Strategies for Optimizing the Production of Proteins and Peptides with Multiple Disulfide Bonds
Abstract
:1. Introduction
2. Host Strains for the Overexpression of Target Proteins
3. Location of Expression
4. Vector Selection for Expression
5. Signal Peptide in Fusion Expression
6. Co-Expression of Chaperones
7. Soluble and Insoluble Expression
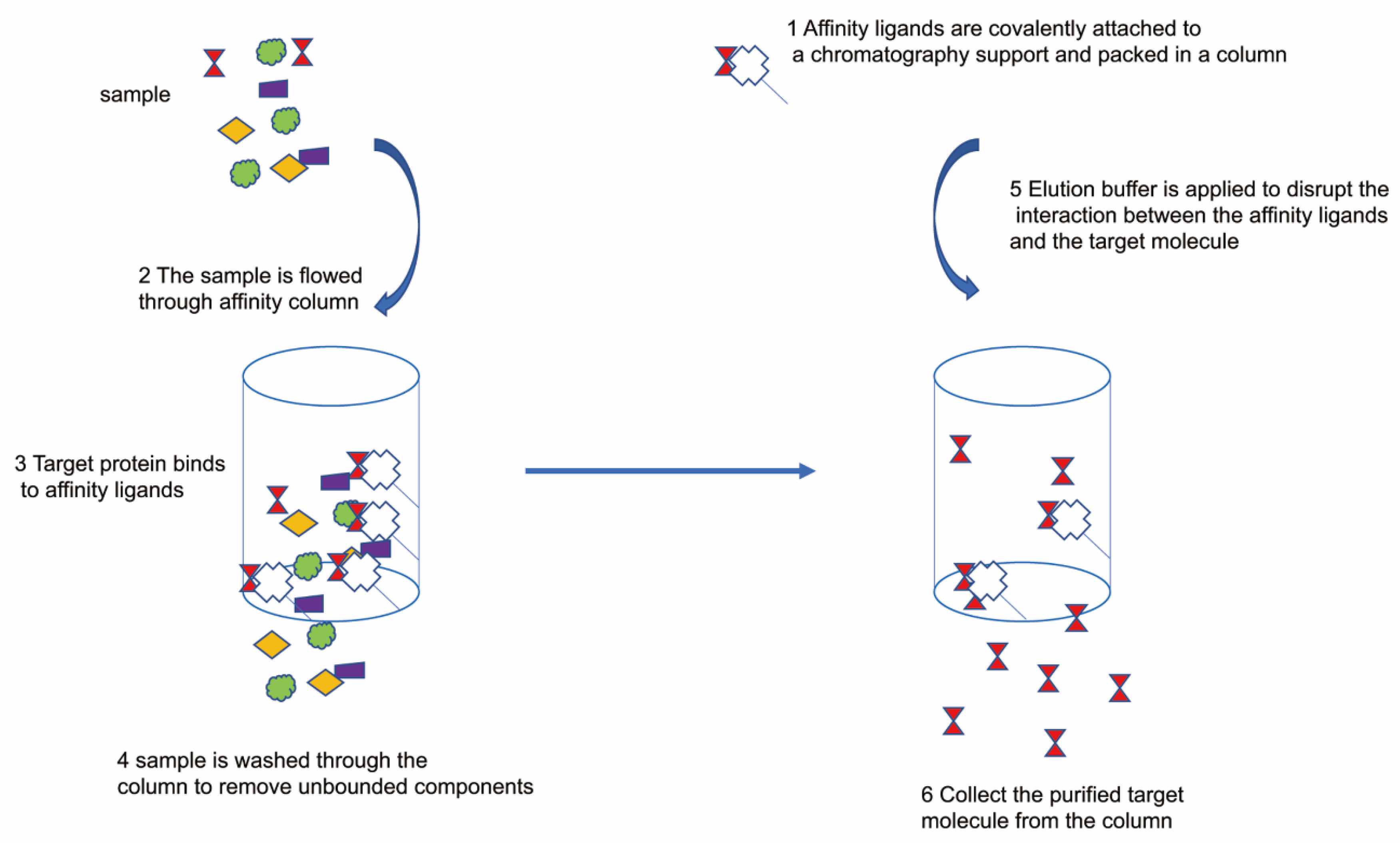
8. Purification
9. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Basagoudanavar, S.H.; Hosamani, M.; Muthuchelvan, D.; Singh, R.P.; Santhamani, R.; Sreenivasa, B.P.; Saravanan, P.; Pandey, A.B.; Singh, R.K.; Venkataramanan, R. Baculovirus expression and purification of peste-des-petits-ruminants virus nucleocapsid protein and its application in diagnostic assay. Biol. J. Int. Assoc. Biol. Stand. 2018, 55, 38–42. [Google Scholar] [CrossRef] [PubMed]
- Faramarzi, T.; Mobasheri, M.; Yoosefy, A.; Valadkhani, Z. Expression and purification of truncated recombinant b8/1 protein of echinococcus granulosus for diagnosis of hydatid infection in human. Acta Trop. 2019, 191, 139–145. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Wang, Z.; Yang, Z.; Li, Z.; Wang, P.; Dong, W. Eukaryotic expression, purification of human igecepsilon2-4 protein and its target fishing. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi Chin. J. Cell. Mol. Immunol. 2018, 34, 205–210. [Google Scholar]
- Jeong, I.S.; Lee, S.; Bonkhofer, F.; Tolley, J.; Fukudome, A.; Nagashima, Y.; May, K.; Rips, S.; Lee, S.Y.; Gallois, P.; et al. Purification and characterization of arabidopsis thaliana oligosaccharyltransferase complexes from the native host: A protein super-expression system for structural studies. Plant J. Cell Mol. Biol. 2018, 94, 131–145. [Google Scholar] [CrossRef] [Green Version]
- Mahesh, G.; Jaiswal, P.; Dey, S.; Sengupta, J.; Mukherjee, S. Cloning, expression, purification and characterization of oligomeric states of the native 5ht2a g-protein-coupled receptor. Protein Pept. Lett. 2018, 25, 390–397. [Google Scholar] [CrossRef] [PubMed]
- Ng, E.Y.; Loh, Y.R.; Li, Y.; Li, Q.; Kang, C. Expression, purification of zika virus membrane protein-ns2b in detergent micelles for nmr studies. Protein Expr. Purif. 2019, 154, 1–6. [Google Scholar] [CrossRef]
- Richardson, D.; Itkonen, J.; Nievas, J.; Urtti, A.; Casteleijn, M.G. Accelerated pharmaceutical protein development with integrated cell free expression, purification, and bioconjugation. Sci. Rep. 2018, 8, 11967. [Google Scholar] [CrossRef]
- Ortega, C.; Prieto, D.; Abreu, C.; Oppezzo, P.; Correa, A. Multi-compartment and multi-host vector suite for recombinant protein expression and purification. Front. Microbiol. 2018, 9, 1384. [Google Scholar] [CrossRef] [Green Version]
- Lu, J.; Liang, W.; Wei, K.; Li, J.; Li, J.; Wang, J.; Gao, W. Induction of signal molecules and expression of functional genes after pichia pastoris stimulation in glycyrrhiza uralensis fisch adventitious roots. J. Food Biochem. 2019, 43, e12798. [Google Scholar] [CrossRef] [PubMed]
- Lopes, C.; Dos Santos, N.V.; Dupont, J.; Pedrolli, D.B.; Valentini, S.R.; de Carvalho Santos-Ebinuma, V.; Pereira, J.F.B. Improving the cost effectiveness of enhanced green fluorescent protein production using recombinant escherichia coli bl21 (de3): Decreasing the expression inducer concentration. Biotechnol. Appl. Biochem. 2019, 66, 527–536. [Google Scholar] [CrossRef]
- Olson, L.J.; Dahms, N.M. Cloning, expression, and purification of the glycosylated transmembrane protein, cation-dependent mannose 6-phosphate receptor, from sf9 cells using the baculovirus system. Methods Mol. Biol. 2018, 1722, 105–116. [Google Scholar] [PubMed]
- Ramos-Benitez, M.J.; Lopez-Cruz, L.M.; Aguayo, V.; Ruiz-Jimenez, C.; Espino, A.M. Cell-free expression, purification and immunoreactivity assessment of recombinant fasciola hepatica saposin-like protein-2. Mol. Biol. Rep. 2018, 45, 1551–1556. [Google Scholar] [CrossRef] [PubMed]
- Rajput, V.B.; Karthikeyan, M.; Ramasamy, S. Zebrafish acid ceramidase: Expression in pichia pastoris gs115and biochemical characterization. Int. J. Biol. Macromol. 2019, 122, 587–593. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Halperin, S.A.; Lee, S.F. Expression, purification, and functional analysis of an antigen-targeting fusion protein composed of cd40 ligand and the c-terminal fragment of ovalbumin. Protein Expr. Purif. 2018, 142, 37–44. [Google Scholar] [CrossRef]
- Parra, N.C.; Mansilla, R.; Aedo, G.; Vispo, N.S.; Gonzalez-Horta, E.E.; Gonzalez-Chavarria, I.; Castillo, C.; Camacho, F.; Sanchez, O. Expression and characterization of human vascular endothelial growth factor produced in siha cells transduced with adenoviral vector. Protein J. 2019, 38, 693–703. [Google Scholar] [CrossRef]
- Subramanyam, S.; Spies, M. Expression, purification, and biochemical evaluation of human rad51 protein. Methods Enzymol. 2018, 600, 157–178. [Google Scholar]
- Sui, Y.; Fu, X.; Wang, Y.; Hu, W.; Zhang, T.; Liu, W.; Jiang, L.; Xing, S.; Fu, X.; Xu, X. Expression, purification and characterization of a catalytic domain of human protein tyrosine phosphatase non-receptor 12 (ptpn12) in escherichia coli with fkbp-type ppiase as a chaperon. Protein Expr. Purif. 2018, 142, 45–52. [Google Scholar] [CrossRef]
- Ouephanit, C.; Boonvitthya, N.; Bozonnet, S.; Chulalaksananukul, W. High-level heterologous expression of endo-1,4-beta-xylanase from penicillium citrinum in pichia pastoris x-33 directed through codon optimization and optimized expression. Molecules 2019, 24, 3515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allonso, D.; Pereira, I.B.; Alves, A.M.; Kurtenbach, E.; Mohana-Borges, R. Expression of soluble, glycosylated and correctly folded dengue virus ns1 protein in pichia pastoris. Protein Expr. Purif. 2019, 162, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Bobik, T.V.; Popov, R.Y.; Aliev, T.K.; Mokrushina, Y.A.; Shamborant, O.G.; Khurs, E.N.; Knorre, V.D.; Smirnov, I.V.; Gabibov, A.G. Production of recombinant human transferrin in eukaryotic pichia pastoris expression system. Bull. Exp. Biol. Med. 2019, 167, 335–338. [Google Scholar] [CrossRef] [PubMed]
- Arbulu, S.; Jimenez, J.J.; Gutiez, L.; Feito, J.; Cintas, L.M.; Herranz, C.; Hernandez, P.E. Cloning and expression of synthetic genes encoding native, hybrid- and bacteriocin-derived chimeras from mature class iia bacteriocins, by pichia pastoris (syn. Komagataella spp.). Food Res. Int. 2019, 121, 888–899. [Google Scholar] [CrossRef]
- Tagliavia, M.; Nicosia, A. Advanced strategies for food-grade protein production: A new E. Coli/lactic acid bacteria shuttle vector for improved cloning and food-grade expression. Microorganisms 2019, 7, 116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, H.H.; Zhang, A.X.; Zhang, Y.; Zhu, D.Y. Cloning, expression, and purification of a recombinant tat-ha-nr2b9c peptide. Protein Expr. Purif. 2012, 85, 239–245. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Dong, S.; Zheng, J.; Li, D.; Li, F.; Luo, Z. Expression, stabilization and purification of membrane proteins via diverse protein synthesis systems and detergents involving cell-free associated with self-assembly peptide surfactants. Biotechnol. Adv. 2014, 32, 564–574. [Google Scholar] [CrossRef] [PubMed]
- Zorko, M.; Japelj, B.; Hafner-Bratkovic, I.; Jerala, R. Expression, purification and structural studies of a short antimicrobial peptide. Biochim. Biophys. Acta 2009, 1788, 314–323. [Google Scholar] [CrossRef] [Green Version]
- Zhu, B.; Pu, Q.; Chen, N.; Chen, S. Expression, purification, and bone-inducing activity of recombinant human bone morphogenetic protein-3 mature peptide. Chin. J. Biotechnol. 1999, 15, 153–158. [Google Scholar]
- Zhou, L.; Lin, Q.; Li, B.; Li, N.; Zhang, S. Expression and purification the antimicrobial peptide cm4 in escherichia coli. Biotechnol. Lett. 2009, 31, 437–441. [Google Scholar] [CrossRef]
- Zheng, P.; Gao, F.; Deng, K.; Gong, W.; Sun, Z. Expression, purification and preliminary x-ray crystallographic analysis of arf1-gdp in complex with dimeric p23 peptide. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2013, 69, 1155–1158. [Google Scholar] [CrossRef] [Green Version]
- Zheng, H.L.; Li, H.; Sun, Y.S.; Yang, Z.Y.; Yu, Q. Parathyroid hormone-related peptide (pthrp): Prokaryotic expression, purification, and preparation of a polyclonal antibody. Genet. Mol. Res. GMR 2014, 13, 6448–6454. [Google Scholar] [CrossRef]
- Zheng, C.F.; Simcox, T.; Xu, L.; Vaillancourt, P. A new expression vector for high level protein production, one step purification and direct isotopic labeling of calmodulin-binding peptide fusion proteins. Gene 1997, 186, 55–60. [Google Scholar] [CrossRef]
- Zhang, Z.Z.; Yang, S.S.; Dou, H.; Mao, J.F.; Li, K.S. Expression, purification, and c-terminal amidation of recombinant human glucagon-like peptide-1. Protein Expr. Purif. 2004, 36, 292–299. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.Y.; Zhang, J.C.; Zhao, H. Expression and purification of human amelogenin mature peptide in escherichia coli. Zhonghua Kou Qiang Yi Xue Za Zhi = Zhonghua Kouqiang Yixue Zazhi Chin. J. Stomatol. 2009, 44, 279–281. [Google Scholar]
- Zhang, X.G.; Wang, W.N.; Zhang, C.S.; Li, K.; Ma, G.D.; Li, J.Y. Expression and purification of delta sleep-inducing peptide fused with protein transduction domain and human serum albumin in pichia pastoris. Protein Pept. Lett. 2017, 24, 668–675. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Yu, H.; Song, C.; Lin, X.; Chen, B.; Tan, C.; Cao, G.; Wang, Z. Expression, purification, and characterization of recombinant human beta-amyloid42 peptide in escherichia coli. Protein Expr. Purif. 2009, 64, 55–62. [Google Scholar] [CrossRef]
- Zhang, H.; Huang, P.F.; Meng, E.; Li, W.Y.; Zhou, L.; Zhu, L.Y.; Wu, L.; Li, M.J.; Liang, S.P.; Zhang, D.Y. An efficient strategy for heterologous expression and purification of active peptide hainantoxin-iv. PLoS ONE 2015, 10, e0117099. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.; Cai, Q.; Li, Z.; Zhai, C.; Zheng, Y. Construction of fusion expression vector of human-derived neurotrophin-6 gene encoding mature peptide and purification of its expressed product. Sheng Wu Yi Xue Gong Cheng Xue Za Zhi J. Biomed. Eng. Shengwu Yixue Gongchengxue Zazhi 2005, 22, 1241–1244. [Google Scholar]
- Yang, K.; Su, Y.; Li, J.; Sun, J.; Yang, Y. Expression and purification of the antimicrobial peptide cecropin ad by fusion with cationic elastin-like polypeptides. Protein Expr. Purif. 2012, 85, 200–203. [Google Scholar] [CrossRef]
- Xu, X.; Jin, F.; Yu, X.; Ji, S.; Wang, J.; Cheng, H.; Wang, C.; Zhang, W. Expression and purification of a recombinant antibacterial peptide, cecropin, from escherichia coli. Protein Expr. Purif. 2007, 53, 293–301. [Google Scholar] [CrossRef]
- Xu, J.; Zhu, Z.; Duan, P.; Li, W.; Zhang, Y.; Wu, J.; Hu, Z.; Roque, R.S.; Liu, J. Cloning, expression, and purification of a highly immunogenic recombinant gonadotropin-releasing hormone (gnrh) chimeric peptide. Protein Expr. Purif. 2006, 50, 163–170. [Google Scholar] [CrossRef]
- Xu, C.S.; Tao, K.; Zheng, D.X.; Zheng, C.X.; Liu, S.L. Isolation and purification of a small peptide with activity of increasing e-receptor expression from calf thymus. Immunol. Investig. 1985, 14, 355–365. [Google Scholar]
- Xing, L.; Xu, W.; Zhou, B.; Chen, Y.; Lin, Z. Facile expression and purification of the antimicrobial peptide histatin 1 with a cleavable self-aggregating tag (csat) in escherichia coli. Protein Expr. Purif. 2013, 88, 248–253. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.G.; Han, F.F.; Luan, C.; Zhang, H.W.; Feng, J.; Choi, Y.J.; Groleau, D.; Wang, Y.Z. High-yield soluble expression and simple purification of the antimicrobial peptide og2 using the intein system in escherichia coli. Biomed. Res. Int. 2013, 2013, 754319. [Google Scholar] [CrossRef] [PubMed]
- Xie, P.; Wang, Y.X.; Gao, R.K.; Zhou, L.J. Optimization of expression and purification protocol for human scfv antibody against beta-amyloid peptide. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi Chin. J. Cell. Mol. Immunol. 2012, 28, 718–721. [Google Scholar]
- Xia, L.; Liu, Z.; Ma, J.; Sun, S.; Yang, J.; Zhang, F. Expression, purification and characterization of cecropin antibacterial peptide from bombyx mori in saccharomyces cerevisiae. Protein Expr. Purif. 2013, 90, 47–54. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, B.; Hanif, Q.; Xubiao, W.; Lulu, Z.; Shahid, M.; Dayong, S.; Rijun, Z. Expression and purification of hybrid ll-37talpha1 peptide in pichia pastoris and evaluation of its immunomodulatory and anti-inflammatory activities by lps neutralization. Front. Immunol. 2019, 10, 1365. [Google Scholar] [CrossRef]
- Baeshen, M.N.; Bouback, T.A.; Alzubaidi, M.A.; Bora, R.S.; Alotaibi, M.A.; Alabbas, O.T.; Alshahrani, S.M.; Aljohani, A.A.; Munshi, R.A.; Al-Hejin, A.; et al. Expression and purification of c-peptide containing insulin using pichia pastoris expression system. Biomed Res. Int. 2016, 2016, 3423685. [Google Scholar] [CrossRef] [Green Version]
- Bang, S.K.; Kang, C.S.; Han, M.D.; Bang, I.S. Expression of recombinant hybrid peptide hinnavin ii/alpha-melanocyte-stimulating hormone in escherichia coli: Purification and characterization. J. Microbiol. 2010, 48, 24–29. [Google Scholar] [CrossRef]
- Cao, P.; Yu, J.; Lu, W.; Cai, X.; Wang, Z.; Gu, Z.; Zhang, J.; Ye, T.; Wang, M. Expression and purification of an antitumor-analgesic peptide from the venom of mesobuthus martensii karsch by small ubiquitin-related modifier fusion in escherichia coli. Biotechnol. Prog. 2010, 26, 1240–1244. [Google Scholar] [CrossRef]
- Cao, W.; Zhou, Y.; Ma, Y.; Luo, Q.; Wei, D. Expression and purification of antimicrobial peptide adenoregulin with c-amidated terminus in escherichia coli. Protein Expr. Purif. 2005, 40, 404–410. [Google Scholar] [CrossRef]
- Cao, Z.; Wang, W.; Xiao, X.; Chen, K.; Liang, X.; Yu, D. High-level expression and purification of an analgesic peptide from buthus martensii karch. Protein Pept. Lett. 2007, 14, 247–251. [Google Scholar] [CrossRef]
- Rahman, M.A.; Brot, N.; Weissbach, H. High level expression and purification of peptide methionine sulfoxide reductase in escherichia coli. Cell. Mol. Biol. 1992, 38, 529–542. [Google Scholar] [PubMed]
- Peng, H.; Yang, M.; Huang, W.S.; Ding, J.; Qu, H.D.; Cai, J.J.; Zhang, N.; Wang, K.J. Soluble expression and purification of a crab antimicrobial peptide scygonadin in different expression plasmids and analysis of its antimicrobial activity. Protein Expr. Purif. 2010, 70, 109–115. [Google Scholar] [CrossRef] [Green Version]
- Pei, Z.; Sun, X.; Tang, Y.; Wang, K.; Gao, Y.; Ma, H. Cloning, expression, and purification of a new antimicrobial peptide gene from musca domestica larva. Gene 2014, 549, 41–45. [Google Scholar] [CrossRef] [PubMed]
- Park, T.J.; Kim, J.S.; Choi, S.S.; Kim, Y. Cloning, expression, isotope labeling, purification, and characterization of bovine antimicrobial peptide, lactophoricin in escherichia coli. Protein Expr. Purif. 2009, 65, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Oh, K.S.; Na, D.K.; Kweon, M.H.; Sung, H.C. Expression and purification of the anticomplementary peptide sh-crit-ed1 (formerly sh-tor-ed1) as a tetramultimer in escherichia coli. Protein Expr. Purif. 2003, 27, 202–209. [Google Scholar] [CrossRef]
- Nguyen, H.Q.; Vu, V.H.; Le, P.D.; Chu, H.M. High-level expression, purification and properties of an endochitinase gene without signal peptide from lecanicillium lecanii 43h in pichia pastoris. Mol. Biol. Rep. 2018, 45, 1067–1075. [Google Scholar] [CrossRef]
- Nagata-Uchiyama, M.; Yaguchi, M.; Hirano, Y.; Ueda, T. Expression and purification of uniformly (15)n-labeled amyloid beta peptide 1-40 in escherichia coli. Protein Pept. Lett. 2007, 14, 788–792. [Google Scholar] [CrossRef]
- Nadaud, P.S.; Sarkar, M.; Wu, B.; MacPhee, C.E.; Magliery, T.J.; Jaroniec, C.P. Expression and purification of a recombinant amyloidogenic peptide from transthyretin for solid-state nmr spectroscopy. Protein Expr. Purif. 2010, 70, 101–108. [Google Scholar] [CrossRef]
- Mulder, K.C.; de Lima, L.A.; Aguiar, P.S.; Carneiro, F.C.; Franco, O.L.; Dias, S.C.; Parachin, N.S. Production of a modified peptide clavanin in pichia pastoris: Cloning, expression, purification and in vitro activities. AMB Express 2015, 5, 129. [Google Scholar] [CrossRef] [Green Version]
- Moon, W.J.; Hwang, D.K.; Park, E.J.; Kim, Y.M.; Chae, Y.K. Recombinant expression, isotope labeling, refolding, and purification of an antimicrobial peptide, piscidin. Protein Expr. Purif. 2007, 51, 141–146. [Google Scholar] [CrossRef]
- Mo, Q.; Fu, A.; Lin, Z.; Wang, W.; Gong, L.; Li, W. Expression and purification of antimicrobial peptide ap2 using sumo fusion partner technology in escherichia coli. Lett. Appl. Microbiol. 2018, 67, 606–613. [Google Scholar] [CrossRef]
- Misono, K.S.; Sivasubramanian, N.; Berkner, K.; Zhang, X. Expression and purification of the extracellular ligand-binding domain of the atrial natriuretic peptide (anp) receptor: Monovalent binding with anp induces 2:2 complexes. Biochemistry 1999, 38, 516–523. [Google Scholar] [CrossRef] [PubMed]
- Meiyalaghan, S.; Latimer, J.M.; Kralicek, A.V.; Shaw, M.L.; Lewis, J.G.; Conner, A.J.; Barrell, P.J. Expression and purification of the antimicrobial peptide gsl1 in bacteria for raising antibodies. BMC Res. Notes 2014, 7, 777. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mehrnejad, F.; Naderi-Manesh, H.; Ranjbar, B.; Maroufi, B.; Asoodeh, A.; Doustdar, F. Pcr-based gene synthesis, molecular cloning, high level expression, purification, and characterization of novel antimicrobial peptide, brevinin-2r, in escherichia coli. Appl. Biochem. Biotechnol. 2008, 149, 109–118. [Google Scholar] [CrossRef] [PubMed]
- Masterson, L.R.; Bortone, N.; Yu, T.; Ha, K.N.; Gaffarogullari, E.C.; Nguyen, O.; Veglia, G. Expression and purification of isotopically labeled peptide inhibitors and substrates of camp-dependant protein kinase a for nmr analysis. Protein Expr. Purif. 2009, 64, 231–236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, C.; Gao, M.; Liu, W.; Zhu, J.; Tian, H.; Gao, X.; Yao, W. Intein-mediated expression and purification of an analog of glucagon-like peptide-1 in escherichia coli. Protein Pept. Lett. 2010, 17, 1245–1250. [Google Scholar] [CrossRef]
- Wei, Q.; Kim, Y.S.; Seo, J.H.; Jang, W.S.; Lee, I.H.; Cha, H.J. Facilitation of expression and purification of an antimicrobial peptide by fusion with baculoviral polyhedrin in escherichia coli. Appl. Environ. Microbiol. 2005, 71, 5038–5043. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Wang, W.; Shao, Z.; Gao, B.; Li, J.; Ma, J.; Li, J.; Che, H.; Zhang, W. Eukaryotic expression and purification of anti-epilepsy peptide of buthus martensii karsch and its protein interactions. Mol. Cell. Biochem. 2009, 330, 97–104. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, L.; Cui, Y.; Song, Y.B.; Liu, Y.F.; Zhang, R.; Wu, C.F.; Zhang, J.H. Purification, characterization and functional expression of a new peptide with an analgesic effect from chinese scorpion buthus martensii karsch (bmk agp-sypu1). Biomed. Chromatogr. BMC 2011, 25, 801–807. [Google Scholar] [CrossRef]
- Wang, X.L.; Ma, S.N.; Yuan, Y.H.; Ding, Y.; Li, D.S. Expression and purification recombinant antihypertensive peptide ameliorates hypertension in rats with spontaneous hypertension. Protein Expr. Purif. 2015, 113, 30–34. [Google Scholar] [CrossRef]
- Wang, Q.; Zhu, F.; Xin, Y.; Liu, J.; Luo, L.; Yin, Z. Expression and purification of antimicrobial peptide buforin iib in escherichia coli. Biotechnol. Lett. 2011, 33, 2121–2126. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.; Wang, S.; Shen, M.; Chen, F.; Zou, Z.; Ran, X.; Cheng, T.; Su, Y.; Wang, J. High level expression and purification of bioactive human alpha-defensin 5 mature peptide in pichia pastoris. Appl. Microbiol. Biotechnol. 2009, 84, 877–884. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Zhao, L.; Zhu, L.; Chen, Z.; Li, H. Expression and purification of chimeric peptide comprising egfr b-cell epitope and measles virus fusion protein t-cell epitope in escherichia coli. Protein Expr. Purif. 2013, 88, 7–12. [Google Scholar] [CrossRef] [PubMed]
- Walsh, D.M.; Thulin, E.; Minogue, A.M.; Gustavsson, N.; Pang, E.; Teplow, D.B.; Linse, S. A facile method for expression and purification of the alzheimer’s disease-associated amyloid beta-peptide. FEBS J. 2009, 276, 1266–1281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tu, W.; Li, T.; Wang, Q.; Cai, K.; Gao, X.; Wang, H. A simple method for expression and purification of shiga toxin 1 (stx1) with biological activities by using a single-promoter vector and native signal peptide. Biotechnol. Appl. Biochem. 2016, 63, 539–545. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tran, D.Q.; Andersson, J.; Hardwick, D.; Bebris, L.; Illei, G.G.; Shevach, E.M. Selective expression of latency-associated peptide (lap) and il-1 receptor type i/ii (cd121a/cd121b) on activated human foxp3+ regulatory t cells allows for their purification from expansion cultures. Blood 2009, 113, 5125–5133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Theis, S.; Doring, F.; Daniel, H. Expression of the myc/his-tagged human peptide transporter hpept1 in yeast for protein purification and functional analysis. Protein Expr. Purif. 2001, 22, 436–442. [Google Scholar] [CrossRef]
- Tao, Y.; Zhao, D.M.; Wen, Y. Expression, purification and antibacterial activity of the channel catfish hepcidin mature peptide. Protein Expr. Purif. 2014, 94, 73–78. [Google Scholar] [CrossRef]
- Sun, Y.L.; Liu, Y.S.; Yang, H.J.; Wang, C.S.; Ma, J.Z. Study on the expression, purification and the anti-endotoxin activity of human endotoxin binding peptide and its mutant. Zhonghua Shao Shang Za Zhi Zhonghua Shaoshang Zazhi Chin. J. Burn. 2006, 22, 291–295. [Google Scholar]
- Wu, Y.; Ma, Y.; Li, L.; Yang, X. Molecular modification, expression and purification of new subtype antioxidant peptide from pinctada fucata by recombinant escherichia coli to improve antioxidant-activity. J. Food Sci. Technol. 2018, 55, 4266–4275. [Google Scholar] [CrossRef]
- Sun, Y.; Li, Q.; Li, Z.; Zhang, Y.; Zhao, J.; Wang, L. Molecular cloning, expression, purification, and functional characterization of palustrin-2ce, an antimicrobial peptide of rana chensinensis. Biosci. Biotechnol. Biochem. 2012, 76, 157–162. [Google Scholar] [CrossRef] [Green Version]
- Stevens, C.M.; Paetzel, M. Purification of a tat leader peptide by co-expression with its chaperone. Protein Expr. Purif. 2012, 84, 167–172. [Google Scholar] [CrossRef] [PubMed]
- Saez, N.J.; Nozach, H.; Blemont, M.; Vincentelli, R. High throughput quantitative expression screening and purification applied to recombinant disulfide-rich venom proteins produced in e. Coli. J. Vis. Exp. JoVE 2014, 89, e51464. [Google Scholar] [CrossRef]
- Klint, J.K.; Senff, S.; Saez, N.J.; Seshadri, R.; Lau, H.Y.; Bende, N.S.; Undheim, E.A.; Rash, L.D.; Mobli, M.; King, G.F. Production of recombinant disulfide-rich venom peptides for structural and functional analysis via expression in the periplasm of e. Coli. PLoS ONE 2013, 8, e63865. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sha, C.; Yu, X.W.; Lin, N.X.; Zhang, M.; Xu, Y. Enhancement of lipase r27rcl production in pichia pastoris by regulating gene dosage and co-expression with chaperone protein disulfide isomerase. Enzym. Microb. Technol. 2013, 53, 438–443. [Google Scholar] [CrossRef] [PubMed]
- Laboissiere, M.C.; Chivers, P.T.; Raines, R.T. Production of rat protein disulfide isomerase in saccharomyces cerevisiae. Protein Expr. Purif. 1995, 6, 700–706. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, S.K.; Tiendrebeogo, R.W.; Chourasia, B.K.; Kana, I.H.; Singh, S.; Theisen, M. Lactococcus lactis provides an efficient platform for production of disulfide-rich recombinant proteins from plasmodium falciparum. Microb. Cell Factories 2018, 17, 55. [Google Scholar] [CrossRef] [PubMed]
- Srinivasulu, B.; Syvitski, R.; Seo, J.K.; Mattatall, N.R.; Knickle, L.C.; Douglas, S.E. Expression, purification and structural characterization of recombinant hepcidin, an antimicrobial peptide identified in japanese flounder, paralichthys olivaceus. Protein Expr. Purif. 2008, 61, 36–44. [Google Scholar] [CrossRef]
- Solov’eva, O.V.; Kakuev, D.L.; Gibanova, N.V.; Lipkin, V.M. Deletion mutants of the natriuretic peptide receptor type b: Expression of cdna, purification and characteristics of proteins. Bioorganicheskaia Khimiia 2000, 26, 433–441. [Google Scholar] [CrossRef]
- Shen, L.; Guo, Z.Y.; Chen, Y.; Liu, L.Y.; Feng, Y.M. Expression, purification, characterization of amphioxus insulin-like peptide and preparation of polyclonal antibody to it. Sheng Wu Hua Xue Yu Sheng Wu Wu Li Xue Bao Acta Biochim. Biophys. Sin. 2001, 33, 629–633. [Google Scholar]
- Sharpe, S.; Yau, W.M.; Tycko, R. Expression and purification of a recombinant peptide from the alzheimer’s beta-amyloid protein for solid-state nmr. Protein Expr. Purif. 2005, 42, 200–210. [Google Scholar] [CrossRef] [PubMed]
- Sharon, M.; Gorlach, M.; Levy, R.; Hayek, Y.; Anglister, J. Expression, purification, and isotope labeling of a gp120 v3 peptide and production of a fab from a hiv-1 neutralizing antibody for nmr studies. Protein Expr. Purif. 2002, 24, 374–383. [Google Scholar] [CrossRef] [PubMed]
- Shanmuganathan, A.; Bishop, A.C.; French, K.C.; McCallum, S.A.; Makhatadze, G.I. Bacterial expression and purification of the amyloidogenic peptide papf39 for multidimensional nmr spectroscopy. Protein Expr. Purif. 2013, 88, 196–200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanchez-Cach, L.A.; Ortiz-Garcia, M.M.; Minero-Garcia, Y.; Munoz-Sanchez, J.A.; Hernandez-Sotomayor, S.T.; Suarez-Solis, V.M.; De Los Santos-Briones, C. Isolation of cdna encoding the catalytic site of phosphatidylinositol-specific phospholipase c from coffea arabica l.: Recombinant expression and peptide purification. Plant Signal. Behav. 2008, 3, 913–916. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riahi, N.; Cappadocia, L.; Henry, O.; Omichinski, J.; De Crescenzo, G. Soluble expression, purification and functional characterization of a coil peptide composed of a positively charged and hydrophobic motif. Amino Acids 2016, 48, 567–577. [Google Scholar] [CrossRef] [PubMed]
- Rao, S.; Zang, X.; Yang, Z.; Gao, L.; Yin, Y.; Fang, W. Soluble expression and purification of the recombinant bioactive peptide precursor bpp-1 in escherichia coli using a celp-sumo dual fusion system. Protein Expr. Purif. 2016, 118, 113–119. [Google Scholar] [CrossRef]
- Ramos, R.; Moreira, S.; Rodrigues, A.; Gama, M.; Domingues, L. Recombinant expression and purification of the antimicrobial peptide magainin-2. Biotechnol. Prog. 2013, 29, 17–22. [Google Scholar] [CrossRef] [Green Version]
- Lv, Z.; Ou, Y.; Li, Q.; Zhang, W.; Ye, B.; Wu, W. Expression, purification and bioactivities analysis of recombinant active peptide from shark liver. Mar. Drugs 2009, 7, 258–267. [Google Scholar] [CrossRef] [Green Version]
- Long, F.; Cho, W.; Ishii, Y. Expression and purification of 15n- and 13c-isotope labeled 40-residue human alzheimer’s beta-amyloid peptide for nmr-based structural analysis. Protein Expr. Purif. 2011, 79, 16–24. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Wang, J.; Yang, J.; Wan, C.; Wang, X.; Sun, H. Recombinant expression, purification and characterization of antimicrobial peptide orbk in escherichia coli. Protein Expr. Purif. 2014, 95, 182–187. [Google Scholar] [CrossRef]
- Li, Y.; Ren, S.; Gong, W. Cloning, high-level expression, purification and crystallization of peptide deformylase from leptospira interrogans. Acta Crystallogr. Sect. D Biol. Crystallogr. 2002, 58, 846–848. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, X.; Wang, G. Cloning, expression, isotope labeling, and purification of human antimicrobial peptide ll-37 in escherichia coli for nmr studies. Protein Expr. Purif. 2006, 47, 498–505. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Wang, J.X.; Zhao, X.F.; Kang, C.J.; Liu, N.; Xiang, J.H.; Li, F.H.; Sueda, S.; Kondo, H. High level expression, purification, and characterization of the shrimp antimicrobial peptide, ch-penaeidin, in pichia pastoris. Protein Expr. Purif. 2005, 39, 144–151. [Google Scholar] [CrossRef]
- Li, J.; Zheng, L.; Li, P.; Wang, F. Intein-mediated expression, purification, and characterization of thymosin alpha1-thymopentin fusion peptide in escherichia coli. Protein Expr. Purif. 2012, 84, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Le, L.T.M.; Nyengaard, J.R.; Golas, M.M.; Sander, B. Vectors for expression of signal peptide-dependent proteins in baculovirus/insect cell systems and their application to expression and purification of the high-affinity immunoglobulin gamma fc receptor i in complex with its gamma chain. Mol. Biotechnol. 2018, 60, 31–40. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.V.; Lott, J.S.; Johnson, R.D.; Arcus, V.L. Expression and purification of an adenylation domain from a eukaryotic nonribosomal peptide synthetase: Using structural genomics tools for a challenging target. Protein Expr. Purif. 2010, 74, 162–168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, B.C.; Zhang, S.Q.; Dan, W.B.; Chen, Y.Q.; Cao, P. Expression in escherichia coli and purification of bioactive antibacterial peptide abp-cm4 from the chinese silk worm, bombyx mori. Biotechnol. Lett. 2007, 29, 1031–1036. [Google Scholar] [CrossRef]
- Kumar, P.; Vahedi-Faridi, A.; Merino, E.; Lopez de Castro, J.A.; Volz, A.; Ziegler, A.; Saenger, W.; Uchanska-Ziegler, B. Expression, purification and preliminary x-ray crystallographic analysis of the human major histocompatibility antigen hla-b*1402 in complex with a viral peptide and with a self-peptide. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2007, 63, 631–634. [Google Scholar] [CrossRef] [Green Version]
- Kuddus, M.R.; Rumi, F.; Tsutsumi, M.; Takahashi, R.; Yamano, M.; Kamiya, M.; Kikukawa, T.; Demura, M.; Aizawa, T. Expression, purification and characterization of the recombinant cysteine-rich antimicrobial peptide snakin-1 in pichia pastoris. Protein Expr. Purif. 2016, 122, 15–22. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, T.; Li, Y.; Li, L.; Wang, Y.; Yang, B.; Wang, Y. High-level expression of thermomyces dupontii thermo-alkaline lipase in pichia pastoris under the control of different promoters. 3 Biotech 2019, 9, 33. [Google Scholar] [CrossRef]
- Verdu, C.; Sanchez, E.; Ortega, C.; Hidalgo, A.; Berenguer, J.; Mencia, M. A modular vector toolkit with a tailored set of thermosensors to regulate gene expression in thermus thermophilus. ACS Omega 2019, 4, 14626–14632. [Google Scholar] [CrossRef] [Green Version]
- Velastegui, E.; Theron, C.; Berrios, J.; Fickers, P. Downregulation by organic nitrogen of aox1 promoter used for controlled expression of foreign genes in the yeast pichia pastoris. Yeast 2019, 36, 297–304. [Google Scholar] [CrossRef] [PubMed]
- Varela, G.M.; Stroppa, M.M.; Garcia, B.A. Daily variations in the expression of genes related to insecticide resistance in the chagas disease vector triatoma infestans (hemiptera: Reduviidae). Am. J. Trop. Med. Hyg. 2019, 100, 1482–1485. [Google Scholar] [CrossRef] [PubMed]
- Trabelsi, S.; Sahnoun, M.; Elgharbi, F.; Ameri, R.; Ben Mabrouk, S.; Mezghani, M.; Hmida-Sayari, A.; Bejar, S. Aspergillus oryzae s2 amya amylase expression in pichia pastoris: Production, purification and novel properties. Mol. Biol. Rep. 2019, 46, 921–932. [Google Scholar] [CrossRef] [PubMed]
- Theron, C.W.; Berrios, J.; Steels, S.; Telek, S.; Lecler, R.; Rodriguez, C.; Fickers, P. Expression of recombinant enhanced green fluorescent protein provides insight into foreign gene-expression differences between mut+ and muts strains of pichia pastoris. Yeast 2019, 36, 285–296. [Google Scholar] [CrossRef]
- Duong, T.T.; Lim, J.; Vasireddy, V.; Papp, T.; Nguyen, H.; Leo, L.; Pan, J.; Zhou, S.; Chen, H.I.; Bennett, J.; et al. Comparative aav-egfp transgene expression using vector serotypes 1-9, 7m8, and 8b in human pluripotent stem cells, rpes, and human and rat cortical neurons. Stem Cells Int. 2019, 2019, 7281912. [Google Scholar] [CrossRef]
- Palanikumar, I.; Katla, S.; Tahara, N.; Yui, M.; Zhang, R.; Ebihara, A.; Sivaprakasam, S. Heterologous expression, purification, and functional characterization of recombinant ovine angiotensinogen in the methylotrophic yeast pichia pastoris. Biotechnol. Prog. 2019, 35, e2866. [Google Scholar] [CrossRef]
- Huang, L.; Peng, F.; Wei, Y.; He, W.; Zhao, S.; Wang, J.; Zhang, Y.; Zhao, H.; Deng, W. A novel method to investigate the effects of gene mutations at the cellular level using a dual expression lentiviral vector. Biosci. Rep. 2019, 39, BSR20182383. [Google Scholar] [CrossRef] [Green Version]
- Jervis, A.J.; Carbonell, P.; Taylor, S.; Sung, R.; Dunstan, M.S.; Robinson, C.J.; Breitling, R.; Takano, E.; Scrutton, N.S. Selprom: A queryable and predictive expression vector selection tool for escherichia coli. ACS Synth. Biol. 2019, 8, 1478–1483. [Google Scholar] [CrossRef] [Green Version]
- Vasques, R.M.; Correa, R.F.T.; da Silva, L.A.; Blawid, R.; Nagata, T.; Ribeiro, B.M.; Ardisson-Araujo, D.M.P. Assembly of tomato blistering mosaic virus-like particles using a baculovirus expression vector system. Arch. Virol. 2019, 164, 1753–1760. [Google Scholar] [CrossRef]
- Tran, H.H.; Chen, B.; Chen, H.; Menassa, R.; Hao, X.; Bernards, M.; Huner, N.P.A.; Wang, A. Development of a cucumber green mottle mosaic virus-based expression vector for the production in cucumber of neutralizing epitopes against a devastating animal virus. J. Virol. Methods 2019, 269, 18–25. [Google Scholar] [CrossRef] [PubMed]
- Fontaine, F.; Fuchs, R.T.; Storz, G. Membrane localization of small proteins in escherichia coli. J Biol. Chem. 2011, 286, 32464–32474. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, Z.; Lu, M.; Ma, Y.; Kwag, D.G.; Kim, S.H.; Park, J.M.; Nam, B.H.; Kim, Y.O.; An, C.M.; Li, H.; et al. Production of disulfide bond-rich peptides by fusion expression using small transmembrane proteins of escherichia coli. Amino Acids 2015, 47, 579–587. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, S.; Amaishi, Y.; Goto, Y.; Ikeda, H.; Fujiwara, H.; Kuzushima, K.; Yasukawa, M.; Shiku, H.; Mineno, J. A promising vector for tcr gene therapy: Differential effect of sirna, 2a peptide, and disulfide bond on the introduced tcr expression. Mol. Ther. Nucl. Acids 2012, 1, e63. [Google Scholar] [CrossRef]
- Nguyen, V.D.; Hatahet, F.; Salo, K.E.H.; Enlund, E.; Zhang, C.; Ruddock, L.W. Pre-expression of a sulfhydryl oxidase significantly increases the yields of eukaryotic disulfide bond containing proteins expressed in the cytoplasm of e-coli. Microb. Cell Fact. 2011, 10, 1–13. [Google Scholar] [CrossRef] [Green Version]
- De Marco, A. Strategies for successful recombinant expression of disulfide bond-dependent proteins in escherichia coli. Microb. Cell Fact. 2009, 8, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Elsner, H.A.; Horn, P.A.; Schoenemann, C.; Altermann, W.W.; Blasczyk, R. Aberrant expression of hla-b*3565q is associated with a disrupted disulfide bond. Immunogenetics 2006, 58, 929–931. [Google Scholar] [CrossRef]
- Wang, X.; Yuan, C.; Huang, B.; Fan, J.; Feng, Y.; Li, A.J.; Zhang, B.; Lei, Y.; Ye, Z.; Zhao, L.; et al. Developing a versatile shotgun cloning strategy for single-vector-based multiplex expression of short interfering rnas (sirnas) in mammalian cells. ACS Synth. Biol. 2019, 8, 2092–2105. [Google Scholar] [CrossRef]
- Mavridis, K.; Wipf, N.; Medves, S.; Erquiaga, I.; Muller, P.; Vontas, J. Rapid multiplex gene expression assays for monitoring metabolic resistance in the major malaria vector anopheles gambiae. Parasites Vectors 2019, 12, 9. [Google Scholar] [CrossRef] [Green Version]
- Li, T.; Liu, B.; Mao, W.; Gao, R.; Wu, J.; Deng, Y.; Shen, Y.; Liu, K.; Cao, J. Prostaglandin e2 promotes nitric oxide synthase 2, platelet-activating factor receptor, and matrix metalloproteinase-2 expression in escherichia coli-challenged ex vivo endometrial explants via the prostaglandin e2 receptor 4/protein kinase a signaling pathway. Theriogenology 2019, 134, 65–73. [Google Scholar]
- Pazmino-Ibarra, V.; Mengual-Marti, A.; Targovnik, A.M.; Herrero, S. Improvement of baculovirus as protein expression vector and as biopesticide by crispr/cas9 editing. Biotechnol. Bioeng. 2019, 116, 2823–2833. [Google Scholar] [CrossRef] [PubMed]
- Smolander, J.; Dehmer, M.; Emmert-Streib, F. Comparing deep belief networks with support vector machines for classifying gene expression data from complex disorders. FEBS Open Bio 2019, 9, 1232–1248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yao, N.; Liu, J.Y.; Tian, Y.Y.; Dong, Y.Y.; Liu, X.M.; Li, H.Y. Cloning and expression analysis of ctbhlh transcription factor gene from carthamus tinctorius and construction of plant expression vector. Zhongguo Zhong Yao Za Zhi Zhongguo Zhongyao Zazhi China J. Chin. Mater. Med. 2019, 44, 278–282. [Google Scholar]
- Zhao, J.Z.; Xu, L.M.; Liu, M.; Cao, Y.S.; Yin, J.S.; Liu, H.B.; Lu, T.Y.; Zhang, Z.Y. Identification of the optimal insertion site for expression of a foreign gene in an infectious hematopoietic necrosis virus vector. Arch. Virol. 2019, 164, 2505–2513. [Google Scholar] [CrossRef]
- Zhu, X.H.; Peng, H.S.; Jiang, Y.L.; Wu, S.H.; Tang, S.Y.; Liu, Y.H. Construction of mouse ccr3 gene rnai lentivirus vector and its expression on mast cells]. Lin Chuang Er Bi Yan Hou Tou Jing Wai Ke Za Zhi J. Clin. Otorhinolaryngol. Head Neck Surg. 2019, 33, 628–634. [Google Scholar]
- Xu, X.; Ding, H.; Liu, J.; Xia, W.; Deng, J.; Chen, Y.; Wang, J.; Shao, Y.; Chen, D.; Ye, X. Construction of eukaryotic expression vector for human platelet cd36 gene 220c>t and 429+4insg variants and analysis of their expressions in hek293t cells. Zhonghua Yi Xue Yi Chuan Xue Za Zhi Zhonghua Yixue Yichuanxue Zazhi Chin. J. Med Genet. 2019, 36, 124–127. [Google Scholar]
- Zhao, L.; Hu, X.; Li, Y.; Wang, Z.; Wang, X. Construction of a novel escherichia coli expression system: Relocation of lpxa from chromosome to a constitutive expression vector. Appl. Microbiol. Biotechnol. 2019, 103, 7177–7189. [Google Scholar] [CrossRef]
- Sandro, Q.; Relizani, K.; Benchaouir, R. Aav production using baculovirus expression vector system. Methods Mol. Biol. 2019, 1937, 91–99. [Google Scholar]
- Takagi, S.; Tsutsumi, N.; Terui, Y.; Kong, X.; Yurimoto, H.; Sakai, Y. Engineering the expression system for komagataella phaffii (pichia pastoris): An attempt to develop a methanol-free expression system. FEMS Yeast Res. 2019, 19, foz059. [Google Scholar] [CrossRef] [Green Version]
- Zhao, D.; Huang, Z.; Liu, J.; Ma, L.; He, J. Expression, purification, and characterization of n-terminal his-tagged proteins with mutations in zinc finger 3 of zinc finger protein znf191(243-368). Prep. Biochem. Biotechnol. 2018, 48, 914–919. [Google Scholar] [CrossRef]
- Zhang, S.B.; Zhang, W.J.; Zhai, H.C.; Lv, Y.Y.; Cai, J.P.; Jia, F.; Wang, J.S.; Hu, Y.S. Expression of a wheat beta-1,3-glucanase in pichia pastoris and its inhibitory effect on fungi commonly associated with wheat kernel. Protein Expr. Purif. 2019, 154, 134–139. [Google Scholar] [CrossRef] [PubMed]
- Li, X.D.; Zhou, L.J.; Zhao, C.; Lu, L.; Niu, N.N.; Han, J.X.; Zhao, K.H. Optimization of expression of orange carotenoid protein in escherichia coli. Protein Expr. Purif. 2019, 156, 66–71. [Google Scholar] [CrossRef] [PubMed]
- Zheng, W.; Yang, L.; Cai, C.; Ni, J.; Liu, B. Expression, purification and characterization of a novel double-sites mutant of the single-chain sweet-tasting protein monellin (mnei) with both improved sweetness and stability. Protein Expr. Purif. 2018, 143, 52–56. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.Y.; Lan, X.; Li, X.J.; Huang, L.J.; Zhang, Y.J.; Wang, Z. High-level expression and characterization of a stereoselective lipase from aspergillus oryzae in pichia pastoris. Protein Expr. Purif. 2019, 155, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Wang, Q.; Xu, Z.; Liu, R.; Cui, F. Distinct replication and gene expression strategies of the rice stripe virus in vector insects and host plants. J. Gen. Virol. 2019, 100, 877–888. [Google Scholar] [CrossRef]
- Zhao, R.Q. Expression, purification and characterization of the plant snf1-related protein kinase 1 from escherichia coli. Protein Expr. Purif. 2019, 162, 24–31. [Google Scholar] [CrossRef]
- Shi, X.; Cordero, T.; Garrigues, S.; Marcos, J.F.; Daros, J.A.; Coca, M. Efficient production of antifungal proteins in plants using a new transient expression vector derived from tobacco mosaic virus. Plant Biotechnol. J. 2019, 17, 1069–1080. [Google Scholar] [CrossRef]
- Seyfi, R.; Babaeipour, V.; Mofid, M.R.; Kahaki, F.A. Expression and production of recombinant scorpine as a potassium channel blocker protein in escherichia coli. Biotechnol. Appl. Biochem. 2019, 66, 119–129. [Google Scholar] [CrossRef] [Green Version]
- Retnoningrum, D.S.; Santika, I.W.M.; Kesuma, S.; Ekowati, S.A.; Riani, C. Construction and characterization of a medium copy number expression vector carrying auto-inducible dps promoter to overproduce a bacterial superoxide dismutase in escherichia coli. Mol. Biotechnol. 2019, 61, 231–240. [Google Scholar] [CrossRef]
- Peng, B.; Xue, G.; Xu, D.; Feng, Z.; Chen, J.; Huang, M.; Lu, H.; Gong, L. Expression and purification of recombinant serine protease domain of human coagulation factor xii in pichia pastoris. Biosci. Biotechnol. Biochem. 2019, 83, 1815–1821. [Google Scholar] [CrossRef]
- Amroudie, M.N.; Ataei, F. Experimental and theoretical study of ibc domain from human ip3r2; molecular cloning, bacterial expression and protein purification. Int. J. Biol. Macromol. 2019, 124, 1321–1327. [Google Scholar] [CrossRef] [PubMed]
- Arjmand, S.; Tavasoli, Z.; Ranaei Siadat, S.O.; Saeidi, B.; Tavana, H. Enhancing chimeric hydrophobin ii-vascular endothelial growth factor a165 expression in pichia pastoris and its efficient purification using hydrophobin counterpart. Int. J. Biol. Macromol. 2019, 139, 1028–1034. [Google Scholar] [CrossRef] [PubMed]
- Ballesteros-Briones, M.C.; Martisova, E.; Casales, E.; Silva-Pilipich, N.; Bunuales, M.; Galindo, J.; Mancheno, U.; Gorraiz, M.; Lasarte, J.J.; Kochan, G.; et al. Short-term local expression of a pd-l1 blocking antibody from a self-replicating rna vector induces potent antitumor responses. Mol. Ther. J. Am. Soc. Gene Ther. 2019, 27, 1892–1905. [Google Scholar] [CrossRef] [PubMed]
- Barkhordari, F.; Sohrabi, N.; Davami, F.; Mahboudi, F.; Garoosi, Y.T. Cloning, expression and characterization of a her2-alpha luffin fusion protein in escherichia coli. Prep. Biochem. Biotechnol. 2019, 49, 759–766. [Google Scholar] [CrossRef]
- Bryant, W.B.; Mills, M.K.; Olson, B.J.; Michel, K. Small rna-seq analysis reveals mirna expression dynamics across tissues in the malaria vector, anopheles gambiae. G3 2019, 9, 1507–1517. [Google Scholar] [CrossRef] [Green Version]
- Camara, E.; Monforte, S.; Albiol, J.; Ferrer, P. Deregulation of methanol metabolism reverts transcriptional limitations of recombinant pichia pastoris (komagataella spp) with multiple expression cassettes under control of the aox1 promoter. Biotechnol. Bioeng. 2019, 116, 1710–1720. [Google Scholar] [CrossRef]
- Carmignotto, G.P.; Azzoni, A.R. On the expression of recombinant cas9 protein in e. Coli bl21(de3) and bl21(de3) rosetta strains. J. Biotechnol. 2019, 306, 62–70. [Google Scholar] [CrossRef]
- Huang, S.F.; Liu, D.B.; Zeng, J.M.; Xiao, Q.; Luo, M.; Zhang, W.P.; Tao, K.; Wen, J.P.; Huang, Z.G.; Feng, W.L. Cloning, expression, purification and functional characterization of the oligomerization domain of bcr-abl oncoprotein fused to the cytoplasmic transduction peptide. Protein Expr. Purif. 2009, 64, 167–178. [Google Scholar] [CrossRef]
- Berkmen, M. Production of disulfide-bonded proteins in escherichia coli. Protein Expr. Purif. 2012, 82, 240–251. [Google Scholar] [CrossRef] [Green Version]
- Santos, B.D.; Morones-Ramirez, J.R.; Balderas-Renteria, I.; Casillas-Vega, N.G.; Galbraith, D.W.; Zarate, X. Optimizing periplasmic expression in escherichia coli for the production of recombinant proteins tagged with the small metal-binding protein smbp. Mol. Biotechnol. 2019, 61, 451–460. [Google Scholar] [CrossRef]
- Wei, X.; Wu, R.; Zhang, L.; Ahmad, B.; Si, D.; Zhang, R. Expression, purification, and characterization of a novel hybrid peptide with potent antibacterial activity. Molecules 2018, 23, 1491. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, Y.; Han, X.; Miao, Y. An effective recombinant protein expression and purification system in saccharomyces cerevisiae. Curr. Protoc. Mol. Biol. 2018, 123, e62. [Google Scholar] [CrossRef]
- Jahangirizadeh, Z.; Ghafouri, H.; Sajedi, R.H.; Sarikhan, S.; Taghdir, M.; Sariri, R. Molecular cloning, prokaryotic expression, purification, structural studies and functional implications of heat shock protein 70 (hsp70) from rutilus frisii kutum. Int. J. Biol. Macromol. 2018, 108, 798–807. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Yuan, D.; Huang, L.J. Development of a gateway-compatible two-component expression vector system for plants. Transgenic Res. 2019, 28, 561–572. [Google Scholar] [CrossRef]
- Li, Q.; Chang, Z.; Oliveira, G.; Xiong, M.; Smith, L.M.; Frey, B.L.; Welham, N.V. Protein turnover during in vitro tissue engineering. Biomaterials 2016, 81, 104–113. [Google Scholar] [CrossRef] [Green Version]
- Kikuchi, M.; Iwabuchi, S.; Kikkou, T.; Noguchi, K.; Odaka, M.; Yohda, M.; Kawata, M.; Sato, C.; Matsumoto, O. Expression, purification, crystallization and preliminary crystallographic analysis of hepatitis b virus core protein dimerized via a peptide linker containing an egfp insertion. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2013, 69, 942–945. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karakas-Sen, A.; Narbad, A. Heterologous expression and purification of nisa, the precursor peptide of lantibiotic nisin from lactococcus lactis. Acta Biol. Hung. 2012, 63, 301–310. [Google Scholar] [CrossRef] [PubMed]
- Johnson, T.; Koria, P. Expression and purification of neurotrophin-elastin-like peptide fusion proteins for neural regeneration. BioDrugs Clin. Immunother. Biopharm. Gene Ther. 2016, 30, 117–127. [Google Scholar] [CrossRef]
- Huo, Y.; Wang, F.; Sun, B.; Yin, L.R.; Zhang, P.P.; Zhang, Y.J.; Zhang, B.M. Prokaryotic expression and purification of antimicrobial peptide ll-37 and the inhibiting effect against candida albicans. Zhonghua Fu Chan Ke Za Zhi 2016, 51, 120–125. [Google Scholar]
- Huang, S.F.; Liu, D.B.; Zeng, J.M.; Yuan, Y.; Xiao, Q.; Sun, C.M.; Li, C.L.; Tao, K.; Wen, J.P.; Huang, Z.G.; et al. Cloning, expression, purification, distribution and kinetics characterization of the bacterial beta-galactosidase fused to the cytoplasmic transduction peptide in vitro and in vivo. Protein Expr. Purif. 2009, 68, 167–176. [Google Scholar] [CrossRef]
- Hu, F.; Ke, T.; Li, X.; Mao, P.H.; Jin, X.; Hui, F.L.; Ma, X.D.; Ma, L.X. Expression and purification of an antimicrobial peptide by fusion with elastin-like polypeptides in escherichia coli. Appl. Biochem. Biotechnol. 2010, 160, 2377–2387. [Google Scholar] [CrossRef] [PubMed]
- He, Q.; Fu, A.Y.; Li, T.J. Expression and one-step purification of the antimicrobial peptide cathelicidin-bf using the intein system in bacillus subtilis. J. Ind. Microbiol. Biotechnol. 2015, 42, 647–653. [Google Scholar] [CrossRef] [PubMed]
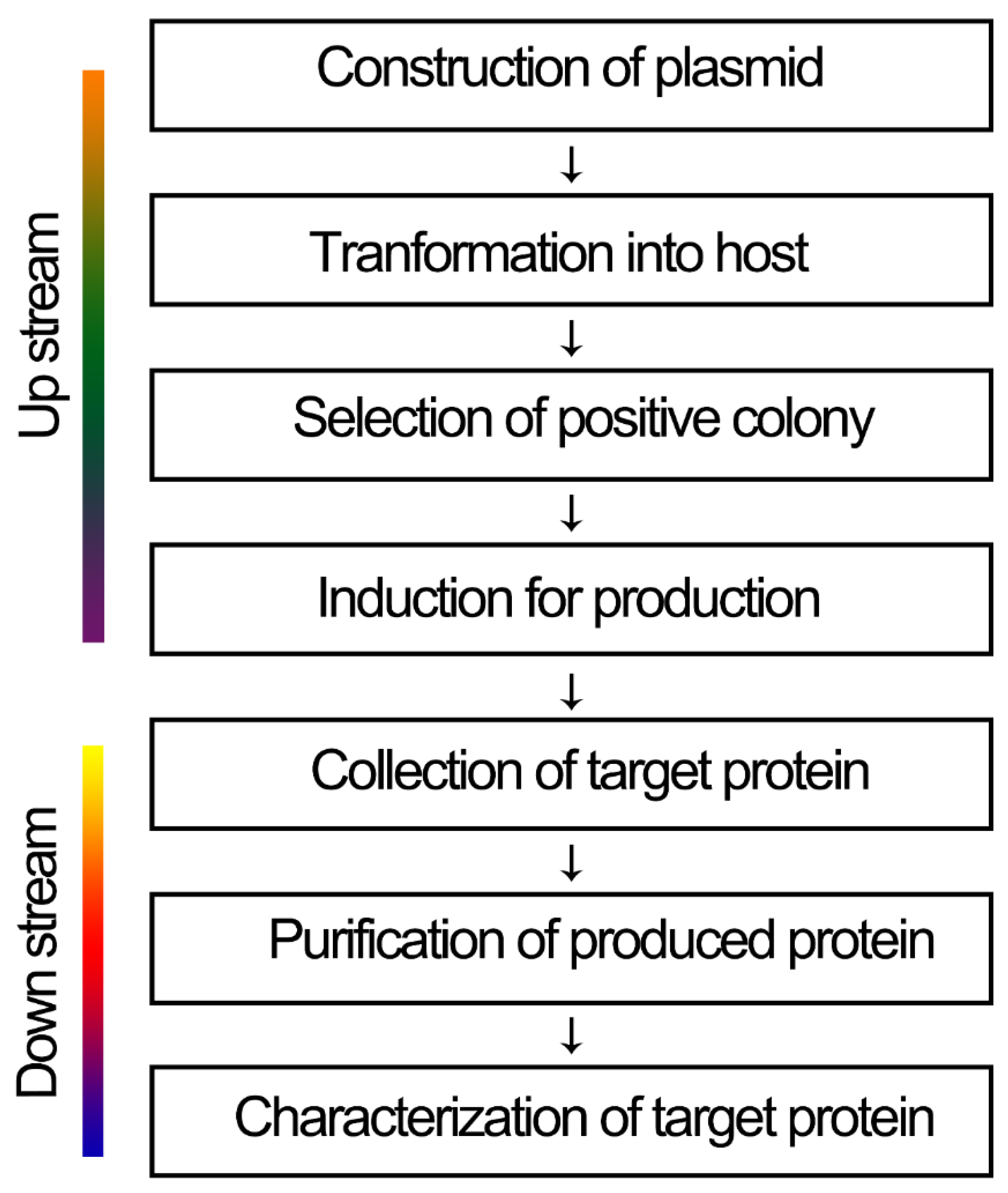
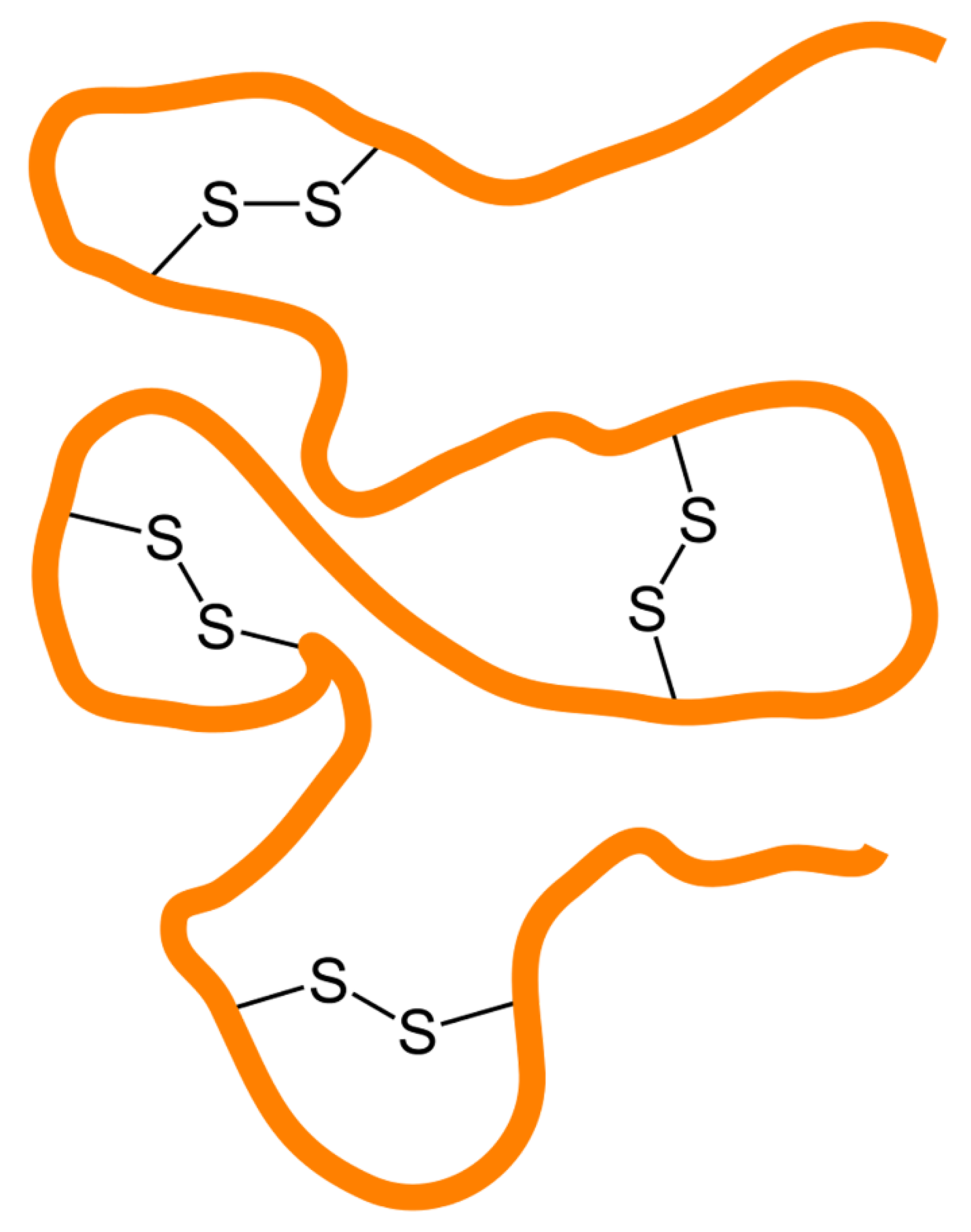
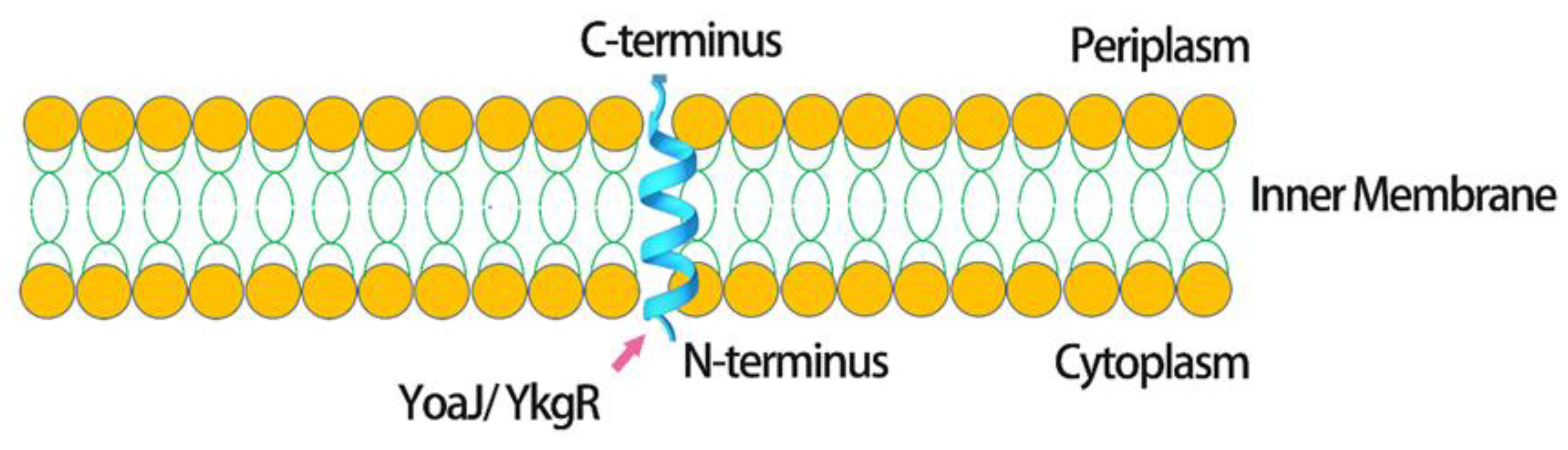
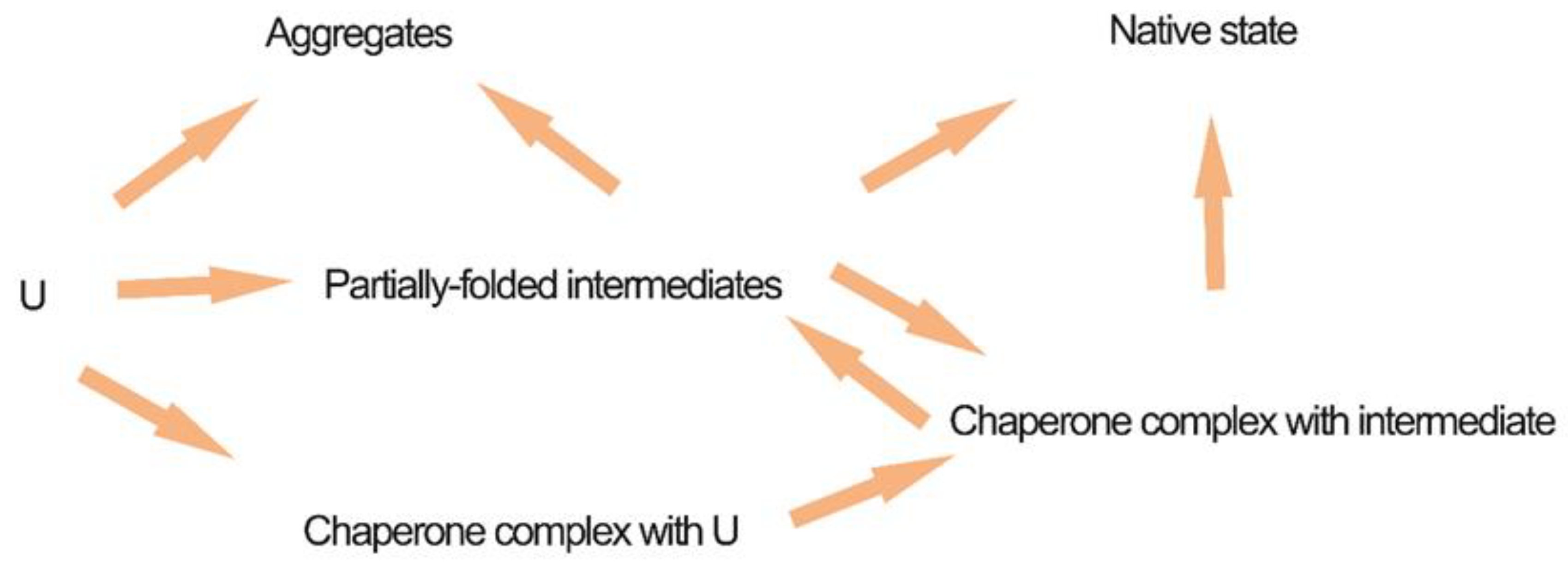

| Advantages | Disadvantages |
|---|---|
| Rapid expression | Proteins with disulfide bonds are difficult to express |
| High yields | Production of unglycosylated proteins |
| Ease of culture and genome modifications | Proteins with endotoxins are produced |
| Inexpensive | Acetate formation results in cell toxicity |
| Rapid mass production and cost-effective | Proteins produced as inclusion bodies are inactive; thus, refolding is required. |
| Advantages | Disadvantages |
|---|---|
| High yield | N or O-linked glycosylation pattern (different from higher eukaryote) Hypermannosylation Proteolytic degradation |
| Stable production strains | |
| Durability | |
| Cost-effective | |
| High-density growth | |
| High productivity | |
| Suitable for isotopically labeled protein production | |
| Rapid growth in chemically defined media | |
| Product processing similar to mammalian cells | |
| Can handle S–S-rich proteins | |
| Can assist in protein folding | |
| Can glycosylate proteins |
| Host | Recombinant Protein | Reference |
|---|---|---|
| E. coli | venom proteins | [83,84] |
| P. pastoris S. cerevisiae | lipase r27RC rat protein disulfide isomerase | [85] [86] |
| L. lactis | Merozoite antigens | [87] |
| Advantage | Disadvantage | |
|---|---|---|
| Cytoplasmic | Higher expression level, simple plasmid construction | Inclusion body may be formed, unfavorable conditions for S-S bond formation, higher proteolysis |
| Periplasmic | Less proteolysis, improved folding, simple purification | Inclusion body may be formed, the signal does not always facilitate export |
| Extracellular | Least extensive proteolysis, simpler purification (fewer protein species), improved folding | Usually no secretion is observed, purification may be complex (protein dilution) |
| Vector | Induction (IPTG) | Level of Expression | Key Feature |
|---|---|---|---|
| lac | 0.2 mM | Low to moderate levels; 15–30% of total cell proteins | Weak, regulated, suitable for gene products at very low intracellular level; comparatively expensive induction |
| Trc and tac | 0.2 mM | Moderately high | High-level, but lower than T7 system; regulated expression still possible; comparatively expensive induction; high basal level |
| T7 RNA polymerase | 0.2 mM | Very high; 40–50% of total cell proteins | Utilizes T7 RNA polymerase; high-level inducible overexpression; T7lac system for tight control of induction needed for more toxic clones; relatively expensive induction; basal level depends on used strain (pLys) |
| Phage promoter PL | Shifting the temperature from 30 to 42 °C (45 °C) | Moderately high | Temperature-sensitive host required; lower likelihood of “leaky” non-induced expression; basal level, high basal level by temperatures below 30 °C |
| Peptide | Disulfide Number |
|---|---|
| Beta-defensin | 3 |
| hepcidin | 3/4 |
| DkTx | 6 |
| ASPA | 3 |
| Fusion Protein | Yield (mg/L) | Disulfide Bond Number | Tag Size (kDa) | Ref |
|---|---|---|---|---|
| YkgR-BD | ~25 | 3 | 4 | [123] |
| YkgR-Hep | ~17 | 4 | 4 | [123] |
| YkgR-DkTx | ~8 | 6 | 4 | [123] |
| YoaJ-ASPA | ~5 | 3 | 2.69 | [123] |
| YoaJ-BD | ~10 | 3 | 2.69 | [123] |
| DsbA-scFv | 0.3 | 23.5 | [124] | |
| MBP-nanobodies | 2.2 | 42 | [125] | |
| DsbC-Huwentoxin | 0.7 | 23.5 | [126] | |
| pelB-human growth hormone | 1.4 | 2.31 | [127] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, Y.; Lee, C.-J.; Park, J.-S. Strategies for Optimizing the Production of Proteins and Peptides with Multiple Disulfide Bonds. Antibiotics 2020, 9, 541. https://doi.org/10.3390/antibiotics9090541
Ma Y, Lee C-J, Park J-S. Strategies for Optimizing the Production of Proteins and Peptides with Multiple Disulfide Bonds. Antibiotics. 2020; 9(9):541. https://doi.org/10.3390/antibiotics9090541
Chicago/Turabian StyleMa, Yunqi, Chang-Joo Lee, and Jang-Su Park. 2020. "Strategies for Optimizing the Production of Proteins and Peptides with Multiple Disulfide Bonds" Antibiotics 9, no. 9: 541. https://doi.org/10.3390/antibiotics9090541
APA StyleMa, Y., Lee, C.-J., & Park, J.-S. (2020). Strategies for Optimizing the Production of Proteins and Peptides with Multiple Disulfide Bonds. Antibiotics, 9(9), 541. https://doi.org/10.3390/antibiotics9090541






