Studying the Gene Expression of Penicillium rubens Under the Effect of Eight Essential Oils
Abstract
1. Introduction
2. Results
2.1. Inhibition of Mycelial Growth Using EOs Vapors
2.2. Impact of Essential Oils on Genome-Wide Gene Expression
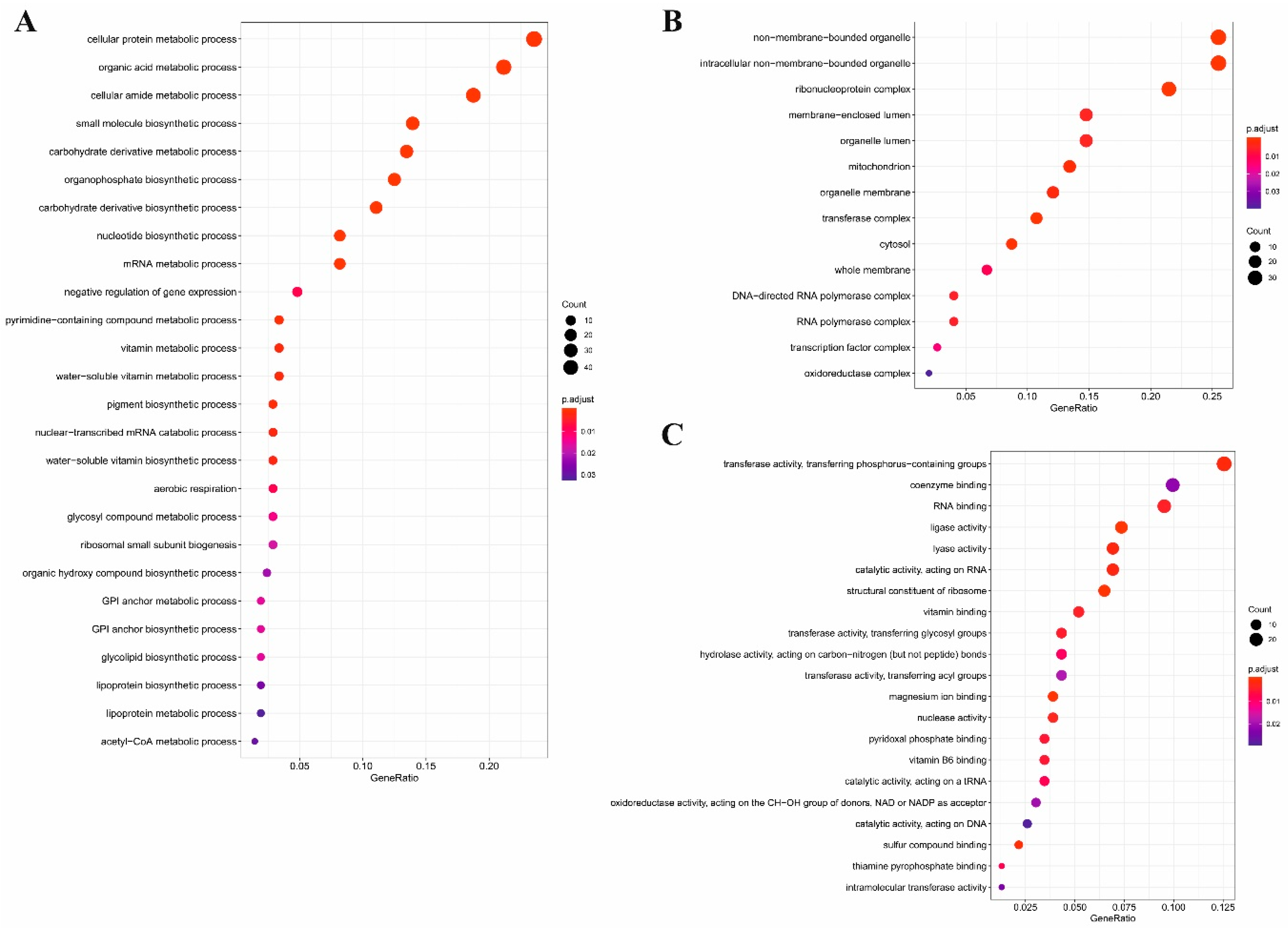
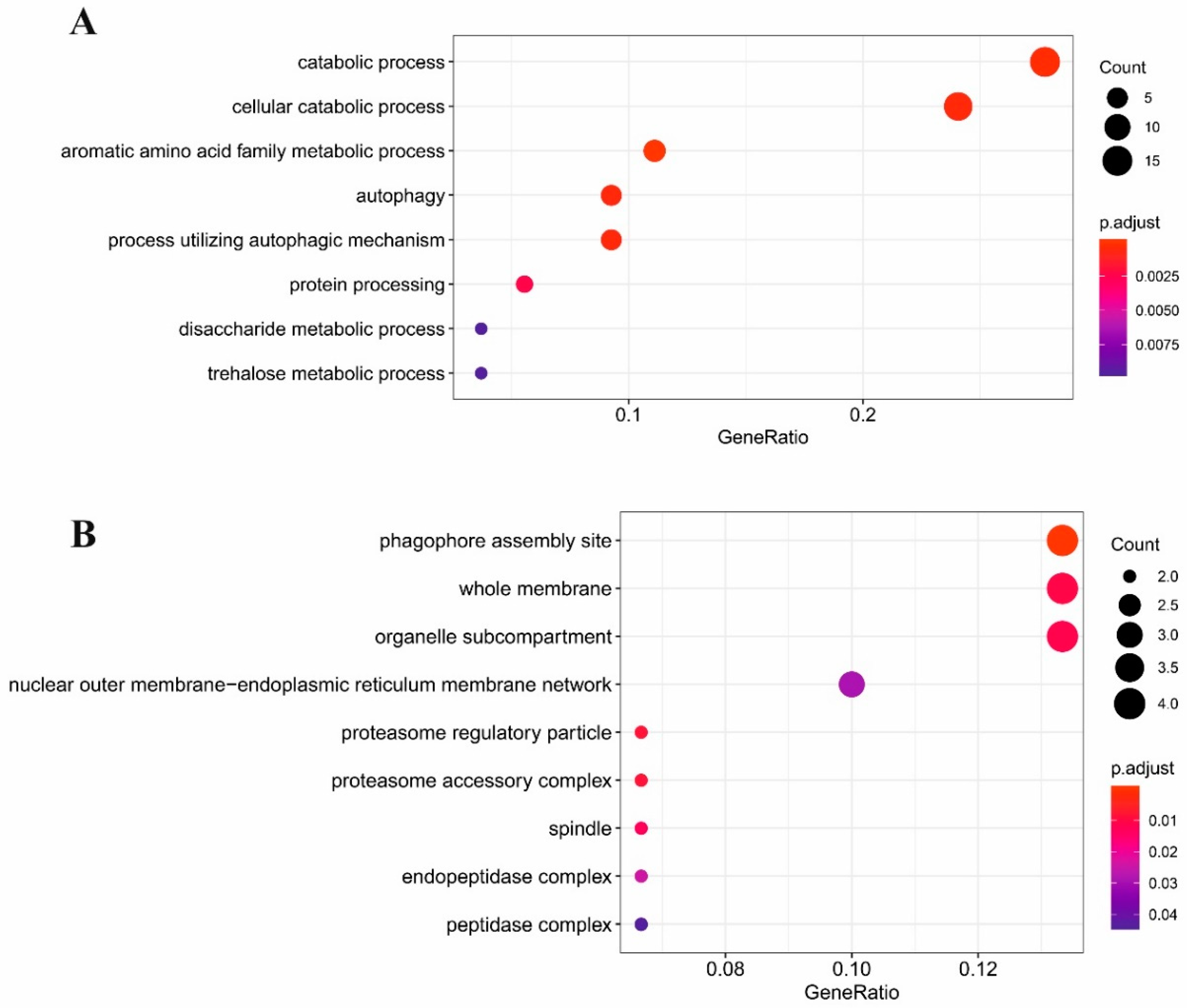
2.3. Gene Ontology Analysis
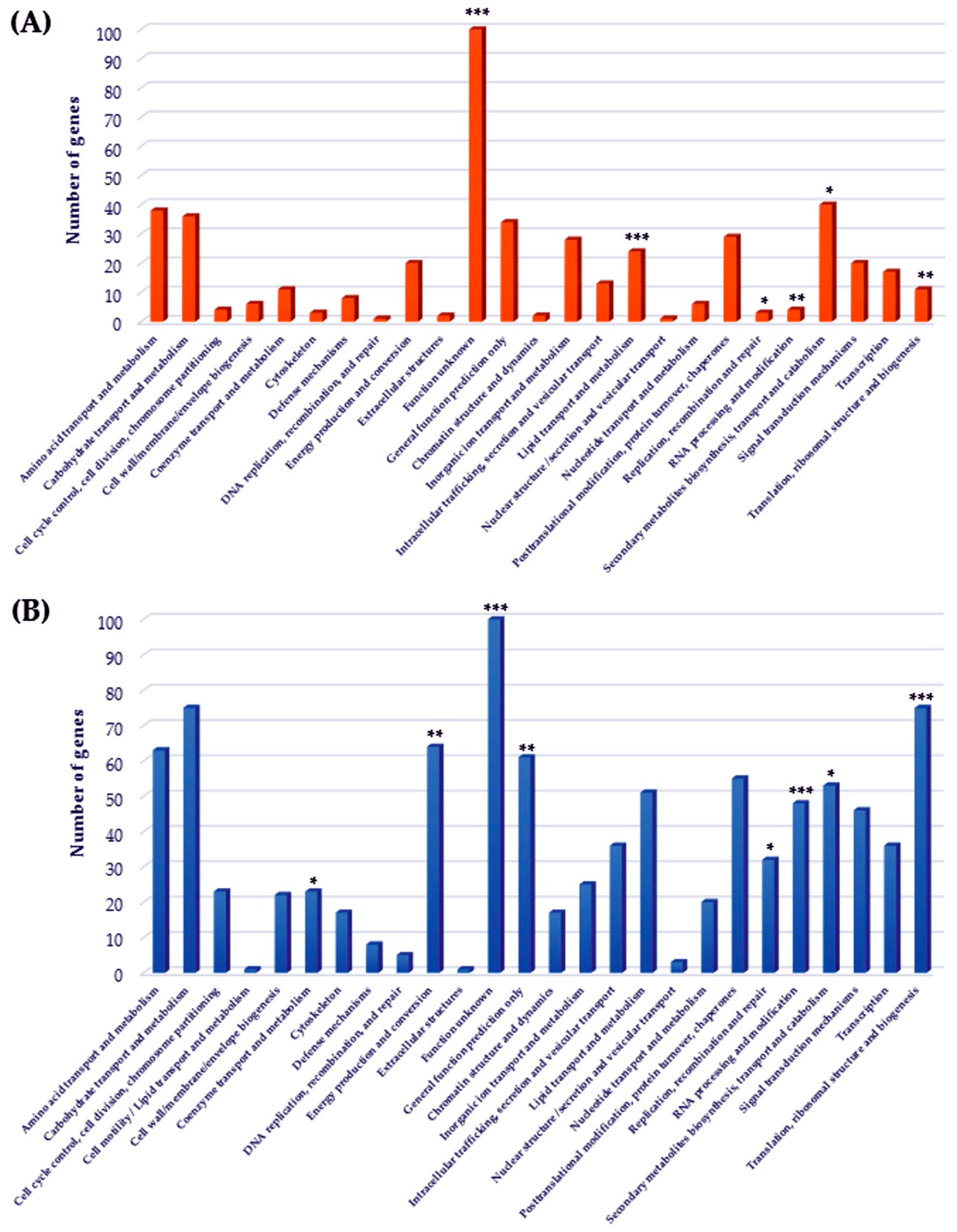
2.4. Functional Classes Analysis
2.5. Metabolic Pathway Analysis
2.5.1. Genes Involved in Protein Synthesis
2.5.2. Transcriptional Modifications of Genes Encoding Carbohydrates
2.5.3. Changed Expression Level of Genes Involved in Fat Metabolism
2.5.4. Gene Related to Secondary Metabolites
3. Discussion
4. Materials and Methods
4.1. Essential Oils
4.2. Fungal Strain and Fungicide Activity of the Vapor Phase of EOs
4.3. RNA Isolation and Quality Control
4.4. Microarray Analysis of Gene Expression
4.5. Image and Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Prestinaci, F.; Pezzotti, P.; Pantosti, A. Antimicrobial resistance: A global multifaceted phenomenon. Pathog. Glob. Health 2015, 109, 309–318. [Google Scholar] [CrossRef] [PubMed]
- Yap, P.S.X.; Yiap, B.C.; Ping, H.C.; Lim, S.H.E. Essential oils, a new horizon in combating bacterial antibiotic resistance. Open Microbiol. J. 2014, 8, 6–14. [Google Scholar] [CrossRef]
- Schmidt, E. Production of Essential Oils. In Handbook of Essential Oils. Science, Technology and Applications; Baser, K.H., Buchbauer, G., Eds.; CRC Press: Boca Raton, FL, USA, 2010; pp. 83–119. [Google Scholar]
- Bilia, A.R.; Guccione, C.; Isacchi, B.; Righeschi, C.; Firenzuoli, F.; Bergonzi, M.C. Essential Oils Loaded in Nanosystems: A Developing Strategy for a Successful Therapeutic Approach. Evid Based Complement Altern. Med. 2014, 651593. [Google Scholar] [CrossRef] [PubMed]
- Opara, E.I.; Chohan, M. Culinary Herbs and Spices: Their Bioactive Properties, the Contribution of Polyphenols and the Challenges in Deducing Their True Health Benefits. Int. J. Mol. Sci. 2014, 15, 19183–19202. [Google Scholar] [CrossRef] [PubMed]
- Nieto, G. Biological Activities of Three Essential Oils of the Lamiaceae Family. Medicines 2017, 4, 63. [Google Scholar] [CrossRef]
- Carson, C.F.; Hammer, K.A.; Riley, T.V. Melaleuca alternifolia (Tea Tree) oil: A review of antimicrobial and other medicinal properties. Clin. Microbiol. Rev. 2006, 19, 50–62. [Google Scholar] [CrossRef]
- Han, X.; Parker, T.L. Anti-inflammatory activity of clove (Eugenia caryophyllata) essential oil on human dermal fibroblasts. Pharm. Biol. 2017, 55, 1619–1622. [Google Scholar] [CrossRef]
- Rao, P.V.; Gan, S.H. Cinnamon: AMultifaceted Medicinal Plant. Evid Based Complement Altern. Med. 2014, 642942. [Google Scholar] [CrossRef]
- Guenther, E. The Essential Oils; Van Nostrand, Inc.: Princeton, NJ, USA, 1950; p. 703. [Google Scholar]
- Akhila, A. Essential Oil-Bearing Grasses: The Genus Cymbopogon; CRC Press: Boca Raton, FL, USA, 2010; p. 262. [Google Scholar]
- Chao, S.C.; Young, D.G.; Oberg, C.J. Screening for inhibitory activity of essential oils on selected bacteria, fungi and viruses. J. Essent. Oil Res. 2000, 12, 639–649. [Google Scholar] [CrossRef]
- Sharma, P.R.; Mondhe, D.M.; Muthiah, S.; Pal, H.C.; Shahi, A.K.; Saxena, A.K.; Qazi, G.N. Anticancer activity of an essential oil from cymbopogon flexuosus. Chem. Biol. Interact. 2009, 179, 160–168. [Google Scholar] [CrossRef]
- Hudson, J.; Kuo, M.; Vimalanathan, S. The Antimicrobial Properties of Cedar Leaf (Thuja plicata) Oil; A Safe and Efficient Decontamination Agent for Buildings. Int. J. Environ. Res. Public Health 2011, 8, 4477–4487. [Google Scholar] [CrossRef] [PubMed]
- Shams-Ghahfarokhi, M.; Aghaei-Gharehbolagh, S.; Aslani, N.; Razzaghi-Abyaneh, M. Investigation on distribution of airborne fungi in outdoor environment in Tehran, Iran. J. Environ. Health Sci. Eng. 2014, 12, 54. [Google Scholar] [CrossRef] [PubMed]
- Burge, H. Bioaerosols: Prevalence and health effects in the indoor environment. J. Allergy Clin. Immun. 1990, 86, 687–701. [Google Scholar] [CrossRef]
- Rogawansamy, S.; Gaskin, S.; Taylor, M.; Pisaniello, D. An Evaluation of Antifungal Agents for the Treatment of Fungal Contamination in Indoor Air Environments. Int. J. Environ. Public Health 2015, 12, 6319–6332. [Google Scholar] [CrossRef]
- Cooley, J.D.; Wong, W.C.; Jumper, C.A.; Straus, D.C. Correlation between the prevalence of certain fungi and sick building syndrome. Occup. Environ. Med. 1998, 55, 579–584. [Google Scholar] [CrossRef]
- Visagie, C.M.; Houbraken, J.; Frisvad, J.C.; Hong, S.-B.; Klaassen, C.H.W.; Perrone, G.; Seifert, K.A.; Varga, J.; Yaguchi, T.; Samson, R.A. Identification and nomenclature of the genus Penicillium. Stud. Mycol. 2014, 78, 343–371. [Google Scholar] [CrossRef]
- Wilson, S.C.; Straus, D.C. The presence of fungi associated with sick building syndrome in North American zoological institutions. J. Zoo Wildl. Med. 2002, 33, 322–327. [Google Scholar] [CrossRef]
- Mehra, T.; Köberle, M.; Braunsdorf, C.; Mailänder-Sanchez, D.; Borelli, C.; Schaller, M. Alternative approaches to antifungal therapies. Exp. Dermatol. 2012, 21, 778–782. [Google Scholar] [CrossRef]
- Glegg, G.A.; Richards, J.P. Chemicals in household products: Problems with solutions. Environ. Manag. 2007, 40, 889–901. [Google Scholar] [CrossRef]
- Whiley, H.; Gaskin, S.; Schroder, T.; Ross, K. Antifungal properties of essential oils for improvement of indoor air quality: A review. Rev. Envirion. Health 2018, 33, 63–76. [Google Scholar] [CrossRef]
- Reyes-Jurado, F.; Navarro-Cruz, A.R.; Ochoa-Velasco, C.E.; Palou, E.; López-Malo, A.; Ávila-Sosa, R. Essential oils in vapor phase as alternative antimicrobials: A review. Crit. Rev. Food Sci. Nutr. 2019, 18, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Dorman, H.J.D.; Deans, S.G. Antibacterial agents from plants: Antibacterial activity of plant volatile oils. J. Appl. Microbiol. 2000, 88, 308–316. [Google Scholar] [CrossRef] [PubMed]
- Pandey, A.K.; Kumar, P.; Singh, P.; Tripathi, N.N.; Bajpai, V.K. Essential Oils: Sources of Antimicrobials and Food Preservatives. Front. Microbiol. 2016, 7, 2161. [Google Scholar] [CrossRef] [PubMed]
- Cox, S.D.; Mann, C.M.I.; Markham, J.L.; Bell, H.C.; Gustafson, J.E.; Warmington, J.R.; Wyllie, S.G. The mode of antimicrobial action of the essential oil of Melaleuca alternifolia (tea tree oil). J. Appl. Microbiol. 2000, 88, 170–175. [Google Scholar] [CrossRef] [PubMed]
- Sealfon, S.C.; Chu, T.T. RNA and DNA Microarrays. Methods Mol. Biol. 2011, 671, 3–34. [Google Scholar] [CrossRef] [PubMed]
- Bajaj, I.; Veiga, T.; van Dissel, D.; Pronk, J.T.; Daran, J.M. Functional characterization of a Penicillium chrysogenum mutanase gene induced upon co-cultivation with Bacillus subtilis. BMC Microbiol. 2014, 14, 114. [Google Scholar] [CrossRef]
- Lowe, R.; Shirley, N.; Bleackley, M.; Dolan, S.; Shafee, T. Transcriptomics technologies. PLoS Comput. Biol. 2017, 18, 13. [Google Scholar] [CrossRef]
- Salo, O.V.; Ries, M.; Medema, M.H.; Lankhorst, P.P.; Vreeken, R.J.; Bovenberg, R.A.L.; Driessen, J.M. Genomic mutational analysis of the impact of the classical strain improvement program of β-lactam producing Penicillium chrysogenum. BMC Genom. 2015, 16, 937. [Google Scholar] [CrossRef]
- Hyldgaard, M.; Mygind, T.; Meyer, R.L. Essential oils in food preservation: Mode of action, synergies, and interactions with food matrix components. Front. Microbiol. 2012, 3, 12. [Google Scholar] [CrossRef]
- Nazzaro, F.; Frantianni, F.; Coppola, R.; Feo, V. Essential oils and antifugal activity. Pharmaceuticals 2017, 10, 4. [Google Scholar] [CrossRef]
- Rajendran, V.; Kalita, P.; Shukla, H.; Kumar, A.; Tripathi, T. Aminoacyl-tRNA synthetases: Structure, function and drug discovery. Int. J. Biol. Macromol. 2018, 111, 400–414. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Crismaru, C.G.; Salo, O.; Bovenberg, R.A.L.; Driessen, A.J.M. Impact of classical strain improvement of Penicillium rubens on amino acid metabolism during β-lactam production. Appl. Environ. Microbiol. 2020, 86, e01561-19. [Google Scholar] [CrossRef] [PubMed]
- Rabha, J.; Jha, D.K. Metabolic Diversity of Penicillium. In Developments in Microbial Biotechnology and Bioengineering-Penicillium System Properties and Applications; Gupta, V.K., Rodriguez-Couto, S., Eds.; Elsevier: Amsterdam, The Netherlands, 2018; pp. 217–234. [Google Scholar]
- Capuder, M.; Šolar, T.; Benčina, M.; Legiša, M. Highly active, citrate inhibition resistant form of Aspergillus niger 6-phosphofructo-1-kinase encoded by a modified pfkA gene. J. Biotechnol. 2009, 144, 51–57. [Google Scholar] [CrossRef] [PubMed]
- Link, T.; Lohaus, G.; Heiser, I.; Mendgen, K.; Hahn, M.; Voegele, R.T. Characterization of a novel NADP+-dependent D-arabitol dehydrogenase from the plant pathogen Uromyces fabae. Biochem. J. 2005, 389, 289–295. [Google Scholar] [CrossRef]
- Martens-Uzunova, E.S.; Schaap, P.J. An evolutionary conserved D-galacturonic acid metabolic pathway operates across filamentous fungi capable of pectin degradation. Fungal. Genet. Biol. 2008, 45, 1449–1457. [Google Scholar] [CrossRef]
- Chinaglia, S.; Chiarelli, L.R.; Maggi, M.; Rodolfi, M.; Valentini, G.; Picco, A.M. Biochemistry of lipolytic enzymes secreted by Penicillium solitum and Cladosporium cladosporioides. Biosci. Biotechnol. Biochem. 2014, 78, 245–254. [Google Scholar] [CrossRef]
- Zhong, Z.; Norvienyeku, J.; Yu, J.; Chen, M.; Cai, R.; Hong, Y.; Chen, L.; Zhang, D.; Wang, B.; Zhou, J.; et al. Two different subcellular-localized Acetoacetyl-CoA acetyltransferases differentiate diverse functions in Magnaporthe oryzae. Fungal. Genet. Biol. 2015, 83, 58–67. [Google Scholar] [CrossRef]
- Coleman, R.A.; Lee, D.P. Enzymes of triacylglycerol synthesis and their regulation. Prog. Lipid. Res. 2004, 43, 134–176. [Google Scholar] [CrossRef]
- Gao, Q.; Shang, Y.; Huang, W.; Wang, C. Glycerol-3-phosphate acyltransferase contributes to triacylglycerol biosynthesis, lipid droplet formation, and host invasion in Metarhizium robertsii. Appl. Environ. Microbiol. 2013, 79, 7646–7653. [Google Scholar] [CrossRef]
- Van der Berg, M.A.; Albang, R.; Albermann, K.; Badger, J.H.; Daran, J.M.; Driessen, A.J.; Garcia-Estrada, C.; Fedorova, N.D.; Harris, D.M.; Heijne, W.H.; et al. Genome sequencing and analysis of the filamentous fungus Penicillium chrysogenum. Nat. Biotechnol. 2008, 26, 1161–1168. [Google Scholar] [CrossRef]
- Slámová, K.; Bojarová, P.; Petrásková, L.; Křen, V. β-N-Acetylhexosaminidase: What’s in a name? Biotechnol. Adv. 2010, 28, 682–693. [Google Scholar] [CrossRef]
- Desbois, S.; John, U.P.; Perugini, A. Dihydrodipicolinate synthase is absent in fungi. Biochimie 2018, 152, 73–84. [Google Scholar] [CrossRef] [PubMed]
- Domigan, L.J.; Scally, S.W.; Fogg, M.J.; Hutton, C.A.; Perugini, M.A.; Dobson, R.C.J.; Muscroft-Taylor, A.C.; Gerrard, J.A.; Devenish, S.R.A. Characterisation of dihydrodipicolinate synthase (DHDPS) from Bacillus anthracis. Biochim. Biophysic. Acta 2009, 1794, 1510–1516. [Google Scholar] [CrossRef]
- Ren, W.; Tao, J.; Shi, D.; Chen, W.; Chen, C. Involvement of a dihydopicolinate synthase gene (FaDHDPS1) in fugal development, pathogenesis and stress responses in Fusarium asiaticum. BMC Microbiol. 2018, 18, 128. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2013; Available online: http://www.R-project.org/ (accessed on 14 April 2020).
- Yu, G.; Wang, L.; Han, Y.; He, Q. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Statist. Soc. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Gu, Z.; Eils, R.; Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef]
- Gu, Z.; Gu, L.; Eils, R.; Schlesner, M.; Brors, B. Circlize Implements and enhances circular visualization in R. Bioinformatics 2014, 30, 2811–2812. [Google Scholar] [CrossRef]
- Luo, W.; Brouwer, C. Pathview: An R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics 2013, 29, 1830–1831. [Google Scholar] [CrossRef]
| Essential Oil | No. Upregulated | No. Downregulated | No. of Genes Detected in at Least 4 Assayed EOs (n ≥ 4) |
|---|---|---|---|
| Thuja plicata | 1730 | 1477 | 1430 upregulated 833 downregulated |
| Cinnamomum cassia | 1760 | 835 | |
| Eugenia caryophyllata | 1818 | 1728 | |
| Cymbopogon flexuosus | 1831 | 1043 | |
| Melaleuca alternifolia | 1121 | 702 | |
| Origanum vulgare | 1609 | 1221 | |
| Mentha piperita | 2419 | 1684 | |
| Thymus vulgaris | 1520 | 1004 |
| Essential Oil | Concentration |
|---|---|
| Thuja plicata (TP) | 0.025 µg/mL |
| Cinnamomum cassia (CC) | 0.05 µg/mL |
| Eugenia caryophyllata (EC) | 0.05 µg/mL |
| Cymbopogon flexuosus (CF) | 0.025 µg/mL |
| Melaleuca alternifolia (MA) | 0.05 µg/mL |
| Origanum vulgare (OV) | 0.01 µg/mL |
| Mentha piperita (MP) | 0.025 µg/mL |
| Thymus vulgaris (TV) | 0.05 µg/mL |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kisová, Z.; Šoltýsová, A.; Bučková, M.; Beke, G.; Puškárová, A.; Pangallo, D. Studying the Gene Expression of Penicillium rubens Under the Effect of Eight Essential Oils. Antibiotics 2020, 9, 343. https://doi.org/10.3390/antibiotics9060343
Kisová Z, Šoltýsová A, Bučková M, Beke G, Puškárová A, Pangallo D. Studying the Gene Expression of Penicillium rubens Under the Effect of Eight Essential Oils. Antibiotics. 2020; 9(6):343. https://doi.org/10.3390/antibiotics9060343
Chicago/Turabian StyleKisová, Zuzana, Andrea Šoltýsová, Mária Bučková, Gábor Beke, Andrea Puškárová, and Domenico Pangallo. 2020. "Studying the Gene Expression of Penicillium rubens Under the Effect of Eight Essential Oils" Antibiotics 9, no. 6: 343. https://doi.org/10.3390/antibiotics9060343
APA StyleKisová, Z., Šoltýsová, A., Bučková, M., Beke, G., Puškárová, A., & Pangallo, D. (2020). Studying the Gene Expression of Penicillium rubens Under the Effect of Eight Essential Oils. Antibiotics, 9(6), 343. https://doi.org/10.3390/antibiotics9060343






