Surface Chemistry, Crystal Structure, Size and Topography Role in the Albumin Adsorption Process on TiO2 Anatase Crystallographic Faces and Its 3D-Nanocrystal: A Molecular Dynamics Study †
Abstract
1. Introduction
2. Materials and Methods
3. Results and Discussion
- (i)
- at first, the initial energy minimization considering different initial trial geometries of interaction albumin fragment-specific solid TiO2 surface (see Section 3.1.1);
- (ii)
- the MD runs at constant temperature until the potential energy, the van der Waals contribute and the conformational changes achieve an equilibrium state, starting from both the initial geometry with the lowest potential energy and the initial metastable geometry displaying the highest initial potential energy (see Section 3.1.2);
- (iii)
- the final geometry optimizations of the final configuration assumed by the adsorbed protein fragment at the end of the MD run (see Section 3.1.2).
3.1. Adsorption of HSA Fragment on Different TiO2 Crystallographic Anatase Faces
3.1.1. Initial Adsorption on Different TiO2 Anatase Faces
3.1.2. Final Adsorption on Different TiO2 Anatase Faces
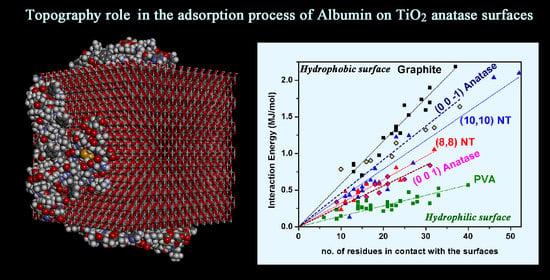
3.2. Adsorption on Ideal TiO2 Anatase 3D Nanocrystal
4. Conclusions
Supplementary Materials
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
Abbreviations
| MM | Molecular mechanics |
| MD | Molecular dynamics |
| HSA | Human serum albumin |
| PDB | Protein Data Bank |
| 3D | Three-dimensional |
| SWCNT | Single-walled carbon nanotube |
| PVA | Polyvinyl alcohol |
| H-bond | Hydrogen bond |
| Eint | Interaction energy |
| Estrain | Strain energy |
| Eintrinsic | Intrinsic energy |
| Rg | Radius of gyration |
| HOPG | Highly oriented pyrolytic graphite |
| SASA | Solvent-accessible surface area |
References
- Silva-Bermudez, P.; Rodil, S.E. An overview of protein adsorption on metal oxide coatings for biomedical implants. Surf. Coat. Technol. 2013, 233, 147–158. [Google Scholar] [CrossRef]
- Hoffman, A.S. Biomaterials in the nano-era. Chin. Sci. Bull. 2013, 58, 4337–4341. [Google Scholar] [CrossRef][Green Version]
- Ratner, B.D. Biomaterials: Been There, Done That, and Evolving into the Future. Annu. Rev. Biomed. Eng. 2019, 21, 171–191. [Google Scholar] [CrossRef] [PubMed]
- Ratner, B.D.; Bryant, S.J. Biomaterials: Where We Have Been and Where We Are Going. Annu. Rev. Biomed. Eng. 2004, 6, 41–75. [Google Scholar] [CrossRef] [PubMed]
- Wei, Q.; Becherer, T.; Angioletti-Uberti, S.; Dzubiella, J.; Wischke, C.; Neffe, A.T.; Lendlein, A.; Ballauff, M.; Haag, R. Protein Interactions with Polymer Coatings and Biomaterials. Angew. Chem. Int. Ed. 2014, 53, 8004–8031. [Google Scholar] [CrossRef] [PubMed]
- Young, T.H.; Lin, D.T.; Chen, L.Y. Human monocyte adhesion and activation on crystalline polymers with different morphology and wettability in vitro. J. Biomed. Mater. Res. 2000, 50, 490–498. [Google Scholar] [CrossRef]
- Mei, Y.; Gerecht, S.; Taylor, M.; Urquhart, A.J.; Bogatyrev, S.R.; Cho, S.-W.; Davies, M.C.; Alexander, M.R.; Langer, R.S.; Anderson, D.G. Mapping the Interactions among Biomaterials, Adsorbed Proteins, and Human Embryonic Stem Cells. Adv. Mater. 2009, 21, 2781–2786. [Google Scholar] [CrossRef]
- González-García, C.; Cantini, M.; Ballester-Beltrán, J.; Altankov, G.; Salmerón-Sánchez, M. The strength of the protein-material interaction determines cell fate. Acta Biomater. 2018, 77, 74–84. [Google Scholar] [CrossRef]
- Chen, H.; Yuan, L.; Song, W.; Wu, Z.; Li, D. Biocompatible polymer materials: Role of protein-surface interactions. Prog. Polym. Sci. 2008, 33, 1059–1087. [Google Scholar] [CrossRef]
- Veiseh, O.; Kievit, F.M.; Ellenbogen, R.G.; Zhang, M. Cancer Cell Invasion: Treatment and Monitoring Opportunities in Nanomedicine. Adv. Drug Deliv. Rev. 2011, 63, 582–596. [Google Scholar] [CrossRef]
- Bencina, M.; Iglic, A.; Mozetic, M.; Junkar, I. Crystallized TiO2 Nanosurfaces in Biomedical Applications. Nanomaterials 2020, 10, 1121. [Google Scholar] [CrossRef]
- Kulkarni, M.; Mazare, A.; Gongadze, E.; Perutkova, S.; Kralj-Iglic, V.; Milosev, I.; Schmuki, P.; Iglic, A.; Mozetic, M. Titanium nanostructures for biomedical applications. Nanotechnology 2015, 26, 062002. [Google Scholar] [CrossRef] [PubMed]
- Yeo, E.L.L.; Thong, P.S.P.; Soo, K.C.; Kah, J.C.Y. Protein corona in drug delivery for multimodal cancer therapy in vivo. Nanoscale 2018, 10, 2461–2472. [Google Scholar] [CrossRef] [PubMed]
- Shin, H.; Jo, S.; Mikos, A.G. Biomimetic materials for tissue engineering. Biomaterials 2003, 24, 209–221. [Google Scholar] [CrossRef]
- Gomes, S.; Leonor, I.B.; Mano, J.F.; Reis, R.L.; Kaplan, D.L. Natural and genetically engineered proteins for tissue engineering. Prog. Polym. Sci. 2012, 37, 1–17. [Google Scholar] [CrossRef]
- Kurinomaru, T.; Inagaki, A.; Hoshi, M.; Nakamura, C.; Yamazoe, H. Protein microswimmers capable of delivering cells for tissue engineering applications. Mater. Horizons 2020, 7, 877–884. [Google Scholar] [CrossRef]
- Liu, F.; Liu, C.; Zheng, B.W.; He, J.; Liu, J.; Chen, C.; Lee, I.S.; Wang, X.H.; Liu, Y. Synergistic Effects on Incorporation of beta-Tricalcium Phosphate and Graphene Oxide Nanoparticles to Silk Fibroin/Soy Protein Isolate Scaffolds for Bone Tissue Engineering. Polymers 2020, 12, 69. [Google Scholar] [CrossRef]
- Sugiyama, N.; Masuda, T.; Shinoda, K.; Nakamura, A.; Tomita, M.; Ishihama, Y. Phosphopeptide enrichment by aliphatic hydroxyl acid-modified metal oxide chromatography for nano-LC-MS/MS in proteomics applications. Mol. Cell Proteom. 2007, 6, 1103–1109. [Google Scholar] [CrossRef]
- Othman, Z.; Pastor, B.C.; van Rijt, S.; Habibovic, P. Understanding interactions between biomaterials and biological systems using proteomics. Biomaterials 2018, 167, 191–204. [Google Scholar] [CrossRef]
- Cormack, A.N.; Tilocca, A. Structure and biological activity of glasses and ceramics. Philos. Trans. R. Soc. A Math. Phys. Eng. Sci. Philos. 2012, 370, 1271–1280. [Google Scholar] [CrossRef]
- Cho, D.H.; Xie, T.; Truong, J.; Stoner, A.C.; Hahm, J.I. Recent advances towards single biomolecule level understanding of protein adsorption phenomena unique to nanoscale polymer surfaces with chemical variations. Nano Res. 2020, 13, 1295–1317. [Google Scholar] [CrossRef]
- Htwe, E.E.; Nakama, Y.; Yamamoto, Y.; Tanaka, H.; Imanaka, H.; Ishida, N.; Imamura, K. Adsorption characteristics of various proteins on a metal surface in the presence of an external electric potential. Colloids Surf. B. Biointerfaces 2018, 166, 262–268. [Google Scholar] [CrossRef]
- Huang, T.T.; Gomez, R.; Geng, T.; Bashir, R.; Bhunia, A.K.; Robinson, J.P.; Ladisch, M.R. Composite surface for blocking bacterial adsorption on protein biochips. Biotechnol. Bioeng. 2003, 81, 618–624. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Handschuh-Wang, S.; Yang, Y.; Zhuang, H.; Schlemper, C.; Wesner, D.; Schonherr, H.; Zhang, W.J.; Jiang, X. Controlled Surface Chemistry of Diamond/beta-SiC Composite Films for Preferential Protein Adsorption. Langmuir 2014, 30, 1089–1099. [Google Scholar] [CrossRef] [PubMed]
- Matsuno, H.; Yokoyama, A.; Watari, F.; Uo, M.; Kawasaki, T. Biocompatibility and osteogenesis of refractory metal implants, titanium, hafnium, niobium, tantalum and rhenium. Biomaterials 2001, 22, 1253–1262. [Google Scholar] [CrossRef]
- Craciun, A.M.; Focsan, M.; Magyari, K.; Vulpoi, A.; Pap, Z. Surface Plasmon Resonance or Biocompatibility-Key Properties for Determining the Applicability of Noble Metal Nanoparticles. Materials 2017, 10, 836. [Google Scholar] [CrossRef] [PubMed]
- McGinley, E.L.; Moran, G.P.; Fleming, G.J.P. Base-metal dental casting alloy biocompatibility assessment using a human-derived three-dimensional oral mucosal model. Acta Biomater. 2012, 8, 432–438. [Google Scholar] [CrossRef]
- Zadorozhnyy, V.Y.; Kozak, D.S.; Shi, X.; Wada, T.; Louzguine-Luzgin, D.V.; Kato, H. Mechanical properties, electrochemical behavior and biocompatibility of the Ti-based low-alloys containing a minor fraction of noble metals. J. Alloys Compd. 2018, 732, 915–921. [Google Scholar] [CrossRef]
- Li, Y.H.; Yang, C.; Zhao, H.D.; Qu, S.G.; Li, X.Q.; Li, Y.Y. New Developments of Ti-Based Alloys for Biomedical Applications. Materials 2014, 7, 1709–1800. [Google Scholar] [CrossRef] [PubMed]
- Stankic, S.; Suman, S.; Haque, F.; Vidic, J. Pure and multi metal oxide nanoparticles: Synthesis, antibacterial and cytotoxic properties. J. Nanobiotechnol. 2016, 14, 1–20. [Google Scholar] [CrossRef]
- Visai, L.; De Nardo, L.; Punta, C.; Melone, L.; Cigada, A.; Imbriani, M.; Arciola, C.R. Titanium oxide antibacterial surfaces in biomedical devices. Int. J. Artif. Organs. 2011, 34, 929–946. [Google Scholar] [CrossRef]
- De Nardo, L.; Raffaini, G.; Ganazzoli, F.; Chiesa, R. Metal surface oxidation and surface interactions. In Surface Modification of Biomaterials: Methods, Analysis and Applications; Williams, R., Ed.; Woodhead Publishing: Cambridge, UK, 2010; pp. 102–142, WOS: 000291542800006. [Google Scholar]
- De Nardo, L.; Raffaini, G.; Ebramzadeh, E.; Ganazzoli, F. Titanium oxide modeling and design for innovative biomedical surfaces: A concise review. Int. J. Artif. Organs 2012, 35, 629–641. [Google Scholar] [CrossRef] [PubMed]
- Almaguer-Flores, A.; Silva-Bermudez, P.; Galicia, R.; Rodil, S.E. Bacterial adhesion on amorphous and crystalline metal oxide coatings. Mater. Sci. Eng. C 2015, 57, 88–99. [Google Scholar] [CrossRef]
- Sit, I.; Xu, Z.Z.; Grassian, V.H. Plasma protein adsorption on TiO2 nanoparticles: Impact of surface adsorption on temperature-dependent structural changes. Polyhedron 2019, 171, 147–154. [Google Scholar] [CrossRef]
- Rudramurthy, G.R.; Swamy, M.K. Potential applications of engineered nanoparticles in medicine and biology: An update. J. Biol. Inorg. Chem. 2018, 23, 1185–1204. [Google Scholar] [CrossRef] [PubMed]
- Thakur, S.; Hashim, N.; Neogi, S.; Ray, A.K. Size-dependent adsorption and conformational changes induced in bovine serum albumin (BSA) on exposure to titanium dioxide (TiO2) nanoparticles. Sep. Sci. Technol. 2017, 52, 421–434. [Google Scholar] [CrossRef]
- Giacomelli, C.E.; Avena, M.J.; DePauli, C.P. Adsorption of bovine serum albumin onto TiO2 particles. J. Colloid Interface Sci. 1997, 188, 387–395. [Google Scholar] [CrossRef]
- Bakri, A.S.; Sandan, M.Z.; Adriyanto, F.; Raship, N.A.; Said, N.D.M.; Abdullah, S.A.; Rahim, M.S. Effect of Annealing Temperature of Titanium Dioxide Thin Films on Structural and Electrical Properties. In AIP Conference Proceedings; AIP Publishing LLC: Melville, NY, USA, 2017; Volume 1788, pp. 030030-1–030030-8. [Google Scholar] [CrossRef]
- Catauro, M.; Tranquillo, E.; Dal Poggetto, G.; Pasquali, M.; Dell’Era, A.; Ciprioti, S.V. Influence of the Heat Treatment on the Particles Size and on the Crystalline Phase of TiO2 Synthesized by the Sol-Gel Method. Materials 2018, 11, 2364. [Google Scholar] [CrossRef]
- Catauro, M.; Dal Poggetto, G.; Risoluti, R.; Ciprioti, S.V. Thermal, chemical and antimicrobial characterization of bioactive titania synthesized by sol-gel method. J. Therm. Anal. Calorim. 2020, 142, 1767–1774. [Google Scholar] [CrossRef]
- Giordano, C.; Saino, E.; Rimondini, L.; Pedeferri, M.P.; Visai, L.; Cigada, A.; Chiesa, R. Electrochemically induced anatase inhibits bacterial colonization on Titanium Grade 2 and Ti6Al4V alloy for dental and orthopedic devices. Colloids Surf. B Biointerfaces 2011, 88, 648–655. [Google Scholar] [CrossRef]
- Kulkarni, M.; Mazare, A.; Park, J.; Gongadze, E.; Killian, M.S.; Kralj, S.; von der Mark, K.; Iglic, A.; Schmuki, P. Protein interactions with layers of TiO2 nanotube and nanopore arrays: Morphology and surface charge influence. Acta Biomater. 2016, 45, 357–366. [Google Scholar] [CrossRef]
- Raffaini, G.; Ganazzoli, F. Surface Topography Effects in Protein Adsorption on Nanostructured Carbon Allotropes. Langmuir 2013, 29, 4883–4893. [Google Scholar] [CrossRef] [PubMed]
- Raffaini, G.; Ganazzoli, F. Surface ordering of proteins adsorbed on graphite. J. Phys. Chem. B 2004, 108, 13850–13854. [Google Scholar] [CrossRef]
- Raffaini, G.; Ganazzoli, F. Separation of chiral nanotubes with an opposite handedness by chiral oligopeptide adsorption: A molecular dynamics study. J. Chromatogr. A 2015, 1425, 221–230. [Google Scholar] [CrossRef]
- Ganazzoli, F.; Raffaini, G. Classical atomistic simulations of protein adsorption on carbon nanomaterials. Curr. Opin. Colloid Interface Sci. 2019, 41, 11–26. [Google Scholar] [CrossRef]
- Raffaini, G.; Ganazzoli, F. Protein adsorption on the hydrophilic surface of a glassy polymer: A computer simulation study. Phys. Chem. Chem. Phys 2006, 8, 2765–2772. [Google Scholar] [CrossRef]
- Bronze-Uhle, E.S.; Dias, L.F.G.; Trino, L.D.; Matos, A.A.; de Oliveira, R.C.; Lisboa, P.N. Physicochemical characterization of albumin immobilized on different TiO2 surfaces for use in implant materials. Colloids Surf. A Physicochem. Eng. Asp. 2019, 564, 39–50. [Google Scholar] [CrossRef]
- Pegueroles, M.; Tonda-Turo, C.; Planell, J.A.; Gil, F.J.; Aparicio, C. Adsorption of Fibronectin, Fibrinogen, and Albumin on TiO2: Time-Resolved Kinetics, Structural Changes, and Competition Study. Biointerface 2012, 7, 48. [Google Scholar] [CrossRef]
- Sousa, S.R.; Moradas-Ferreira, P.; Saramago, B.; Melo, L.V.; Barbosa, M.A. Human serum albumin adsorption on TiO2 from single protein solutions and from plasma. Langmuir 2004, 20, 9745–9754. [Google Scholar] [CrossRef]
- Zhao, F.H.; Chen, Y.M.; Hu, Y.; Lu, X.G.; Xiong, S.B.; Wu, B.Y.; Guo, Y.Q.; Huang, P.; Yang, B.C. Conformation changes of albumin and lysozyme on electrospun TiO2 nanofibers and its effects on MSC behaviors. Colloids Surf. B 2020, 185, 110604. [Google Scholar] [CrossRef]
- Giacomelli, C.E.; Esplandiu, M.J.; Ortiz, P.I.; Avena, M.J.; De Pauli, C.P. Ellipsometric study of bovine serum albumin adsorbed onto Ti/TiO2 electrodes. J. Colloid Interface Sci. 1999, 218, 404–411. [Google Scholar] [CrossRef]
- Kathiravan, A.; Anandan, S.; Renganathan, R. Interaction of colloidal TiO2 with human serum albumin: A fluorescence quenching study. Colloids Surf. A Physicochem. Eng. Asp. 2009, 333, 91–95. [Google Scholar] [CrossRef]
- Wu, Y.Q.; Zhang, H.M.; Wang, Y.Q. Conformational and functional changes of bovine serum albumin induced by TiO2 nanoparticles binding. J. Mol. Liq. 2017, 343, 358–368. [Google Scholar] [CrossRef]
- Sun, T.; Liu, L.S.; Sun, Y.; Tan, C.L.; Yao, F.; Liang, X.H.; Wang, Y.; Yang, Y.H.; Hu, X.Y.; Fan, J. Synthesis and Characterization of TiO2 Nanoparticles: Applications in Research on the Interaction of Colloidal TiO2 with Human Serum Albumin by Fluorescence Spectroscopy. Anal. Sci. 2012, 28, 491–496. [Google Scholar] [CrossRef] [PubMed]
- Vergaro, V.; Carlucci, C.; Cascione, M.; Lorusso, C.; Conciauro, F.; Scremin, B.F.; Congedo, P.M.; Cannazza, G.; Citti, C.; Ciccarella, G. Interaction between Human Serum Albumin and Different Anatase TiO2 Nanoparticles: A Nano-bio Interface Study. Nanomater. Nanotechnol. 2015, 5, 30. [Google Scholar] [CrossRef]
- Xu, Z.Z.; Grassian, V.H. Bovine Serum Albumin Adsorption on TiO2 Nanoparticle Surfaces: Effects of pH and Coadsorption of Phosphate on Protein-Surface Interactions and Protein Structure. J. Phys. Chem. C 2017, 121, 21763–21771. [Google Scholar] [CrossRef]
- Raffaini, G.; Elli, S.; Ganazzoli, F. Computer simulation of bulk mechanical properties and surface hydration of biomaterials. J. Biomed. Mater. Res. Part A 2006, 77, 618–626. [Google Scholar] [CrossRef] [PubMed]
- Raffaini, G.; Ganazzoli, F. Surface Hydration of Polymeric (Bio)Materials: A Molecular Dynamics Simulation Study. J. Biomed. Mater. Res. Part A 2010, 92, 1382–1391. [Google Scholar] [CrossRef]
- Ozboyaci, M.; Kokh, D.B.; Corni, S.; Wade, R.C. Modeling and simulation of protein-surface interactions: Achievements and challenges. Q. Rev. Biophys. 2016, 49, e4. [Google Scholar] [CrossRef] [PubMed]
- Raffaini, G.; Ganazzoli, F. Protein adsorption on biomaterial and nanomaterial surfaces: A molecular modeling approach to study non-covalent interactions. J. Appl. Biomater. Biomech. 2010, 8, 135–145. [Google Scholar] [CrossRef] [PubMed]
- Raffaini, G.; Ganazzoli, F. Understanding the performance of biomaterials through molecular modeling: Crossing the bridge between their intrinsic properties and the surface adsorption of proteins. Macromol. Biosci. 2007, 7, 552–566. [Google Scholar] [CrossRef]
- YazdanYar, A.; Aschauer, U.; Bowen, P. Interaction of biologically relevant ions and organic molecules with titanium oxide (rutile) surfaces: A review on molecular dynamics studies. Colloids Surf. B 2018, 161, 563–577. [Google Scholar] [CrossRef]
- Raffaini, G.; Ganazzoli, F. Simulation study of the interaction of some albumin subdomains with a flat graphite surface. Langmuir 2003, 19, 3403–3412. [Google Scholar] [CrossRef]
- Raffaini, G.; Ganazzoli, F. Adsorption of charged albumin subdomains on a graphite surface. J. Biomed. Mater. Res. A 2006, 76, 638–645. [Google Scholar] [CrossRef]
- Raffaini, G.; Ganazzoli, F. Molecular modelling of protein adsorption on the surface of titanium dioxide polymorphs. Philos. Trans. R. Soc. A 2012, 370, 1444–1462. [Google Scholar] [CrossRef]
- Raffaini, G.; Melone, L.; Punta, C. Understanding the topography effects on competitive adsorption on a nanosized anatase crystal: A molecular dynamics study. Chem. Commun. 2013, 49, 7581–7583. [Google Scholar] [CrossRef] [PubMed]
- Fiorati, A.; Gambarotti, C.; Melone, L.; Pastori, N.; Punta, C.; Raffaini, G.; Truscello, A. Advanced Synthetic Techniques. In Green Synthetic Approaches for Biologically Relevant Heterocycles, 2nd ed.; Brahmachari, G., Ed.; Elsevier: Amsterdam, The Netherlands, 2020; Volume 1, pp. 189–206. ISBN 9780128205860. Available online: https://www.elsevier.com/books-and-journals (accessed on 8 March 2021).
- Mashatooki, M.H.; Ebrahimzadeh, A.R.; Sardroodi, J.J.; Abbasi, A. Investigation of TiO2 anatase (101), (100) and (110) facets as immobilizer for a potential anticancer RNA aptamer: A classical molecular dynamics simulation. Mol. Simul. 2019, 45, 849–858. [Google Scholar] [CrossRef]
- Yan, L.; Chen, H.Z.; Jing, C.Y. TiO2 Facets Shaped by Concentration-Dependent Surface Diffusion of Dopamine. J. Phys. Chem. Lett. 2019, 10, 898–903. [Google Scholar] [CrossRef] [PubMed]
- Materials Studio BIOVIA; Discovery Studio and Accelrys Inc. InsightII 2000; Accelrys Inc.: San Diego, CA, USA, 2000; Available online: http://www.accelrys.com (accessed on 4 April 2021).
- Young, D. Computational Chemistry: A Practical Guide for Applying Techniques to Real World Problems; John Wiley & Sons, Inc.: New York, NY, USA, 2001. [Google Scholar]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Raffaini, G.; Ganazzoli, F. Molecular dynamics simulation of the adsorption of a fibronectin module on a graphite surface. Langmuir 2004, 20, 3371–3378. [Google Scholar] [CrossRef]
- Barinov, N.A.; Prokhorov, V.V.; Dubrovin, E.V.; Klinov, D.V. AFM visualization at a single-molecule level of denaturated states of proteins on graphite. Colloids Surf. B 2016, 146, 777–784. [Google Scholar] [CrossRef]
- Catauro, M.; Barrino, F.; Dal Poggetto, G.; Milazzo, M.; Blanco, I.; Ciprioti, S.V. Structure, drug absorption, bioactive and antibacterial properties of sol-gel SiO2/ZrO2 materials. Ceram. Int. 2020, 46, 29459–29465. [Google Scholar] [CrossRef]
- Catauro, M.; Pagliuca, C.; Lisi, L.; Ruoppolo, G. Synthesis of alkoxide-derived V-Nb catalysts prepared by sol-gel route. Thermochim. Acta 2002, 381, 65–72. [Google Scholar] [CrossRef]
- Catauro, M.; Bollino, F.; Papale, F.; Ferrara, C.; Mustarelli, P. Silica-polyethylene glycol hybrids synthesized by sol-gel: Biocompatibility improvement of titanium implants by coating. Mater. Sci. Eng. C 2015, 55, 118–125. [Google Scholar] [CrossRef]
- Catauro, M.; Verardi, D.; Melisi, D.; Belotti, F.; Mustarelli, P. Novel sol-gel organic-inorganic hybrid materials for drug delivery. J. Appl. Biomater. Funct. Mater. 2010, 8, 42–51. [Google Scholar]
- Catauro, M.; Blanco, I.; De Santis, R.; Russo, T.; Crescente, G. Synthesis of Glass Nanocomposite Powders: Structure, Thermal, and Antibacterial Study. Macromol. Symp. 2021, 395, 2000200. [Google Scholar] [CrossRef]

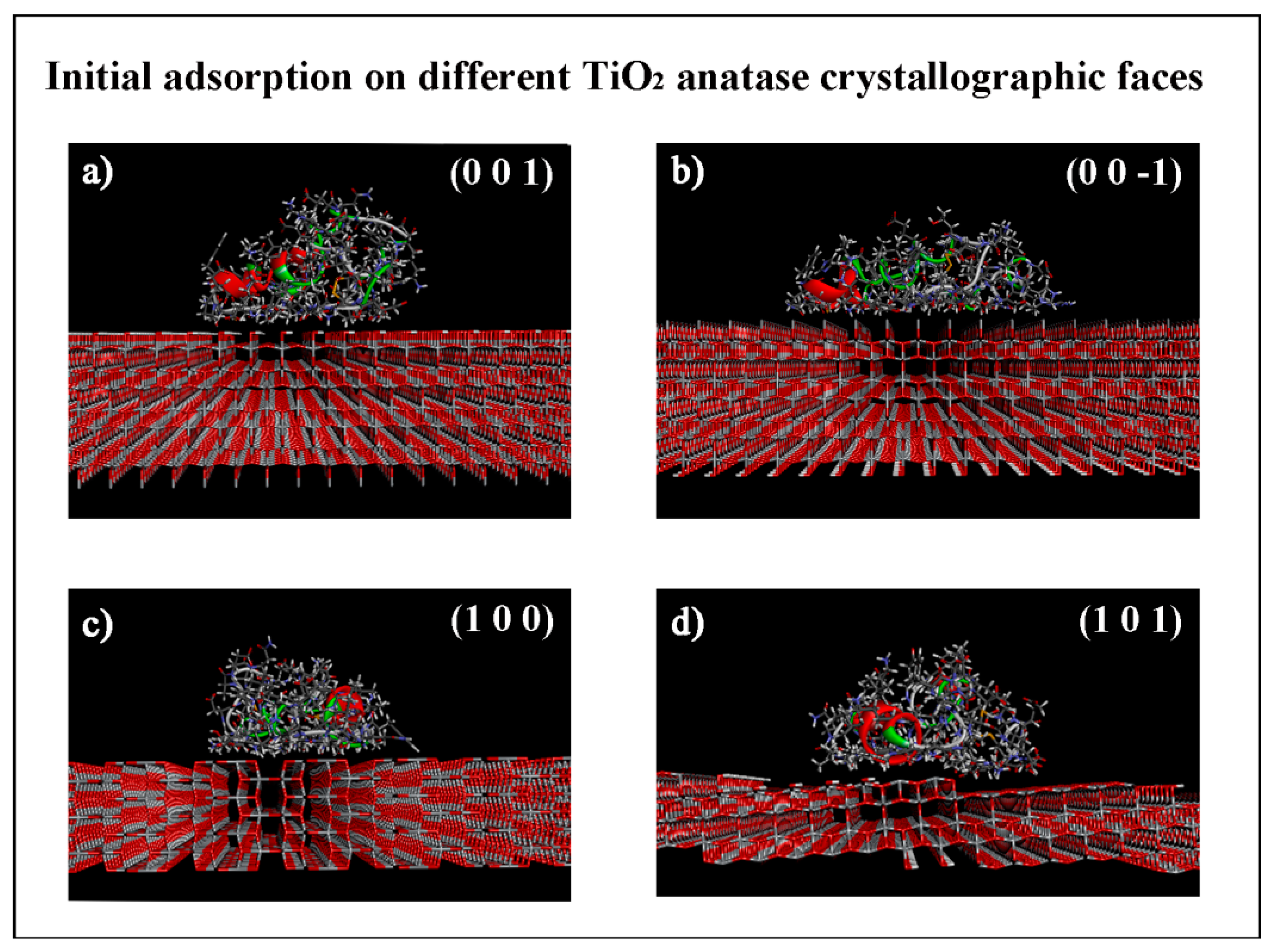
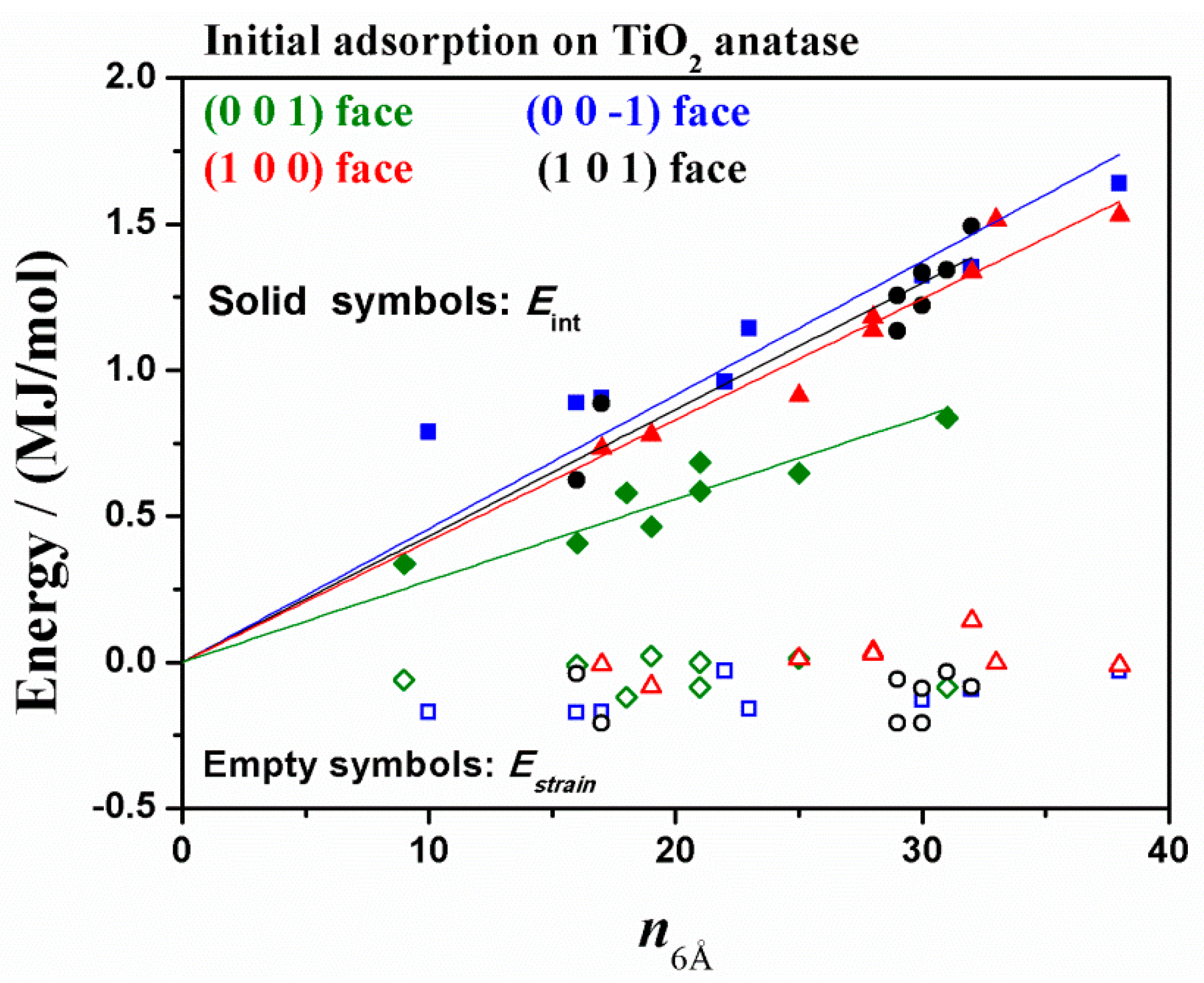
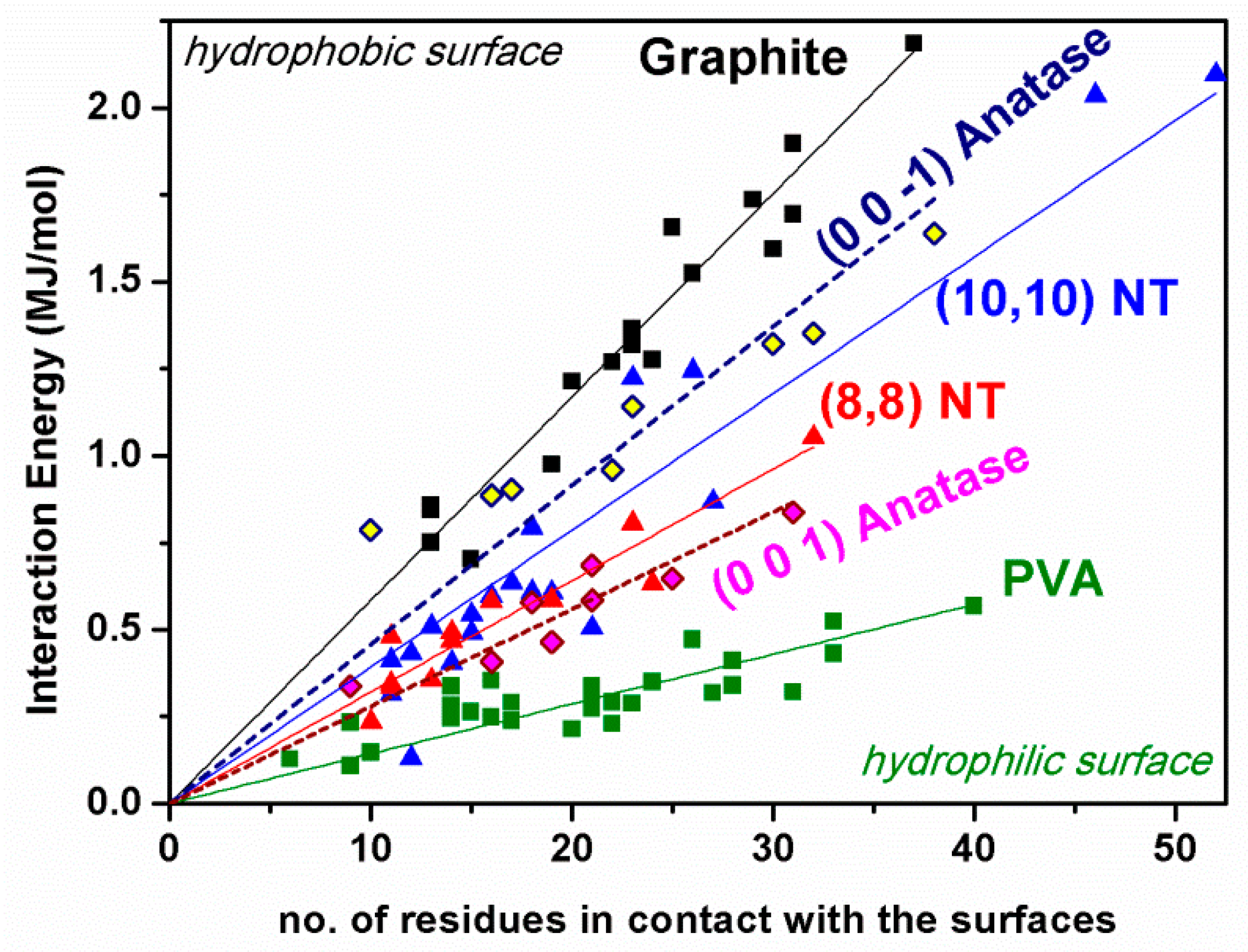
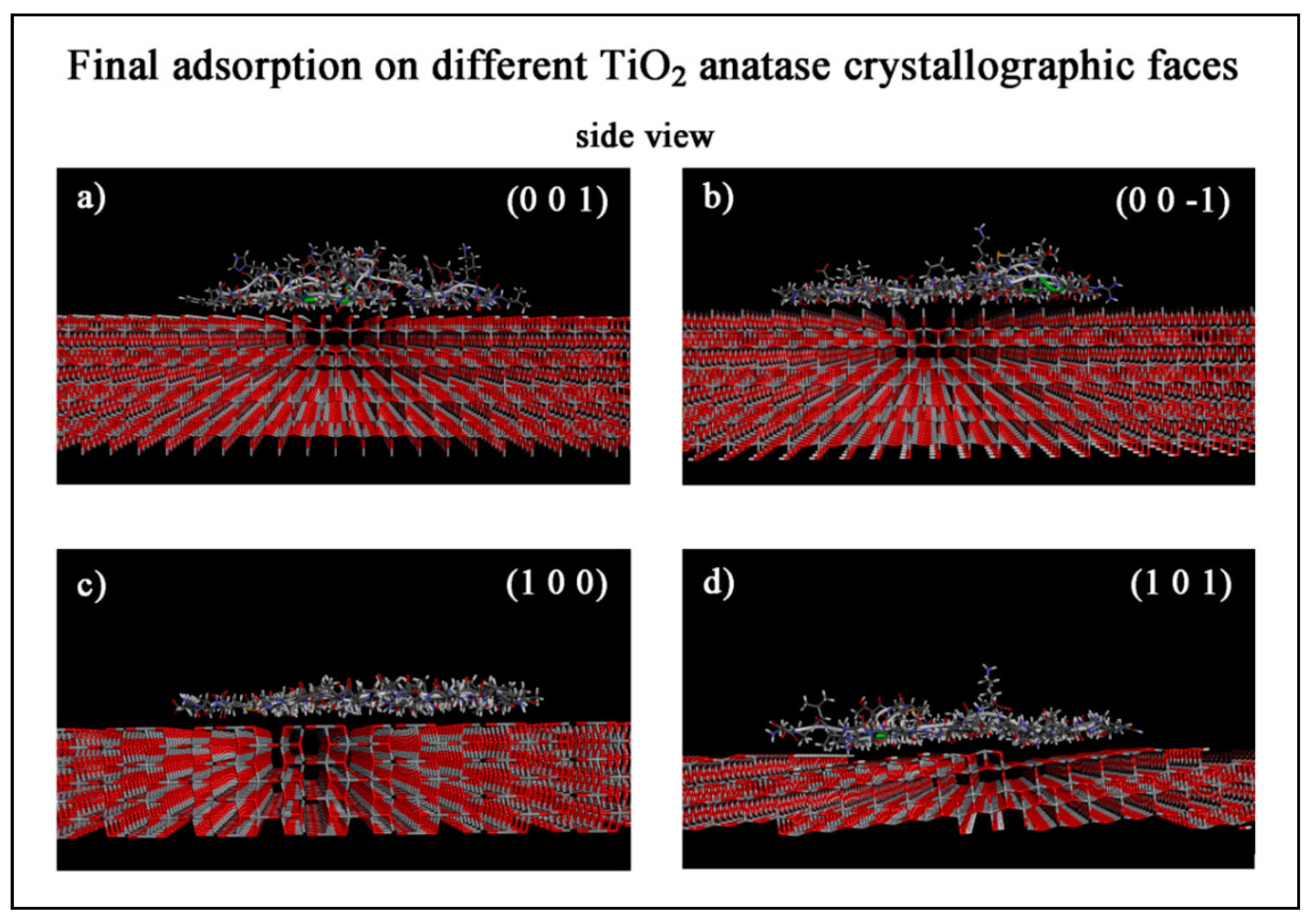



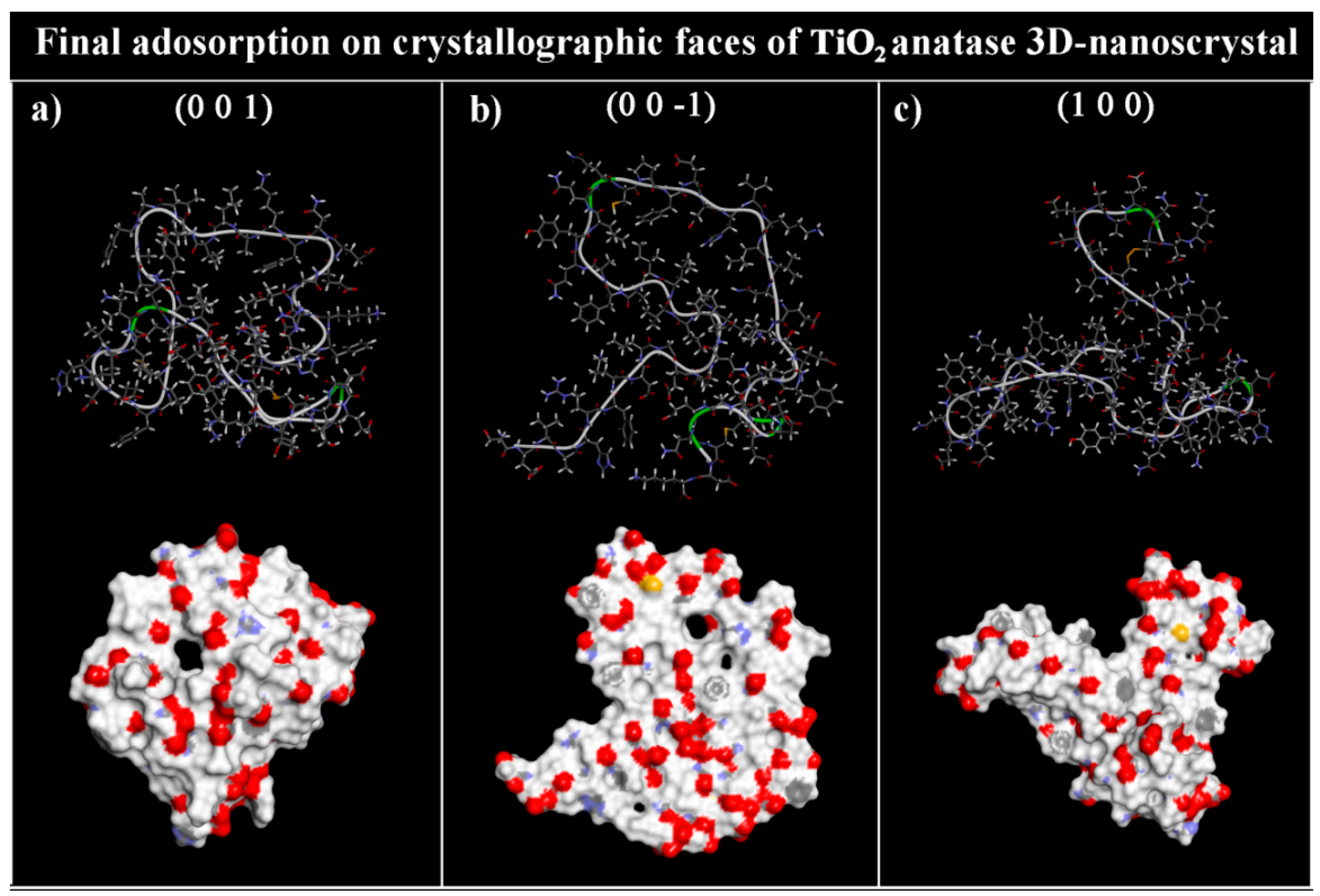
| TiO2 Anatase Crystallographic Face | Initial Eint (kJ/mol) | Estrain (kJ/mol) | Final Eint (kJ/mol) | Estrain (kJ/mol) | Eintrinsic (kJ/mol) | Rg (Å) | Volume (Å3) | SASA (Å2) |
|---|---|---|---|---|---|---|---|---|
| (0 0 1) | 837.6 | −86.3 | 2000.0 | −561.6 | 54.1 | 14.2 | 7133 | 3699 |
| (0 0 −1) | 1639.2 | −32.7 | 2738.5 | −104.4 | 54.8 | 20.0 | 7055 | 4541 |
| (1 0 0) | 1529.8 | 12.2 | 3103.0 | 228.0 | 51.7 | 21.3 | 6854 | 5005 |
| (1 0 1) | 1491.2 | −84.3 | 2861.2 | −8.158 | 54.0 | 17.6 | 6961 | 4613 |
| TiO2 Anatase Crystallographic Face | Initial Eint (kJ/mol) | Estrain (kJ/mol) | Final Eint (kJ/mol) | Estrain (kJ/mol) | Eintrinsic (kJ/mol) | Rg (Å) | Volume (Å3) | SASA (Å2) |
|---|---|---|---|---|---|---|---|---|
| (0 0 1) | 336.6 | −60.6 | 1959.8 | −578.7 | 56.0 | 13.5 | 7433 | 3466 |
| (0 0 −1) | 787.2 | −173.0 | 2574.4 | −311.4 | 59.9 | 15.4 | 7290 | 3873 |
| (1 0 0) | 731.2 | −7.98 | 2716.0 | −209.5 | 59.0 | 15.1 | 7127 | 4060 |
| (1 0 1) | 622.4 | −39.8 | 2853.0 | −75.65 | 54.9 | 16.8 | 7012 | 4506 |
| Crystallographic TiO2 Face | Rg (Å) | Volume (Å3) | Solvent-Accessible Surface Area (Å2) |
|---|---|---|---|
| (0 0 1) | 13.7 | 7388 | 3668 |
| (0 0 −1) | 16.4 | 7053 | 4473 |
| (1 0 0) | 16.3 | 7137 | 4119 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Raffaini, G. Surface Chemistry, Crystal Structure, Size and Topography Role in the Albumin Adsorption Process on TiO2 Anatase Crystallographic Faces and Its 3D-Nanocrystal: A Molecular Dynamics Study. Coatings 2021, 11, 420. https://doi.org/10.3390/coatings11040420
Raffaini G. Surface Chemistry, Crystal Structure, Size and Topography Role in the Albumin Adsorption Process on TiO2 Anatase Crystallographic Faces and Its 3D-Nanocrystal: A Molecular Dynamics Study. Coatings. 2021; 11(4):420. https://doi.org/10.3390/coatings11040420
Chicago/Turabian StyleRaffaini, Giuseppina. 2021. "Surface Chemistry, Crystal Structure, Size and Topography Role in the Albumin Adsorption Process on TiO2 Anatase Crystallographic Faces and Its 3D-Nanocrystal: A Molecular Dynamics Study" Coatings 11, no. 4: 420. https://doi.org/10.3390/coatings11040420
APA StyleRaffaini, G. (2021). Surface Chemistry, Crystal Structure, Size and Topography Role in the Albumin Adsorption Process on TiO2 Anatase Crystallographic Faces and Its 3D-Nanocrystal: A Molecular Dynamics Study. Coatings, 11(4), 420. https://doi.org/10.3390/coatings11040420