Bacteriophage-Induced Lipopolysaccharide Mutations in Escherichia coli Lead to Hypersensitivity to Food Grade Surfactant Sodium Dodecyl Sulfate
Abstract
:1. Introduction
2. Results
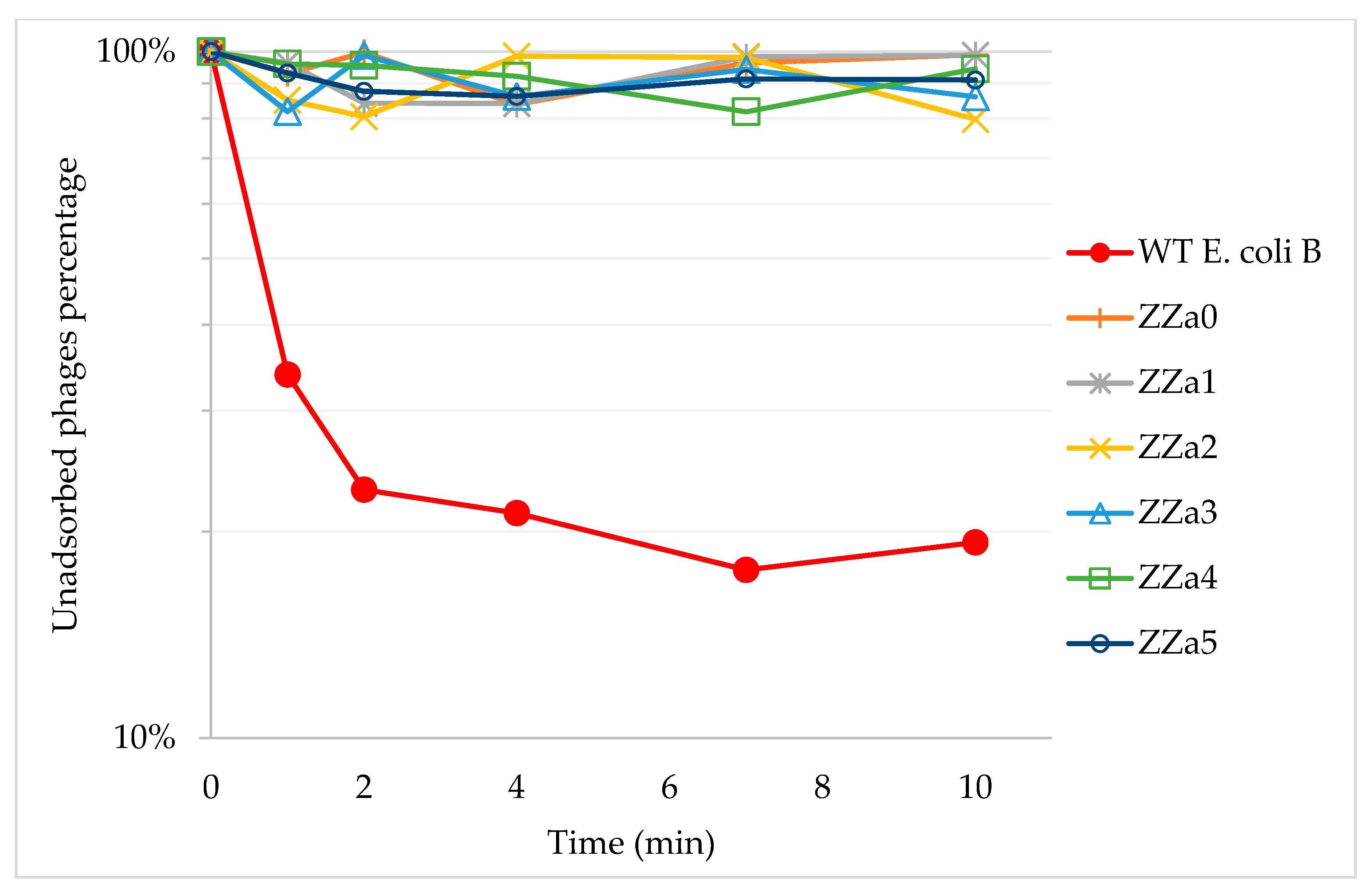
2.1. Phage T4 Resistant Mutant Isolation and Phage Adsorption Assays
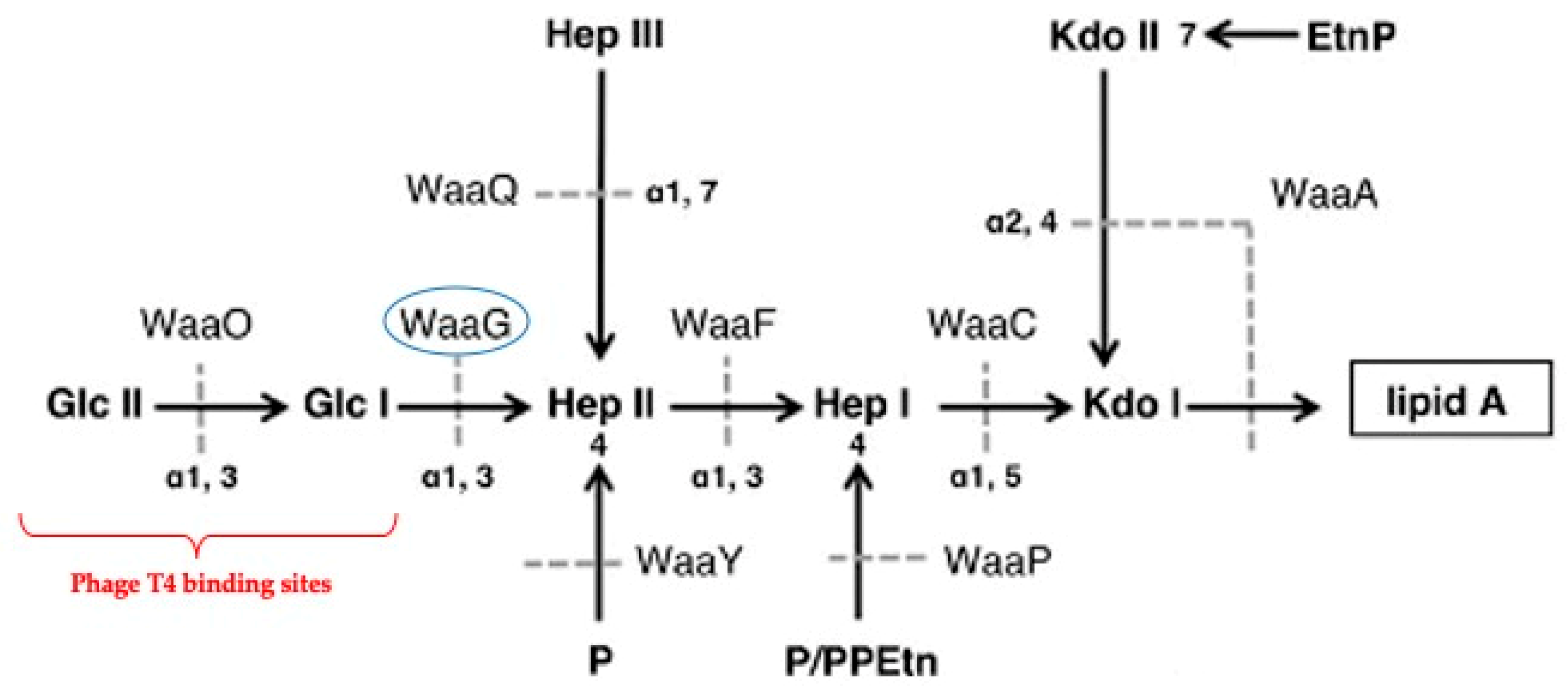
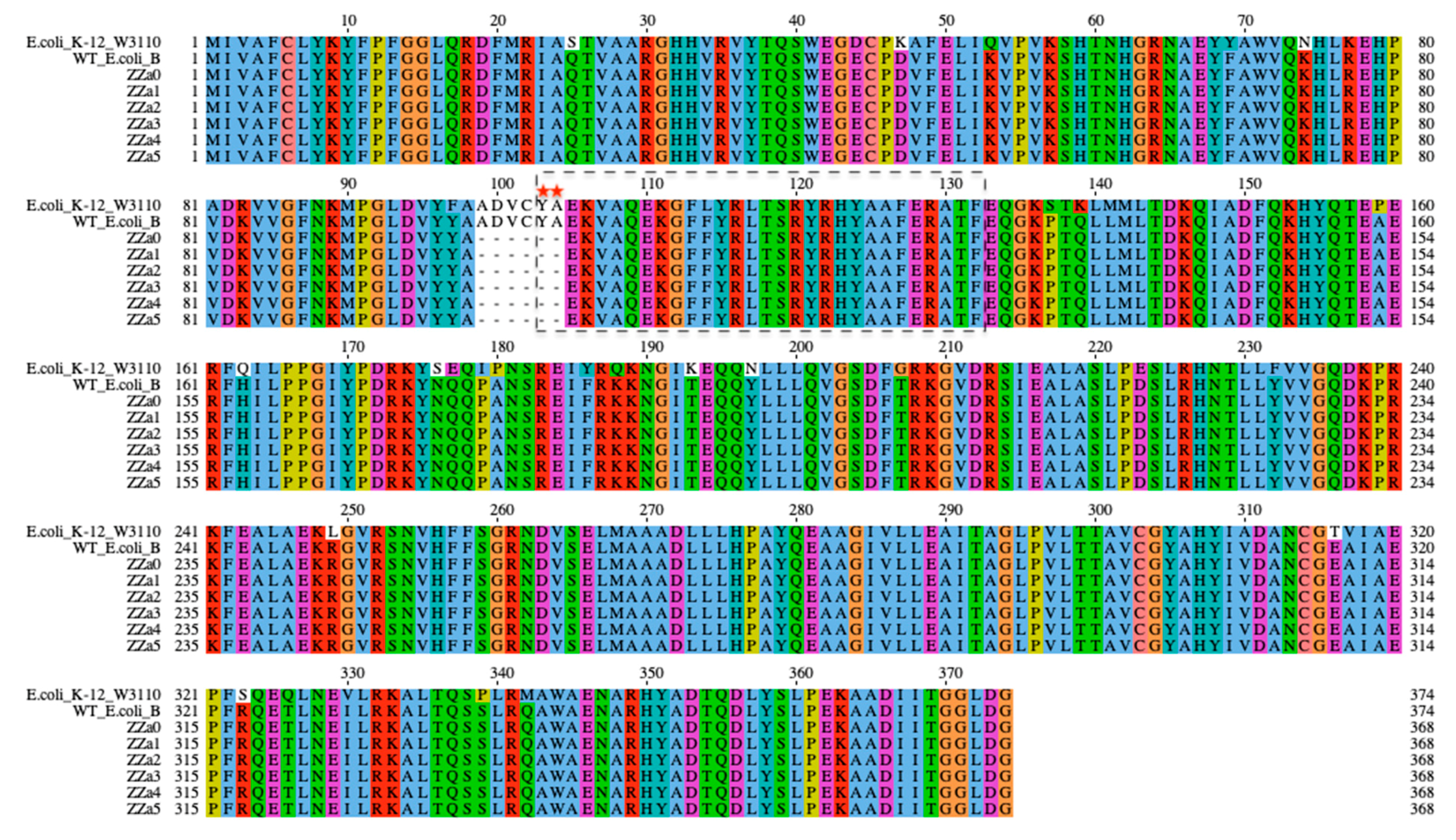
2.2. Identification of Genetic Mutations Associated with phage Resistance
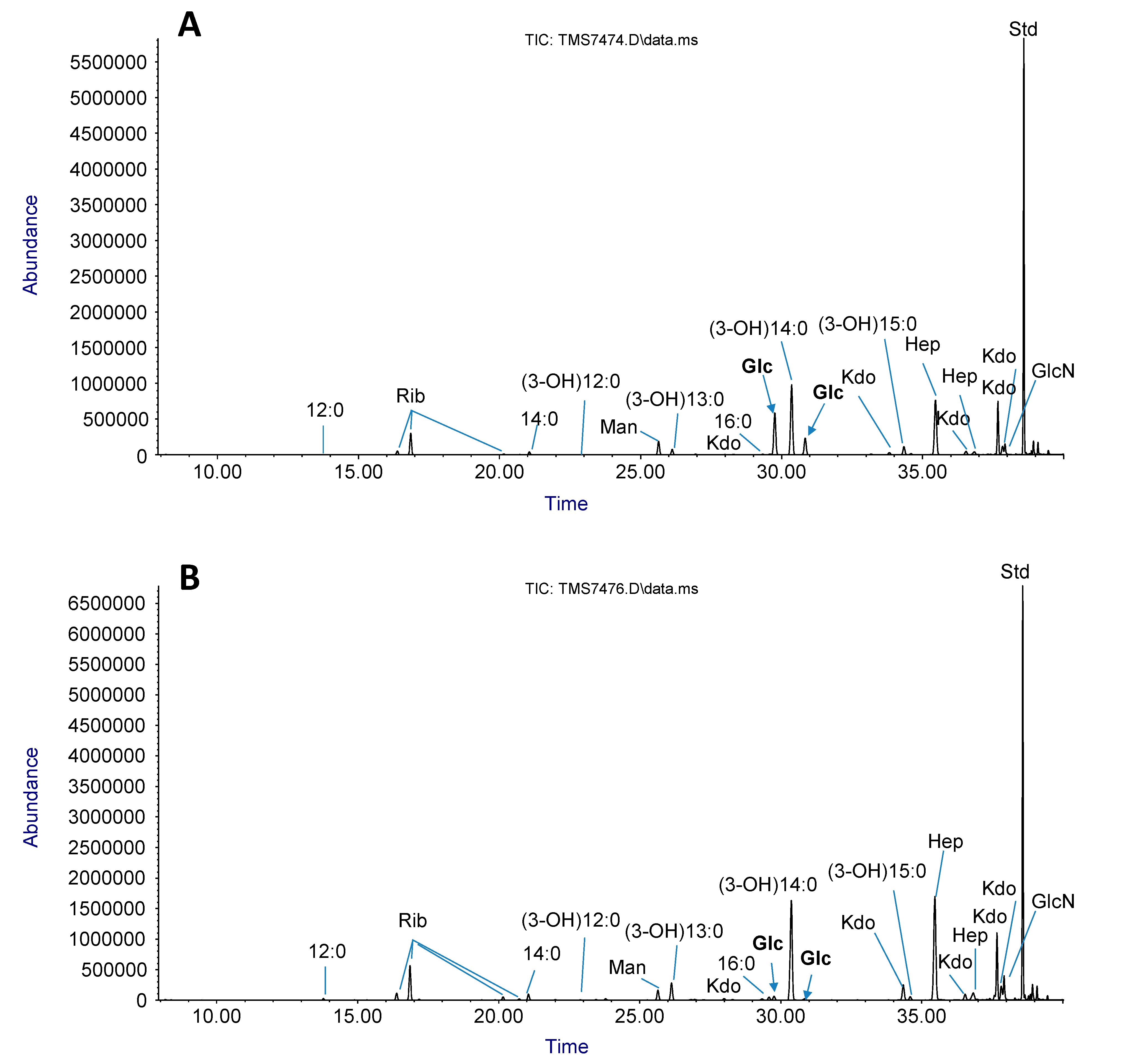
2.3. LPS sugar Composition Analysis
2.4. Minimum Inhibitory Concentration (MIC) Assay
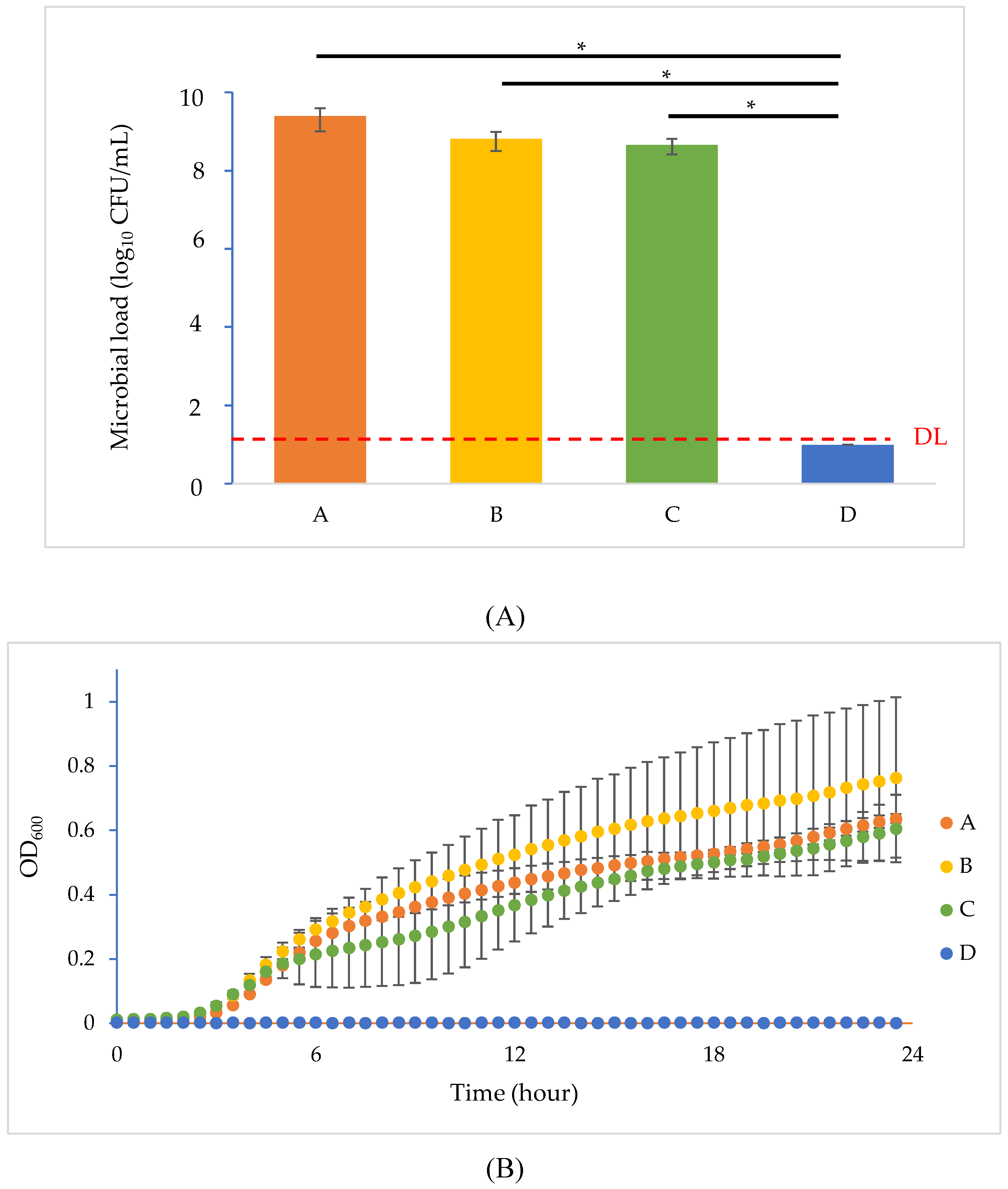
2.5. Phage Resistance Inhibition Test
3. Discussion
4. Materials and Methods
4.1. Bacterial Strain and Growth Condition
4.2. Bacteriophage T4 Preparation
4.3. BIMs Isolation and Confirmation
4.4. Phage Adsorption to Bacterial Cells Measurement
4.5. DNA Extraction, Whole Genome Sequencing and Bioinformatic Analysis
4.6. Bioinformatic Analysis
4.7. Isolation of LPS and Glycosyl Composition Analysis
4.8. Minimum Inhibitory Concentration Assays
4.9. Phage Resistance Inhibition Test
4.10. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- O’Neill, J. Tackling Drug-Resistant Infections Globally: Final Report and Recommendations. The Review on Antimicrobial Resistance. 2016. Available online: https://apo.org.au/node/63983 (accessed on 19 May 2016).
- Council of Canadian Academies. When Antibiotics Fail. The Expert Panel on the Potential Socio-Economic Impacts of Antimicrobial Resistance in Canada; Council of Canadian Academies: Ottawa, ON, Canada, 2019. [Google Scholar]
- Global Action Plan on Antimicrobial Resistance. Microbe Mag. 2015, 10, 354–355. [CrossRef]
- World Health Organization. National Antimicrobial Resistance Surveillance Systems and Participation in the Global Antimicrobial Resistance Surveillance System (GLASS): A Guide to Planning, Implementation, and Monitoring and Evaluation; World Health Organization: Geneva, Switzerland, 2016. [Google Scholar]
- World Health Organization. Antimicrobial Resistance: Global Report on Surveillance; World Health Organization: Geneva, Switzerland, 2014. [Google Scholar]
- Poirel, L.; Madec, J.-Y.; Lupo, A.; Schink, A.-K.; Kieffer, N.; Nordmann, P.; Schwarz, S. Antimicrobial Resistance in Escherichia coli. Antimicrob. Resist. Bact. Livest. Companion Anim. 2018, 6, 289–316. [Google Scholar] [CrossRef] [Green Version]
- Manyi-Loh, C.E.; Mamphweli, S.N.; Meyer, E.; Okoh, A.I. Antibiotic Use in Agriculture and Its Consequential Resistance in Environmental Sources: Potential Public Health Implications. Molecules 2018, 23, 795. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shepherd, A.K.; Pottinger, P.S. Management of Urinary Tract Infections in the Era of Increasing Antimicrobial Resistance. Med. Clin. N. Am. 2013, 97, 737–757. [Google Scholar] [CrossRef]
- Pitout, J.D.D. Extraintestinal Pathogenic Escherichia coli: A Combination of Virulence with Antibiotic Resistance. Front. Microbiol. 2012, 3, 9. [Google Scholar] [CrossRef] [Green Version]
- Gao, L.; Hu, J.; Zhang, X.; Wei, L.; Li, S.; Miao, Z.; Chai, T. Application of swine manure on agricultural fields contributes to extended-spectrum β-lactamase-producing Escherichia coli spread in Tai’an, China. Front. Microbiol. 2015, 6, 313. [Google Scholar] [CrossRef] [Green Version]
- Rasheed, M.U.; Thajuddin, N.; Ahamed, P.; Teklemariam, Z.; Jamil, K. Antimicrobial Drug Resistance in Strains of Escherichia coli Isolated from Food Sources. Rev. Inst. Med. Trop Sao Paulo 2014, 56, 341–346. [Google Scholar] [CrossRef]
- Bonyadian, M.; Moshtaghi, H.; Mohamadtaghipour, L. Antibiotic Resistance of Verotoxigenic Escherichia coli Isolated from Vegetables. Biol. J. Microorg. 2017, 5, 9–16. [Google Scholar]
- Alvarez-Uria, G.; Gandra, S.; Mandal, S.; Laxminarayan, R. Global forecast of antimicrobial resistance in invasive isolates of Escherichia coli and Klebsiella pneumoniae. Int. J. Infect. Dis. 2018, 68, 50–53. [Google Scholar] [CrossRef] [Green Version]
- McDanel, J.; Schweizer, M.L.; Crabb, V.; Nelson, R.; Samore, M.; Khader, K.; Blevins, A.; Diekema, D.J.; Chiang, H.-Y.; Nair, R.; et al. Incidence of Extended-Spectrum β-Lactamase (ESBL)-Producing Escherichia coli and Klebsiella Infections in the United States: A Systematic Literature Review. Infect. Control. Hosp. Epidemiol. 2017, 38, 1209–1215. [Google Scholar] [CrossRef]
- Lin, D.M.; Koskella, B.; Lin, H.C. Phage therapy: An alternative to antibiotics in the age of multi-drug resistance. World J. Gastrointest. Pharmacol. Ther. 2017, 8, 162–173. [Google Scholar] [CrossRef]
- Ghosh, C.; Sarkar, P.; Issa, R.; Haldar, J. Alternatives to Conventional Antibiotics in the Era of Antimicrobial Resistance. Trends Microbiol. 2019, 27, 323–338. [Google Scholar] [CrossRef] [PubMed]
- Bergh, Ø.; Børsheim, K.Y.; Bratbak, G.; Heldal, M. High abundance of viruses found in aquatic environments. Nature 1989, 340, 467–468. [Google Scholar] [CrossRef] [PubMed]
- Moye, Z.D.; Woolston, J.; Sulakvelidze, A. Bacteriophage Applications for Food Production and Processing. Viruses 2018, 10, 205. [Google Scholar] [CrossRef] [Green Version]
- Greer, G.G. Bacteriophage Control of Foodborne Bacteria†. J. Food Prot. 2005, 68, 1102–1111. [Google Scholar] [CrossRef] [PubMed]
- Chan, B.K.; Sistrom, M.; Wertz, J.E.; Kortright, K.E.; Narayan, D.; Turner, P.E. Phage selection restores antibiotic sensitivity in MDR Pseudomonas aeruginosa. Sci. Rep. 2016, 6, 26717. [Google Scholar] [CrossRef] [PubMed]
- Valério, N.; Oliveira, C.; Jesus, V.; Branco, T.; Pereira, C.; Moreirinha, C.; Almeida, A. Effects of single and combined use of bacteriophages and antibiotics to inactivate Escherichia coli. Virus Res. 2017, 240, 8–17. [Google Scholar] [CrossRef] [PubMed]
- Prehm, P.; Jann, B.; Jann, K.; Schmidt, G.; Stirm, S. On a bacteriophage T3 and T4 receptor region within the cell wall lipopolysaccharide of Escherichia coli B. J. Mol. Boil. 1976, 101, 277–281. [Google Scholar] [CrossRef]
- Washizaki, A.; Yonesaki, T.; Otsuka, Y. Characterization of the interactions between Escherichia coli receptors, LPS and OmpC, and bacteriophage T4 long tail fibers. Microbiology 2016, 5, 1003–1015. [Google Scholar] [CrossRef] [Green Version]
- Coutinho, P.M.; Deleury, E.; Davies, G.; Henrissat, B. An Evolving Hierarchical Family Classification for Glycosyltransferases. J. Mol. Boil. 2003, 328, 307–317. [Google Scholar] [CrossRef]
- Lairson, L.L.; Henrissat, B.; Davies, G.; Withers, S. Glycosyltransferases: Structures, Functions, and Mechanisms. Annu. Rev. Biochem. 2008, 77, 521–555. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Comeau, A.M.; Tétart, F.; Trojet, S.N.; Prère, M.-F.; Krisch, H.M. Phage-Antibiotic Synergy (PAS): β-Lactam and Quinolone Antibiotics Stimulate Virulent Phage Growth. PLoS ONE 2007, 2, e799. [Google Scholar] [CrossRef] [PubMed]
- Karam, J.D.; Drake, J.W. Molecular Biology of Bacteriophage; American Society for Microbiology: Washington DC, USA, 1994; Volume 4. [Google Scholar]
- Kasman, L.M.; Kasman, A.; Westwater, C.; Dolan, J.; Schmidt, M.G.; Norris, J.S. Overcoming the Phage Replication Threshold: A Mathematical Model with Implications for Phage Therapy. J. Virol. 2002, 76, 5557–5564. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- D’Herelle, F. Sur le rôle du microbe filtrant bactériophage dans la dysentérie bacillaire. CR Acad. Sci. 1918, 167, 970. [Google Scholar]
- Weidel, W. Phage Receptor Systems of Escherichia coli B. In Proceedings of the Cold Spring Harbor Symposia on Quantitative Biology, Cold Spring Harbor, NY, USA, 5 June 1953; pp. 155–157. [Google Scholar]
- Brenner, S. The adsorption of bacteriophages by sensitive and resistant cells of Escherichia coli strain B. Proc. R. Soc. Lond. Ser. B Boil. Sci. 1955, 144, 93–99. [Google Scholar] [CrossRef]
- Silva, J.B.; Storms, Z.; Sauvageau, D. Host receptors for bacteriophage adsorption. FEMS Microbiol. Lett. 2016, 363, fnw002. [Google Scholar] [CrossRef] [Green Version]
- Yoon, S.H.; Han, M.-J.; Jeong, H.; Lee, C.H.; Xia, X.-X.; Lee, D.-H.; Shim, J.H.; Lee, S.Y.; Oh, T.-K.; Kim, J.F. Comparative multi-omics systems analysis of Escherichia coli strains B and K-12. Genome Boil. 2012, 13, R37. [Google Scholar] [CrossRef] [Green Version]
- Yethon, J.A.; Vinogradov, E.; Perry, M.B.; Whitfield, C. Mutation of the Lipopolysaccharide Core Glycosyltransferase Encoded by waaG Destabilizes the Outer Membrane of Escherichia coli by Interfering with Core Phosphorylation. J. Bacteriol. 2000, 182, 5620–5623. [Google Scholar] [CrossRef] [Green Version]
- Tamaki, S.; Sato, T.; Matsuhashi, M. Role of Lipopolysaccharides in Antibiotic Resistance and Bacteriophage Adsorption of Escherichia coli K-12. J. Bacteriol. 1971, 105, 968–975. [Google Scholar] [CrossRef] [Green Version]
- Chang, V.; Chen, L.Y.; Wang, A.; Yuan, X. The Effect of Lipopolysaccharide Core Structure Defects on Transformation Efficiency in Isogenic Escherichia coli BW25113 rfaG, rfaP, and rfaC Mutants. JEMI 2010, 14, 101–107. [Google Scholar]
- Liebau, J.; Pettersson, P.; Szpryngiel, S.; Mäler, L. Membrane Interaction of the Glycosyltransferase WaaG. Biophys. J. 2015, 109, 552–563. [Google Scholar] [CrossRef] [Green Version]
- Albesa-Jové, D.; Giganti, D.; Jackson, M.C.; Alzari, P.M.; Guerin, M.E. Structure-function relationships of membrane-associated GT-B glycosyltransferases. Glycobiology 2013, 24, 108–124. [Google Scholar] [CrossRef] [Green Version]
- Beloin, C.; Roux, A.; Ghigo, J.-M. Escherichia coli Biofilms. In Inducible Lymphoid Organs; Springer Science and Business Media LLC Heidelberg: Berlin, Germany, 2008; Volume 322, pp. 249–289. [Google Scholar]
- Yi, W.; Liu, X.; Li, Y.; Li, J.; Xia, C.; Zhou, G.; Zhang, W.; Zhao, W.; Chen, X.; Wang, P.G. Remodeling bacterial polysaccharides by metabolic pathway engineering. Proc. Natl. Acad. Sci. USA 2009, 106, 4207–4212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nikaido, H. Molecular Basis of Bacterial Outer Membrane Permeability Revisited. Microbiol. Mol. Boil. Rev. 2003, 67, 593–656. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scanlan, P.D.; Bischofberger, A.M.; Hall, A.R. Modification of Escherichia coli-bacteriophage interactions by surfactants and antibiotics in vitro. FEMS Microbiol. Ecol. 2016, 93, 93. [Google Scholar] [CrossRef] [Green Version]
- Gabig-Cimińska, M.; Herman-Antosiewicz, A.; Kwiatkowska, M.; Los, M.; Thomas, M.S.; Węgrzyn, G. The cell surface protein Ag43 facilitates phage infection of Escherichia coli in the presence of bile salts and carbohydrates. Microbiology 2002, 148, 1533–1542. [Google Scholar] [CrossRef] [Green Version]
- Canadian Food Inspection Agency. Permitted Synonyms for Food Additives Table. Available online: https://www.inspection.gc.ca/food-label-requirements/labelling/industry/food-additives/permitted-synonyms/eng/1369857665232/1369857767799 (accessed on 16 April 2020).
- Food and Drug Administration. Code of Federal Regulations Title 21. Available online: https://www.accessdata.fda.gov/scripts/cdrh/cfdocs/cfcfr/CFRSearch.cfm?fr=172.822 (accessed on 16 April 2020).
- Zhong, Z.; Goodridge, L. Characterization of Phage AR1 Resistant Escherichia coli O157:H7 Mutants. 2020; Manuscript in Preparation. [Google Scholar]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1989. [Google Scholar]
- Kropinski, A.M.; Mazzocco, A.; Waddell, T.E.; Lingohr, E.; Johnson, R.P. Enumeration of Bacteriophages by Double Agar Overlay Plaque Assay. In Methods in Molecular Biology; Springer Science and Business Media LLC: Berlin, Germany, 2009; Volume 501, pp. 69–76. [Google Scholar]
- Clokie, M.R.; Kropinski, A.M.; Lavigne, R. Bacteriophages; Springer: Berlin, Germany, 2009. [Google Scholar]
- Kropinski, A.M. Measurement of the Rate of Attachment of Bacteriophage to Cells. In Methods in Molecular Biology; Springer Science and Business Media LLC: Berlin, Germany, 2009; Volume 501, pp. 151–155. [Google Scholar]
- Krueger, A.P. The Sorption of Bacteriophage by Living and Dead Susceptible Bacteria. J. Gen. Physiol. 1931, 14, 493–516. [Google Scholar] [CrossRef] [Green Version]
- Tritt, A.; Eisen, J.A.; Facciotti, M.T.; E Darling, A. An Integrated Pipeline for de Novo Assembly of Microbial Genomes. PLoS ONE 2012, 7, e42304. [Google Scholar] [CrossRef] [Green Version]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.Y.; Glass, E.; Kubal, M.; et al. The RAST Server: Rapid Annotations using Subsystems Technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef] [Green Version]
- Madeira, F.; Park, Y.; Lee, J.; Buso, N.; Gur, T.; Madhusoodanan, N.; Basutkar, P.; Tivey, A.R.N.; Potter, S.C.; Finn, R.D.; et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019, 47, W636–W641. [Google Scholar] [CrossRef] [Green Version]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.A.; Clamp, M.; Barton, G.J. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zankari, E.; Hasman, H.; Cosentino, S.; Vestergaard, M.; Rasmussen, S.; Lund, O.; Aarestrup, F.M.; Larsen, M.V. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012, 67, 2640–2644. [Google Scholar] [CrossRef] [PubMed]
- Westphal, O.; Jann, K. Bacterial Lipopolysaccharides Extraction with Phenol-Water and Further Applications of the Procedure. Methods Carbohydr. Chem. 1965, 5, 83–91. [Google Scholar]
- Bhat, U.R.; Forsberg, L.S.; Carlson, R.W. Structure of lipid A component of Rhizobium leguminosarum bv. phaseoli lipopolysaccharide. Unique nonphosphorylated lipid A containing 2-amino-2-deoxygluconate, galacturonate, and glucosamine. J. Boil. Chem. 1994, 269, 14402–14410. [Google Scholar]
- York, W.S.; Darvill, A.G.; McNeil, M.; Stevenson, T.T.; Albersheim, P. Isolation and Characterization of Plant Cell Walls and Cell Wall Components. In RNA Interference; Elsevier BV: Amsterdam, The Netherlands, 1986; Volume 118, pp. 3–40. [Google Scholar]
| Strain | LPS Glycosyl Composition [Mole %] | |||||
|---|---|---|---|---|---|---|
| Rib | Man | Glc | Hep | GlcN | Kdo * | |
| WT E. coli B | 12.2 | 4.3 | 17.8 | 62.2 | 3.5 | + |
| ZZa3 | 12.8 | 2.1 | 1 | 78.5 | 5.7 | + |
| Substrates | WT | ZZa0 | ZZa1 | ZZa2 | ZZa3 | ZZa4 | ZZa5 |
|---|---|---|---|---|---|---|---|
| Ampicillin (µg/mL) | 4 | 2 | 2 | 2 | 2 | 1 | 1 |
| Kanamycin (µg/mL) | 32 | 16 | 16 | 16 | 16 | 16 | 16 |
| Novobiocin (µg/mL) | >200 | >200 | >200 | >200 | >200 | >200 | >200 |
| Polymyxin B (µg/mL) | 64 | 32 | 32 | 32 | 32 | 32 | 32 |
| EDTA (mg/mL) | 50 | 25 | 25 | 25 | 25 | 25 | 25 |
| Sodium Cholate (mg/mL) | >128 | 128 | 128 | 128 | 128 | 128 | 128 |
| SDS (mg/mL) | >200 | 3 | 1.5 | 3 | 3 | 3 | 1.5 |
| Setting A | Setting B | Setting C | Setting D | |
|---|---|---|---|---|
| E. coli B (106 CFU) | + | + | + | + |
| Phage T4 (107 PFU) | − | + | − | + |
| SDS (10 mg/mL) | − | − | + | + |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhong, Z.; Emond-Rheault, J.-G.; Bhandare, S.; Lévesque, R.; Goodridge, L. Bacteriophage-Induced Lipopolysaccharide Mutations in Escherichia coli Lead to Hypersensitivity to Food Grade Surfactant Sodium Dodecyl Sulfate. Antibiotics 2020, 9, 552. https://doi.org/10.3390/antibiotics9090552
Zhong Z, Emond-Rheault J-G, Bhandare S, Lévesque R, Goodridge L. Bacteriophage-Induced Lipopolysaccharide Mutations in Escherichia coli Lead to Hypersensitivity to Food Grade Surfactant Sodium Dodecyl Sulfate. Antibiotics. 2020; 9(9):552. https://doi.org/10.3390/antibiotics9090552
Chicago/Turabian StyleZhong, Zeyan, Jean-Guillaume Emond-Rheault, Sudhakar Bhandare, Roger Lévesque, and Lawrence Goodridge. 2020. "Bacteriophage-Induced Lipopolysaccharide Mutations in Escherichia coli Lead to Hypersensitivity to Food Grade Surfactant Sodium Dodecyl Sulfate" Antibiotics 9, no. 9: 552. https://doi.org/10.3390/antibiotics9090552





