Derivates of the Antifungal Peptide Cm-p5 Inhibit Development of Candida auris Biofilms In Vitro
Abstract
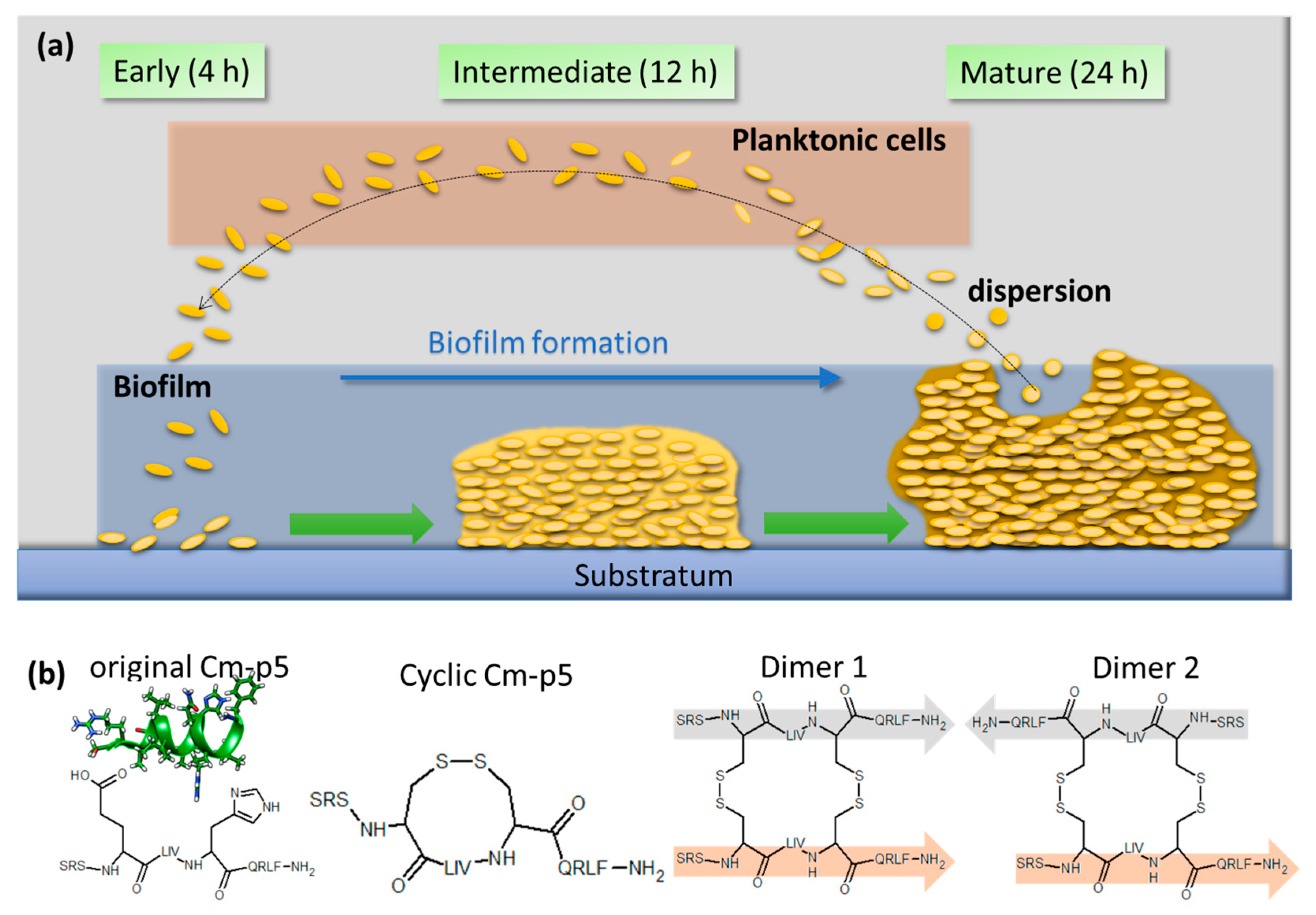
1. Introduction
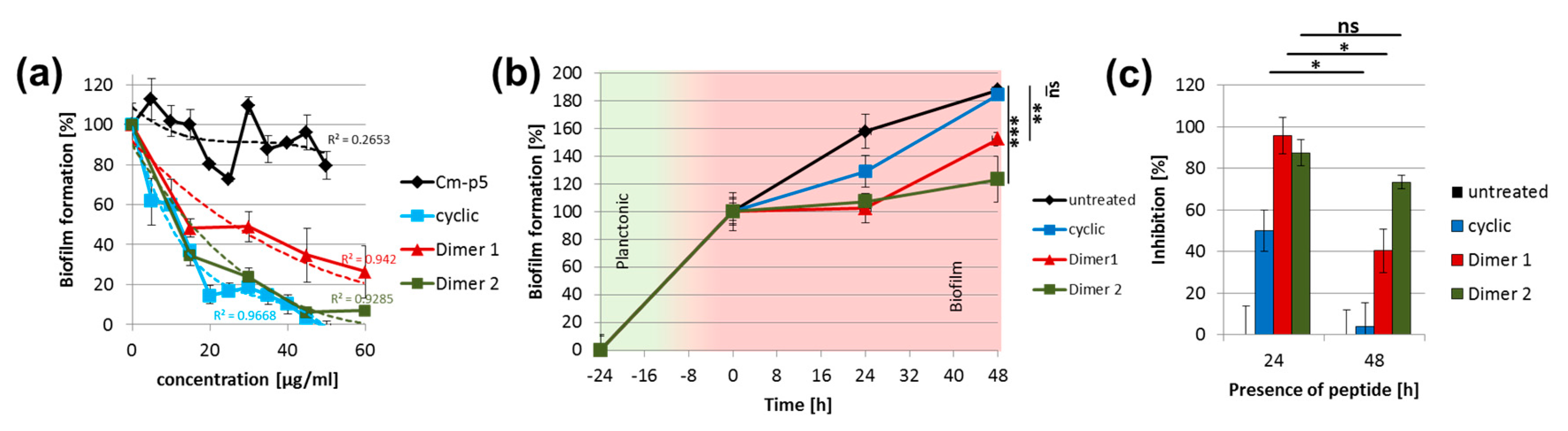
2. Results and Discussion
3. Materials and Methods
3.1. Cultivation of C. auris
3.2. Biofilm Formation and Quantification
3.3. Effects of Cm-p5 Derivatives on Mature Biofilms
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Nadell, C.D.; Bassler, B.L.; Levin, S.A. Observing bacteria through the lens of social evolution. J. Biol. 2008, 7, 27. [Google Scholar] [CrossRef] [PubMed]
- Licking, E. Getting a grip on bacterial slime. Bus. Week 1999, 13, 98–100. [Google Scholar]
- Davies, D. Understanding biofilm resistance to antibacterial agents. Nat. Rev. Drug Discov. 2003, 2, 114–122. [Google Scholar] [CrossRef] [PubMed]
- Fox, E.P.; Nobile, C.J. A sticky situation: Untangling the transcriptional network controlling biofilm development in Candida albicans. Transcription 2012, 3, 315–322. [Google Scholar] [CrossRef] [PubMed]
- Papon, N.; Courdavault, V.; Clastre, M.; Bennett, R.J. Emerging and Emerged Pathogenic Candida Species: Beyond the Candida albicans Paradigm. PLoS Pathog. 2013, 9. [Google Scholar] [CrossRef] [PubMed]
- Satoh, K.; Makimura, K.; Hasumi, Y.; Nishiyama, Y.; Uchida, K.; Yamaguchi, H. Candida auris sp. nov., a novel ascomycetous yeast isolated from the external ear canal of an inpatient in a Japanese hospital. Microbiol. Immunol. 2009, 53, 41–44. [Google Scholar] [CrossRef]
- Morales-López, S.E.; Parra-Giraldo, C.M.; Ceballos-Garzón, A.; Martínez, H.P.; Rodríguez, G.J.; Álvarez-Moreno, C.A.; Rodríguez, J.Y. Invasive infections with multidrug-resistant yeast Candida auris, Colombia. Emerg. Infect. Dis. 2017, 23, 162–164. [Google Scholar] [CrossRef]
- Lockhart, S.R.; Etienne, K.A.; Vallabhaneni, S.; Farooqi, J.; Chowdhary, A.; Govender, N.P.; Colombo, A.L.; Calvo, B.; Cuomo, C.A.; Desjardins, C.A.; et al. Simultaneous emergence of multidrug-resistant candida auris on 3 continents confirmed by whole-genome sequencing and epidemiological analyses. Clin. Infect. Dis. 2017, 64, 134–140. [Google Scholar] [CrossRef]
- Osei Sekyere, J. Candida auris: A systematic review and meta-analysis of current updates on an emerging multidrug-resistant pathogen. Microbiologyopen 2018, 7, e00578. [Google Scholar] [CrossRef]
- Forbes. Available online: https://www.forbes.com/sites/judystone/2017/08/24/candida-auris-a-new-fungal-superbug-emerging-as-a-global-threat/ (accessed on 14 May 2020).
- Ben-Ami, R.; Berman, J.; Novikov, A.; Bash, E.; Shachor-Meyouhas, Y.; Zakin, S.; Maor, Y.; Tarabia, J.; Schechner, V.; Adler, A.; et al. Multidrug-resistant candida haemulonii and C. Auris, tel aviv, Israel. Emerg. Infect. Dis. 2017, 23, 195–203. [Google Scholar] [CrossRef]
- Kean, R.; Delaney, C.; Sherry, L.; Borman, A.; Johnson, E.M.; Richardson, M.D.; Rautemaa-Richardson, R.; Williams, C.; Ramage, G. Transcriptome Assembly and Profiling of Candida auris Reveals Novel Insights into Biofilm-Mediated Resistance. mSphere 2018, 3. [Google Scholar] [CrossRef] [PubMed]
- Ciociola, T.; Giovati, L.; Conti, S.; Magliani, W.; Santinoli, C.; Polonelli, L. Natural and synthetic peptides with antifungal activity. Future Med. Chem. 2016, 8, 1413–1433. [Google Scholar] [CrossRef] [PubMed]
- Rautenbach, M.; Troskie, A.M.; Vosloo, J.A. Antifungal peptides: To be or not to be membrane active. Biochimie 2016, 130, 132–145. [Google Scholar] [CrossRef] [PubMed]
- López-Abarrategui, C.; McBeth, C.; Mandal, S.M.; Sun, Z.J.; Heffron, G.; Alba-Menéndez, A.; Migliolo, L.; Reyes-Acosta, O.; García-Villarino, M.; Nolasco, D.O.; et al. Cm-p5: An antifungal hydrophilic peptide derived from the coastal mollusk Cenchritis muricatus (Gastropoda: Littorinidae). FASEB J. 2015, 29, 3315–3325. [Google Scholar] [CrossRef]
- Kubiczek, D.; Flaig, C.; Raber, H.; Dietz, S.; Kissmann, A.K.; Heerde, T.; Bodenberger, N.; Wittgens, A.; González-Garcia, M.; Kang, F.; et al. A Cerberus-Inspired Anti-Infective Multicomponent Gatekeeper Hydrogel against Infections with the Emerging “Superbug” Yeast Candida auris. Macromol. Biosci. 2020, 20. [Google Scholar] [CrossRef]
- Morales-Vicente, F.E.; González-Garcia, M.; Diaz Pico, E.; Moreno-Castillo, E.; Garay, H.E.; Rosi, P.E.; Jimenez, A.M.; Campos-Delgado, J.A.; Rivera, D.G.; Chinea, G.; et al. Design of a Helical-Stabilized, Cyclic, and Nontoxic Analogue of the Peptide Cm-p5 with Improved Antifungal Activity. ACS Omega 2019, 4, 19081–19095. [Google Scholar] [CrossRef]
- Høiby, N.; Ciofu, O.; Johansen, H.K.; Song, Z.J.; Moser, C.; Jensen, P.Ø.; Molin, S.; Givskov, M.; Tolker-Nielsen, T.; Bjarnsholt, T. The clinical impact of bacterial biofilms. Int. J. Oral Sci. 2011, 3, 55–65. [Google Scholar] [CrossRef]
- Welsh, R.M.; Bentz, M.L.; Shams, A.; Houston, H.; Lyons, A.; Rose, L.J.; Litvintseva, A.P. Survival, persistence, and isolation of the emerging multidrug-resistant pathogenic yeast Candida auris on a plastic health care surface. J. Clin. Microbiol. 2017, 55, 2996–3005. [Google Scholar] [CrossRef]
- Sherry, L.; Ramage, G.; Kean, R.; Borman, A.; Johnson, E.M.; Richardson, M.D.; Rautemaa-Richardson, R. Biofilm-forming capability of highly virulent, multidrug-resistant Candida auris. Emerg. Infect. Dis. 2017, 23, 328–331. [Google Scholar] [CrossRef]
- O’Toole, G.; Kaplan, H.B.; Kolter, R. Biofilm Formation as Microbial Development. Annu. Rev. Microbiol. 2000, 54, 49–79. [Google Scholar] [CrossRef]
- Villar, C.C.; Kashleva, H.; Nobile, C.J.; Mitchell, A.P.; Dongari-Bagtzoglou, A. Mucosal tissue invasion by Candida albicans is associated with E-cadherin degradation, mediated by transcription factor Rim101p and protease Sap5p. Infect. Immun. 2007, 75, 2126–2135. [Google Scholar] [CrossRef] [PubMed]
- Winter, M.B.; Salcedo, E.C.; Lohse, M.B.; Hartooni, N.; Gulati, M.; Sanchez, H.; Takagi, J.; Hube, B.; Andes, D.R.; Johnson, A.D.; et al. Global identification of biofilm-specific proteolysis in Candida albicans. MBio 2016, 7. [Google Scholar] [CrossRef]
- Jose, A.; Coco, B.J.; Milligan, S.; Young, B.; Lappin, D.F.; Bagg, J.; Murray, C.; Ramage, G. Reducing the incidence of denture stomatitis: Are denture cleansers sufficient? J. Prosthodont. 2010, 19, 252–257. [Google Scholar] [CrossRef] [PubMed]
- O’Toole, G.A. Microtiter dish Biofilm formation assay. J. Vis. Exp. 2010. [Google Scholar] [CrossRef] [PubMed]
- Negri, M.; Gonçalves, V.; Silva, S.; Henriques, M.; Azeredo, J.; Oliveira, R. Crystal violet staining to quantity Candida adhesion to epithelial cells. Br. J. Biomed. Sci. 2010, 67, 120–125. [Google Scholar] [CrossRef] [PubMed]
- Sherry, L.; Rajendran, R.; Lappin, D.F.; Borghi, E.; Perdoni, F.; Falleni, M.; Tosi, D.; Smith, K.; Williams, C.; Jones, B.; et al. Biofilms formed by Candida albicans bloodstream isolates display phenotypic and transcriptional heterogeneity that are associated with resistance and pathogenicity. BMC Microbiol. 2014, 14, 182. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kubiczek, D.; Raber, H.; Gonzalez-García, M.; Morales-Vicente, F.; Staendker, L.; Otero-Gonzalez, A.J.; Rosenau, F. Derivates of the Antifungal Peptide Cm-p5 Inhibit Development of Candida auris Biofilms In Vitro. Antibiotics 2020, 9, 363. https://doi.org/10.3390/antibiotics9070363
Kubiczek D, Raber H, Gonzalez-García M, Morales-Vicente F, Staendker L, Otero-Gonzalez AJ, Rosenau F. Derivates of the Antifungal Peptide Cm-p5 Inhibit Development of Candida auris Biofilms In Vitro. Antibiotics. 2020; 9(7):363. https://doi.org/10.3390/antibiotics9070363
Chicago/Turabian StyleKubiczek, Dennis, Heinz Raber, Melaine Gonzalez-García, Fidel Morales-Vicente, Ludger Staendker, Anselmo J. Otero-Gonzalez, and Frank Rosenau. 2020. "Derivates of the Antifungal Peptide Cm-p5 Inhibit Development of Candida auris Biofilms In Vitro" Antibiotics 9, no. 7: 363. https://doi.org/10.3390/antibiotics9070363
APA StyleKubiczek, D., Raber, H., Gonzalez-García, M., Morales-Vicente, F., Staendker, L., Otero-Gonzalez, A. J., & Rosenau, F. (2020). Derivates of the Antifungal Peptide Cm-p5 Inhibit Development of Candida auris Biofilms In Vitro. Antibiotics, 9(7), 363. https://doi.org/10.3390/antibiotics9070363





