the HIV-1 packaging signal. Virology 1995, 212, 718–723. [Google Scholar] [CrossRef]
- Brandt, S.; Blissenbach, M.; Grewe, B.; Konietzny, R.; Grunwald, T.; Uberla, K. Rev proteins of human and simian immunodeficiency virus enhance RNA encapsidation. PLoS Pathog. 2007, 3, e54. [Google Scholar] [CrossRef]
- Blissenbach, M.; Grewe, B.; Hoffmann, B.; Brandt, S.; Uberla, K. Nuclear RNA export and packaging functions of HIV-1 Rev revisited. J. Virol. 2010, 84, 6598–6604. [Google Scholar] [CrossRef]
- Cochrane, A. How does the journey affect the message(RNA)? RNA Biol. 2009, 6, 169–170. [Google Scholar] [CrossRef]
- Cockrell, A.S.; van, P.H.; Santistevan, N.; Ma, H.; Kafri, T. The HIV-1 Rev/RRE system is required for HIV-1 5' UTR cis elements to augment encapsidation of heterologous RNA into HIV-1 viral particles. Retrovirology 2011, 8, 51. [Google Scholar] [CrossRef]
- Lever, A.M. HIV-1 RNA packaging. Adv. Pharmacol. 2007, 55, 1–32. [Google Scholar] [CrossRef]
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Jeang, K.-T. Multi-Faceted Post-Transcriptional Functions of HIV-1 Rev. Biology 2012, 1, 165-174. https://doi.org/10.3390/biology1020165
Jeang K-T. Multi-Faceted Post-Transcriptional Functions of HIV-1 Rev. Biology. 2012; 1(2):165-174. https://doi.org/10.3390/biology1020165
Chicago/Turabian StyleJeang, Kuan-Teh. 2012. "Multi-Faceted Post-Transcriptional Functions of HIV-1 Rev" Biology 1, no. 2: 165-174. https://doi.org/10.3390/biology1020165
APA StyleJeang, K.-T. (2012). Multi-Faceted Post-Transcriptional Functions of HIV-1 Rev. Biology, 1(2), 165-174. https://doi.org/10.3390/biology1020165



