An Active Gelatin Coating Containing Eugenol and Vacuum Delays the Decay of Chinese Seabass (Lateolabrax maculatus) Fillets during Cold Storage: A Microbiome Perspective
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials and Reagents
2.2. Preparation of the Active Gelatin–Eugenol Coating Solution
2.3. Preparation of the Seabass and Sample Treatments
2.4. Determination of the Total Volatile Basic Nitrogen
2.5. Characterization of the Microbiota Based on Culture-Dependent Methods
2.6. Characterization of the Microbiota Based on Culture-Independent Methods
2.6.1. DNA Extraction and PCR Amplification
2.6.2. Illumina MiSeq Sequencing and Data Processing
2.7. Data Processing and Taxonomic Classification
2.8. Statistical Analysis
3. Results and Discussions
3.1. TVB-N Values
3.2. Analysis of High-Throughput Sequencing Data
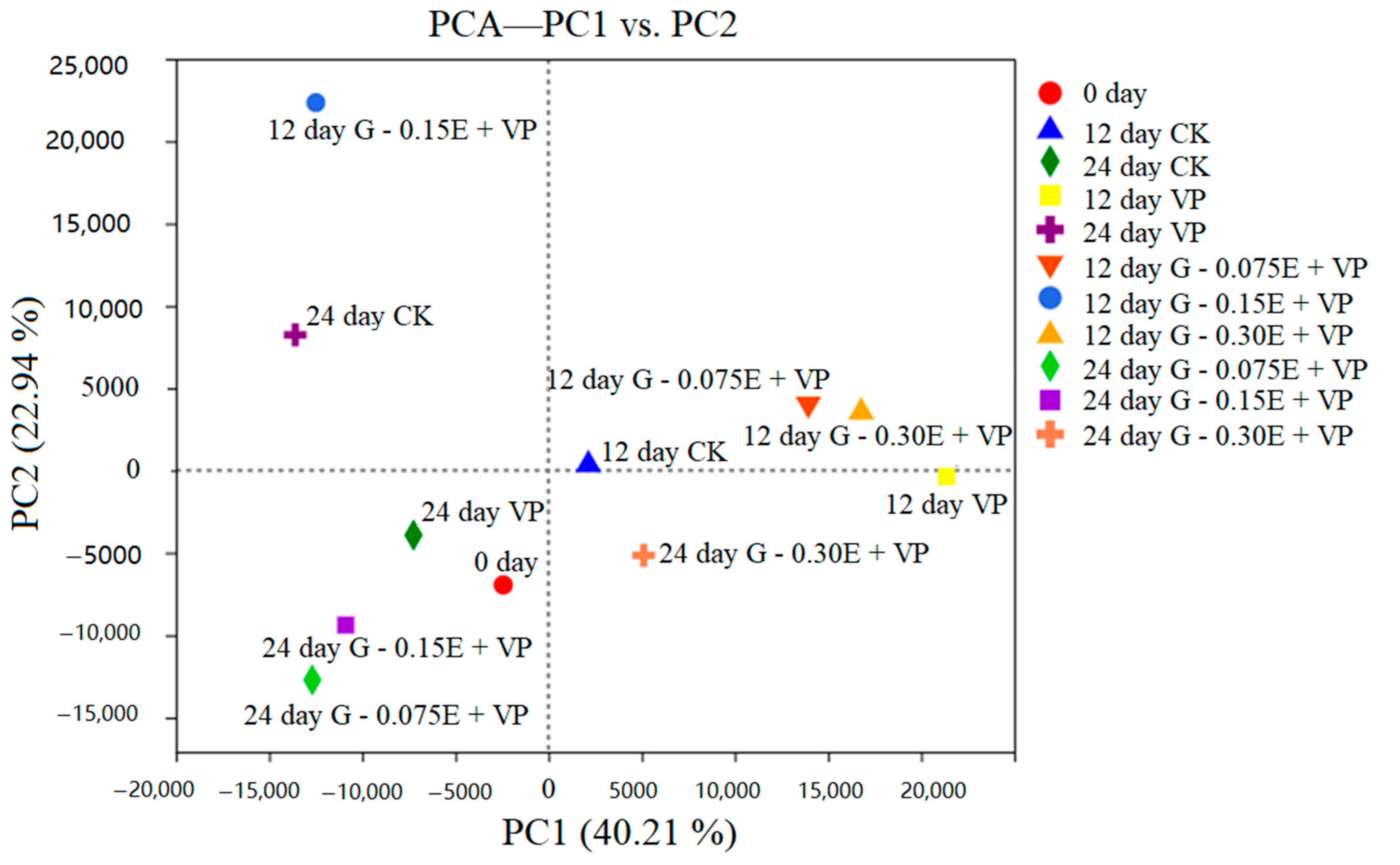
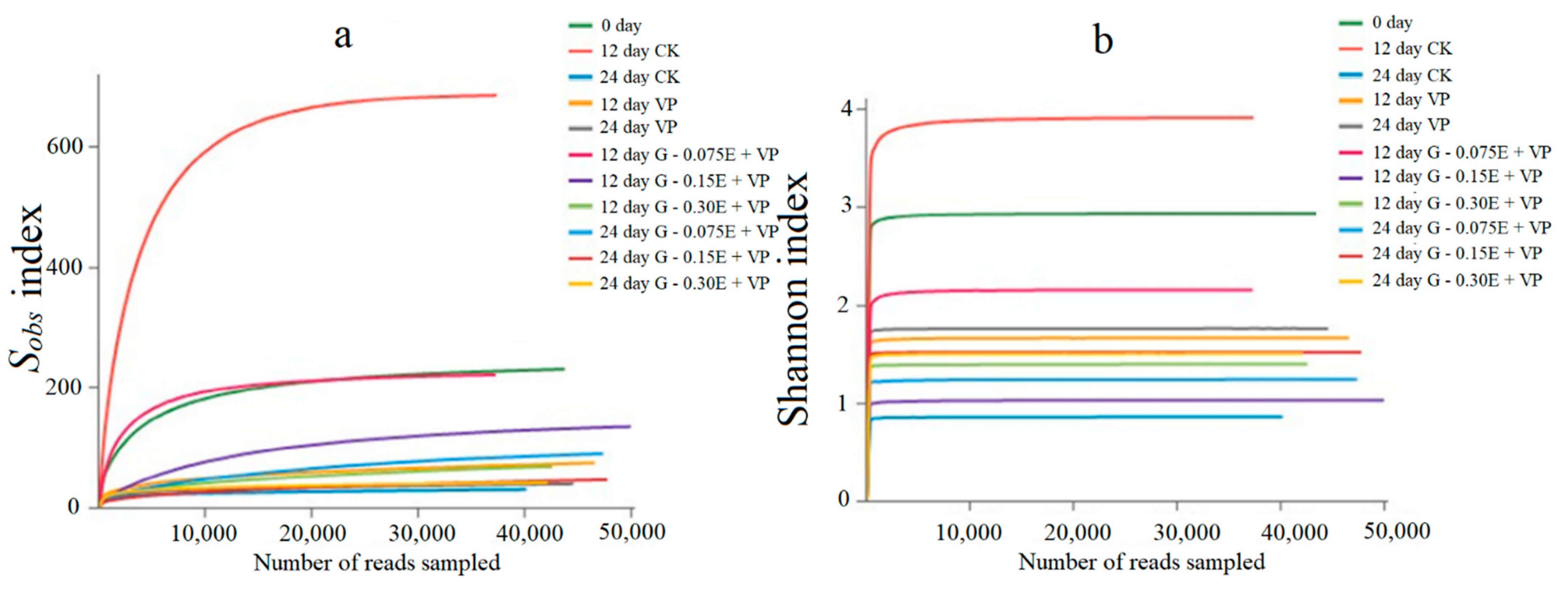
3.3. Bacterial Diversity Analysis
3.4. Analysis of the Species Community Composition
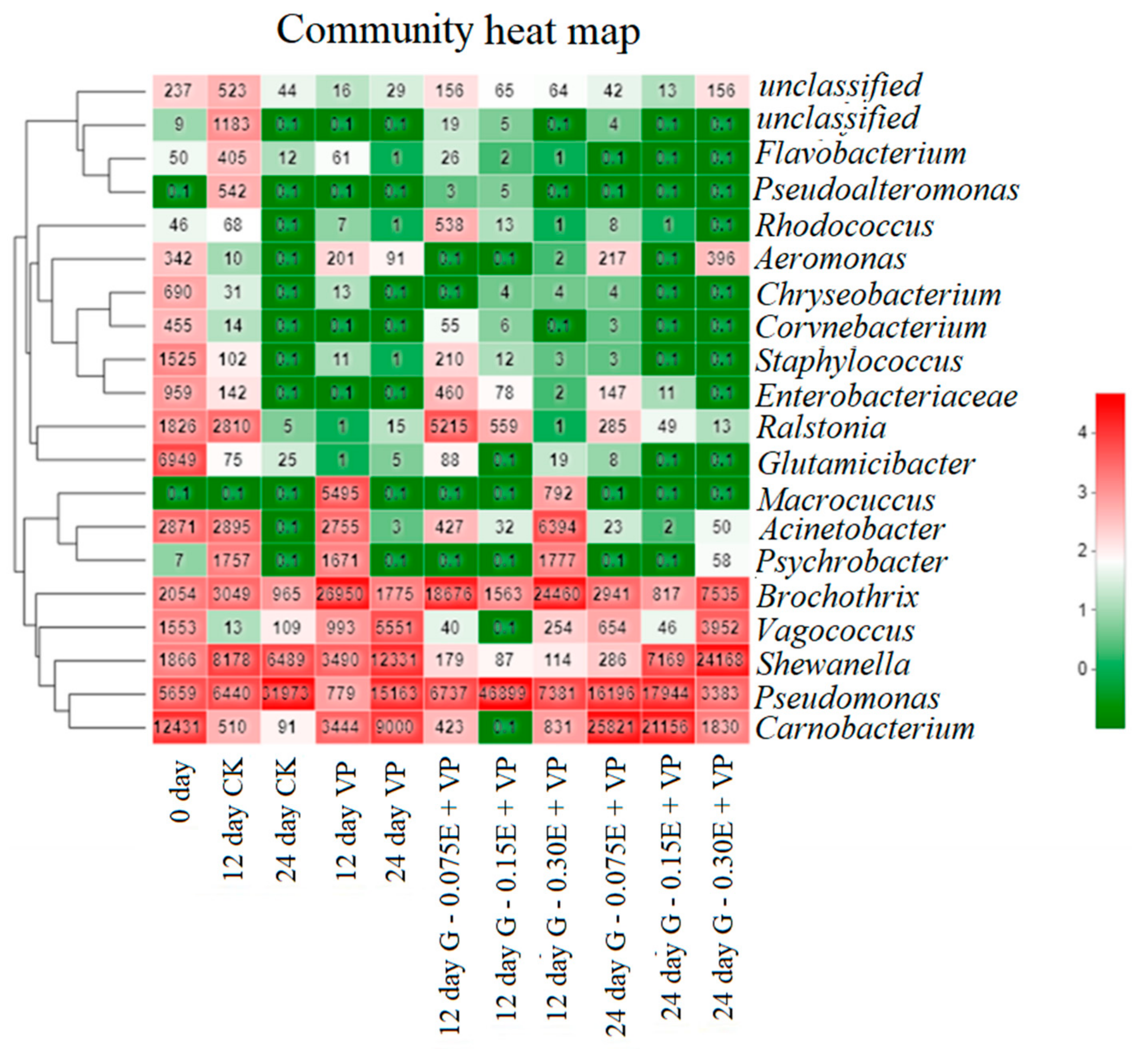
3.5. Microbial Community Succession Heat Map
3.6. Culture-Dependent Analyses of the Microbiota
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, W.H.; Dong, H.B.; Sun, Y.X.; Cao, M.; Duan, Y.F.; Li, H.; Liu, Q.S.; Gu, Q.H.; Zhang, J.S. The efficacy of eugenol and tricaine methanesulphonate as anaesthetics for juvenile Chinese sea bass (Lateolabrax maculatus) during simulated transport. J. Appl. Ichthyol. 2019, 35, 551–557. [Google Scholar] [CrossRef]
- Chaijan, S.; Panpipat, W.; Panya, A.; Cheong, L.Z.; Chaijan, M. Preservation of chilled Asian sea bass (Lates calcarifer) steak by whey protein isolate coating containing polyphenol extract from ginger, lemongrass, or green tea. Food Control 2020, 118, 107400. [Google Scholar] [CrossRef]
- Martínez, O.; Salmerón, J.; Epelde, L.; Vicente, M.S.; de Vega, C. Quality enhancement of smoked sea bass (Dicentrarchus labrax) fillets by adding resveratrol and coating with chitosan and alginate edible films. Food Control 2018, 85, 168–176. [Google Scholar] [CrossRef]
- Cai, L.; Cao, A.; Bai, F.; Li, J. Effect of ε-polylysine in combination with alginate coating treatment on physicochemical and microbial characteristics of Japanese sea bass (Lateolabrax japonicas) during refrigerated storage. LWT Food Sci. Technol. 2015, 62, 1053–1059. [Google Scholar] [CrossRef]
- Teixeira, B.; Fidalgo, L.G.; Mendes, R.; Costa, G.D.; Cordeiro, C.; Marques, A.; Saraiva, J.A.; Nunes, M.L. Effect of high pressure processing in the quality of sea bass (Dicentrarchus labrax) fillets: Pressurization rate, pressure level and holding time. Innov. Food Sci. Emerg. Technol. 2014, 22, 31–39. [Google Scholar] [CrossRef]
- Qiu, X.; Chen, S.; Liu, G.; Yang, Q. Quality enhancement in the Japanese sea bass (Lateolabrax japonicas) fillets stored at 4 °C by chitosan coating incorporated with citric acid or licorice extract. Food Chem. 2014, 162, 156–160. [Google Scholar] [CrossRef]
- Sun, L.; Sun, J.; Thavaraj, P.; Yang, X.; Guo, Y. Effects of thinned young apple polyphenols on the quality of grass carp (Ctenopharyngodon idellus) surimi during cold storage. Food Chem. 2017, 224, 372–381. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, D.; Lv, J.; Li, Q.; Kong, C.; Luo, Y. Effect of cinnamon essential oil on bacterial diversity and shelf-life in vacuum-packaged common carp (Cyprinus carpio) during refrigerated storage. Int. J. Food Microbiol. 2017, 249, 1–8. [Google Scholar] [CrossRef]
- Yu, D.; Wu, L.; Regenstein, J.; Jiang, Q.; Yang, F.; Xu, Y.; Xia, W. Recent advances in quality retention of non-frozen fish and fishery products: A review. Crit. Rev. Food Sci. Nutr. 2019, 60, 1–13. [Google Scholar] [CrossRef]
- Carrión-Granda, X.; Fernández-Pan, I.; Rovira, J.; Maté, J. Effect of antimicrobial edible coatings and modified atmosphere packaging on the microbiological quality of cold stored hake (Merluccius merluccius) fillets. J. Food Qual. 2018, 2018, 1–12. [Google Scholar] [CrossRef]
- Zhao, J.; Lv, W.; Wang, J.; Li, J.; Liu, X.; Zhu, J. Effects of tea polyphenols on the post-mortem integrity of large yellow croaker (Pseudosciaena crocea) fillet proteins. Food Chem. 2013, 141, 2666–2674. [Google Scholar] [CrossRef] [PubMed]
- Alotaibi, S.; Tahergorabi, R. Development of a sweet potato starch-based coating and its effect on quality attributes of shrimp during refrigerated storage. LWT Food Sci. Technol. 2018, 88, 203–209. [Google Scholar] [CrossRef]
- Alparslan, Y.; Baygar, T.N. Effect of chitosan film coating combined with orange peel essential oil on the shelf life of deepwater pink shrimp. Food Bioprocess. Technol. 2017, 10, 842–853. [Google Scholar] [CrossRef]
- Alparslan, Y.; Yapici, H.H.; Metin, C.; Baygar, T.; Gunlu, A.; Baygar, T. Quality assessment of shrimps preserved with orange leaf essential oil incorporated gelatin. LWT Food Sci. Technol. 2016, 72, 457–466. [Google Scholar] [CrossRef]
- Nilsuwan, K.; Benjakul, S.; Prodpran, T.; Caba, K.D.I. Fish gelatin monolayer and bilayer films incorporated with epigallocatechin gallate: Properties and their use as pouches for storage of chicken skin oil. Food Hydrocoll. 2019, 89, 783–791. [Google Scholar] [CrossRef]
- Nilsuwan, K.; Guerrero, P.; Caba, K.D.I.; Benjakul, S.; Prodpran, T. Properties and application of bilayer films based on poly (lactic acid) and fish gelatin containing epigallocatechin gallate fabricated by thermo-compression molding. Food Hydrocoll. 2020, 105, 105792. [Google Scholar] [CrossRef]
- Suderman, N.; Isa, M.I.N.; Sarbon, N.M. The effect of plasticizers on the functional properties of biodegradable gelatin-based film: A review. Food Biosci. 2018, 24, 111–119. [Google Scholar] [CrossRef]
- Kan, J.; Liu, J.; Yong, H.; Liu, Y.; Qin, Y.; Liu, J. Development of active packaging based on chitosan-gelatin blend films functionalized with Chinese hawthorn (Crataegus pinnatifida) fruit extract. Int. J. Biol. Macromol. 2019, 140, 384–392. [Google Scholar] [CrossRef]
- Staroszczyk, H.; Kusznierewicz, B.; Malinowska-Pańczyk, E.; Sinkiewicz, I.; Gottfried, K.; Kołodziejska, I. Fish gelatin films containing aqueous extracts from phenolic-rich fruit pomace. LWT Food Sci. Technol. 2020, 117, 108613. [Google Scholar] [CrossRef]
- Yeddes, W.; Djebali, K.; Wannes, W.A.; Horchani-Naifer, K.; Hammami, M.; Younes, I.; Tounsi, M.S. Gelatin-chitosan-pectin films incorporated with rosemary essential oil: Optimized formulation using mixture design and response surface methodology. Int. J. Biol. Macromol. 2020, 154, 92–103. [Google Scholar] [CrossRef]
- Jamróz, E.; Juszczak, L.; Kucharek, M. Investigation of the physical properties, antioxidant and antimicrobial activity of ternary potato starch-furcellaran-gelatin films incorporated with lavender essential oil. Int. J. Biol. Macromol. 2018, 114, 1094–1101. [Google Scholar] [CrossRef] [PubMed]
- Bermúdez-Oria, A.; Rodríguez-Gutiérrez, G.; Vioque, B.; Rubio-Senent, F.; Fernández-Bolaños, J. Physical and functional properties of pectin-fish gelatin films containing the olive phenols hydroxytyrosol and 3,4-dihydroxyphenylglycol. Carbohydr. Polym. 2017, 178, 368–377. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.; Rhim, J.W. Preparation of antimicrobial and antioxidant gelatin/curcumin composite films for active food packaging application. Colloids Surf. B 2020, 188, 110761. [Google Scholar] [CrossRef] [PubMed]
- Neira, L.M.; Martucci, J.F.; Stejskal, N.; Ruseckaite, R.A. Time-dependent evolution of properties of fish gelatin edible films enriched with carvacrol during storage. Food Hydrocoll. 2019, 94, 304–310. [Google Scholar] [CrossRef]
- Ashrafudoulla, M.; Mizan, M.F.R.; Ha, A.J.; Park, S.H.; Ha, S.D. Antibacterial and antibiofilm mechanism of eugenol against antibiotic resistance Vibrio parahaemolyticus. Food Microbiol. 2020, 91, 103500. [Google Scholar] [CrossRef]
- Qian, W.; Sun, Z.; Wang, T.; Yang, M.; Liu, M.; Zhang, J.; Li, Y. Antimicrobial activity of eugenol against carbapenem-resistant Klebsiella pneumoniae and its effect on biofilms. Microb. Pathog. 2020, 139, 103924. [Google Scholar] [CrossRef]
- Wang, Q.; Zhang, L.; Ding, W. Eugenol nanocapsules embedded with gelatin-chitosan for chilled pork preservation. Int. J. Biol. Macromol. 2020, 158, 837–844. [Google Scholar] [CrossRef]
- Cai, L.; Dai, Y.; Cao, A.; Cao, M. The effects of CS@Fe3O4 nanoparticles combined with microwave or far infrared thawing on microbial diversity of red seabream (Pagrus major) fillets based on high-throughput sequencing. Food Microbiol. 2020, 91, 103511. [Google Scholar] [CrossRef]
- Jagadeesan, B.; Gerner-Smidt, P.; Allard, M.W.; Leuillet, S.; Winkler, A.; Xiao, Y.; Chaffron, S.; Vossen, J.V.D.; Tang, S.; Katase, M.; et al. The use of next generation sequencing for improving food safety: Translation into practice. Food Microbiol. 2019, 79, 96–115. [Google Scholar] [CrossRef]
- Parlapani, F.F.; Michailidou, S.; Pasentsis, K.; Argiriou, A.; Krey, G.; Boziaris, I.S. A meta-barcoding approach to assess and compare the storage temperature-dependent bacterial diversity of gilt-head sea bream (Sparus aurata) originating from fish farms from two geographically distinct areas of Greece. Int. J. Food Microbiol. 2018, 278, 36–43. [Google Scholar] [CrossRef]
- Huang, H.; Sun, W.; Xiong, G.; Shi, L.; Jiao, C.; Wu, W.; Li, X.; Qiao, Y.; Liao, L.; Ding, A.; et al. Effects of HVEF treatment on microbial communities and physicochemical properties of catfish fillets during chilled storage. LWT Food Sci. Technol. 2020, 131, 109667. [Google Scholar] [CrossRef]
- Huang, Z.; Liu, X.; Jia, S.; Zhang, L.; Luo, Y. The effect of essential oils on microbial composition and quality of grass carp (Ctenopharyngodon idellus) fillets during chilled storage. Int. J. Food Microbiol. 2018, 266, 52–59. [Google Scholar] [CrossRef] [PubMed]
- Balti, R.; Mansour, M.B.; Zayoud, N.; Balc’h, R.L.; Brodu, N.; Arhaliass, A.; Massé, A. Active exopolysaccharides based edible coatings enriched with red seaweed (Gracilaria gracilis) extract to improve shrimp preservation during refrigerated storage. Food Biosci. 2020, 34, 100522. [Google Scholar] [CrossRef]
- Jia, S.; Huang, Z.; Lei, Y.; Zhang, L.; Li, Y.; Luo, Y. Application of Illumina-MiSeq high throughput sequencing and culture-dependent techniques for the identification of microbiota of silver carp (Hypophthalmichthys molitrix) treated by tea polyphenols. Food Microbiol. 2018, 76, 52–61. [Google Scholar] [CrossRef]
- Zang, J.; Xu, Y.; Xia, W.; Yu, D.; Gao, P.; Jiang, Q.; Yang, F. Dynamics and diversity of microbial community succession during fermentation of Suan yu, a Chinese traditional fermented fish, determined by high throughput sequencing. Food Res. Int. 2018, 111, 565–573. [Google Scholar] [CrossRef]
- Rong, C.; Ling, Z.; Huihui, S.; Qi, L. Characterization of microbial community in high-pressure treated oysters by high-throughput sequencing technology. Innov. Food Sci. Emerg. Technol. 2018, 45, 241–248. [Google Scholar] [CrossRef]
- Zhou, Q.; Li, P.; Fang, S.; Mei, J.; Xie, J. Preservative effects of gelatin active coating containing eugenol and higher CO2 concentration modified atmosphere packaging on Chinese sea bass (Lateolabrax maculatus) during superchilling (−0.9 °C) storage. Molecules 2020, 25, 871. [Google Scholar] [CrossRef]
- Zhou, Q.; Li, P.; Fang, S.; Liu, W.; Mei, J.; Xie, J. Preservative effects of gelatin active coating enriched with eugenol emulsion on Chinese seabass (Lateolabrax maculatus) during superchilling (−0.9 °C) storage. Coatings 2019, 9, 489. [Google Scholar] [CrossRef]
- Sun, X.; Guo, X.; Ji, M.; Wu, J.; Zhu, W.; Wang, J.; Cheng, C.; Chen, L.; Zhang, Q. Preservative effects of fish gelatin coating enriched with CUR/βCD emulsion on grass carp (Ctenopharyngodon idellus) fillets during storage at 4 °C. Food Chem. 2019, 272, 643–652. [Google Scholar] [CrossRef]
- Wang, W.; Li, M.; Li, H.; Liu, X.; Guo, T.; Zhang, G.; Xiong, Y. A renewable intelligent colorimetric indicator based on polyaniline for detecting freshness of tilapia. Packag. Technol. Sci. 2018, 31, 133–140. [Google Scholar] [CrossRef]
- Liu, X.; Li, D.; Li, K.; Luo, Y. Monitoring bacterial communities in ε-Polylysine-treated bighead carp (Aristichthys nobilis) fillets using culture-dependent and culture-independent techniques. Food Microbiol. 2018, 76, 257–266. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhang, Y.; Li, D.; Luo, Y. Characterization of the microbiota in lightly salted bighead carp (Aristichthys nobilis) fillets stored at 4 °C. Food Microbiol. 2017, 62, 106–111. [Google Scholar] [CrossRef] [PubMed]
- EzBioCloud Database Home Page. Available online: http://www.eztaxon.org (accessed on 28 January 2021).
- UPARSE OTU Clustering. Available online: http://drive5.com/uparse (accessed on 28 January 2021).
- RDP Classifier Home Page. Available online: http://rdp.cme.msu.edu/ (accessed on 28 January 2021).
- Fast Tree-Comparison Tools. Available online: http://www.microbesonline.org/fasttree (accessed on 28 January 2021).
- Liu, X.; Huang, Z.; Jia, S.; Zhang, J.; Li, K.; Luo, Y. The roles of bacteria in the biochemical changes of chill-stored bighead carp (Aristichthys nobilis): Proteins degradation, biogenic amines accumulation, volatiles production, and nucleotides catabolism. Food Chem. 2018, 255, 174–181. [Google Scholar] [CrossRef] [PubMed]
- Aponte, M.; Anastasio, A.; Marrone, R.; Mercogliano, R.; Peruzy, M.F.; Murru, N. Impact of gaseous ozone coupled to passive refrigeration system to maximize shelf-life and quality of four different fresh fish products. LWT Food Sci. Technol. 2018, 93, 412–419. [Google Scholar] [CrossRef]
- Ojagh, S.M.; Rezaei, M.; Razavi, S.H.; Hosseini, S.M.H. Effect of chitosan coatings enriched with cinnamon oil on the quality of refrigerated rainbow trout. Food Chem. 2010, 120, 193–198. [Google Scholar] [CrossRef]
- Cai, L.; Wu, X.; Li, X.; Zhong, K.; Li, Y.; Li, J. Effects of different freezing treatments on physicochemical responses and microbial characteristics of Japanese sea bass (Lateolabrax japonicas) fillets during refrigerated storage. LWT Food Sci. Technol. 2014, 59, 122–129. [Google Scholar] [CrossRef]
- Mallon, C.A.; Elsas, J.D.V.; Salles, J.F. Microbial invasions: The process, patterns, and mechanisms. Trends Microbiol. 2015, 23, 719–729. [Google Scholar] [CrossRef]
- Li, P.; Peng, Y.; Mei, J.; Xie, J. Effects of microencapsulated eugenol emulsions on microbiological, chemical and organoleptic qualities of farmed Japanese sea bass (Lateolabrax japonicus) during cold storage. LWT Food Sci. Technol. 2020, 118, 108831. [Google Scholar] [CrossRef]
- Zheng, K.; Xiao, S.; Li, W.; Wang, W.; Chen, H.; Yang, F.; Qin, C. Chitosan-acorn starch-eugenol edible film: Physico-chemical, barrier, antimicrobial, antioxidant and structural properties. Int. J. Biol. Macromol. 2019, 135, 344–352. [Google Scholar] [CrossRef]
- Veeck, A.P.D.L.; Daniel, A.P.; Klein, B.; Quatrin, A.; Rezer, A.P.D.S.; Milani, L.I.G.; Zeppenfeld, C.C.; Cunha, M.A.D.; Heldwein, C.G.; Heinzmann, B.M. Chemical, microbiological, and sensory parameters during the refrigerated storage of silver catfish (Rhamdia quelen) exposed in vivo to the essential oil of Lippia alba. J. Food Sci. Technol. 2018, 55, 1416–1425. [Google Scholar] [CrossRef]
- Volpe, M.G.; Siano, F.; Paolucci, M.; Sacco, A.; Sorrentino, A.; Malinconico, M.; Varricchio, E. Active edible coating effectiveness in shelf-life enhancement of trout (Oncorhynchusmykiss) fillets. LWT Food Sci. Technol. 2015, 60, 615–622. [Google Scholar] [CrossRef]
- Zhao, X.; Wu, J.E.; Chen, L.; Yang, H. Effect of vacuum impregnated fish gelatin and grape seed extract on metabolite profiles of tilapia (Oreochromis niloticus) fillets during storage. Food Chem. 2019, 293, 418–428. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, A.; Rodiles, A.; Merrifield, D.L.; Chandra, G.; Ghosh, K. Exploring intestinal microbiome composition in three Indian major carps under polyculture system: A high-throughput sequencing based approach. Aquaculture 2020, 524, 735206. [Google Scholar] [CrossRef]
- Huyben, D.; Vidaković, A.; Werner Hallgren, S.; Langeland, M. High-throughput sequencing of gut microbiota in rainbow trout (Oncorhynchus mykiss) fed larval and pre-pupae stages of black soldier fly (Hermetia illucens). Aquaculture 2019, 500, 485–491. [Google Scholar] [CrossRef]
- Odeyemi, O.A.; Burke, C.M.; Bolch, C.C.J.; Stanley, R. Spoilage microbial community profiling by 16S rRNA amplicon sequencing of modified atmosphere packaged live mussels stored at 4 °C. Food Res. Int. 2019, 121, 568–576. [Google Scholar] [CrossRef]
- Jia, S.; Liu, X.; Huang, Z.; Li, Y.; Zhang, L.; Luo, Y. Effects of chitosan oligosaccharides on microbiota composition of silver carp (Hypophthalmichthys molitrix) determined by culture-dependent and independent methods during chilled storage. Int. J. Food Microbiol. 2018, 268, 81–91. [Google Scholar] [CrossRef]
- Olatunde, O.O.; Benjakul, S.; Vongkamjan, K. Combined effect of ethanolic coconut husk extract and modified atmospheric packaging (MAP) in extending the shelf life of asian sea bass slices. J. Aquat. Food Prod. Technol. 2019, 28, 689–702. [Google Scholar] [CrossRef]
- Singh, S.; Lee, M.; Gaikwad, K.K.; Lee, Y.S. Antibacterial and amine scavenging properties of silver-silica composite for post-harvest storage of fresh fish. Food Bioprod. Process. 2018, 107, 61–69. [Google Scholar] [CrossRef]
- Höll, L.; Behr, J.; Vogel, R.F. Identification and growth dynamics of meat spoilage microorganisms in modified atmosphere packaged poultry meat by MALDI-TOF MS. Food Microbiol. 2016, 60, 84–91. [Google Scholar] [CrossRef]
- Li, Y.; Huang, J.; Yuan, C.; Ding, T.; Chen, S.; Hu, Y. Developing a new spoilage potential algorithm and identifying spoilage volatiles in small yellow croaker (Larimichthys polyactis) under vacuum packaging condition. LWT Food Sci. Technol. 2019, 106, 209–217. [Google Scholar] [CrossRef]
- Jeyakumar, G.E.; Lawrence, R. Mechanisms of bactericidal action of Eugenol against Escherichia coli. J. Herb. Med. 2020, in press. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, Y.; Zhu, X.; Cao, P.; Wei, S.; Lu, Y. Antibacterial and antibiofilm activities of eugenol from essential oil of Syzygium aromaticum (L.) Merr. & L. M. Perry (clove) leaf against periodontal pathogen Porphyromonas gingivalis. Microb. Pathog. 2017, 113, 396–402. [Google Scholar] [PubMed]
- Odeyemi, O.A.; Burke, C.M.; Bolch, C.C.J.; Stanley, R. Seafood spoilage microbiota and associated volatile organic compounds at different storage temperatures and packaging conditions. Int. J. Food Microbiol. 2018, 280, 87–99. [Google Scholar] [CrossRef] [PubMed]
- Mallappa, R.H.; Singh, D.K.; Rokana, N.; Pradhan, D.; Batish, V.K.; Grover, S. Screening and selection of probiotic Lactobacillus strains of Indian gut origin based on assessment of desired probiotic attributes combined with principal component and heatmap analysis. LWT Food Sci. Technol. 2019, 105, 272–281. [Google Scholar] [CrossRef]
- Huang, Z.; Liu, X.; Jia, S.; Luo, Y. Antimicrobial effects of cinnamon bark oil on microbial composition and quality of grass carp (Ctenopharyngodon idellus) fillets during chilled storage. Food Control 2017, 82, 316–324. [Google Scholar] [CrossRef]
- Liu, M.; Liu, Y.; Cao, M.J.; Liu, G.M.; Chen, Q.; Sun, L.; Chen, H. Antibacterial activity and mechanisms of depolymerized fucoidans isolated from Laminaria japonica. Carbohydr. Polym. 2017, 172, 294–305. [Google Scholar] [CrossRef]
- Ghosh, V.; Mukherjee, A.; Chandrasekaran, N. Ultrasonic emulsification of food-grade nanoemulsion formulation and evaluation of its bactericidal activity. Ultrason. Sonochem. 2013, 20, 338–344. [Google Scholar] [CrossRef] [PubMed]
- Sangare, L.; Chen, W.B.; Wang, C.L.; Chen, X.B.; Wu, M.H.; Zhang, X.; Zang, J.Y. Structural insights into the conformational change of Staphylococcus aureus NreA at C-terminus. Biotechnol. Lett. 2020, 42, 787–795. [Google Scholar] [CrossRef]
- Baptista, R.F.; Lemos, M.; Teixeira, C.E.; Vital, H.C.; Carneiro, C.S.; Mársico, E.T.; Conte, C.A.; Mano, S.B. Microbiological quality and biogenic amines in ready-to-eat grilled chicken fillets under vacuum packing, freezing, and high-dose irradiation. Poult. Sci. 2014, 93, 1571–1577. [Google Scholar] [CrossRef]
- Silbande, A.; Adenet, S.; Chopin, C.; Cornet, J.; Smith-Ravin, J.; Rochefort, K.; Leroi, F. Effect of vacuum and modified atmosphere packaging on the microbiological, chemical and sensory properties of tropical red drum (Sciaenops ocellatus) fillets stored at 4 °C. Int. J. Food Microbiol. 2018, 266, 31–41. [Google Scholar] [CrossRef]
- Zhao, R.; Rong, S.; Sun, G.; Liu, S.; Li, B.; Cao, Y.; Li, Y. Cutoff Ostwald ripening stability of eugenol-in-water emulsion by co-stabilization method and antibacterial activity evaluation. Food Hydrocoll. 2020, 107, 105925. [Google Scholar] [CrossRef]

| TVB-N (mg N/100 g) | CK | VP | G-0.075E + VP | G-0.15E + VP | G-0.30E + VP | |
|---|---|---|---|---|---|---|
| Time | ||||||
| 0 day | 9.11 ± 0.31 | 9.11 ± 0.31 | 9.11 ± 0.31 | 9.11 ± 0.31 | 9.11 ± 0.31 | |
| 12th day | 13.24 ± 0.44 | 12.29 ± 0.50 | 11.84 ± 0.29 | 11.46 ± 0.17 | 11.08 ± 0.30 | |
| 24th day | 26.61 ± 0.60 | 22.69 ± 0.58 | 19.73 ± 0.41 | 16.54 ± 0.35 | 14.28 ± 0.37 | |
| Sample\Estimators | Total Tags | OTUs | Average Length | ACE | Chao1 | Coverage | Shannon | Simpson | Sobs |
|---|---|---|---|---|---|---|---|---|---|
| 0 day | 44,432.00 | 146.70 | 424.00 | 236.20 | 239.20 | 1.00 | 2.93 | 0.11 | 229.00 |
| 12th day CK | 40,338.00 | 12.15 | 424.00 | 686.62 | 687.79 | 1.00 | 3.91 | 0.06 | 684.00 |
| 24th day CK | 45,713.00 | 6.57 | 429.00 | 44.99 | 37.50 | 1.00 | 0.85 | 0.62 | 30.00 |
| 12th day VP | 50,269.00 | 10.45 | 429.00 | 98.49 | 104.00 | 1.00 | 1.66 | 0.36 | 74.00 |
| 24th day VP | 50,944.00 | 6.00 | 429.00 | 48.18 | 67.00 | 1.00 | 1.76 | 0.22 | 39.00 |
| 12th day G-0.075E + VP | 37,492.00 | 35.68 | 427.00 | 223.20 | 222.12 | 1.00 | 2.15 | 0.29 | 220.00 |
| 12th day G-0.15E + VP | 50,858.00 | 22.22 | 429.00 | 148.84 | 159.20 | 1.00 | 1.02 | 0.46 | 134.00 |
| 12th day G-0.30E + VP | 44,114.00 | 15.61 | 429.00 | 119.78 | 85.77 | 1.00 | 1.39 | 0.39 | 68.00 |
| 24th day G-0.075E + VP | 48,208.00 | 8.85 | 429.00 | 110.16 | 99.12 | 1.00 | 1.24 | 0.41 | 89.00 |
| 24th day G-0.15E + VP | 51,070.00 | 7.93 | 429.00 | 106.28 | 84.25 | 1.00 | 1.52 | 0.28 | 46.00 |
| 24th day G-0.30E + VP | 49,811.00 | 7.11 | 429.00 | 56.68 | 46.25 | 1.00 | 1.50 | 0.37 | 41.00 |
| Number | Results | Similarity | Accession No. | |
|---|---|---|---|---|
| 1 | Pseudomonas | Pseudomonas fragi | 99.58% | NR_024946.1 |
| 2 | Pseudomonas | Pseudomonas lactis | 100.00% | NR_156986.1 |
| 3 | Pseudomonas | Pseudomonas psychrophila | 99.31% | NR_028619.1 |
| 4 | Brochothrix | Brochothrix thermosphacta | 100.00% | NR_113587.1 |
| 5 | Pseudomonas | Pseudomonas migulae | 99.52% | NR_024927.1 |
| 6 | Brachybacterium | Brachybacterium rhamnosum | 99.93% | NR_042109.1 |
| 7 | Shewanella | Shewanella baltica | 98.57% | NR_025267.1 |
| 8 | Pseudomonas | Pseudomonas libanensis | 99.51% | NR_024901.1 |
| 9 | Pseudomonas | Pseudomonas weihenstephanensis | 99.31% | NR_148764.1 |
| 10 | Pseudomonas | Pseudomonas helleri | 99.37% | NR_148763.1 |
| 11 | Carnobacterium | Carnobacterium divergens | 99.45% | NR_113798.1 |
| 12 | Pseudomonas | Pseudomonas veronii | 99.51% | NR_028706.1 |
| 13 | Carnobacterium | Carnobacterium maltaromaticum | 98.31% | NR_044710.2 |
| 14 | Agrobacterium | Agromyces indicus | 99.65% | NR_108908.1 |
| 15 | Microbacterium | Microbacterium trichothecenolyticum | 98.72% | NR_044937.1 |
| 16 | Pseudomonas | Pseudomonas paralactis | 99.86% | NR_156987.1 |
| 17 | Staphylococcus | Staphylococcus edaphicus | 99.80% | NR_074999.2 |
| 0d Species identification | Number | 12th day CK Species identification | Number | 24th day CK Species identification | Number | 12th day VP Species identification | Number | 24th day VP Species identification | Number | 12th day G-0.075E + VP Species identification | Number | 12th day G-0.15E + VP Species identification | Number | 12th day G-0.30E + VP Species identification | Number | 24th day G-0.075E + VP Species identification | Number | 24th day G-0.15E + VP Species identification | Number | 24th day G-0.30E + VP Species identification | Number |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pseudomonas | 1 | Pseudomonas | 2 | Pseudomonas | 1 | Pseudomonas | 5 | Pseudomonas | 5 | Pseudomonas | 10 | Pseudomonas | 2 | Pseudomonas | 2 | Pseudomonas | 2 | Pseudomonas | 2 | Pseudomonas | 8 |
| – | 5 | – | 16 | – | 2 | Shewanella | 7 | Shewanella | 7 | Shewanella | 7 | – | 8 | – | 10 | – | 8 | – | 8 | – | 12 |
| – | 10 | Shewanella | 7 | – | 3 | Carnobacterium | 11 | Carnobacterium | 11 | Carnobacterium | 11 | – | 9 | – | 12 | Carnobacterium | 13 | – | 9 | Shewanella | 7 |
| Brochothrix | 4 | Brochothrix | 4 | – | 9 | – | 13 | – | 13 | Brochothrix | 4 | – | 10 | Brochothrix | 4 | Brochothrix | 4 | Shewanella | 7 | Brochothrix | 4 |
| Shewanella | 7 | Brachybacterium | 6 | – | 10 | Brochothrix | 4 | Staphylococcus | 17 | Brachybacterium | 6 | Brochothrix | 4 | Carnobacterium | 11 | – | – | Carnobacterium | 11 | Carnobacterium | 13 |
| Staphylococcus | 17 | – | – | Shewanella | 7 | Brachybacterium | 6 | Microbacterium | 15 | Agrobacterium | 14 | Carnobacterium | 11 | – | 13 | – | – | – | 13 | – | – |
| Carnobacterium | 11 | – | – | – | – | Microbacterium | 15 | – | – | – | – | – | 13 | – | – | – | – | – | – | – | – |
| – | 13 | – | – | – | – | Staphylococcus | 17 | – | – | – | – | Brachybacterium | 6 | – | – | – | – | – | – | – | – |
| Microbacterium | 15 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | ||
| Brachybacterium | 6 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | |||
| Agrobacterium | 14 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, X.; Zhou, Q.; Qiu, W.; Mei, J.; Xie, J. An Active Gelatin Coating Containing Eugenol and Vacuum Delays the Decay of Chinese Seabass (Lateolabrax maculatus) Fillets during Cold Storage: A Microbiome Perspective. Coatings 2021, 11, 147. https://doi.org/10.3390/coatings11020147
Ma X, Zhou Q, Qiu W, Mei J, Xie J. An Active Gelatin Coating Containing Eugenol and Vacuum Delays the Decay of Chinese Seabass (Lateolabrax maculatus) Fillets during Cold Storage: A Microbiome Perspective. Coatings. 2021; 11(2):147. https://doi.org/10.3390/coatings11020147
Chicago/Turabian StyleMa, Xuan, Qianqian Zhou, Weiqiang Qiu, Jun Mei, and Jing Xie. 2021. "An Active Gelatin Coating Containing Eugenol and Vacuum Delays the Decay of Chinese Seabass (Lateolabrax maculatus) Fillets during Cold Storage: A Microbiome Perspective" Coatings 11, no. 2: 147. https://doi.org/10.3390/coatings11020147
APA StyleMa, X., Zhou, Q., Qiu, W., Mei, J., & Xie, J. (2021). An Active Gelatin Coating Containing Eugenol and Vacuum Delays the Decay of Chinese Seabass (Lateolabrax maculatus) Fillets during Cold Storage: A Microbiome Perspective. Coatings, 11(2), 147. https://doi.org/10.3390/coatings11020147









