1. Introduction
Escherichia coli is ubiquitous in nature, often found in soil, water, food, human, and animal intestinal tracts. However,
E. coli can also act as a pathogen in a wide range of conditions, from enteric diseases to extraintestinal infections.
E. coli strains which cause enteric diseases or diarrhea are known as diarrheagenic
E. coli (DEC) and are divided into six distinct pathotypes based on clinical, epidemiological, and molecular criteria [
1]. Among them, enteropathogenic
E. coli (EPEC) is the predominant cause of diarrhea in developing countries [
1,
2,
3,
4,
5] and enterohemorrhagic
E. coli (EHEC) is attributed to the foodborne outbreaks in developed countries and can cause bloody diarrhea due to the production of Shiga toxins (
stxs) and is known as Shiga toxin-producing
E. coli (STEC). Among the DEC, STEC includes the most dangerous strains and more than 400 serotypes that produce
stxs have been identified, and this term was created since
E. coli species possess the toxin, which is more or less identical to that produced by
Shigella dysentery type I [
6]. STEC possesses a broad range of virulence factors (VFs), which are encoded by chromosomal genes, and they are often located in pathogenicity islands (PAIs) or plasmids, with the production of
stxs being the most crucial resulting in endothelial cell damage and possible hemolytic uremic syndrome (HUS) [
7,
8]. In addition, STEC is responsible for 2.8 million illnesses, and 3890 cases per year of enteric disease in humans, globally [
8]. Shiga toxin is classified into two types, Shiga toxin type 1 (
stx1) and Shiga toxin type 2
(stx2) and subtypes which dominate the pathogenicity of STEC, and it is known to be an important factor in differentiating the severity of illness [
2,
9,
10], but cannot be solely responsible for full pathogenicity, as a vast arsenal of VFs are essential for STEC pathogenicity. Thus, STEC possesses other VFs necessary for infection, such as intimin (
eae), which is an outer membrane encoded by the locus for enterocyte effacement (LEE) and is essential for the intimate adherence of
eae-positive STEC strains to the host’s intestine and, eventually, for the attaching and effacing (A/E) lesions frequently perceived in STEC infections. In addition, several STEC strains contain a number of plasmid-encoded VFs, including enterohemolysin (pO157;
ehxA). The toxin
ehxA is a heat-labile pore-forming toxin that causes hemolysis of host red blood cells and the possession of
ehxA by a STEC strain has been attributed to HUS [
11]. Other plasmid-encoded (pO113) VFs include a subtilase cytotoxin gene (
subA; triggers apoptosis in human cells) and an autoagglutinating adhesin gene (
saa; associated with the absence of LEE) [
12] also recognized as key plasmid-encoded VFs. The gene
subA suppresses the host’s immune system and allows STEC to adhere to enterocytes. Growing evidence suggests that differences in virulence between pathogenic and nonpathogenic bacterial strains can be attributed to VFs in pathogenicity islands [
13]. Several PAIs of STEC, including genes encoding OI-43/48, OI-57, and OI-71, are absent in nonpathogenic
E. coli and are viewed as STEC VFs. These genes have been used in molecular risk assessment research to classify STEC serotypes into various seropathotypes depending on whether a specific serotype in humans has been involved in mild, severe ailment, or no disease at all. Furthermore, numerous genes located on OI-43/48, including
IrgA homolog adhesin (encodes an adhesin;
iha) and
ureC (encode urease resistance), and on OI-71, including
nleA (effector; disrupts protein secretion) are considered appropriate virulence markers in STEC serotypes involved in severe human ailments and outbreaks but their individual role in the pathogenesis of human infection is still poorly understood [
2]. In addition, these genes are mainly found in high risk (HUS) STEC strains and are associated with colonization and survival in the host, and may interfere with signaling pathways during inflammation [
14].
In the last decade, the STEC has gained substantial attention as a global public health concern for various sporadic infections and major outbreaks due to the emergence of resistance to multiple antibiotics as there are fewer, or often no, effective antibiotics available for infections caused by such bacteria. Furthermore, multidrug resistance (MDR) in
E. coli strains has increased in recent years, causing serious problems in healthcare settings worldwide [
1,
15,
16]. MDR represents one of the notable global health challenges of this century and the global increase in its spread is a major issue, particularly in developing countries where there is limited control over the quality, distribution, and use of antibiotics in human medicine, veterinary medicine, and food animal farming [
17]. MDR bacterial infections have increased at an alarming rate due to the tremendous dissemination of antibiotic resistance determinants [
18,
19,
20] and MDR pathogens and infections thereof are expected to cause 10 million deaths per year by 2050 [
21,
22]. Even though the role of commensal bacteria in providing antibiotic resistance has long been recognized, however, these bacteria have not been extensively studied [
18].
E. coli comprises only a small fraction of the human gastrointestinal tract’s bacterial flora, but it is not a significant reservoir. Resistance in commensal
E. coli from healthy patients was first demonstrated over 50 years ago, and several recent studies have demonstrated the high or increased incidence of MDR in commensal
E. coli from healthy children and adults from various countries [
23,
24,
25]. Currently, the use of antibiotics to treat STEC infection in humans is controversial and not recommended in many countries according to the current clinical guidelines, as some antibiotics can induce the production of Shiga toxin [
8,
26,
27]. However, antibiotic resistance is a matter of growing concern due to the wide spread of
E. coli resistance to all antibiotics used in human therapy, and the dispersion of resistance via mobile genetic elements. Additionally, STEC bacteriophages may carry antibiotic resistance-encoding genes that can be transferred to naïve
E. coli which are then transformed into antibiotic-resistant strains [
8,
21].
Hence, bacteria with VFs and antibiotic resistance should be carefully monitored. Currently, there is limited information available on the prevalence of antibiotic resistance in E. coli isolates from humans and food samples. Understanding the prevalence of resistance to antibiotics, especially for critically important antibiotics, in E. coli isolates from human and food samples, will provide the useful information for developing risk management options to mitigate the spread of resistance. This research was performed to investigate the diversity and distribution of the major virulence-associated factors (stx1, stx2, iha, ureC, eae, uidA, nleA, ehxA, subA, and saa) in previously isolated strains from diverse sources including foods, animal carcasses and feces, and humans by using PCR. The second objective was to determine the strains’ potential as human pathogens by employing 17 frequently used antibiotics in clinical practices. This study emphasizes on the importance of preventing the spread of E. coli isolates that harbor both antibiotic resistance and virulence genes and the overall objective of this research was to contribute to STEC surveillance and gain insight into STEC strains’ molecular epidemiology in human diseases.
4. Discussion
The concept of molecular risk assessment [
41] has been used effectively to classify STEC strains into those that are attributed to outbreaks and life-threatening diseases in humans and those causing less severe disease or that are not involved in human disease.
E. coli strains have been characterized by serotype and the presence and subtype of VFs, as well as other toxins and plasmid-associated adherence and virulence factors. Thus, in this study, we investigated the presence of VFs in
E. coli strains to broaden the knowledge of the properties of
E. coli strains isolated from diverse sources. Recent epidemiological studies have revealed that the STEC serotypes O26, O103, O111, O145, and O157 are highly related to human infections (they may account for up to 80% of human STEC infections) [
8] and our results are in agreement with reports from other countries describing the high pathogenic potential of strains associated with these serogroups [
2]. The presence of non-LEE effector genes encoded by O-island O1-71 is highly attributed to strains that were often involved in outbreaks and serious disease in humans [
41,
42,
43]. We observed that
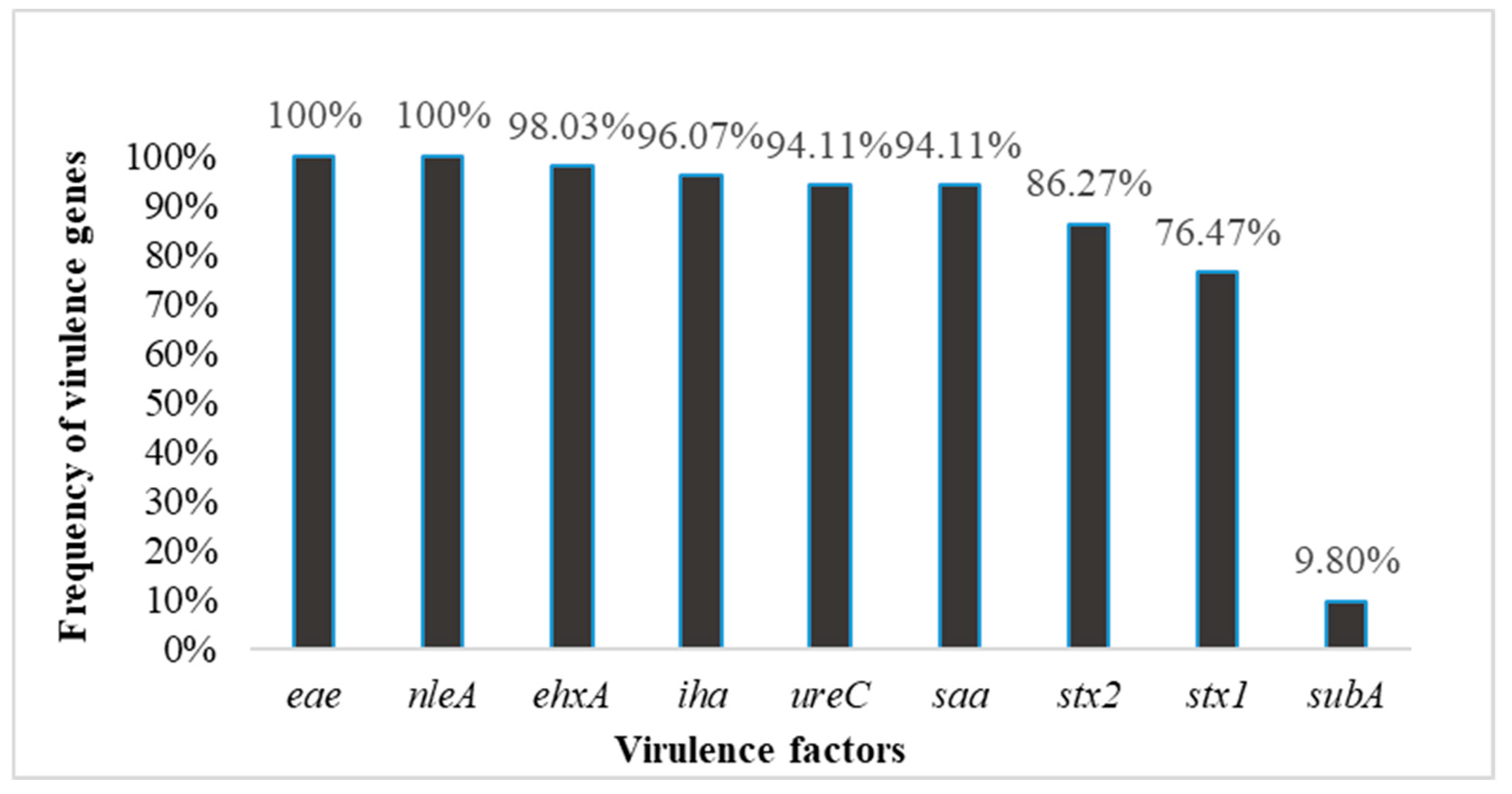
stx2-harboring isolates were more frequent than
stx1-harboring isolates. The dominant combinations of VFs present in the strains studied were
stx1,
stx2, and
eae (76.4% of strains). The distribution pattern of VFs was similar to the STEC strains isolated from domestic animals in Mexico [
44]. In France, the eminent toxin genotype was that of
stx2-carrying STEC strains. From these findings, it seems that STEC strains carrying the
stx1 gene are more often confronted than those carrying
stx2. Possession of OI-43/48, OI-71, and non-LEE effectors genes together with
stx2,
eae and a whole plasmid is the hallmark of highly virulent STEC strains that are frequently associated with outbreaks and serious diseases such as hemorrhagic colitis (HC) and HUS [
8]. Since the carriage of combinations of
stx genes has been correlated with severity of the disease, the
stx gene profile provides us an overview of the pathogenic potential of these STEC strains from diverse sources. STEC strains that carry both
stx2 and the
eae genes were more often associated with severe disease. Interestingly, all of the isolates were harboring the
eae gene, and it is reported that a significant majority of human STEC isolates obtained from HC or HUS patients contained
eae [
45]. The presence of
subA in the
E. coli strains was similar to that observed in some STEC strains isolated from human infections in the USA and Australia [
46]. It was previously reported that the amount of
stx2 production is capable of determining the severity of diseases caused by STEC strains. Results of this characterization have identified that
E. coli strains, defined by the presence of
eae,
subA, and the
nleA genes, may be considered a significant food safety threat. Plasmid-encoded VFs enhance pathogenesis and contribute to the survival of STEC in humans [
47], however, the pathogenic mechanisms of STEC infection are only partially understood. The varying prevalence of various VFs indicates that STEC strains are heterogeneous and it has been hypothesized that the combination of these genes may complement the Shiga toxin effect and enhance its virulence among STEC strains. The majority of strains in our research were positive for key VFs. Most of the strains carried the full complement of OI-43/48 VFs and all non-LEE-encoding effector genes. There is no specific pattern of virulence markers capable of interfering with the pathogenic potential of a given STEC isolate, and the search for a broad set of VFs has become the best strategy for measuring the microbiological and clinical risks that these pathogens may pose.
The 17 most frequently used antibiotics in clinical practice were employed to assess the actual frequency of antibiotic resistance in 51
E. coli strains. Generally, antibiotics are divided mainly into three categories on the basis of their functions (
Table 4). While MDR was seen, however, there was no common pattern of resistance. It is important to note that all strains were resistant to at least three different classes of antibiotic agents and were considered as MDR. The majority of the
E. coli strains of our research showed a high prevalence of resistance against first-line antibiotics (commonly prescribed oral antibiotics) such as ampicillin, penicillin, methicillin, gentamicin, vancomycin, novobiocin, streptomycin, kanamycin, and erythromycin (
Table 4). The similar results of antibiotic resistance of
E. coli strains were reported in developing countries such as Brazil, Turkey, China, and Ghana [
1,
4,
5,
45]. Our data exhibit a high resistance rate in
E. coli strains that is comparable to those reported in previous studies. These results illustrate the growing extent of the misuse of antibiotics in clinical practices. In particular,
E. coli strain resistance to methicillin, penicillin, vancomycin, and novobiocin reached 100%. The resistance rates of these
E. coli strains were higher than reported in developing countries [
3,
48]. These kinds of MDR STEC strains pose a serious threat to human health by affecting treatment against them. It was previously reported that patients infected with STEC should not be treated with antibiotics due to the risk of developing HUS [
26,
49].
E. coli forms part of the human gut’s commensal flora and has been identified as the predominant reservoir of genes for antibiotic resistance [
20]. These resistance genes are stable once acquired and are easily transferable to pathogenic bacteria [
50]. These transfers have effectively changed the etiological and pathogenic character of bacterial species [
51]. The majority of the STEC serotypes found in this study have also been reported in other countries. Depending on comparison by serotype and sequence type with human strains and the prevalence of VFs, the STEC strains could have a higher potential to cause human disease. Documented data showed that STEC strains isolated from human and food samples show a high prevalence of resistance to various types of antibiotics, including aminoglycosides, tetracycline, penicillin, and chloramphenicol. Molecular epidemiological studies have shown that the presence of certain antibiotic resistance genes, including the genes that encode resistance against tetracycline (
tetA and
tetB), ampicillin (
CITM), gentamicin (
aac (3)-IV), chloramphenicol (
cat1 and
cmlA), and aminoglycosides (
aadA1), is the key cause of antibiotic resistance in STEC [
52,
53]. With regard to macrolide antibiotics (erythromycin),
E. coli is an enteric bacterium, which are often non-susceptible due to the presence of chromosomal efflux pumps (
mel) or cellular impermeability [
54,
55]. A number of different mechanisms have been reported for the macrolide resistance of Gram-negative bacteria. These mechanisms include the presence of a number of genes, such as two ester genes (
ere(A) and
ere(B)) [
56,
57,
58], phosphorylase genes (
mph(A),
mph(B), and
mph(D)) [
59], and one rRNA methylase gene (
erm(B)) [
56,
60,
61]. In addition, if they acquire macrolide resistance genes, such as
mef(A) and
mef(B), that may increase their resistance levels further [
49]. We found that the pattern of phenotypic resistance of STEC strains was supported by the genotypic resistance of STEC strains isolated from various samples followed by a high prevalence of antibiotic resistance genes [
52]. Several studies have recorded that the prevalence of antibiotic-resistant
E. coli has increased since 1950 [
62]. An alarming increase in the prevalence of MDR
E. coli strains all over the world has been reported and this is a result of the spread of plasmids and other genetic elements. This has made antibiotic resistance a major public health issue globally [
62].
E. coli is an important food safety and public health concern because of its pathogenicity and potential for MDR.
Lastly, but most importantly, we found that
E. coli strains harbor a high level of VFs in addition to high MDR (
Table 3 and
Table 4). These results explain how
E. coli strains can effectively invade the human body and evade antibiotic treatment. The results of this research demonstrate that MDR
E. coli strains harbor a high frequency of VFs and their VF profiles are highly heterogeneous. These results suggest that use of antibiotics needs to be monitored by the private, public, and agricultural sectors as certain antibiotics can induce the production of
stx and thus encourage the onset of severe disease symptoms in humans. The improper use of antibiotics has become a public health problem worldwide in healthcare settings. Given the importance of
E. coli in food safety and public health, our findings on the prevalence of antibiotic resistance and VFs provide valuable information for risk management strategies to protect public health. The monitoring of the antibiotic resistance of STEC is pivotal due to the likelihood of the horizontal transfer of resistance genes from notorious STEC strains to other pathogens. In addition, the monitoring process will help in developing new treatment approaches and help in establishing effective control strategies that assist in stopping the spread of resistance. Hence, the molecular typing and contentious monitoring of antibiotic resistance could be helpful in developing efficacious control strategies against STEC and in formulating new antibiotics with reduced tendency for antibiotic resistance. The diversity in the prevalence of
stx genes, enterotoxin genes, and other virulence-related genes in this study and the other studies can be attributed to the geographical origin of samples, the sample size, the handling of the collected samples, the number of strains examined, the type of the examined VFs, and the role of the examined VFs in the pathogenesis of the disease. Surveillance data suggest that resistance in
E. coli is consistently highest for antibiotics that have been in use in human and veterinary medicine for the longest period of time [
62]. The past two decades have witnessed major increases in the emergence and spread of MDR bacteria and increasing resistance to newer compounds, such as fluoroquinolones and certain cephalosporins [
62]. Thus, surveillance and control measures need to be intensified to prevent further spread of these strains in the world.
In summary, E. coli is a significant cause of diarrheal and foodborne outbreaks, resulting in severe economic losses. This research demonstrates a high prevalence and heterogeneity of VF profiles among human MDR E. coli strains. The virulotyping revealed that the majority of E. coli strains were positive for stx1, stx2, eae, ehxA, ureC, nleA, and iha but subA was observed in a very small number of isolates. The serotype O26 strains possess the highest number of virulence-associated factors. The majority of isolates were resistant to two or more antibiotics that are commonly used in clinical medicine for the treatment of various bacterial diseases. However, all the 51 isolates were sensitive to imipenem and meropenem and, therefore, these drugs could be the drugs of choice in the treatment of STEC infections. We conclude that appropriate efforts should be focused on surveillance and that control measures to prevent/reduce further the spread of such microorganisms are crucial. However, further research using whole genome sequences would therefore be required to better understand the prevalence of VFs and antibiotic resistance in E. coli strains that may arise in this important human pathogen. The continuous monitoring and screening for MDR foodborne pathogens should be performed. Taken together, this knowledge will provide a better understanding of the risks associated with STEC and will aid in the development of appropriate and tailored intervention strategies.