The Bristol Sponge Microbiome Collection: A Unique Repository of Deep-Sea Microorganisms and Associated Natural Products
Abstract
1. Introduction
2. Results
2.1. Sponge Sample Collection
2.2. Isolation of Strains Producing Active Compounds
2.2.1. Bacterial Culturing and Isolation
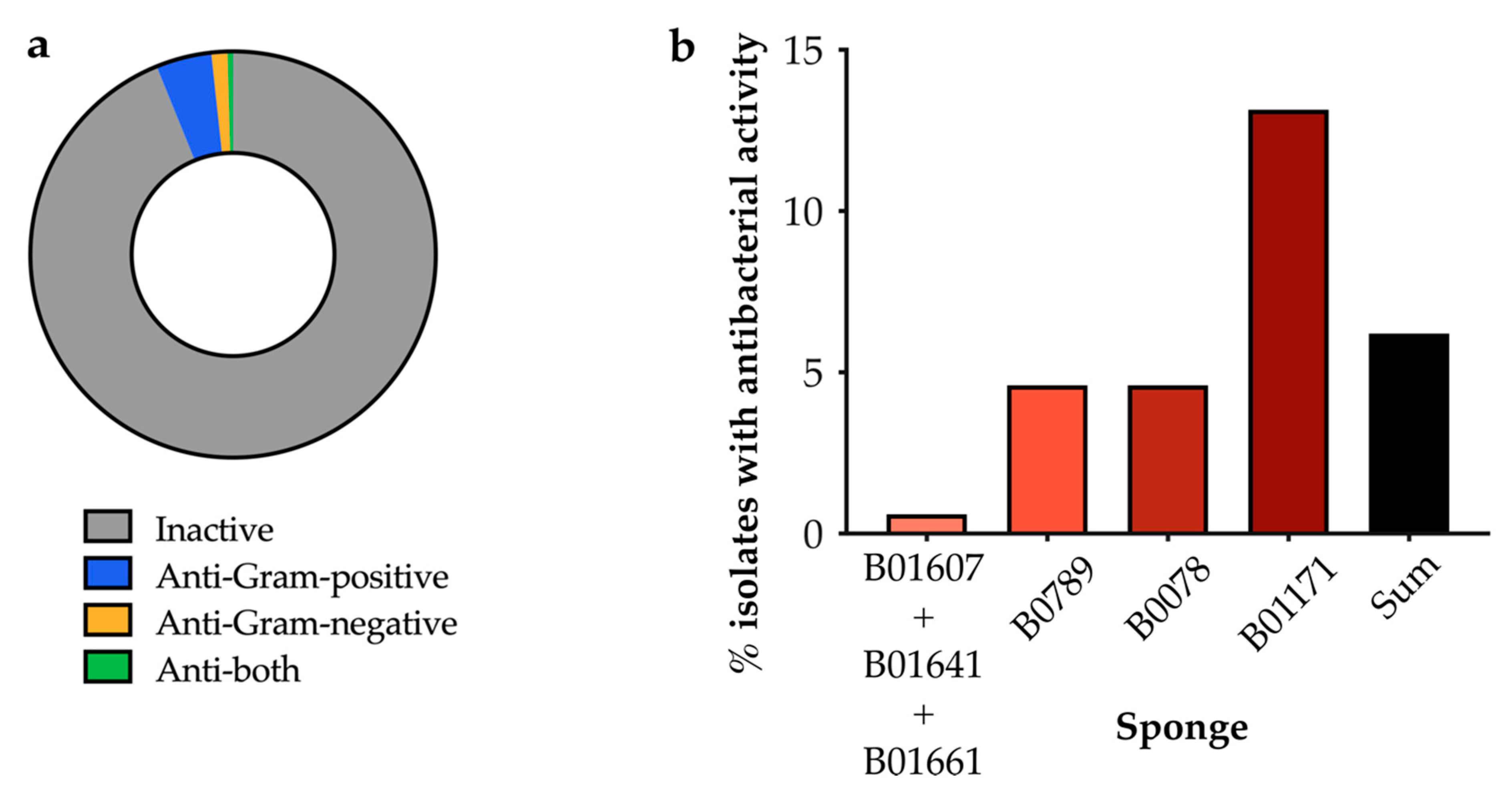
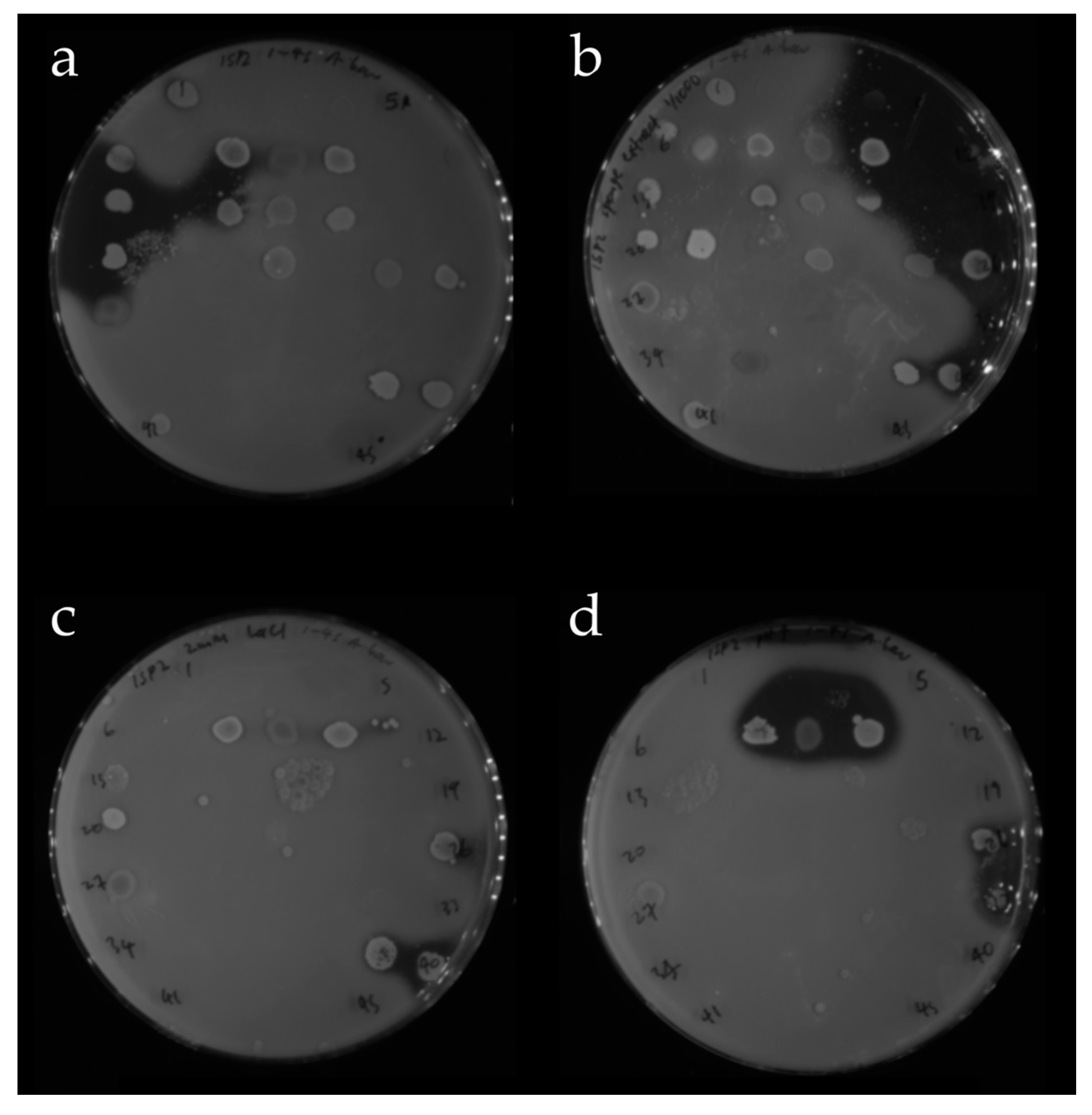
2.2.2. Screening of Isolated Strains for Antibiotic Activity
2.3. Composition of Cultured Bacteria
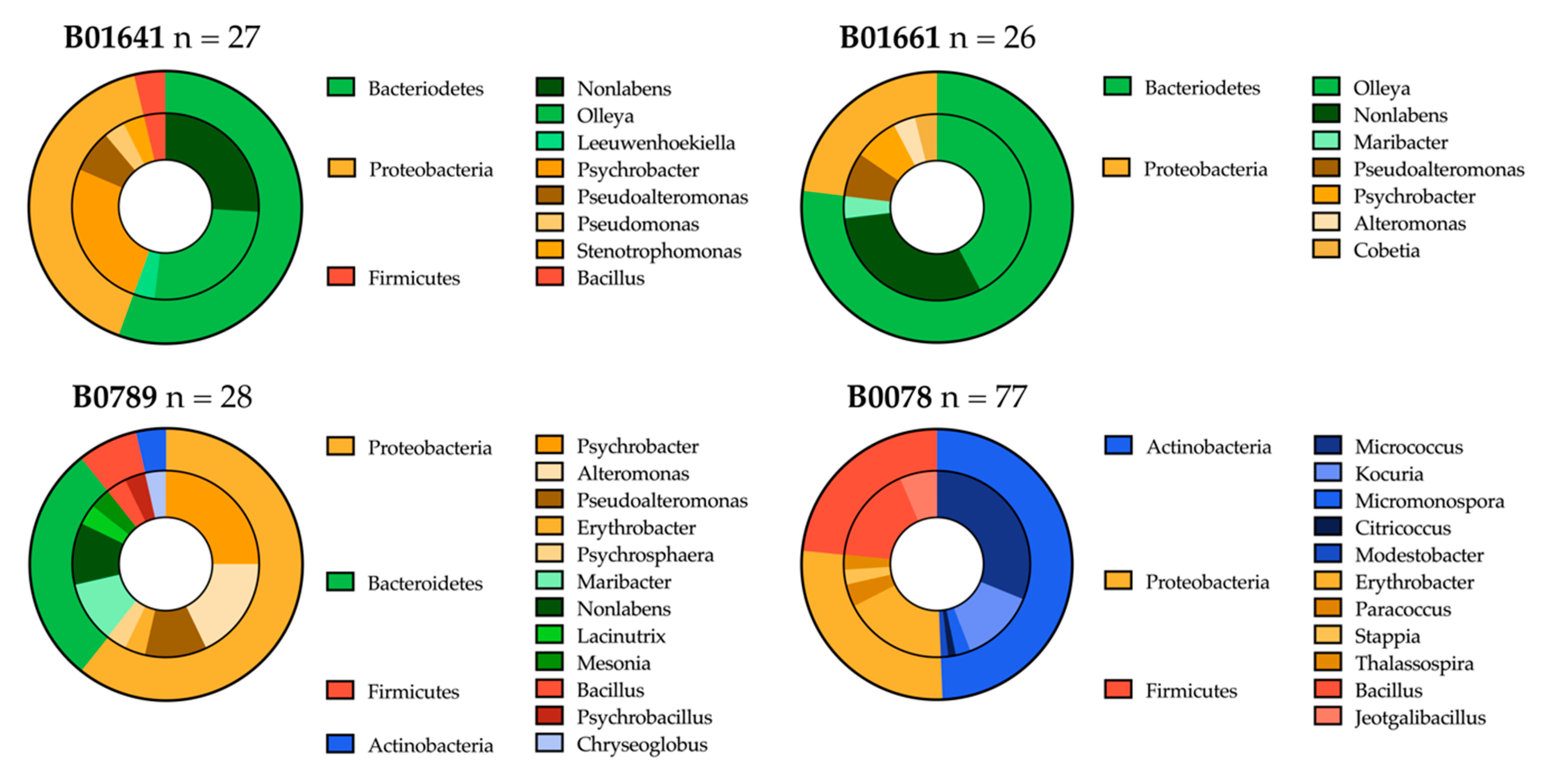
2.3.1. Distribution of Isolated Strains from Sponges B01641, B01661, B0789 and B0078
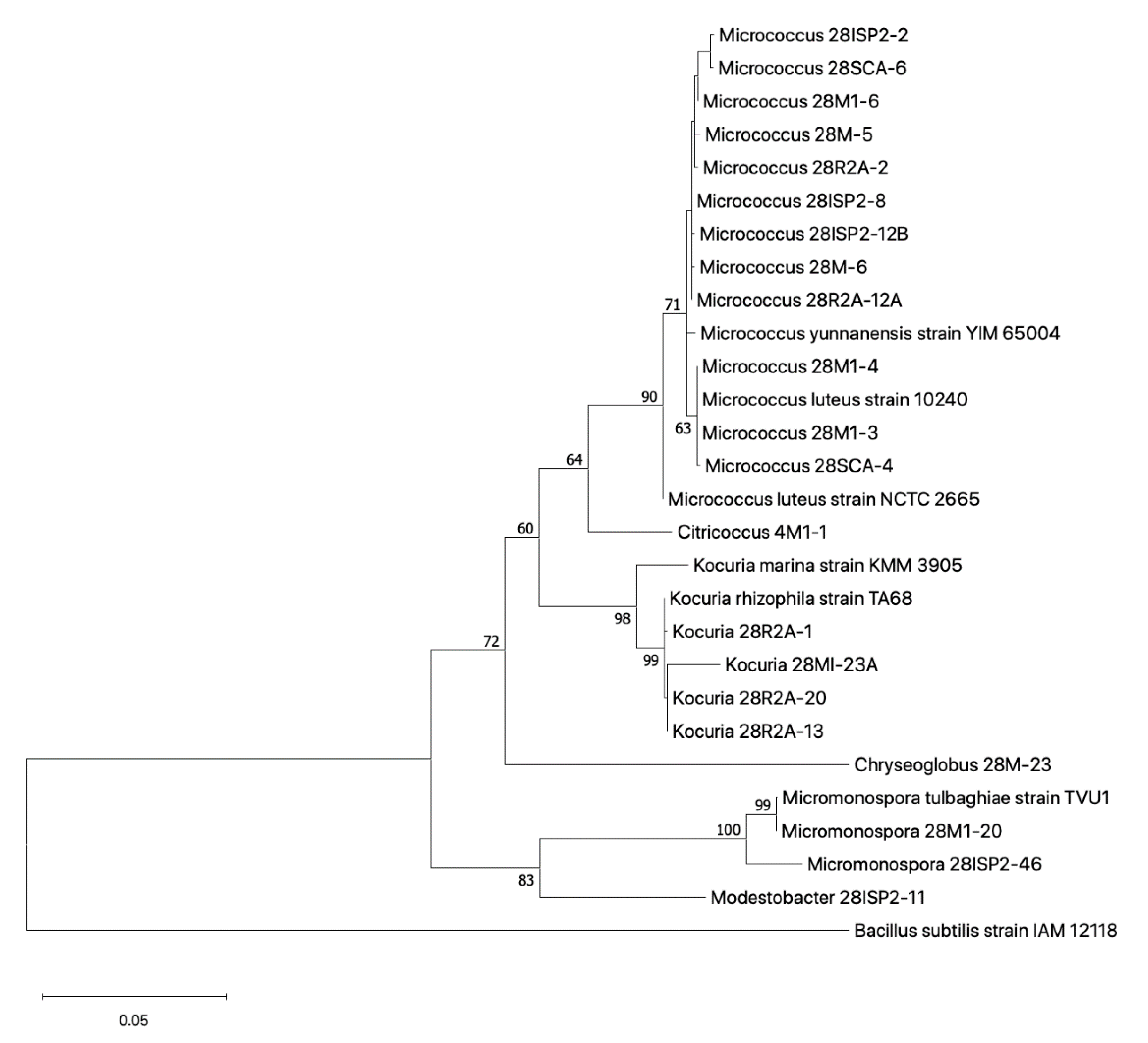
2.3.2. Phylogenetic Relationship of Cultured Actinobacteria
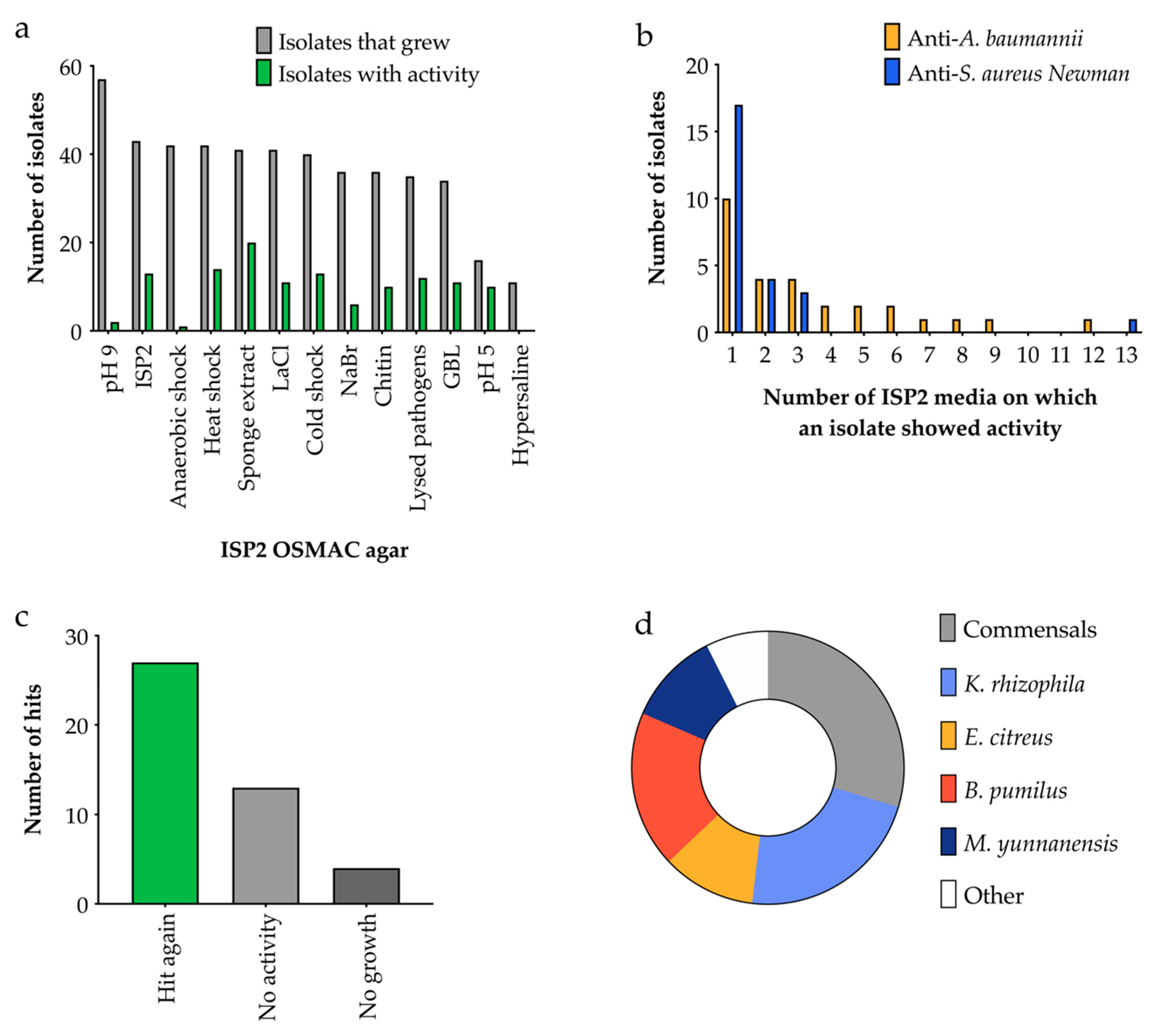
2.4. OSMAC Screen of 90 Isolates
2.4.1. OSMAC Screen Using IPS2 Media
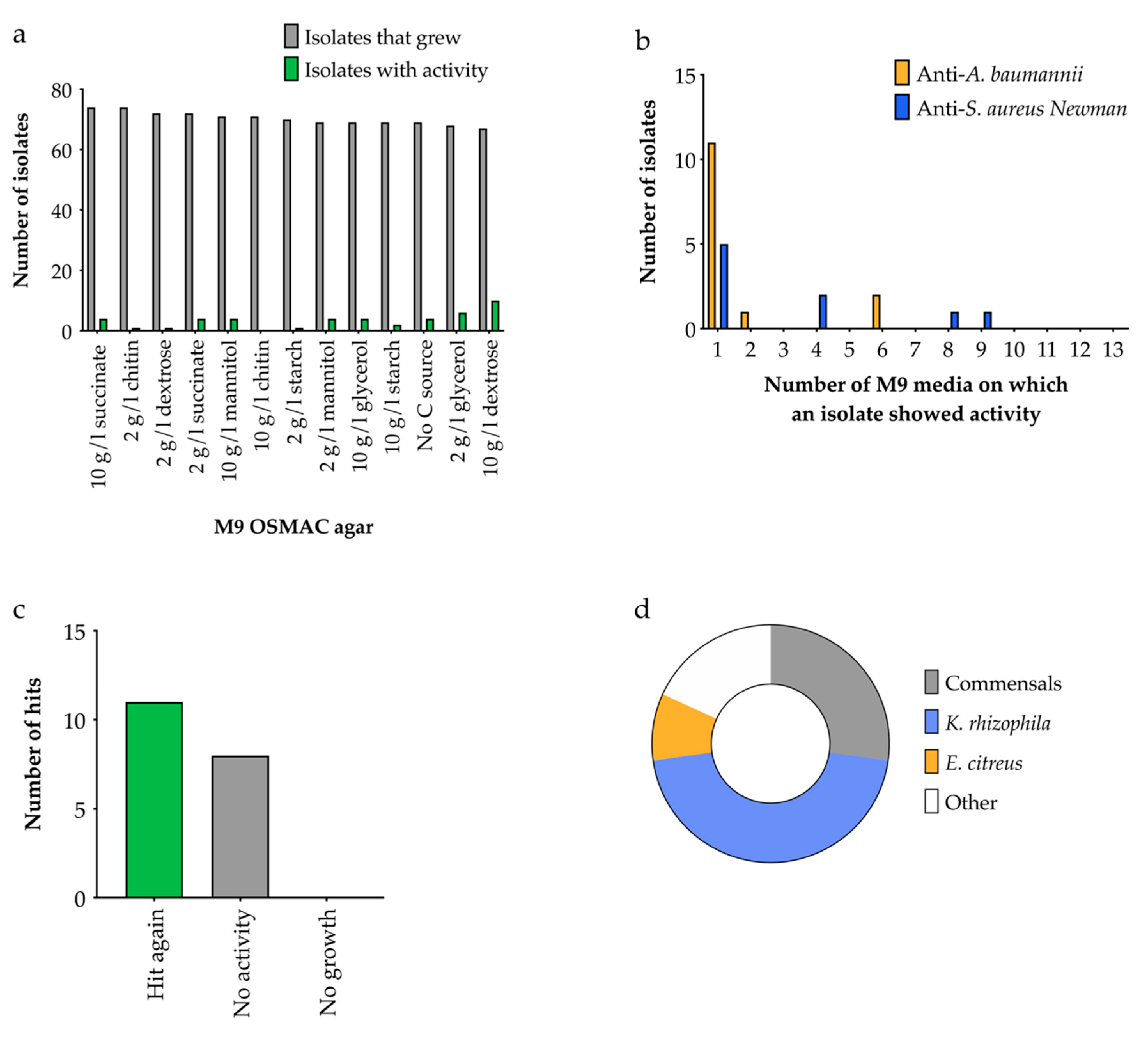
2.4.2. OSMAC Screen Using M9 Media
3. Discussion
4. Materials and Methods
4.1. Sponge Sample Collection
4.2. Sponge Processing and Isolation of Bacterial Strains
4.3. Sponge Extract Preparation
4.4. Soft Agar Overlay Antimicrobial Activity Assay
4.5. 16S rRNA PCR Gene Amplification
4.6. Phylogenetic Analysis
4.7. OSMAC Screening
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Newman, D.J.; Cragg, G.M. Natural Products as Sources of New Drugs from 1981 to 2014. J. Nat. Prod. 2016, 79, 629–661. [Google Scholar] [CrossRef] [PubMed]
- Dias, D.A.; Urban, S.; Roessner, U. A historical overview of natural products in drug discovery. Metabolites 2012, 2, 303–336. [Google Scholar] [CrossRef] [PubMed]
- Harvey, A.L.; Edrada-Ebel, R.; Quinn, R.J. The re-emergence of natural products for drug discovery in the genomics era. Nat. Rev. Drug Discov. 2015, 14, 111–129. [Google Scholar] [CrossRef] [PubMed]
- Li, J.W.H.; Vederas, J.C. Drug Discovery and Natural Products: End of an Era or an Endless Frontier? Science 2009, 325, 161. [Google Scholar] [CrossRef]
- Li, F.; Wang, Y.; Li, D.; Chen, Y.; Dou, Q.P. Are we seeing a resurgence in the use of natural products for new drug discovery? Expert Opin. Drug Discov. 2019, 14, 417–420. [Google Scholar] [CrossRef]
- Fox Ramos, A.E.; Evanno, L.; Poupon, E.; Champy, P.; Beniddir, M.A. Natural products targeting strategies involving molecular networking: Different manners, one goal. Nat. Prod. Rep. 2019, 36, 960–980. [Google Scholar] [CrossRef]
- Zhang, X.; Hindra; Elliot, M.A. Unlocking the trove of metabolic treasures: Activating silent biosynthetic gene clusters in bacteria and fungi. Curr. Opin. Microbiol. 2019, 51, 9–15. [Google Scholar] [CrossRef]
- Bull, A.T.; Goodfellow, M. Dark, rare and inspirational microbial matter in the extremobiosphere: 16 000 m of bioprospecting campaigns. Microbiology 2019, 165, 1252–1264. [Google Scholar] [CrossRef]
- Tortorella, E.; Tedesco, P.; Palma Esposito, F.; January, G.G.; Fani, R.; Jaspars, M.; De Pascale, D. Antibiotics from Deep-Sea Microorganisms: Current Discoveries and Perspectives. Mar. Drugs 2018, 16, 355. [Google Scholar] [CrossRef]
- Skropeta, D.; Wei, L. Recent advances in deep-sea natural products. Nat. Prod. Rep. 2014, 31, 999–1025. [Google Scholar] [CrossRef]
- Cheng, M.M.; Tang, X.L.; Sun, Y.T.; Song, D.Y.; Cheng, Y.J.; Liu, H.; Li, P.L.; Li, G.Q. Biological and Chemical Diversity of Marine Sponge-Derived Microorganisms over the Last Two Decades from 1998 to 2017. Molecules 2020, 25, 853. [Google Scholar] [CrossRef]
- Carroll, A.R.; Copp, B.R.; Davis, R.A.; Keyzers, R.A.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2020, 37, 175–223. [Google Scholar] [CrossRef]
- Schmidt, E.W.; Obraztsova, A.Y.; Davidson, S.K.; Faulkner, D.J.; Haygood, M.G. Identification of the antifungal peptide-containing symbiont of the marine sponge Theonella swinhoei as a novel δ-proteobacterium, “Candidatus Entotheonella palauensis”. Mar. Biol. 2000, 136, 969–977. [Google Scholar] [CrossRef]
- Sayed, A.M.; Hassan, M.H.A.; Alhadrami, H.A.; Hassan, H.M.; Goodfellow, M.; Rateb, M.E. Extreme environments: Microbiology leading to specialized metabolites. J. Appl. Microbiol. 2020, 128, 630–657. [Google Scholar] [CrossRef]
- Sun, C.; Mudassir, S.; Zhang, Z.; Feng, Y.; Chang, Y.; Che, Q.; Gu, Q.; Zhu, T.; Zhang, G.; Li, D. Secondary Metabolites from Deep-sea Derived Microorganisms. Curr. Med. Chem. 2019. [Google Scholar] [CrossRef]
- Jaspars, M.; De Pascale, D.; Andersen, J.H.; Reyes, F.; Crawford, A.D.; Ianora, A. The marine biodiscovery pipeline and ocean medicines of tomorrow. J. Mar. Biol. Assoc. U.K. 2016, 96, 151–158. [Google Scholar] [CrossRef]
- Tormo, J.R.; Oves, D.; Lacret, R.; Moreno, C.; DeLa Cruz, M.; Díaz, C.; González, I.; Martín, J.; Pérez-Victoria, I.; Vicente, F.; et al. Marine natural products drug discovery from actinomycetes at Fundacion MEDINA. Planta. Med. 2015, 81, PK18. [Google Scholar] [CrossRef]
- Lertcanawanichakul, M.; Sawangnop, S. A Comparison of Two Methods Used for Measuring the Antagonistic Activity of Bacillus Species. Walailak J. Sci. Tech. 2011, 5, 161–171. [Google Scholar]
- Elleuch, L.; Shaaban, M.; Smaoui, S.; Mellouli, L.; Karray-Rebai, I.; Fourati-Ben Fguira, L.; Shaaban, K.A.; Laatsch, H. Bioactive secondary metabolites from a new terrestrial Streptomyces sp. TN262. Appl. Biochem. Biotechnol. 2010, 162, 579–593. [Google Scholar] [CrossRef]
- Anand, T.P.; Bhat, A.W.; Shouche, Y.S.; Roy, U.; Siddharth, J.; Sarma, S.P. Antimicrobial activity of marine bacteria associated with sponges from the waters off the coast of South East India. Microbiol. Res. 2006, 161, 252–262. [Google Scholar] [CrossRef]
- El-Hawary, S.S.; Sayed, A.M.; Mohammed, R.; Khanfar, M.A.; Rateb, M.E.; Mohammed, T.A.; Hajjar, D.; Hassan, H.M.; Gulder, T.A.M.; Abdelmohsen, U.R. New Pim-1 Kinase Inhibitor From the Co-culture of Two Sponge-Associated Actinomycetes. Front. Chem. 2018, 6, 538. [Google Scholar] [CrossRef] [PubMed]
- Dalisay, D.S.; Williams, D.E.; Wang, X.L.; Centko, R.; Chen, J.; Andersen, R.J. Marine Sediment-Derived Streptomyces Bacteria from British Columbia, Canada Are a Promising Microbiota Resource for the Discovery of Antimicrobial Natural Products. PLoS ONE 2013, 8, e77078. [Google Scholar] [CrossRef] [PubMed]
- Bredholt, H.; Fjaervik, E.; Johnsen, G.; Zotchev, S.B. Actinomycetes from sediments in the Trondheim fjord, Norway: Diversity and biological activity. Mar. Drugs 2008, 6, 12–24. [Google Scholar] [CrossRef] [PubMed]
- Kamjam, M.; Sivalingam, P.; Deng, Z.; Hong, K. Deep Sea Actinomycetes and Their Secondary Metabolites. Front. Microbiol. 2017, 8, 760. [Google Scholar] [CrossRef] [PubMed]
- Bode, H.B.; Bethe, B.; Höfs, R.; Zeeck, A. Big effects from small changes: Possible ways to explore nature’s chemical diversity. Chembiochem 2002, 3, 619–627. [Google Scholar] [CrossRef]
- Romano, S.; Jackson, S.A.; Patry, S.; Dobson, A.D.W. Extending the “One Strain Many Compounds” (OSMAC) Principle to Marine Microorganisms. Mar. Drugs 2018, 16, 244. [Google Scholar] [CrossRef]
- Shirling, E.B.; Gottlieb, D. Methods for characterization of Streptomyces species1. Int. J. Syst. Evol. Microbiol. 1966, 16, 313–340. [Google Scholar] [CrossRef]
- Millero, F.J.; Feistel, R.; Wright, D.G.; McDougall, T.J. The composition of Standard Seawater and the definition of the Reference-Composition Salinity Scale. Deep Sea Res. Part. I Oceanogr. Res. Pap. 2008, 55, 50–72. [Google Scholar] [CrossRef]
- Sung, A.A.; Gromek, S.M.; Balunas, M.J. Upregulation and Identification of Antibiotic Activity of a Marine-Derived Streptomyces sp. via Co-Cultures with Human Pathogens. Mar. Drugs 2017, 15, 250. [Google Scholar] [CrossRef]
- Kai, W.; Peisheng, Y.; Rui, M.; Wenwen, J.; Zongze, S. Diversity of culturable bacteria in deep-sea water from the South Atlantic Ocean. Bioengineered 2017, 8, 572–584. [Google Scholar] [CrossRef]
- Palomo, S.; González, I.; De la Cruz, M.; Martín, J.; Tormo, J.R.; Anderson, M.; Hill, R.T.; Vicente, F.; Reyes, F.; Genilloud, O. Sponge-derived Kocuria and Micrococcus spp. as sources of the new thiazolyl peptide antibiotic kocurin. Mar. Drugs 2013, 11, 1071–1086. [Google Scholar] [CrossRef] [PubMed]
- Pye, C.R.; Bertin, M.J.; Lokey, R.S.; Gerwick, W.H.; Linington, R.G. Retrospective analysis of natural products provides insights for future discovery trends. Proc. Natl. Acad. Sci. USA 2017, 114, 5601–5606. [Google Scholar] [CrossRef] [PubMed]
- Esteves, A.I.; Amer, N.; Nguyen, M.; Thomas, T. Sample Processing Impacts the Viability and Cultivability of the Sponge Microbiome. Front. Microbiol. 2016, 7, 499. [Google Scholar] [CrossRef] [PubMed]
- Sipkema, D.; Schippers, K.; Maalcke, W.J.; Yang, Y.; Salim, S.; Blanch, H.W. Multiple approaches to enhance the cultivability of bacteria associated with the marine sponge Haliclona (gellius) sp. Appl. Environ. Microbiol. 2011, 77, 2130–2140. [Google Scholar] [CrossRef] [PubMed]
- Wilson, M.C.; Mori, T.; Rückert, C.; Uria, A.R.; Helf, M.J.; Takada, K.; Gernert, C.; Steffens, U.A.E.; Heycke, N.; Schmitt, S.; et al. An environmental bacterial taxon with a large and distinct metabolic repertoire. Nature 2014, 506, 58–62. [Google Scholar] [CrossRef] [PubMed]
- Baltz, R.H. Natural product drug discovery in the genomic era: Realities, conjectures, misconceptions, and opportunities. J. Ind. Microbiol. Biotechnol. 2019, 46, 281–299. [Google Scholar] [CrossRef]
- Sun, W.; Zhang, F.; He, L.; Karthik, L.; Li, Z. Actinomycetes from the South China Sea sponges: Isolation, diversity, and potential for aromatic polyketides discovery. Front. Microbiol. 2015, 6, 1048. [Google Scholar] [CrossRef]
- Indraningrat, A.A.; Smidt, H.; Sipkema, D. Bioprospecting Sponge-Associated Microbes for Antimicrobial Compounds. Mar. Drugs 2016, 14, 87. [Google Scholar] [CrossRef]
- Baik, K.S.; Park, S.C.; Kim, H.J.; Lee, K.H.; Seong, C.N. Chryseoglobus frigidaquae gen. nov., sp. nov., a novel member of the family Microbacteriaceae. Int. J. Syst. Evol. Microbiol. 2010, 60, 1311–1316. [Google Scholar] [CrossRef][Green Version]
- Stierle, A.C.; Cardellina, J.H., 2nd; Singleton, F.L. A marine Micrococcus produces metabolites ascribed to the sponge Tedania ignis. Experientia 1988, 44, 1021. [Google Scholar] [CrossRef]
- Subramani, R.; Sipkema, D. Marine Rare Actinomycetes: A Promising Source of Structurally Diverse and Unique Novel Natural Products. Mar. Drugs 2019, 17, 249. [Google Scholar] [CrossRef]
- Liu, Y.; Lai, Q.; Dong, C.; Sun, F.; Wang, L.; Li, G.; Shao, Z. Phylogenetic diversity of the Bacillus pumilus group and the marine ecotype revealed by multilocus sequence analysis. PLoS ONE 2013, 8, e80097. [Google Scholar] [CrossRef] [PubMed]
- Bérdy, J. Bioactive microbial metabolites. J. Antibiot. (Tokyo) 2005, 58, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Boumehira, A.Z.; El-Enshasy, H.A.; Hacène, H.; Elsayed, E.A.; Aziz, R.; Park, E.Y. Recent progress on the development of antibiotics from the genus Micromonospora. Biotechnol. Bioproc. E. 2016, 21, 199–223. [Google Scholar] [CrossRef]
- Li, Y.; Li, Z.; Yamanaka, K.; Xu, Y.; Zhang, W.; Vlamakis, H.; Kolter, R.; Moore, B.S.; Qian, P.-Y. Directed natural product biosynthesis gene cluster capture and expression in the model bacterium Bacillus subtilis. Sci. Rep. 2015, 5, 9383. [Google Scholar] [CrossRef]
- Hu, Y.; MacMillan, J.B. Erythrazoles A–B, Cytotoxic Benzothiazoles from a Marine-Derived Erythrobacter sp. Org. Lett. 2011, 13, 6580–6583. [Google Scholar] [CrossRef]
- Felske, A.; Rheims, H.; Wolterink, A.; Stackebrandt, E.; Akkermans, A.D.L. Ribosome analysis reveals prominent activity of an uncultured member of the class Actinobacteria in grassland soils. Microbiology 1997, 143, 2983–2989. [Google Scholar] [CrossRef]
- Stecher, G.; Tamura, K.; Kumar, S. Molecular Evolutionary Genetics Analysis (MEGA) for macOS. Mol. Biol. Evol. 2020, 37, 1237–1239. [Google Scholar] [CrossRef]
- Giubergia, S.; Phippen, C.; Gotfredsen, C.H.; Nielsen, K.F.; Gram, L. Influence of Niche-Specific Nutrients on Secondary Metabolism in Vibrionaceae. Appl. Environ. Microbiol. 2016, 82, 4035–4044. [Google Scholar] [CrossRef]
- Selvin, J.; Joseph, S.; Asha, K.R.T.; Manjusha, W.A.; Sangeetha, V.S.; Jayaseema, D.M.; Antony, M.C.; Denslin Vinitha, A.J. Antibacterial potential of antagonistic Streptomyces sp. isolated from marine sponge Dendrilla nigra. FEMS Microbiol. Ecol. 2004, 50, 117–122. [Google Scholar] [CrossRef]
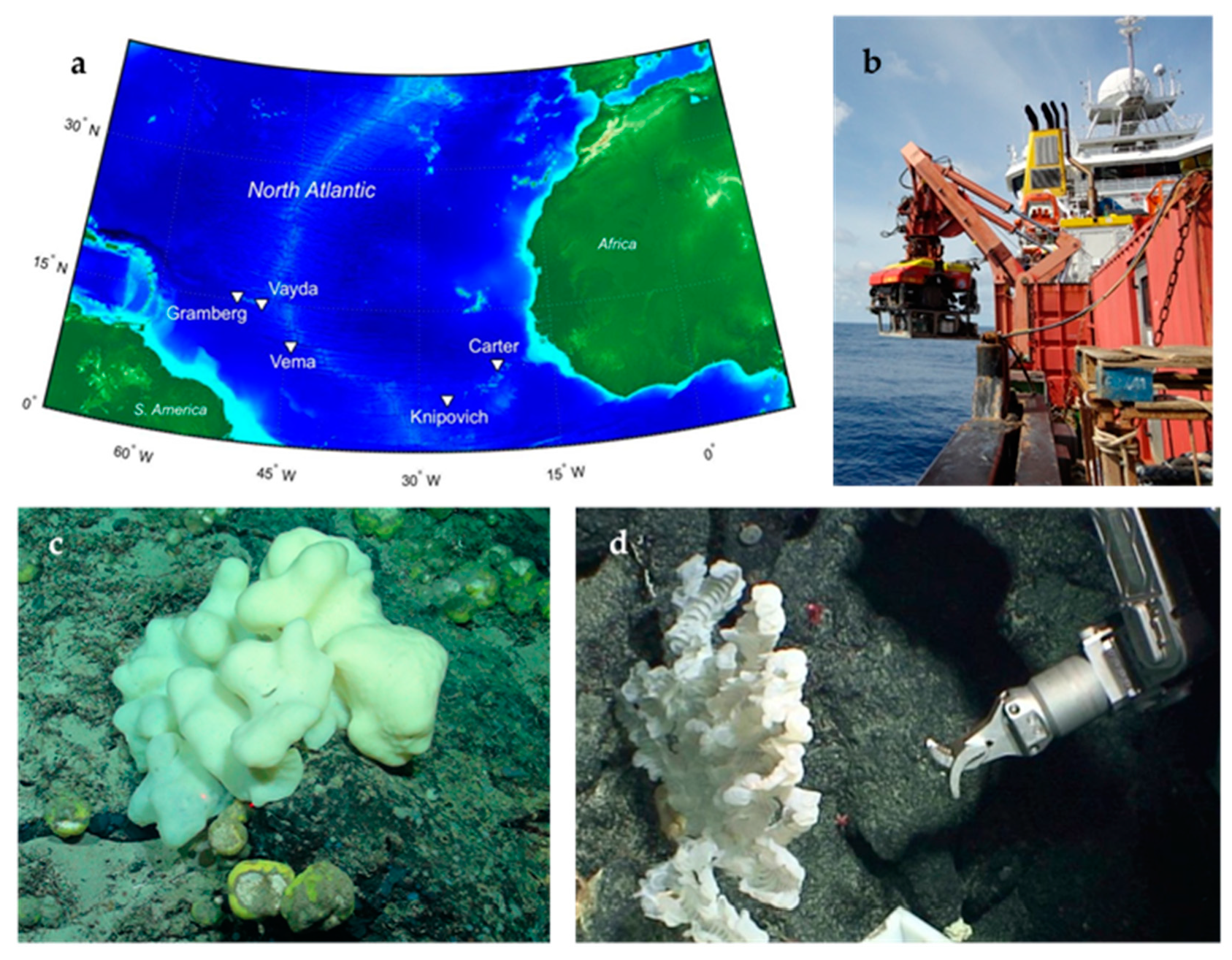
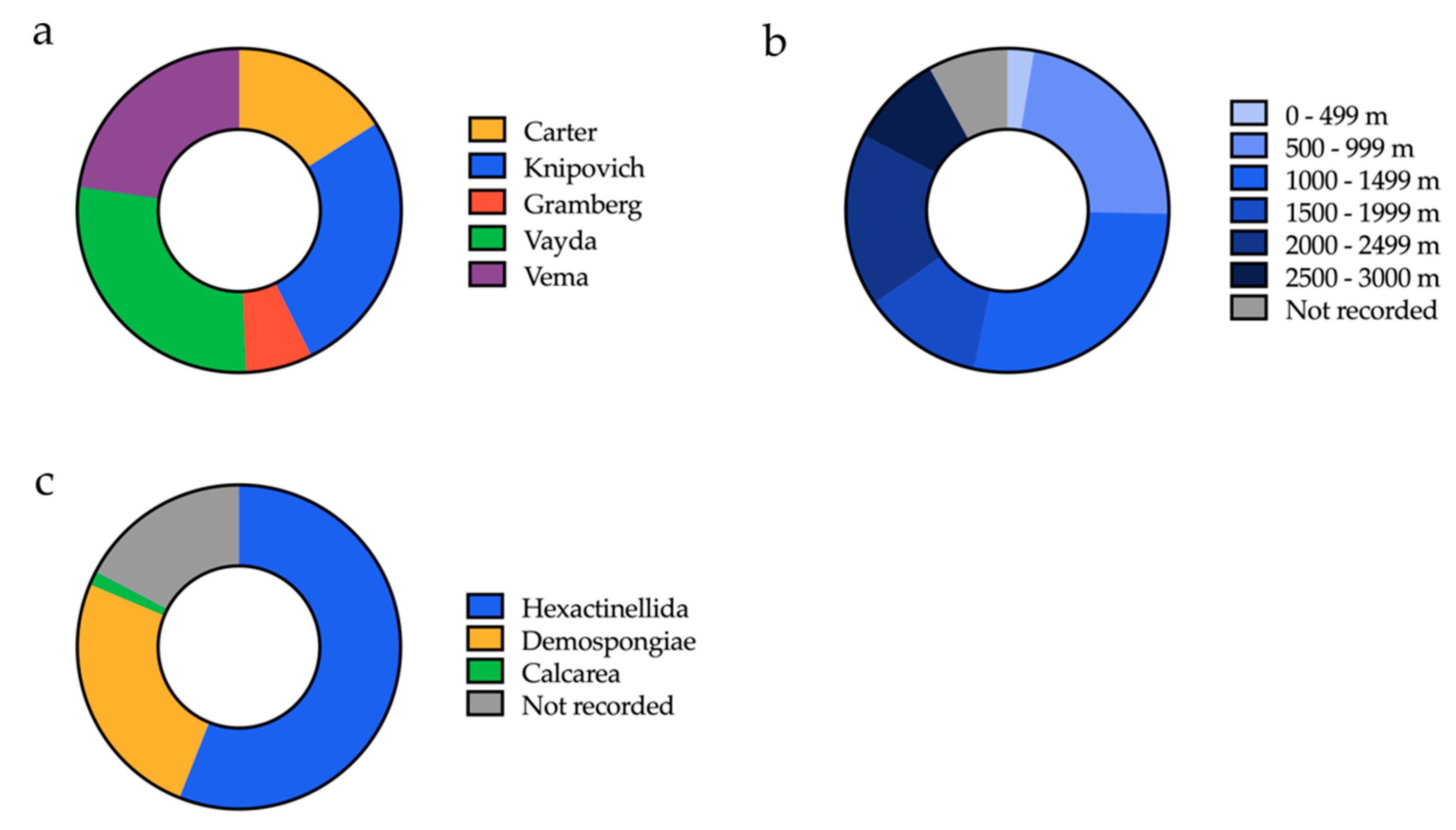

| Order of Processing | Sample ID | Latitude (N) | Longitude (W) | Depth (m) | Sampling Region | Visual ID | Total Bacterial Isolates a |
|---|---|---|---|---|---|---|---|
| 1 | B01607 | 10°96′49′′ | 44°36′58′′ | 2887 | Vema fracture zone | Hexactinellid | 37 |
| 2 | B01641 | 14°53′3′′ | 48°7′27′′ | 868 | Vayda seamount | Hexactinellid | 55 |
| 3 | B01661 | 14°51′47′′ | 48°14′29′′ | 1483 | Vayda seamount | Demosponge | 73 |
| 4 | B0789 | 14°53′25′′ | 48°9′7′′ | 824 | Vayda seamount | Hexactinellid | 136 |
| 5 | B0078 | 5°37′30′′ | 26°57′29′′ | 971 | Knipovich seamount | Demosponge | 182 |
| 6 | B01171 | 5°35′43′′ | 25°58′28′′ | 2257 | Knipovich seamount | Demosponge | 229 |
| Sponge Sample | Agar Type | Colonies Isolated | |
|---|---|---|---|
| 28 °C | 4 °C | ||
| B01607 | AIA-XO | 0 | 0 |
| Marine | 19 | 19 | |
| Peptone-starch | 0 | 0 | |
| TOTAL | 19 | 19 | |
| B01641 | AIA-XO | 17 | 0 |
| Marine | 30 | 9 | |
| Peptone-starch | 1 | 0 | |
| TOTAL | 48 | 9 | |
| B01661 | AIA-XO | 0 | 0 |
| Marine | 52 | 20 | |
| Peptone-starch | 8 | 0 | |
| TOTAL | 60 | 20 | |
| B0789 | 60:40 | 2 | 1 |
| AIA | 16 | 11 | |
| AIA-XO | 4 | - | |
| Basic | 7 | 4 | |
| Marine | 43 | 21 | |
| Peptone-starch | 10 | 18 | |
| TOTAL | 82 | 55 | |
| B0078 | 60:40 | 10 | 0 |
| ISP2 | 50 | 0 | |
| M1 | 25 | 1 | |
| Marine | 23 | 4 | |
| Peptone-starch | 11 | 0 | |
| R2A | 27 | 4 | |
| SCA | 27 | 0 | |
| TOTAL | 173 | 9 | |
| B01171 | 60/40 | 7 | 3 |
| ISP2 | 17 | 5 | |
| ISP2/10 | 7 | 4 | |
| ISP4 | 4 | 3 | |
| M1 | 5 | 2 | |
| MB1/2 | 11 | 10 | |
| MB20 | 9 | 10 | |
| MB20-NO3 | 3 | - | |
| Mucin | 7 | 13 | |
| Peptone-starch | 22 | 7 | |
| R2A | 3 | 5 | |
| SA1 | 8 | 0 | |
| SAC | 12 | 0 | |
| Spicule-extract | 12 | 8 | |
| TOTAL | 127 | 70 | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Williams, S.E.; Stennett, H.L.; Back, C.R.; Tiwari, K.; Ojeda Gomez, J.; Challand, M.R.; Hendry, K.R.; Spencer, J.; Essex-Lopresti, A.E.; Willis, C.L.; et al. The Bristol Sponge Microbiome Collection: A Unique Repository of Deep-Sea Microorganisms and Associated Natural Products. Antibiotics 2020, 9, 509. https://doi.org/10.3390/antibiotics9080509
Williams SE, Stennett HL, Back CR, Tiwari K, Ojeda Gomez J, Challand MR, Hendry KR, Spencer J, Essex-Lopresti AE, Willis CL, et al. The Bristol Sponge Microbiome Collection: A Unique Repository of Deep-Sea Microorganisms and Associated Natural Products. Antibiotics. 2020; 9(8):509. https://doi.org/10.3390/antibiotics9080509
Chicago/Turabian StyleWilliams, Sam E., Henry L. Stennett, Catherine R. Back, Kavita Tiwari, Jorge Ojeda Gomez, Martin R. Challand, Katharine R. Hendry, James Spencer, Angela E. Essex-Lopresti, Christine L. Willis, and et al. 2020. "The Bristol Sponge Microbiome Collection: A Unique Repository of Deep-Sea Microorganisms and Associated Natural Products" Antibiotics 9, no. 8: 509. https://doi.org/10.3390/antibiotics9080509
APA StyleWilliams, S. E., Stennett, H. L., Back, C. R., Tiwari, K., Ojeda Gomez, J., Challand, M. R., Hendry, K. R., Spencer, J., Essex-Lopresti, A. E., Willis, C. L., Curnow, P., & Race, P. R. (2020). The Bristol Sponge Microbiome Collection: A Unique Repository of Deep-Sea Microorganisms and Associated Natural Products. Antibiotics, 9(8), 509. https://doi.org/10.3390/antibiotics9080509