Antimicrobial Resistance and Virulence of Methicillin-Resistant Staphylococcus aureus from Human, Chicken and Environmental Samples within Live Bird Markets in Three Nigerian Cities
Abstract
:1. Introduction
2. Results
2.1. Identification of Staphylococcus aureus
2.2. Antimicrobial Susceptibility
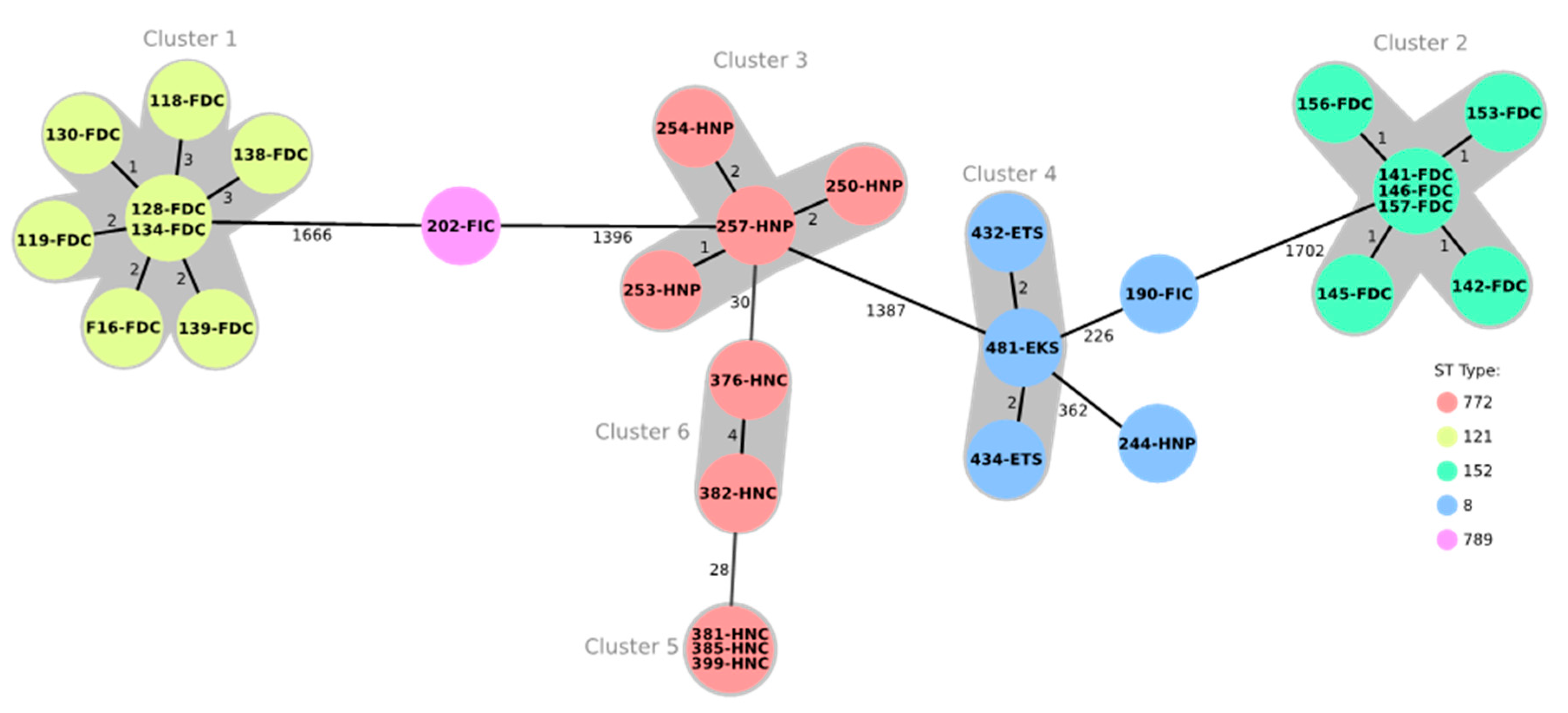
2.3. Multilocus Sequence Typing and Core Genome Multilocus Sequence Typing
2.4. Staphylococcal Cassette Chromosome mec (SCCmec) Typing
2.5. Staphylococcus aureus Protein A (spa) and Direct Repeat Unit (dru) Typing
2.6. Virulence Genes
2.7. Antimicrobial Resistance Genes and Resistance-Mediating Mutations
3. Discussion
4. Materials and Method
4.1. Sampling Locations and Sample Collection
4.2. Isolation and Phenotypic Identification of Staphylococci Including S. aureus
4.3. Antimicrobial Susceptibility Testing
4.4. DNA Extraction and Whole Genome Sequencing
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Rahman, M.M.; Amin, K.B.; Rahman, S.; Khair, A.; Rahman, M.; Hossain, A.; Rahman, A.; Parvez, M.S.; Miura, N.; Alam, M.M. Investigations of methicillin-resistant Staphylococcus aureus among clinical isolates from humans and animals by culture methods and multiplex PCR. BMC Vet. Res. 2018, 14, 300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fritz, S.A.; Hogan, P.G.; Singh, L.N.; Thompson, R.M.; Wallace, M.A.; Whitney, K.; Al-Zubeidi, D.; Burnham, C.A.; Fraser, V.J. Staphylococcus aureus contamination of environmental surfaces in households with children infected with methicillin-resistant S. aureus. JAMA Pediatr. 2014, 168, 1030–1038. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tong, S.Y.C.; Davis, J.S.; Eichenberger, E.; Holland, T.L.; Fowler, G., Jr. Staphylococcus aureus infections: Epidemiology, pathophysiology, clinical manifestation, and management. Clin. Microbiol. Rev. 2015, 28, 603–661. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lakhundi, S.; Zhang, K. Methicillin-resistant Staphylococcus aureus: Molecular characterization, evolution, and epidemiology. Clin. Microbiol. Rev. 2018, 31, e00020-18. [Google Scholar] [CrossRef] [Green Version]
- Price, L.B.; Stegger, M.; Hasman, H.; Aziz, M.; Larsen, J.; Andersen, P.S.; Pearson, T.; Waters, A.E.; Foster, J.T.; Schupp, J.; et al. Staphylococcus aureus CC398: Host adaptation and emergence of methicillin resistance in livestock. mBio 2012, 3, e00305-11. [Google Scholar] [CrossRef] [Green Version]
- Shittu, A.O.; Oyedara, O.; Abegunrin, F.; Okon, K.; Raji, A.; Taiwo, S.; Ogunsola, F.; Onyedibe, K.; Elisha, G. Characterization of methicillin-susceptible and -resistant staphylococci in the clinical setting: A multicentre study in Nigeria. BMC Infect. Dis. 2012, 12, 286. [Google Scholar] [CrossRef] [Green Version]
- Raji, A.; Ojemhen, O.; Umejiburu, U.; Ogunleye, A.; Blanc, D.S.; Basset, P. High genetic diversity of Staphylococcus aureus in a tertiary care hospital in Southwest Nigeria. Diagn. Microbiol. Infect. Dis. 2013, 77, 367–369. [Google Scholar] [CrossRef]
- Nworie, A.; Onyema, A.S.; Okekpa, S.I.; Elom, M.O.; Umoh, N.O.; Usanga, V.U.; Ibiam, G.A.; Ukwah, B.N.; Nwadi, L.C.; Ezeruigbo, C.; et al. A novel methicillin-resistant Staphylococcus aureus t11469 and a poultry endemic strain t002 (ST5) are present in chicken in Ebonyi State, Nigeria. BioMed Res. Int. 2017, 2017, 2936461. [Google Scholar] [CrossRef] [Green Version]
- Kwoji, I.D.; Jauro, S.; Musa, J.A.; Lekko, Y.M.; Salihu, S.I.; Danchuwa, H.A. Phenotypic detection of methicillin-resistant Staphylococcus aureus in village chickens from poultry markets in Maiduguri, Nigeria. J. Adv. Vet. Anim. Res. 2019, 6, 163–167. [Google Scholar] [CrossRef]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing, 29th ed.; CLSI Supplement M100; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2019. [Google Scholar]
- Schwarz, S.; Silley, P.; Simjee, S.; Woodford, N.; van Duijkeren, E.; Johnson, A.P.; Gaastra, W. Editorial: Assessing the antimicrobial susceptibility of bacteria obtained from animals. J. Antimicrob. Chemother. 2010, 65, 601–604. [Google Scholar] [CrossRef]
- CLSI. Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals, 4th ed.; CLSI Supplement VET08; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2018. [Google Scholar]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing, 30th ed.; CLSI Supplement M100; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2020. [Google Scholar]
- Otto, M. MRSA virulence and spread. Cell Microbiol. 2012, 14, 1513–1521. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fisher, E.L.; Otto, M.; Cheung, G.Y.C. Basis of virulence in enterotoxin-mediated staphylococcal food poisoning. Front. Microbiol. 2018, 13, 436. [Google Scholar] [CrossRef] [PubMed]
- Griffith, E.C.; Wallace, M.J.; Wu, Y.; Kumar, G.; Gajewski, S.; Jackson, P.; Phelps, G.A.; Zheng, Z.; Rock, C.O.; Lee, R.E.; et al. The structural and functional basis for recurring sulfa drug resistance mutations in Staphylococcus aureus dihydropteroate synthase. Front. Microbiol. 2018, 9, 1369. [Google Scholar] [CrossRef] [PubMed]
- Fournié, G.; Guitian, J.; Desvaux, S.; Mangtani, P.; Ly, S.; Cong, V.C.; San, S.; Dung, D.; Holl, D.; Pfeiffer, D.U.; et al. Identifying live bird markets with the potential to act as reservoirs of avian influenza A (H5N1) virus: A survey in northern Vietnam and Cambodia. PLoS ONE 2012, 7, e37986. [Google Scholar] [CrossRef] [Green Version]
- Greger, M. The human/animal interface: Emergence and resurgence of zoonotic infectious diseases. Crit. Rev. Microbiol. 2007, 33, 243–299. [Google Scholar] [CrossRef]
- Zhou, X.; Li, Y.; Wang, Y.; Edwards, J.; Guo, F.; Clements, A.C.; Huang, B.; Magalhaes, R.J. The role of live poultry movement and live bird market biosecurity in the epidemiology of influenza A (H7N9): A cross-sectional observational study in four eastern China provinces. J. Infect. 2015, 71, 470–479. [Google Scholar] [CrossRef] [Green Version]
- Otalu, O., Jr.; Kabir, J.; Okolocha, E.C.; Umoh, V.P.; Kwaga, J.K.P.; Owoladun, A.O. Detection of methicillin resistant Staphylococcus aureus in chicken carcasses and live birds in Zaria, Nigeria. FUTA J. Res. Sci. 2015, 1, 132–138. [Google Scholar]
- Suleiman, A.; Zaria, L.T.; Grema, H.A.; Ahmadu, P. Antimicrobial resistant coagulase positive Staphylococcus aureus from chickens in Maiduguri, Nigeria. Sokoto J. Vet. Sci. 2013, 11, 51–55. [Google Scholar] [CrossRef] [Green Version]
- De Boer, E.; Zwartkruis-Nahuis, J.T.; Wit, B.; Huijsdens, X.W.; de Neeling, A.J.; Bosch, T.; van Oosterom, R.A.; Vila, A.; Heuvelink, A.E. Prevalence of methicillin-resistant Staphylococcus aureus in meat. Int. J. Food Microbiol. 2009, 134, 52–56. [Google Scholar] [CrossRef]
- Feßler, A.T.; Kadlec, K.; Hassel, M.; Hauschild, T.; Eidam, C.; Ehricht, R.; Monecke, S.; Schwarz, S. Characterization of methicillin-resistant Staphylococcus aureus isolates from food and food products of poultry origin in Germany. Appl. Environ. Microbiol. 2011, 77, 7151–7157. [Google Scholar] [CrossRef] [Green Version]
- Monecke, S.; Ruppelt, A.; Wendlandt, S.; Schwarz, S.; Slickers, P.; Ehricht, R.; Jäckel, S.C. Genotyping of Staphylococcus aureus isolates from diseased poultry. Vet. Microbiol. 2013, 162, 806–812. [Google Scholar] [CrossRef] [PubMed]
- Wendlandt, S.; Kadlec, K.; Feßler, A.T.; Monecke, S.; Ehricht, R.; van de Giessen, A.W.; Hengeveld, P.D.; Huijsdens, X.; Schwarz, S.; van Duijkeren, E. Resistance phenotypes and genotypes of methicillin-resistant Staphylococcus aureus isolates from broiler chickens at slaughter and abattoir workers. J. Antimicrob. Chemother. 2013, 68, 2458–2463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shittu, A.O.; Okon, K.; Adesida, S.; Oyedara, O.; Witte, W.; Strommenger, B.; Layer, F.; Nübel, U. Antibiotic resistance and molecular epidemiology of Staphylococcus aureus in Nigeria. BMC Microbiol. 2011, 11, 92. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abdulgader, S.A.; Shittu, A.O.; Nicol, M.B.; Kabar, M. Molecular epidemiology of methicillin-resistant Staphylococcus aureus in Africa: A systematic review. Front. Microbiol. 2015, 6, 348. [Google Scholar] [CrossRef]
- Egyir, B.; Guardabassi, L.; Sørum, M.; Nielsen, S.S.; Kolekang, A.; Frimpong, E.; Addo, K.K.; Newman, M.J.; Larsen, A.R. Molecular epidemiology and antimicrobial susceptibility of clinical Staphylococcus aureus from healthcare institutions in Ghana. PLoS ONE 2014, 9, e89716. [Google Scholar] [CrossRef] [Green Version]
- Ghebremedhin, B.; Olugbosi, M.O.; Raji, A.M.; Layer, F.; Bakare, R.A.; König, B.; König, W. Emergence of a community-associated methicillin-resistant Staphylococcus aureus strain with a unique resistance profile in Southwest Nigeria. J. Clin. Microbiol. 2009, 47, 2975–2980. [Google Scholar] [CrossRef] [Green Version]
- Rao, Q.; Shang, W.; Hu, X.; Rao, X. Staphylococcus aureus ST121: A globally disseminated hypervirulent clone. Med. Microbiol. 2015, 64, 1462–1473. [Google Scholar] [CrossRef]
- Brennan, G.I.; Shore, A.C.; Corcoran, S.; Tecklenborg, S.; Coleman, D.C.; O’Connell, B. Emergence of Hospital- and Community-Associated Panton-Valentine Leukocidin-Positive methicillin-resistant Staphylococcus aureus genotype ST772-MRSA-V in Ireland and detailed investigation of an ST772-MRSA-V cluster in a neonatal intensive care unit. J. Clin. Microbiol. 2012, 50, 841–847. [Google Scholar] [CrossRef] [Green Version]
- Monecke, S.; Baier, V.; Coombs, G.W.; Slickers, P.; Ziegler, A.; Ehricht, R. Genome sequencing and molecular characterization of Staphylococcus aureus ST772-MRSA-V, “Bengal Bay Clone”. BMC Res. Note 2013, 6, 548. [Google Scholar]
- Conceição, T.; Coelho, C.; Santos-Silva, I.; de Lencastre, H.; Aires-de-Sousa, M. Epidemiology of methicillin-resistant and -susceptible Staphylococcus aureus in Luanda, Angola: First description of the spread of the MRSA ST5-IVa clone in the African continent. Microb. Drug Resist. 2014, 20, 441–449. [Google Scholar] [CrossRef]
- Chambers, H.F.; DeLeo, F.R. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat. Rev. Microbiol. 2010, 7, 629–641. [Google Scholar] [CrossRef] [PubMed]
- DeLeo, F.R.; Otto, M.; Kreiswirth, B.N.; Chambers, H.F. Community-associated methicillin-resistant Staphylococcus aureus. Lancet 2010, 375, 1557–1568. [Google Scholar] [CrossRef] [Green Version]
- Boerlin, P.; Burnens, A.P.; Frey, J.; Kuhnert, P.; Nicolet, J. Molecular epidemiology and genetic linkage of macrolide and aminoglycoside resistance in Staphylococcus intermedius of canine origin. Vet. Microbiol. 2001, 79, 155–169. [Google Scholar] [CrossRef]
- Soimala, T.; Lübke-Becker, A.; Hanke, D.; Eichhorn, I.; Feßler, A.T.; Schwarz, S.; Eule, J.C. Molecular and phenotypic characterization of methicillin-resistant Staphylococcus pseudintermedius from ocular surfaces of dogs and cats suffering from ophthalmological diseases. Vet. Microbiol. 2020, 244. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.Y. Molecular pathogenesis of Staphylococcus aureus infection. Pediatr. Res. 2009, 65, 71R–77R. [Google Scholar] [CrossRef]
- Argudin, M.A.; Mendoza, M.C.; Rodicio, M.R. Food poisoning and Staphylococcus aureus enterotoxins. Toxins 2010, 2, 1751–1773. [Google Scholar] [CrossRef]
- Pokhrel, R.H.; Aung, M.S.; Thapa, B.; Chaudhary, R.; Mishra, S.K.; Kawaguchiya, M.; Urushibara, N.; Kobayashi, N. Detection of ST772 Panton-Valentine leucocidin-positive methicillin-resistant Staphylococcus aureus (Bengal Bay Clones) and ST22 S. aureus isolates with genetic variant of elastin binding protein in Nepal. New Microbes New Infect. 2016, 11, 20–27. [Google Scholar] [CrossRef] [Green Version]
- McCarthy, A.J.; Lindsay, J.A. The distribution of plasmids that carry virulence and resistance genes in Staphylococcus aureus is lineage associated. BMC Microbiol. 2012, 12, 104. [Google Scholar] [CrossRef] [Green Version]
- McCarthy, A.J.; Witney, A.A.; Lindsay, J.A. Staphylococcus aureus temperate bacteriophage: Carriage and horizontal gene transfer is lineage associated. Front. Cell. Infect. Microbiol. 2012, 2, 6. [Google Scholar] [CrossRef] [Green Version]
- Kraushaar, B.; Ballhausen, B.; Leeser, D.; Tenhagen, B.A.; Käsbohrer, A.; Fetsch, A. Antimicrobial resistances and virulence markers in methicillin-resistant Staphylococcus aureus from broiler and turkey: A molecular view from farm to fork. Vet. Microbiol. 2017, 200, 25–32. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Leopold, S.R.; Goering, R.V.; Witten, A.; Harmsen, D.; Mellmann, A. Bacterial whole-genome sequencing revisited: Portable, scalable, and standardized analysis for typing and detection of virulence and antibiotic resistance genes. J. Clin. Microbiol. 2014, 52, 2365–2370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feldgarden, M.; Brover, V.; Haft, D.H.; Prasad, A.B.; Slotta, D.J.; Tolstoy, I.; Tyson, G.H.; Zhao, S.; Hsu, C.H.; McDermott, P.F.; et al. Validating the AMRFinder tool and resistance gene database by using antimicrobial resistance genotype-phenotype correlations in a collection of isolates. Antimicrob. Agents Chemother. 2019, 63, e00483-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carattoli, A.; Zankari, E.; García-Fernández, A.; Larsen, M.V.; Lund, O.; Villa, L.; Aarestrup, F.M.; Hasman, H. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef] [Green Version]
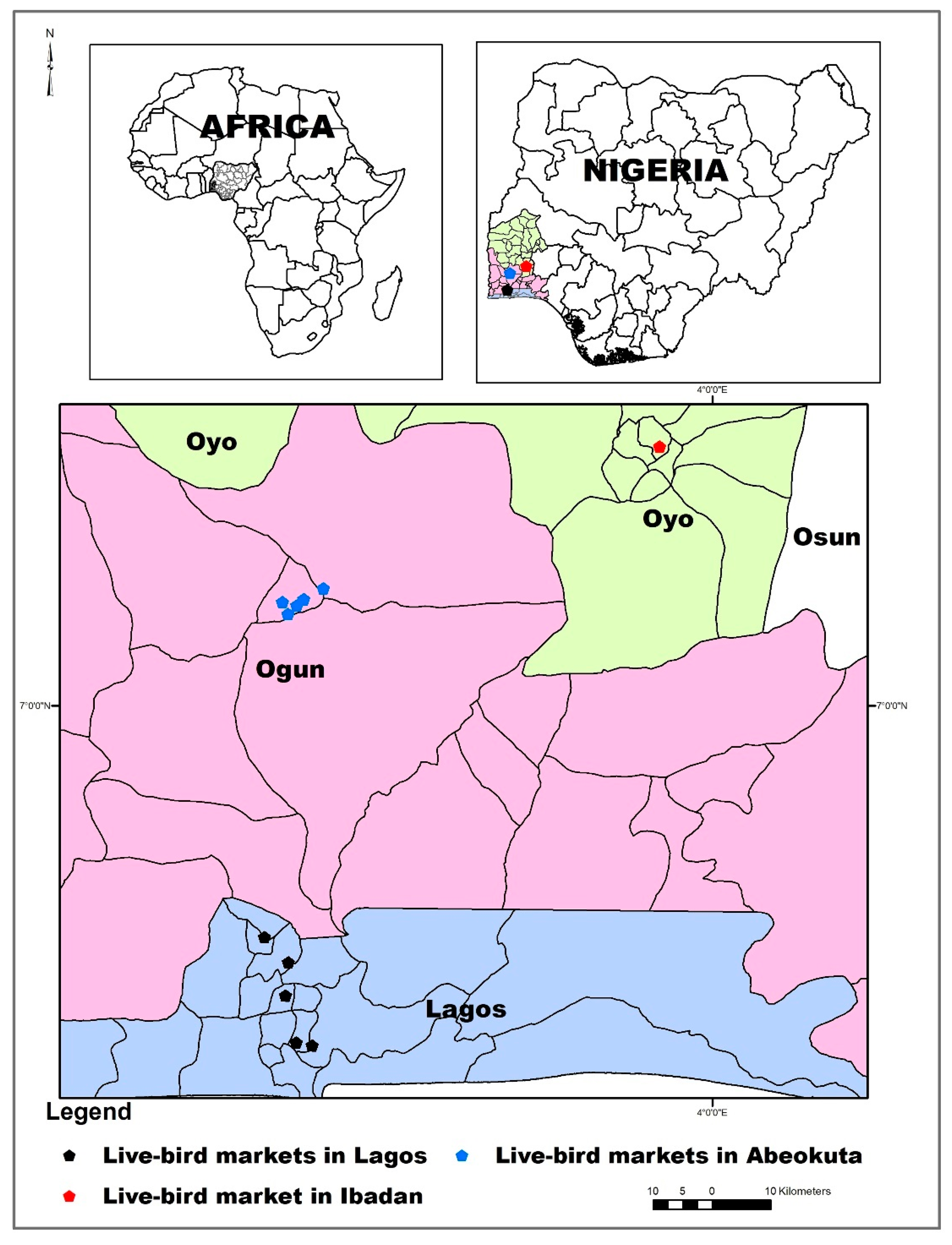
| Sample Categories | Sample Locations and Sample Size | |||
|---|---|---|---|---|
| Abeokuta | Ibadan | Lagos | Total | |
| Freshly dressed chicken | 13 | 15 | 0 | 28 |
| Frozen/imported chicken | 2 | 0 | 0 | 2 |
| Processors | 10 | 0 | 0 | 10 |
| Consumers | 0 | 0 | 8 | 8 |
| Knives | 2 | 0 | 0 | 2 |
| Tables | 0 | 6 | 0 | 6 |
| Total | 27 | 21 | 8 | 56 |
| Antimicrobial Agent (s) | Number of Isolates with MIC Value (mg/L) Is | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0.008 | 0.015 | 0.03 | 0.06 | 0.12 | 0.25 | 0.5 | 1 | 2 | 4 | 8 | 16 | 32 | 64 | 128 | 256 | 512 | |
| Vancomycin (VAN) | - | - | - | - | - | - | 51 | 5 | - | - | - | - | |||||
| Florfenicol (FFC) | - | - | - | - | - | 47 | 9 | - | - | - | - | - | |||||
| Erythromycin (ERY) | - | - | - | - | 16 | 24 | - | - | - | 3 | 12 | 1 | |||||
| Clindamycin (CLI) | - | 3 | 51 | 2 | - | - | - | - | - | - | - | - | |||||
| Streptomycin (STR) | - | - | - | - | 4 | 39 | 12 | 1 | - | - | - | - | |||||
| Gentamicin (GEN) | 1 | 13 | 16 | 6 | 2 | - | - | - | 2 | 6 | 5 | 3 | 2 | ||||
| Neomycin (NEO) | 1 | 15 | 22 | 1 | - | - | - | - | - | - | 17 | ||||||
| Ciprofloxacin (CIP) | - | - | - | - | - | 2 | 28 | 6 | 1 | - | 3 | 14 | 2 | ||||
| Enrofloxacin 1 (ENR) | - | - | - | - | 12 | 18 | 7 | - | - | 13 | 5 | - | 1 | ||||
| Nalidixic acid (NAL) | - | - | - | - | - | - | - | - | 1 | 5 | 27 | 4 | 19 | ||||
| Trimethoprim/sulfamethoxazole (1:19) c (SXT) | - | - | - | - | 1 | 7 | 30 | 12 | 4 | - | 1 | - | 1 | ||||
| Tetracycline (TET) | - | 6 | 12 | 4 | - | - | - | - | 6 | 26 | 1 | 1 | |||||
| Doxycycline (DOX) | - | 13 | 9 | 1 | - | 5 | 23 | 5 | - | - | - | - | |||||
| Linezolid (LZD) | - | - | - | - | - | 14 | 37 | 5 | - | - | - | - | |||||
| Tiamulin (TIA) | - | - | - | 2 | 32 | 22 | - | - | - | - | - | - | |||||
| Quinupristin/dalfopristin (Q/D) | - | - | - | - | 29 | 26 | 1 | - | - | - | |||||||
| ID | Source | Resistance Profile * | spa | dru | SCCmec | MLST |
|---|---|---|---|---|---|---|
| 118 | FDC/Abeokuta | BLA | t314 | Nt | IVa | ST121 |
| 119 | FDC/Abeokuta | BLA-SXT-TET | t314 | Nt | IVa | ST121 |
| 128 | FDC/Abeokuta | BLA-TET | t314 | Nt | IVa | ST121 |
| 130 | FDC/Abeokuta | BLA | t314 | Nt | IVa | ST121 |
| 134 | FDC/Abeokuta | BLA-TET | t314 | Nt | IVa | ST121 |
| 138 | FDC/Abeokuta | BLA | t314 | Nt | IVa | ST121 |
| 139 | FDC/Abeokuta | BLA-TET | t314 | Nt | IVa | ST121 |
| F16 | FDC/Abeokuta | BLA-TET | t314 | Nt | IVa | ST121 |
| 141 | FDC/Ibadan | BLA-SXT-TET | t4690 | dt11a | Vc | ST152 |
| 142 | FDC/Ibadan | BLA-SXT-TET | t4690 | dt11a | Vc | ST152 |
| 145 | FDC/Ibadan | BLA-ERY-TET | t4690 | dt11a | Vc | ST152 |
| 146 | FDC/Ibadan | BLA-SXT-TET | t4690 | dt11a | Vc | ST152 |
| 153 | FDC/Ibadan | BLA-TET | t4690 | dt11a | Vc | ST152 |
| 156 | FDC/Ibadan | BLA-TET | t4690 | dt9aw | Vc | ST152 |
| 157 | FDC/Ibadan | BLA-TET | t4690 | dt11a | Vc | ST152 |
| 190 | FIC/Abeokuta | BLA-CIP-GEN-TET | t1476 | dt10dr ** | Unknown | ST8 |
| 202 | FIC/Abeokuta | BLA-CIP-GEN-TET | t091 | dt11a | V | ST789 |
| 244 | HNP/Abeokuta | BLA-CIP-SXT-TET | t2331 | dt11dw ** | Vc | ST8 |
| 250 | HNP/Abeokuta | BLA-CIP-ERY-GEN | t657 | dt10ao | V | ST772 |
| 253 | HNP/Abeokuta | BLA-CIP-ERY-GEN | t657 | dt10ao | V | ST772 |
| 254 | HNP/Abeokuta | BLA-CIP-ERY-GEN | t657 | dt10ao | V | ST772 |
| 257 | HNP/Abeokuta | BLA-CIP-ERY-GEN | t657 | dt10ao | V | ST772 |
| 376 | HNC/Lagos | BLA-CIP-ERY-GEN | t657 | dt10cj | V | ST772 |
| 381 | HNC/Lagos | BLA-CIP-ERY-GEN | t657 | dt10ao | V | ST772 |
| 382 | HNC/Lagos | BLA-CIP-ERY-GEN | t657 | dt10cj | V | ST772 |
| 385 | HNC/Lagos | BLA-CIP-ERY-GEN | t657 | dt10ao | V | ST772 |
| 399 | HNC/Lagos | BLA-CIP-ERY-GEN | t657 | dt10ao | V | ST772 |
| 432 | ETS/Ibadan | BLA-TET | t12236 | dt11a | Vc | ST8 |
| 434 | ETS/Ibadan | BLA-TET | t12236 | dt11a | Vc | ST8 |
| 481 | EKS/Abeokuta | BLA-TET | t12236 | dt9aw | Vc | ST8 |
| Genes Encoding Virulence Factors | Present |
|---|---|
| cap8 H, I, J, K (capsular polysaccharide) | All ST121 and ST789 isolates |
| chp (chemotaxis inhibitory protein) | Only in one isolate (ST8, t1476) |
| clfA (clumping factor A) | All isolates but one (ST152/t4690) |
| clfB (clumping factor B) | All but four isolates (ST152/t4690, ST8/t2331, ST8/t12236 and ST772/t657) |
| cna (collagen adhesin) | All ST152, ST772 isolates and one ST121 isolate |
| ebp (elastin binding protein) | All ST8, ST152, ST789 and ST772 isolates |
| esaC (Type VII secretion system) | All ST8, ST121, ST152, ST789 isolates |
| esxB (Type VII secretion system) | All ST8, ST121, ST152, ST789 isolates |
| fnbB (fibronectin binding protein) | All but three isolates (ST772) |
| hysA (hyaluronidase) | All ST8, ST121, ST152, ST772 isolates |
| lukF-PV (Panton-Valentine leukocidin) | All ST121, ST152 except one isolate (157), ST772 isolates |
| lukS-PV (Panton-Valentine leukocidin) | All ST121, ST152 except one isolate (157), ST772 isolates |
| lukD (Leukotoxin D) | All ST8, ST121, ST152, ST789 isolates |
| lukE (Leukotoxin E) | All ST8, ST121, ST152, ST789 isolates |
| map (MHC class II analog protein) | All ST8, ST121, ST772, ST789 isolates |
| sak (staphylokinase) | All ST8, ST121, ST152, ST789 isolates |
| sdrC (serine aspartate repeat protein) | All ST8, ST121, ST772, ST789 isolates |
| sdrD (serine aspartate repeat protein) | All ST8, ST121, ST772, ST789 isolates |
| sdrE (serine aspartate repeat protein) | All but one isolate (ST8/t2331) |
| sea (staphylococcal enterotoxin) | All ST772, ST789 and four ST8 isolates |
| seb (staphylococcal enterotoxin) | All ST121 isolates |
| sec (staphylococcal enterotoxin) | Only in five isolates (ST772) |
| sell (staphylococcal enterotoxin-like) | Only in five isolates (ST772) |
| Profile | Resistance Genes Present | MLST/spa/dru Type (Number of Isolates) |
| 1 | mecA-dfrG-tet(K)-tet(38) | ST152/t4690/dt11a (4) or ST152/t4690/dt9aw (1) |
| 2 | mecA-blaZ-dfrG-fosB-tet(38) | ST121/t314/nt (3) |
| 3 | mecA-blaZ-dfrG-tet(K)-tet(38) | ST152/t4690/dt11a (2) |
| 4 | mecA-blaZ-dfrG-fosB-tet(K)-tet(38) | ST8/t12236/dt11a (2), ST8/t12236/dt9aw (1), and ST121/t314/nt (5) |
| 5 | mecA-blaZ-dfrG-fosB-tet(38)-msr(A)-mph(C)-aacA/aphD-aphA3-Δsat4-ΔaadE | ST772/t657/dt10ao (7), ST772/t657/dt10cj (2) |
| 6 | mecA-blaZ-dfrG-fosB-tet(K)-tet(38) | ST8/t2331/dt11dw (1) |
| 7 | mecA-blaZ-dfrG-dfrS1-fosB-tet(K)-tet(38)-aacA/aphD | ST8/t1476/dt10dr (1) |
| 8 | mecA-blaZ-dfrG-tet(K)-tet(38)-aacA/aphD-aphA3-sat4-ΔaadE | ST789/t091/dt11a (1) |
| Sample Categories | Sample Locations and Sample Size | |||
|---|---|---|---|---|
| Abeokuta | Ibadan | Lagos | Total | |
| A. On-farm | ||||
| Live chicken | 68 | - | 74 | 142 |
| B. Live bird markets (meat samples) | ||||
| Freshly dressed chicken | 20 | 5 | 33 | 58 |
| Frozen/imported chicken | 27 | - | 63 | 90 |
| C. Live bird markets (human samples) | ||||
| Processors | 12 | 5 | 15 | 32 |
| Consumers | 34 | - | 245 | 279 |
| D. Live bird markets (environmental samples) | ||||
| Knives | 6 | - | 53 | 59 |
| Tables | 16 | 5 | 53 | 74 |
| Total | 183 | 15 | 536 | 734 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ogundipe, F.O.; Ojo, O.E.; Feßler, A.T.; Hanke, D.; Awoyomi, O.J.; Ojo, D.A.; Akintokun, A.K.; Schwarz, S.; Maurischat, S. Antimicrobial Resistance and Virulence of Methicillin-Resistant Staphylococcus aureus from Human, Chicken and Environmental Samples within Live Bird Markets in Three Nigerian Cities. Antibiotics 2020, 9, 588. https://doi.org/10.3390/antibiotics9090588
Ogundipe FO, Ojo OE, Feßler AT, Hanke D, Awoyomi OJ, Ojo DA, Akintokun AK, Schwarz S, Maurischat S. Antimicrobial Resistance and Virulence of Methicillin-Resistant Staphylococcus aureus from Human, Chicken and Environmental Samples within Live Bird Markets in Three Nigerian Cities. Antibiotics. 2020; 9(9):588. https://doi.org/10.3390/antibiotics9090588
Chicago/Turabian StyleOgundipe, Flora Olubunmi, Olufemi Ernest Ojo, Andrea T. Feßler, Dennis Hanke, Olajoju Jokotola Awoyomi, David Ajiboye Ojo, Aderonke Kofoworola Akintokun, Stefan Schwarz, and Sven Maurischat. 2020. "Antimicrobial Resistance and Virulence of Methicillin-Resistant Staphylococcus aureus from Human, Chicken and Environmental Samples within Live Bird Markets in Three Nigerian Cities" Antibiotics 9, no. 9: 588. https://doi.org/10.3390/antibiotics9090588
APA StyleOgundipe, F. O., Ojo, O. E., Feßler, A. T., Hanke, D., Awoyomi, O. J., Ojo, D. A., Akintokun, A. K., Schwarz, S., & Maurischat, S. (2020). Antimicrobial Resistance and Virulence of Methicillin-Resistant Staphylococcus aureus from Human, Chicken and Environmental Samples within Live Bird Markets in Three Nigerian Cities. Antibiotics, 9(9), 588. https://doi.org/10.3390/antibiotics9090588







