Non-Destructive Monitoring of P. fluorescens and S. epidermidis Biofilm under Different Media by Fourier Transform Infrared Spectroscopy and Other Corroborative Techniques
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Media Used and Their Preparation
2.3. Bacteria Cultivation
2.4. Planktonic P. fluorescens and S. epidermidis Preparation for ATR Analysis
2.5. Spectroscopy Study
2.6. Adhesion Assay
3. Results and Discussion
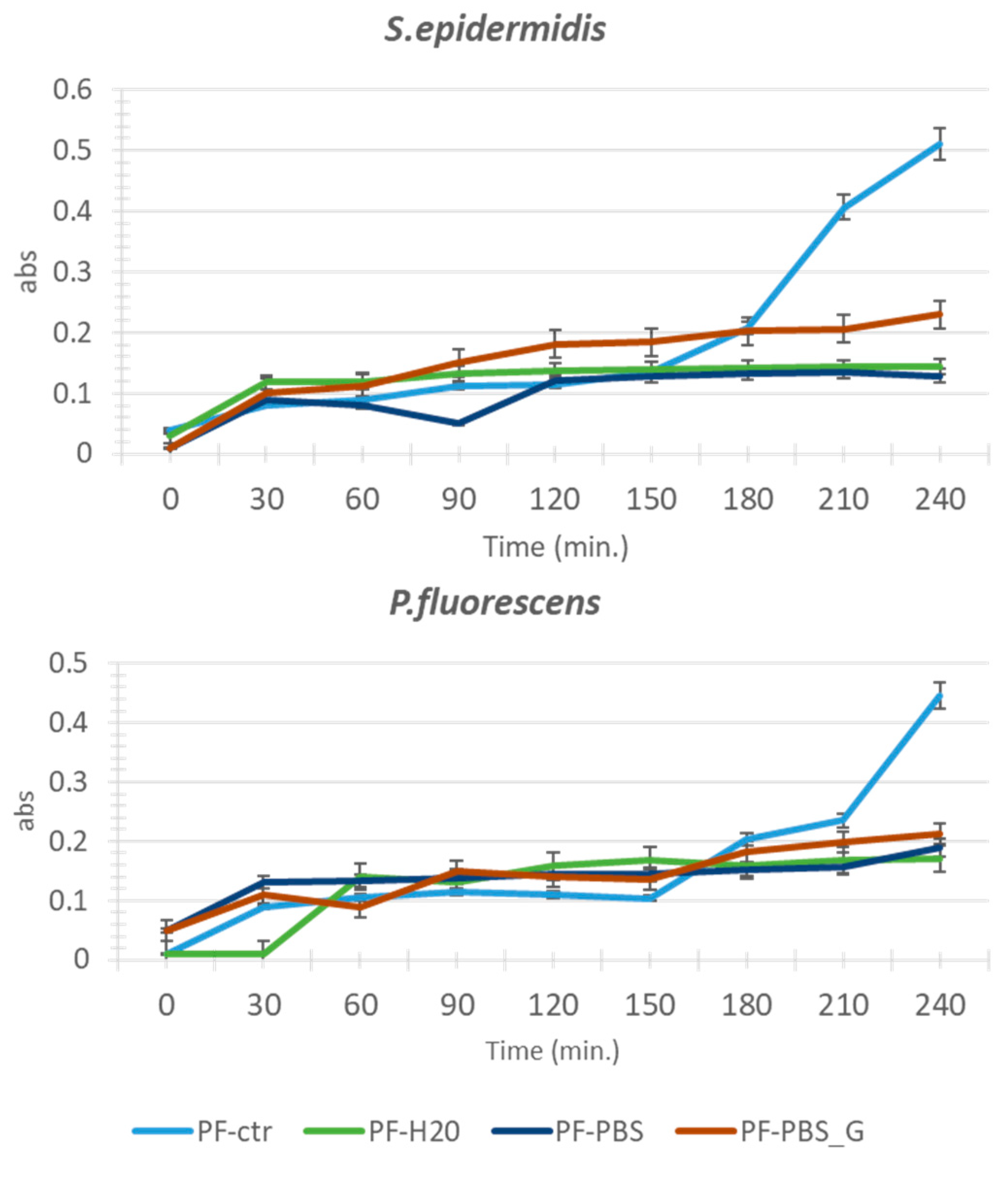
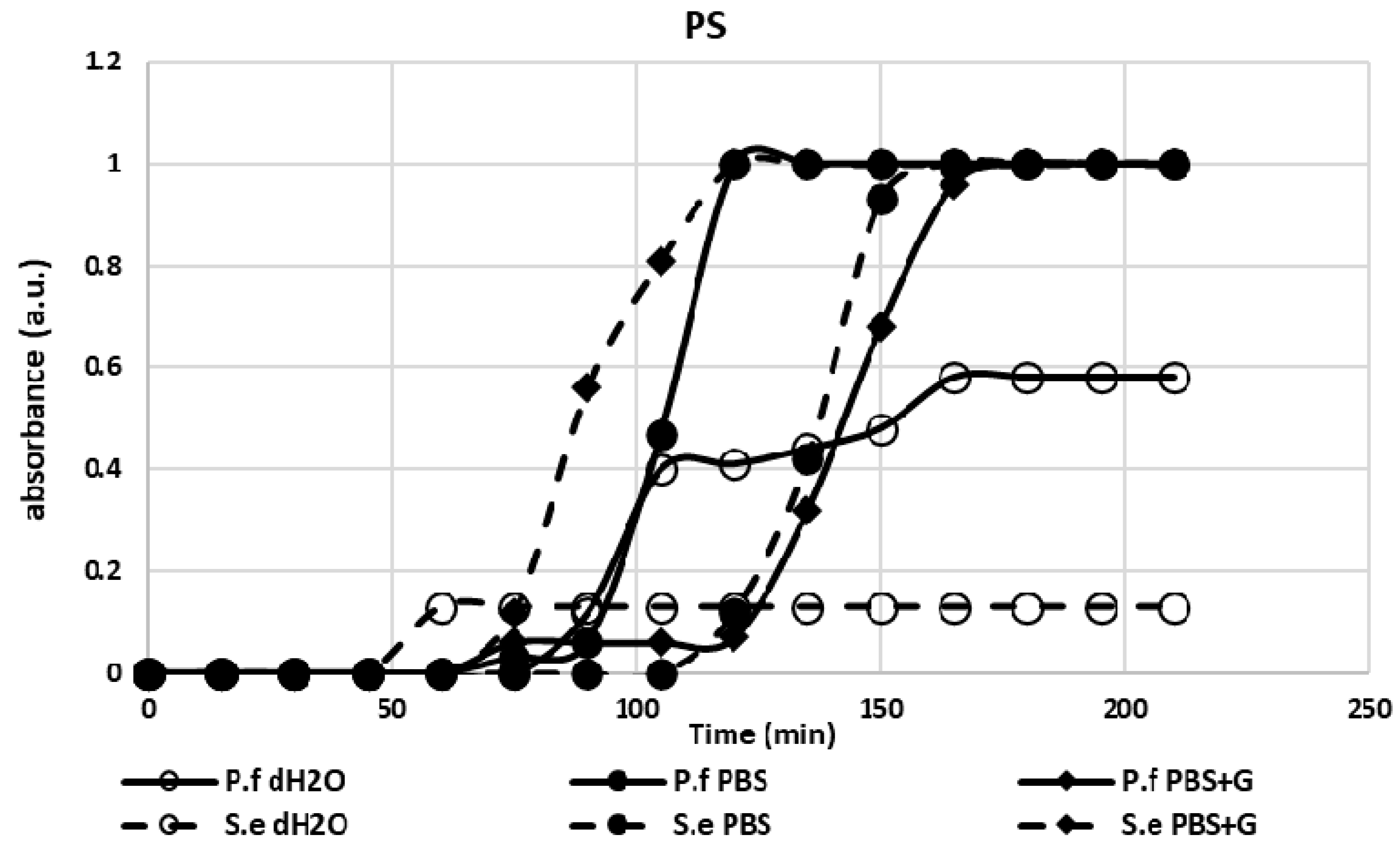
3.1. Adhesion Test
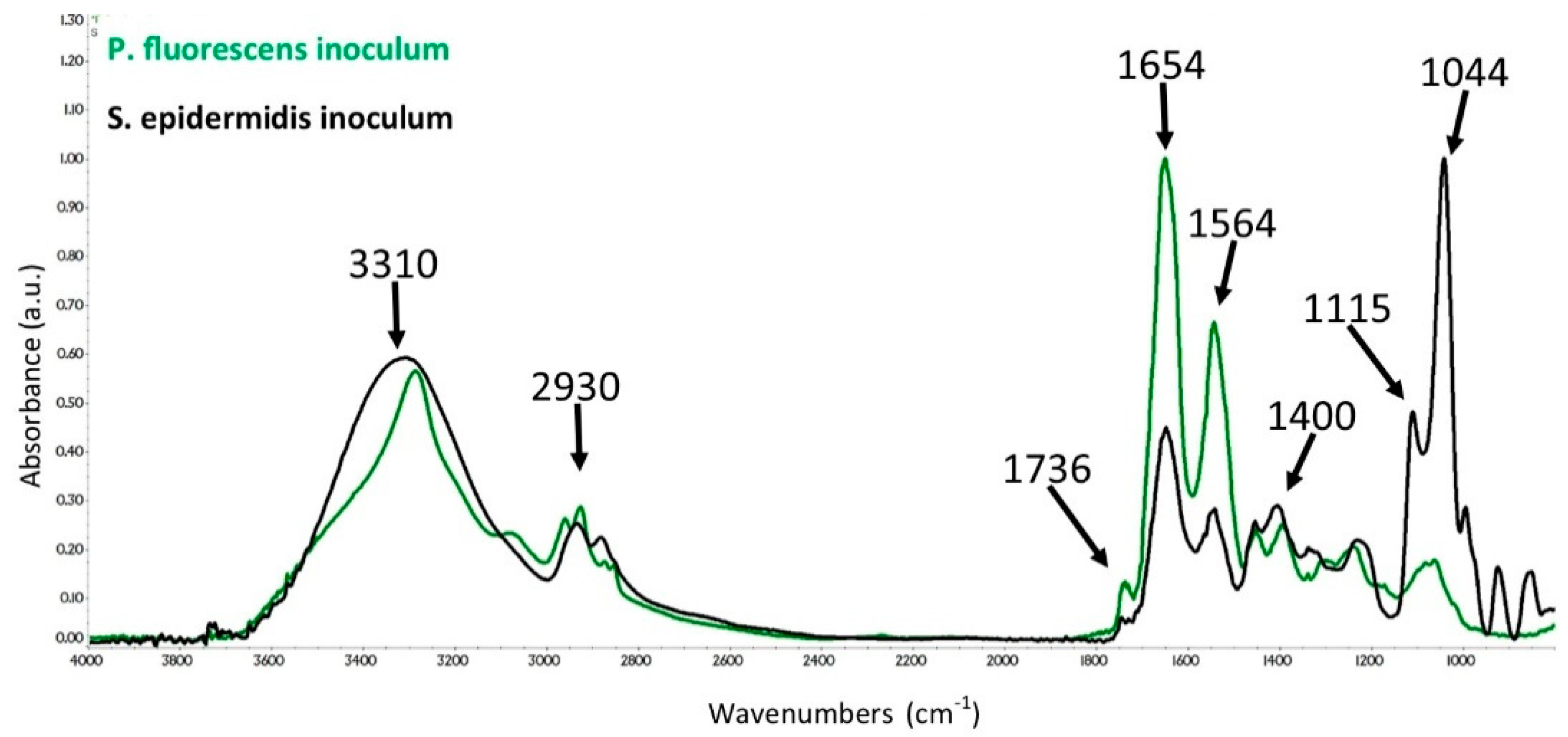
3.2. ATR-FTIR Spectra of SE and PF Bacteria
3.3. Changing of Amide Bands as a Function of Time and Media
3.4. Changing of Polysaccharides Bands as a Function of Time and Media
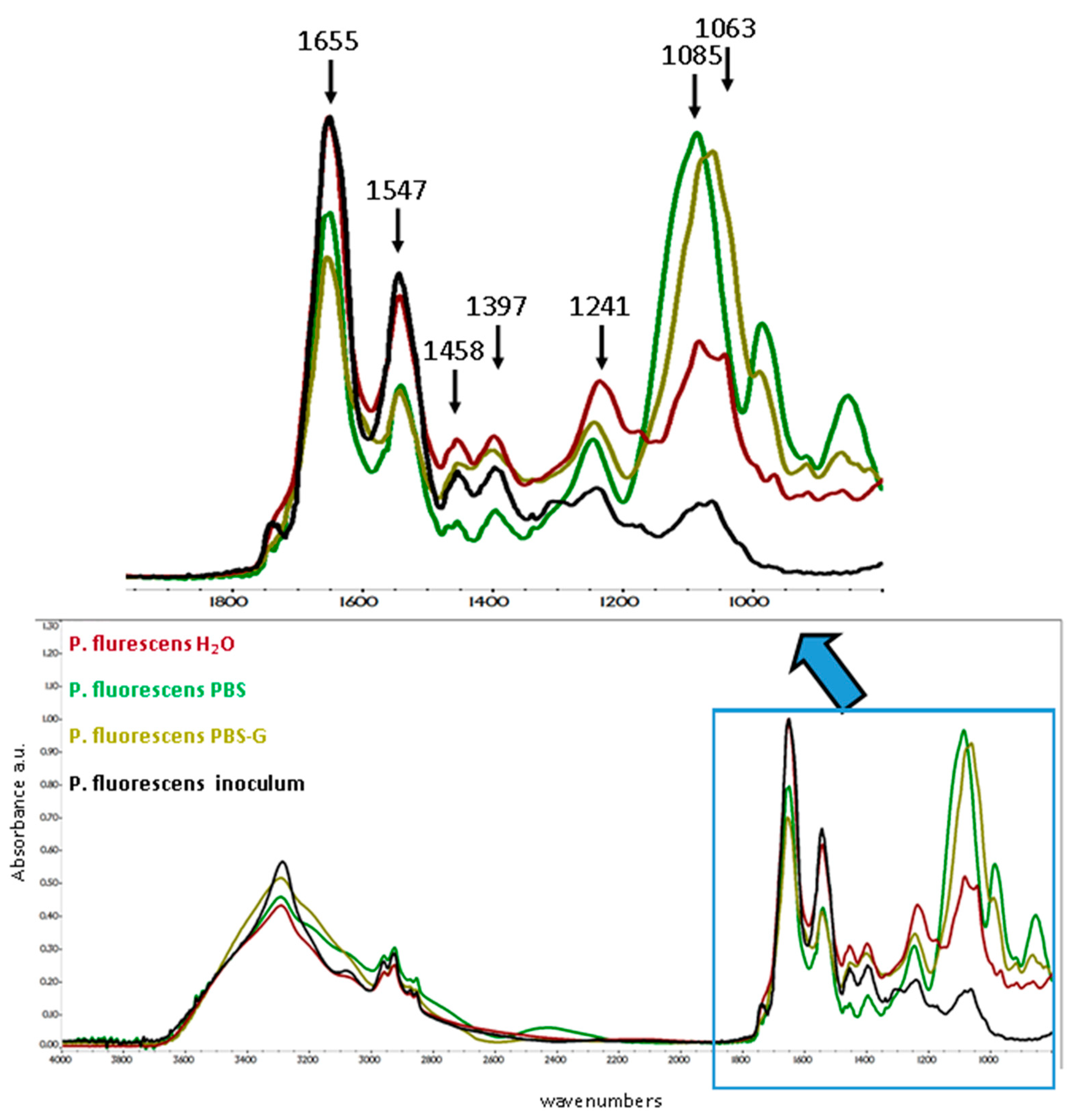
3.5. FTIR Spectra Changes as a Function of Media Composition
3.5.1. S. epidermidis
3.5.2. P. fluorescens
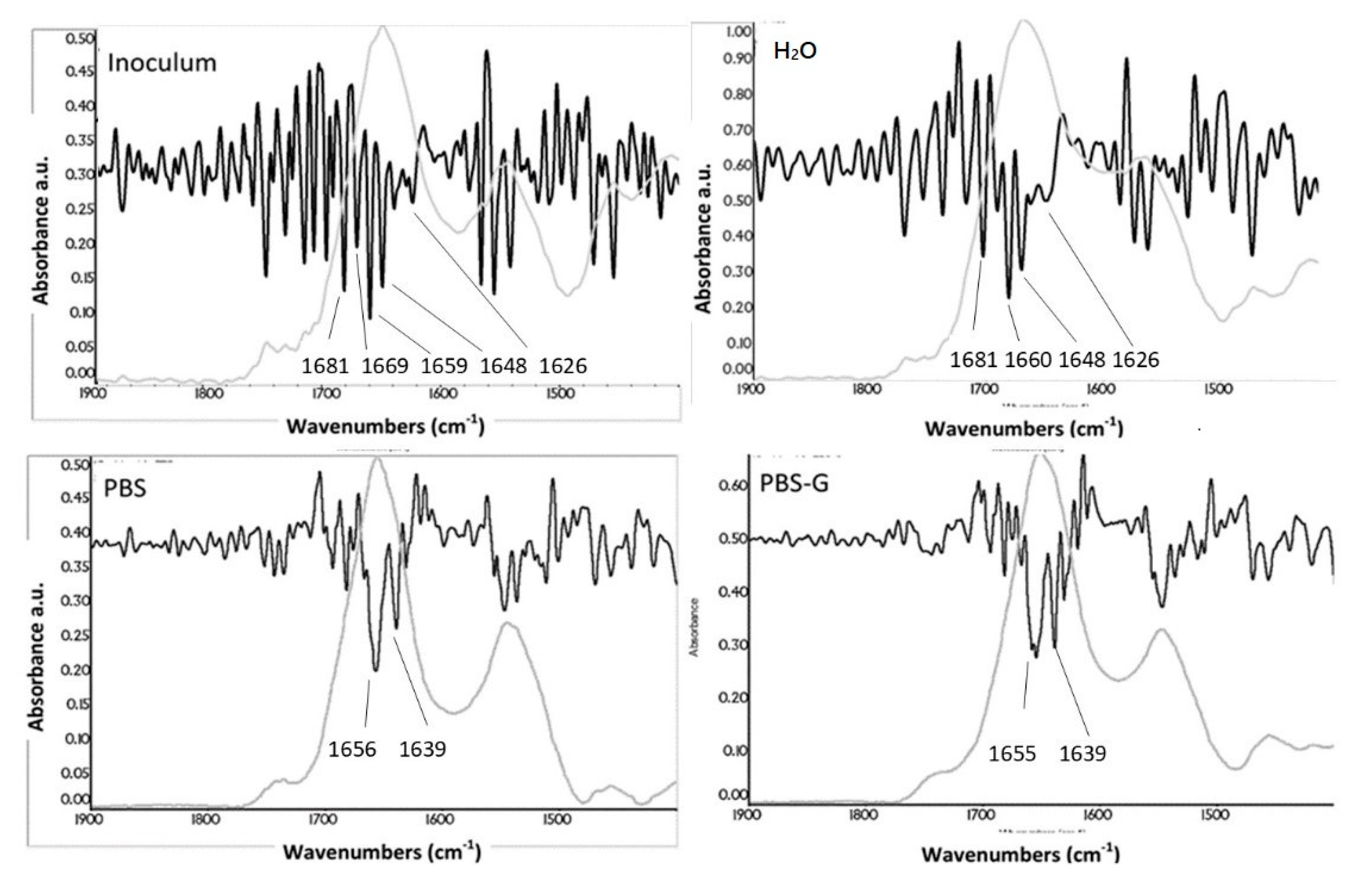
3.6. Secondary-Structure Analysis
3.7. Bacteria Staining Test
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Flemming, H.-C. The perfect slime. Colloids Surf. B Biointerfaces 2011, 86, 251–259. [Google Scholar] [CrossRef]
- Pamp, S.J.; Gjermansen, M.; Tolker-Nielsen, T. The Biofilm Matrix: A Sticky Framework. In Bacterial Biofilm Formation and Adaptation; Givskov, M.C., Ed.; Horizon BioScience: Norfolk, UK, 2007; Volume 34, pp. 37–69. [Google Scholar]
- Watnick, P.; Kolter, R. Biofilm, City of Microbes. J. Bacteriol. 2000, 182, 2675–2679. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flemming, H.-C.; Wingender, J.; Szewzyk, U.; Steinberg, P.; Rice, S.A.; Kjelleberg, S. Biofilms: An emergent form of bacterial life. Nat. Rev. Microbiol. 2016, 14, 563–575. [Google Scholar] [CrossRef] [PubMed]
- Costerton, J.W. Bacterial Biofilms: A Common Cause of Persistent Infections. Science 1999, 284, 1318–1322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bahn, Y.-S.; Xue, C.; Idnurm, A.; Rutherford, J.C.; Heitman, J.; Cardenas, M.E. Sensing the environment: Lessons from fungi. Nat. Rev. Microbiol. 2007, 5, 57–69. [Google Scholar] [CrossRef] [PubMed]
- Yin, W.; Wang, Y.; Liu, L.; He, J. Biofilms: The Microbial “Protective Clothing” in Extreme Environments. Int. J. Mol. Sci. 2019, 20, 3423. [Google Scholar] [CrossRef] [Green Version]
- Harrison, J.J.; Ceri, H.; Turner, R.J. Multimetal resistance and tolerance in microbial biofilms. Nat. Rev. Microbiol. 2007, 5, 928–938. [Google Scholar] [CrossRef]
- Fulaz, S.; Vitale, S.; Quinn, L.; Casey, E. Nanoparticle–Biofilm Interactions: The Role of the EPS Matrix. Trends Microbiol. 2019, 27, 915–926. [Google Scholar] [CrossRef]
- Whitchurch, C.B. Extracellular DNA Required for Bacterial Biofilm Formation. Science 2002, 295, 1487. [Google Scholar] [CrossRef]
- Tolker-Nielsen, T. Biofilm Development. Microbiol. Spectr. 2015, 3. [Google Scholar] [CrossRef] [Green Version]
- Davies, D.G. The Involvement of Cell-to-Cell Signals in the Development of a Bacterial Biofilm. Science 1998, 280, 295–298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Kievit, T.R.; Gillis, R.; Marx, S.; Brown, C.; Iglewski, B.H. Quorum-Sensing Genes in Pseudomonas aeruginosa Biofilms: Their Role and Expression Patterns. Appl. Environ. Microbiol. 2001, 67, 1865–1873. [Google Scholar] [CrossRef] [Green Version]
- Carniello, V.; Peterson, B.W.; van der Mei, H.C.; Busscher, H.J. Physico-chemistry from initial bacterial adhesion to surface-programmed biofilm growth. Adv. Colloid Interface Sci. 2018, 261, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Serra, D.O.; Richter, A.M.; Hengge, R. Cellulose as an Architectural Element in Spatially Structured Escherichia coli Biofilms. J. Bacteriol. 2013, 195, 5540–5554. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rohde, H.; Frankenberger, S.; Zähringer, U.; Mack, D. Structure, function and contribution of polysaccharide intercellular adhesin (PIA) to Staphylococcus epidermidis biofilm formation and pathogenesis of biomaterial-associated infections. Eur. J. Cell Biol. 2010, 89, 103–111. [Google Scholar] [CrossRef] [PubMed]
- McDougald, D.; Rice, S.A.; Barraud, N.; Steinberg, P.D.; Kjelleberg, S. Should we stay or should we go: Mechanisms and ecological consequences for biofilm dispersal. Nat. Rev. Microbiol. 2012, 10, 39–50. [Google Scholar] [CrossRef]
- Franklin, M.J.; Nivens, D.E.; Weadge, J.T.; Howell, P.L. Biosynthesis of the Pseudomonas aeruginosa Extracellular Polysaccharides, Alginate, Pel, and Psl. Front. Microbiol. 2011, 2, 167. [Google Scholar] [CrossRef] [Green Version]
- Colvin, K.M.; Irie, Y.; Tart, C.S.; Urbano, R.; Whitney, J.C.; Ryder, C.; Howell, P.L.; Wozniak, D.J.; Parsek, M.R. The Pel and Psl polysaccharides provide Pseudomonas aeruginosa structural redundancy within the biofilm matrix: Polysaccharides of the P. aeruginosa biofilm matrix. Environ. Microbiol. 2012, 14, 1913–1928. [Google Scholar] [CrossRef] [Green Version]
- Conrad, A.; Kontro, M.; Keinänen, M.M.; Cadoret, A.; Faure, P.; Mansuy-Huault, L.; Block, J.-C. Fatty acids of lipid fractions in extracellular polymeric substances of activated sludge flocs. Lipids 2003, 38, 1093–1105. [Google Scholar] [CrossRef]
- Romero, D.; Vlamakis, H.; Losick, R.; Kolter, R. An accessory protein required for anchoring and assembly of amyloid fibres in B. subtilis biofilms: B. subtilis amyloid fibre accessory protein. Mol. Microbiol. 2011, 80, 1155–1168. [Google Scholar] [CrossRef] [Green Version]
- Grunert, T.; Monahan, A.; Lassnig, C.; Vogl, C.; Müller, M.; Ehling-Schulz, M. Deciphering Host Genotype-Specific Impacts on the Metabolic Fingerprint of Listeria monocytogenes by FTIR Spectroscopy. PLoS ONE 2014, 9, e115959. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sutherland, I. The biofilm matrix—An immobilized but dynamic microbial environment. Trends Microbiol. 2001, 9, 222–227. [Google Scholar] [CrossRef]
- Klausen, M.; Heydorn, A.; Ragas, P.; Lambertsen, L.; Aaes-Jørgensen, A.; Molin, S.; Tolker-Nielsen, T. Biofilm formation by Pseudomonas aeruginosa wild type, flagella and type IV pili mutants: Roles of bacterial motility in the formation of the flat P. aeruginosa biofilm. Mol. Microbiol. 2003, 48, 1511–1524. [Google Scholar] [CrossRef]
- Chen, R.; Guo, C.; Chu, W.; Jiang, N.; Li, H. ATR-FTIR study of Bacillus sp. and Escherichia coli settlements on the bare and Al2O3 coated ZnSe internal reflection element. Chin. Chem. Lett. 2019, 30, 115–119. [Google Scholar] [CrossRef]
- Genkawa, T.; Ahamed, T.; Noguchi, R.; Takigawa, T.; Ozaki, Y. Simple and rapid determination of free fatty acids in brown rice by FTIR spectroscopy in conjunction with a second-derivative treatment. Food Chem. 2016, 191, 7–11. [Google Scholar] [CrossRef] [PubMed]
- Pink, J.; Smith-Palmer, T.; Chisholm, D.; Beveridge, T.J.; Pink, D.A. An FTIR study of Pseudomonas aeruginosa PAO1 biofilm development: Interpretation of ATR–FTIR data in the 1500–1180 cm−1 region. Biofilms 2005, 2, 165–175. [Google Scholar] [CrossRef]
- Pink, J.; Smith-Palmer, T.; Beveridge, T.J.; Pink, D.A. An FTIR study of Pseudomonas aeruginosa PAO1 biofilm growth and dispersion. An improved ATR method for studying biofilms: The C–H stretch spectral region. Biofilms 2004, 1, 157–163. [Google Scholar] [CrossRef]
- Cwalina, B.; Dec, W.; Michalska, J.K.; Jaworska-Kik, M.; Student, S. Initial stage of the biofilm formation on the NiTi and Ti6Al4V surface by the sulphur-oxidizing bacteria and sulphate-reducing bacteria. J. Mater. Sci. Mater. Med. 2017, 28, 173. [Google Scholar] [CrossRef] [Green Version]
- Boualam, M.; Quilès, F.; Mathieu, L.; Block, J.-C. Monitoring the Effect of Organic Matter on Biofilm Growth in Low Nutritive Waters by ATR/FT-IR Spectroscopy. Biofouling 2002, 18, 73–81. [Google Scholar] [CrossRef]
- Van der Kooij, D. The occurrence of Pseudomonas spp. in surface water and in tap water as determined on citrate media. Antonie Leeuwenhoek 1977, 43, 187–197. [Google Scholar] [CrossRef]
- Van der Kooij, D.; Visser, A.; Oranje, J.P. Multiplication of fluorescent pseudomonads at low substrate concentrations in tap water. Antonie Leeuwenhoek 1982, 48, 229–243. [Google Scholar] [CrossRef] [PubMed]
- Pop, C.; Apostu, S.; Rotar, A.M.; Semeniuc, C.A.; Sindic, M.; Mabon, N. FTIR spectroscopic characterization of a new biofilm obtained from kefiran. J. Agroaliment. Process. Technol. 2013, 19, 157–159. [Google Scholar]
- Karadenizli, A.; Kolayli, F.; Ergen, K. A novel application of Fourier-transformed infrared spectroscopy: Classification of slime from staphylococci. Biofouling 2007, 23, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Cappelli, A.; Razzano, V.; Paolino, M.; Grisci, G.; Giuliani, G.; Donati, A.; Mendichi, R.; Samperi, F.; Battiato, S.; Boccia, A.C.; et al. Bithiophene-based polybenzofulvene derivatives with high stacking and hole mobility. Polym. Chem. 2015, 6, 7377–7388. [Google Scholar] [CrossRef]
- Leone, G.; Consumi, M.; Franzi, C.; Tamasi, G.; Lamponi, S.; Donati, A.; Magnani, A.; Rossi, C.; Bonechi, C. Development of liposomal formulations to potentiate natural lovastatin inhibitory activity towards 3-hydroxy-3-methyl-glutaryl coenzyme A (HMG-CoA) reductase. J. Drug Deliv. Sci. Technol. 2018, 43, 107–112. [Google Scholar] [CrossRef]
- Genevaux, P.; Muller, S.; Bauda, P. A rapid screening procedure to identify mini-Tn10 insertion mutants of Escherichia coli K-12 with altered adhesion properties. FEMS Microbiol. Lett. 1996, 142, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Quilès, F.; Humbert, F.; Delille, A. Analysis of changes in attenuated total reflection FTIR fingerprints of Pseudomonas fluorescens from planktonic state to nascent biofilm state. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2010, 75, 610–616. [Google Scholar] [CrossRef]
- Fasasi, Y.A.; Mirjankar, N.; Fasasi, A. Fourier Transform Infrared Spectroscopic Analysis of Protein Secondary Structures Found in Egusi. Am. J. Appl. Ind. Chem. 2015, 1, 1–4. [Google Scholar]
- Bonechi, C.; Donati, A.; Tamasi, G.; Pardini, A.; Rostom, H.; Leone, G.; Lamponi, S.; Consumi, M.; Magnani, A.; Rossi, C. Chemical characterization of liposomes containing nutraceutical compounds: Tyrosol, hydroxytyrosol and oleuropein. Biophys. Chem. 2019, 246, 25–34. [Google Scholar] [CrossRef]
- Pietralik, Z.; Mucha-Kruczynska, I.; Kozak, M. FTIR analysis of protein secondary structure in solid and solution states. Synchrotron Radiation News 2012, 11, 52. [Google Scholar]
- Leone, G.; Consumi, M.; Pepi, S.; Lamponi, S.; Bonechi, C.; Tamasi, G.; Donati, A.; Rossi, C.; Magnani, A. Alginate–gelatin formulation to modify lovastatin release profile from red yeast rice for hypercholesterolemia therapy. Ther. Deliv. 2017, 8, 843–854. [Google Scholar] [CrossRef]
- Leone, G.; Consumi, M.; Lamponi, S.; Bonechi, C.; Tamasi, G.; Donati, A.; Rossi, C.; Magnani, A. Thixotropic PVA hydrogel enclosing a hydrophilic PVP core as nucleus pulposus substitute. Mater. Sci. Eng. C 2019, 98, 696–704. [Google Scholar] [CrossRef] [PubMed]
- Delille, A.; Quilès, F.; Humbert, F. In Situ Monitoring of the Nascent Pseudomonas fluorescens Biofilm Response to Variations in the Dissolved Organic Carbon Level in Low-Nutrient Water by Attenuated Total Reflectance-Fourier Transform Infrared Spectroscopy. AEM 2007, 73, 5782–5788. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, W.; Saxena, A.; Song, B.; Ward, B.B.; Beveridge, T.J.; Myneni, S.C.B. Elucidation of Functional Groups on Gram-Positive and Gram-Negative Bacterial Surfaces Using Infrared Spectroscopy. Langmuir 2004, 20, 11433–11442. [Google Scholar] [CrossRef]
- Jubeen, F.; Liaqat, A.; Amjad, F.; Sultan, M.; Iqbal, S.Z.; Sajid, I.; Khan Niazi, M.B.; Sher, F. Synthesis of 5-Fluorouracil Cocrystals with Novel Organic Acids as Coformers and Anticancer Evaluation against HCT-116 Colorectal Cell Lines. Cryst. Growth Des. 2020, 20, 2406–2414. [Google Scholar] [CrossRef]
- Jubeen, F.; Liaqat, A.; Sultan, M.; Zafar Iqbal, S.; Sajid, I.; Sher, F. Green synthesis and biological evaluation of novel 5-fluorouracil derivatives as potent anticancer agents. Saudi Pharm. J. 2019, 27, 1164–1173. [Google Scholar] [CrossRef] [PubMed]
- McWhirter, M.J.; Bremer, P.J.; McQuillan, A.J. Direct Infrared Spectroscopic Evidence of pH- and Ionic Strength-Induced Changes in Distance of Attached Pseudomonas a eruginosa from ZnSe Surfaces. Langmuir 2002, 18, 1904–1907. [Google Scholar] [CrossRef]
- Lasch, P. Spectral pre-processing for biomedical vibrational spectroscopy and microspectroscopic imaging. Chemom. Intell. Lab. Syst. 2012, 117, 100–114. [Google Scholar] [CrossRef] [Green Version]
- Bunaciu, A.A.; Fleschin, Ş.; Aboul-Enein, H.Y. Evaluation of the Protein Secondary Structures Using Fourier Transform Infrared Spectroscopy. Gazi Univ. J. Sci. 2014, 27, 637–644. [Google Scholar]



| Tentative Assignment of the Main Band to the Relevant Functional Groups | Ref | |||
|---|---|---|---|---|
| Wavenumber (cm−1) | Assignment | Principal Compounds | Main Corresponding Cellular Compounds | |
| 1736 | νC=O | Esters from lipids | Membranes | [30,31,32,33,44] |
| 1713 | νC=O | Esters, carboxylic acids | Nucleoid, ribosomes | |
| 1700–1580 | νC=O, νC=N, νC=C, δNH | DNA, RNA bases | Nucleoid, ribosomes | |
| 1693–1627 | Amide I (νC=O coupled with δN-H), δH2O | Proteins, water (1640cm-1) | Membranes, cytoplasm, flagella, pili, ribosomes | [41] |
| 1568-1531 | Amide II (δN-H coupled with νC-N) | Proteins | Membranes, cytoplasm, flagella, pili, ribosomes | [41] |
| 1468,1455 | δCH2, δCH3 | Lipids | Membranes | [26,27,28] |
| 1400 | νsCOO- | Amino acids, fatty acids chains | Capsule, peptidoglycan | |
| 1317;1281 | τCH2; Amide III(νC-N coupled with δN-H) | Fatty acids chains, proteins | Membranes, cytoplasm, flagella, pili, ribosomes | |
| 1238 | νa PO2- | Phosphodiester, phospholipids, LPS, nucleic acids, ribosomes | Membrane, nucleoids, ribosomes | |
| 1220 | νC-O-C | - | Capsule, storage inclusion | [16,25,33,44] |
| 1200–900 | νC-O, νC-C, δC-O-H, | Polysaccharides | - | |
| - | δC-O-C | - | - | |
| 1172,1153 | νsC-OH, νC-O | Proteins, carbohydrates, esters | - | |
| 1118 | νsCC | Phosphodiester, phosphoilipids | Nucleoids, ribosomes | [20,28,40,41,42,43,44,45,46,47,48] |
| 1086 | νsPO2- | LPS, nucleic acids | Membranes, nucleoid, ribosomes | [28,48] |
| 1058 | νsC-O-C, | Polysaccharides | Capsule, peptidoglycan | [16,33,40,44] |
| - | νsP-O-C (R-O-P-O-R’) | - | - | |
| 1041 | νO-H coupled with δC-O | Polysaccharides | Capsule, peptidoglycan | [42,43,44,45] |
| 1026 | CH2OH | Carbohydrates | Storage inclusion | [28,40,47] |
| 993 | - | Ribose skelet (ARN) | Ribosomes | |
| 970 | νC-C,νP-O-P | RNA backbone | Ribosomes | |
| Inoculum | H2O | PBS | PBS-G | Assignment |
|---|---|---|---|---|
| Frequencies (cm−1) | ||||
| Pseudomonas Fluorescent | ||||
| 1656 | 1656 | 1659 | 1658 | α-helix |
| - | - | 1648 | - | Random coils |
| 1628 | 1638 | 1639 | 1639 | β-sheet |
| Staphylococcus Epidermidis | ||||
| 1681,1669 | 1681 | - | - | β-sheet |
| 1659 | 1660 | 1656 | 1655 | α-helix |
| 1648 | 1648 | - | - | Random coils |
| 1637,1626 | 1626 | 1639 | 1639 | β-sheet |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Consumi, M.; Jankowska, K.; Leone, G.; Rossi, C.; Pardini, A.; Robles, E.; Wright, K.; Brooker, A.; Magnani, A. Non-Destructive Monitoring of P. fluorescens and S. epidermidis Biofilm under Different Media by Fourier Transform Infrared Spectroscopy and Other Corroborative Techniques. Coatings 2020, 10, 930. https://doi.org/10.3390/coatings10100930
Consumi M, Jankowska K, Leone G, Rossi C, Pardini A, Robles E, Wright K, Brooker A, Magnani A. Non-Destructive Monitoring of P. fluorescens and S. epidermidis Biofilm under Different Media by Fourier Transform Infrared Spectroscopy and Other Corroborative Techniques. Coatings. 2020; 10(10):930. https://doi.org/10.3390/coatings10100930
Chicago/Turabian StyleConsumi, Marco, Kamila Jankowska, Gemma Leone, Claudio Rossi, Alessio Pardini, Eric Robles, Kevin Wright, Anju Brooker, and Agnese Magnani. 2020. "Non-Destructive Monitoring of P. fluorescens and S. epidermidis Biofilm under Different Media by Fourier Transform Infrared Spectroscopy and Other Corroborative Techniques" Coatings 10, no. 10: 930. https://doi.org/10.3390/coatings10100930








