Pathogenic Escherichia coli Possess Elevated Growth Rates under Exposure to Sub-Inhibitory Concentrations of Azithromycin
Abstract
1. Introduction
2. Results
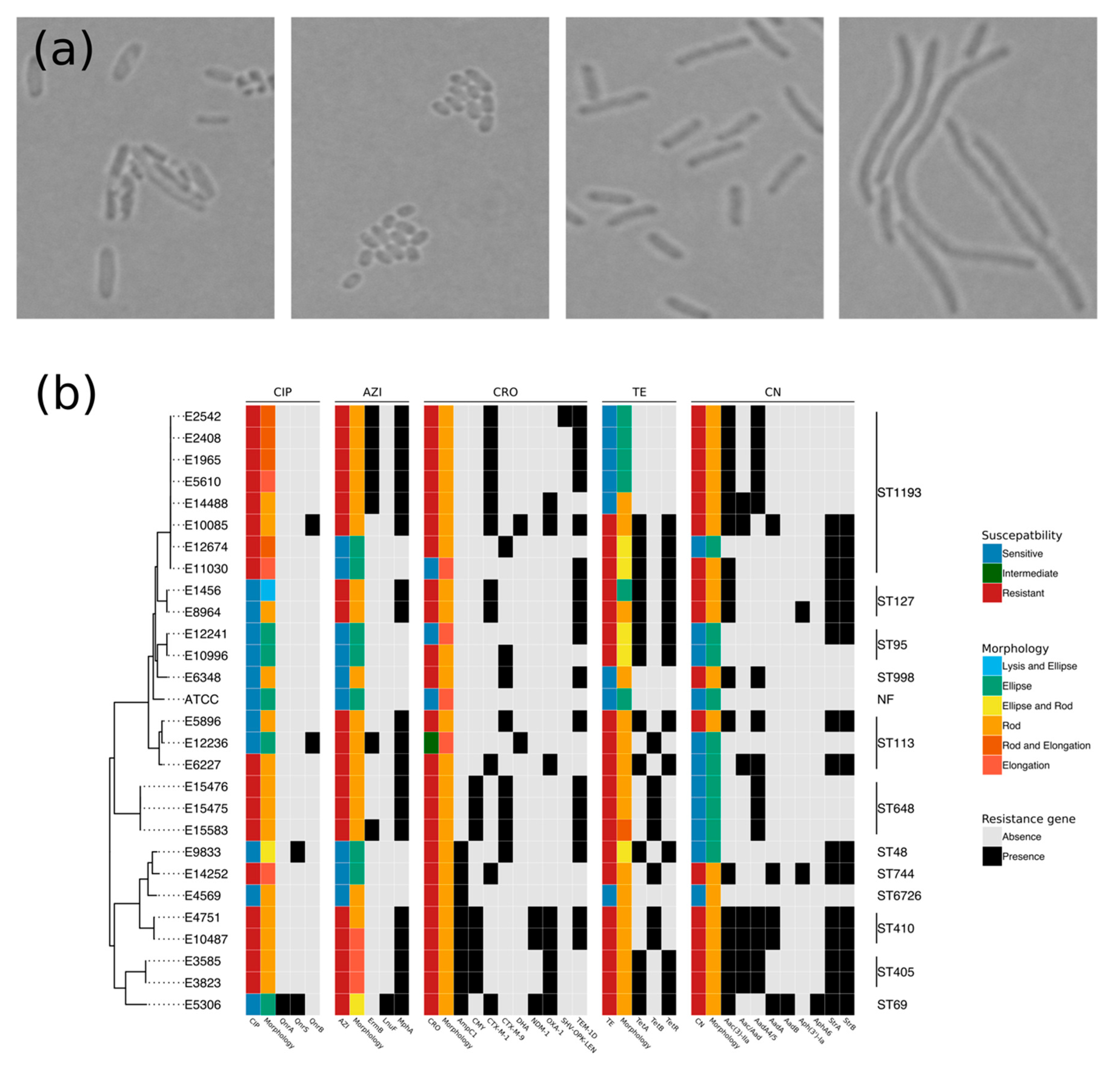
2.1. Resistome and Morphology Associated with AMR
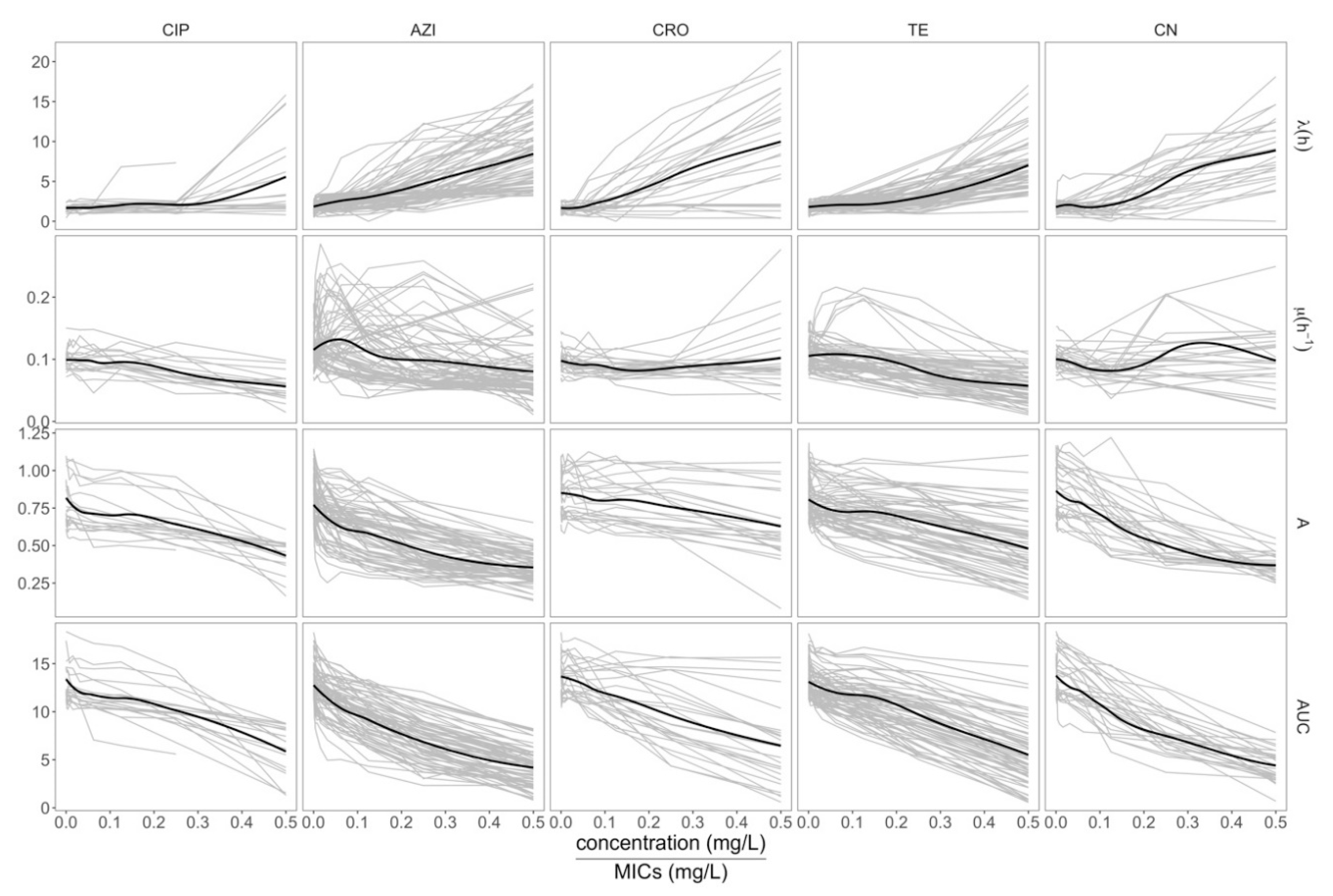
2.2. Assessing the Growth Dynamics of E. coli under Exposure to Antimicrobials
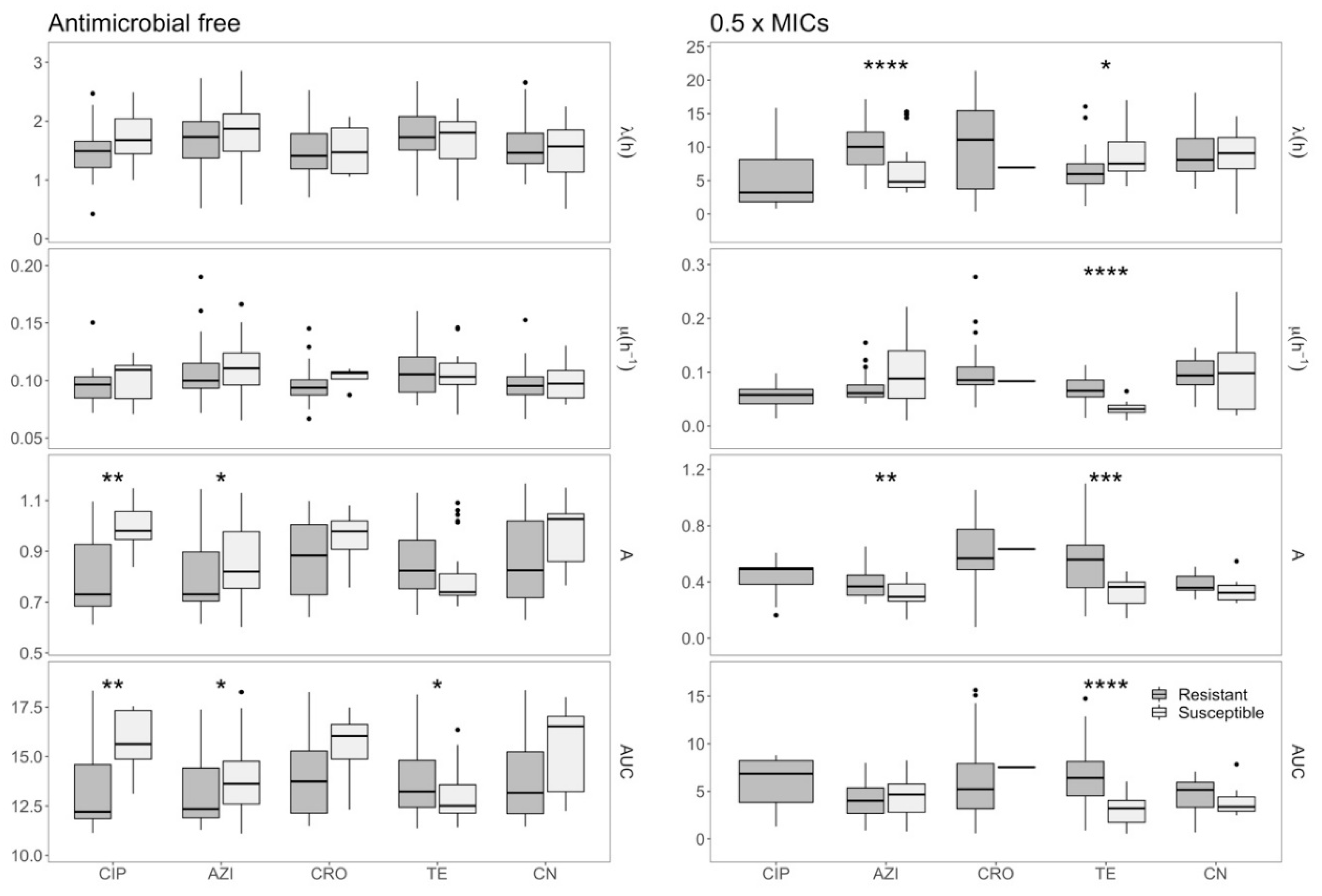
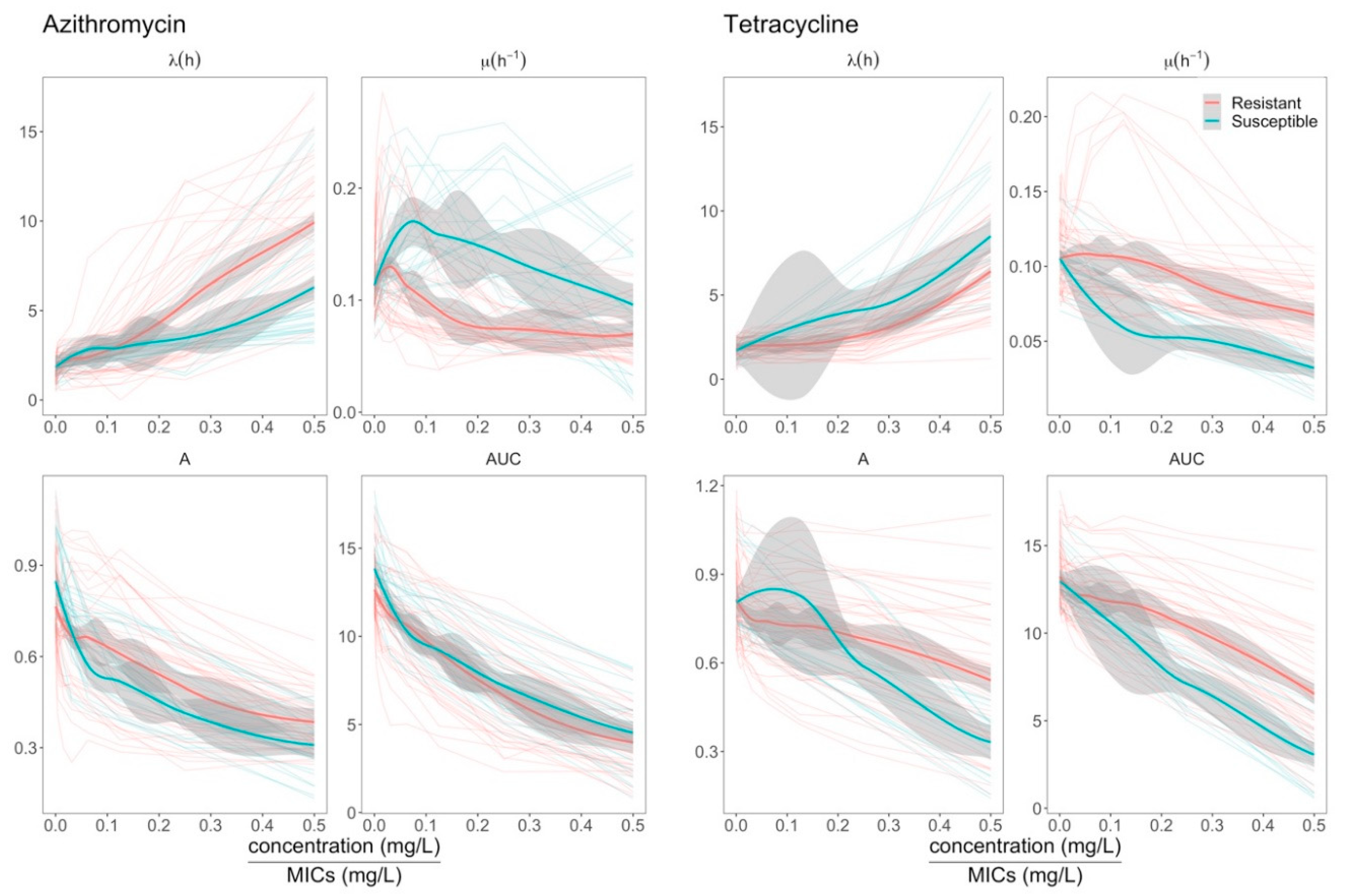
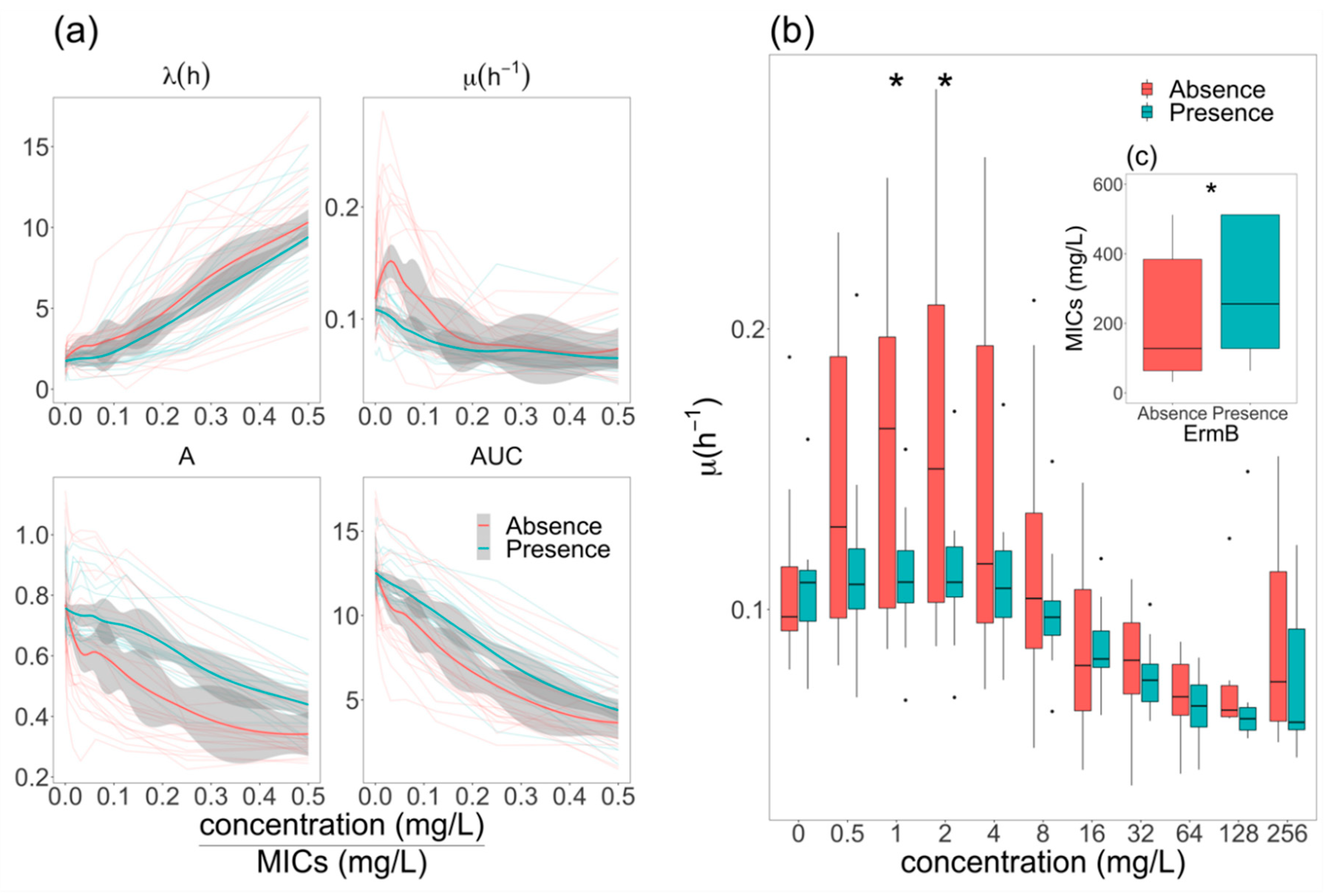
2.3. Effects of Azithromycin and Tetracycline Resistance on Growth Dynamics of E. coli
2.4. Elevation of Growth Rate in ermB- E. coli at Sub-MIC Concentrations of Azithromycin
3. Discussion
4. Materials and Methods
4.1. Organisms and Whole Genome Sequencing
4.2. Monitoring and Modeling Bacterial Growth
4.3. Bright-Field Microscopy
4.4. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- European Centre for Disease Prevention and Control Surveillance of antimicrobial resistance in Europe 2016. In Annual Report of the European Antimicrobial REsistance Surveillance Network (EARS-Net); European Centre for Disease Prevention and Control: Stockholm, Sweden, 2017.
- Lan, N.P.H.; Hien, N.H.; Le Thi Phuong, T.; Thanh, D.P.; Thieu, N.T.V.; Ngoc, D.T.T.; Tuyen, H.T.; Vinh, P.V.; Ellington, M.J.; Thwaites, G.E.; et al. Phenotypic and genotypic characteristics of ESBL and AmpC producing organisms associated with bacteraemia in Ho Chi Minh City, Vietnam. Antimicrob. Resist. Infect. Control. 2017, 6, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Darton, T.C.; Thanh Tuyen, H.; Chung The, H.; Newton, P.N.; Dance, D.A.B.; Phetsouvanh, R.; Davong, V.; Campbell, J.I.; Van Minh Hoang, N.; Thwaites, G.E.; et al. Azithromycin resistance in Shigella spp. in Southeast Asia. Antimicrob. Agents Chemother. 2018, 62, e01748-17. [Google Scholar] [CrossRef] [PubMed]
- Word Health Organization. Antimicrobial Resistance: Global Report on Surveillance 2014; Word Health Organization: Geneva, Switzerland, 2014. [Google Scholar]
- Shrestha, P.; Cooper, B.S.; Coast, J.; Oppong, R.; Do Thi Thuy, N.; Phodha, T.; Celhay, O.; Guerin, P.J.; Wertheim, H.; Lubell, Y. Enumerating the economic cost of antimicrobial resistance per antibiotic consumed to inform the evaluation of interventions affecting their use. Antimicrob. Resist. Infect. Control. 2018, 7, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Hamad, B. The antibiotics market. Nat. Rev. Drug Discov. 2010, 9, 675–676. [Google Scholar] [CrossRef]
- Odoki, M.; Aliero, A.A.; Tibyangye, J.; Nyabayo Maniga, J.; Wampande, E.; Kato, C.D.; Agwu, E.; Bazira, J. Prevalence of Bacterial Urinary Tract Infections and Associated Factors among Patients Attending Hospitals in Bushenyi District, Uganda. Int. J. Microbiol. 2019, 1, 1–8. [Google Scholar] [CrossRef]
- Gonzales, L.; Joffre, E.; Rivera, R.; Sjöling, Å.; Svennerholm, A.M.; Iñiguez, V. Prevalence, seasonality and severity of disease caused by pathogenic Escherichia coli in children with diarrhoea in Bolivia. J. Med. Microbiol. 2013, 62, 1697–1706. [Google Scholar] [CrossRef]
- Khalil, I.A.; Troeger, C.; Blacker, B.F.; Rao, P.C.; Brown, A.; Atherly, D.E.; Brewer, T.G.; Engmann, C.M.; Houpt, E.R.; Kang, G.; et al. Morbidity and mortality due to shigella and enterotoxigenic Escherichia coli diarrhoea: The Global Burden of Disease Study 1990–2016. Lancet Infect. Dis. 2018, 18, 1229–1240. [Google Scholar] [CrossRef]
- Otero, F.; Santiso, R.; Tamayo, M.; Fernández, J.L.; Bou, G.; Lepe, J.A.; McConnell, M.J.; Gosálvez, J.; Cisneros, J.M. Rapid Detection of Antibiotic Resistance in Gram-Negative Bacteria Through Assessment of Changes in Cellular Morphology. Microb. Drug Resist. 2017, 23, 157–162. [Google Scholar] [CrossRef]
- Choi, J.; Yoo, J.; Lee, M.; Kim, E.-G.; Lee, J.S.; Lee, S.; Joo, S.; Song, S.H.; Kim, E.-C.; Lee, J.C.; et al. A rapid antimicrobial susceptibility test based on single-cell morphological analysis. Sci. Transl. Med. 2014, 6, 267ra174. [Google Scholar] [CrossRef]
- Spalding, C.; Keen, E.; Smith, D.J.; Krachler, A.; Jabbari, S. Mathematical modelling of the antibiotic- induced morphological transition of Pseudomonas aeruginosa. PLoS Comput. Biol. 2018, 14, e1006012. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Qiu, Y.; Shi, H.; Yin, H. The importance of lag time extension in determining bacterial resistance to antibiotics. Analyst 2016, 141, 3059–3067. [Google Scholar] [CrossRef] [PubMed]
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef]
- Aldred, K.J.; Kerns, R.J.; Osheroff, N. Mechanism of quinolone action and resistance. Biochemistry 2014, 53, 1565–1574. [Google Scholar] [CrossRef] [PubMed]
- Performance Standards for Antimicrobial Susceptibility Testing, 28th ed.; Clinical And Laboratory Standars Institute: Wayne, PA, USA, 2018.
- Blázquez, J.; Gómez-Gómez, J.M.; Oliver, A.; Juan, C.; Kapur, V.; Martín, S. PBP3 inhibition elicits adaptive responses in Pseudomonas aeruginosa. Mol. Microbiol. 2006, 62, 84–99. [Google Scholar] [CrossRef] [PubMed]
- Yao, Z.; Kahne, D.; Kishony, R. Distinct Single-Cell Morphological Dynamics under Beta-Lactam Antibiotics. Mol. Cell 2012, 48, 705–712. [Google Scholar] [CrossRef] [PubMed]
- Buijs, J.; Dofferhoff, A.S.M.; Mouton, J.W.; Wagenvoort, J.H.T.; Van Der Meer, J.W.M. Concentration-dependency of β-lactam-induced filament formation in Gram-negative bacteria. Clin. Microbiol. Infect. 2008, 14, 344–349. [Google Scholar] [CrossRef]
- Miller, C.; Thomsen, L.E.; Gaggero, C.; Mosseri, R.; Ingmer, H.; Cohen, S.N. SOS response induction by β-lactams and bacterial defense against antibiotic lethality. Science 2004, 305, 1629–1631. [Google Scholar] [CrossRef]
- Ryan, D.M.; Monsey, D. Bacterial filamentation and in vivo efficacy: A comparison of several cephalosporins. J. Antimicrob. Chemother. 1981, 7, 57–63. [Google Scholar] [CrossRef]
- Rolinson, G.N. Effect of β-lactam antibiotics on bacterial cell growth rate. J. Gen. Microbiol. 1980, 120, 317–323. [Google Scholar] [CrossRef]
- Godinez, W.J.; Chan, H.; Hossain, I.; Li, C.; Ranjitkar, S.; Rasper, D.; Simmons, R.L.; Zhang, X.; Feng, B.Y. Morphological Deconvolution of Beta-Lactam Polyspecificity in E. coli. ACS Chem. Biol. 2019, 14, 1217–1226. [Google Scholar] [CrossRef]
- Baltekin, Ö.; Boucharin, A.; Tano, E.; Andersson, D.I.; Elf, J. Antibiotic susceptibility testing in less than 30 min using direct single-cell imaging. Proc. Natl. Acad. Sci. USA 2017, 114, 9170–9175. [Google Scholar] [CrossRef] [PubMed]
- Levy, S.B. The Antibiotic Paradox; Perseus Publishing: Cambridge, MA, USA, 2002. [Google Scholar]
- Daghrir, R.; Drogui, P. Tetracycline antibiotics in the environment: A review. Environ. Chem. Lett. 2013, 11, 209–227. [Google Scholar] [CrossRef]
- Karami, N.; Nowrouzian, F.; Adlerberth, I.; Wold, A.E. Tetracycline resistance in escherichia coli and persistence in the infantile colonic microbiota. Antimicrob. Agents Chemother. 2006, 50, 156–161. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, A.; Zachariasen, C.; Christiansen, L.E.; Græsbøll, K.; Toft, N.; Matthews, L.; Damborg, P.; Agersø, Y.; Olsen, J.E.; Nielsen, S.S. Pharmacodynamic modelling of in vitro activity of tetracycline against a representative, naturally occurring population of porcine Escherichia coli. Acta Vet. Scand. 2015, 57, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Dunai, A.; Spohn, R.; Farkas, Z.; Lázár, V.; Györkei, Á.; Apjok, G.; Boross, G.; Szappanos, B.; Grézal, G.; Faragó, A.; et al. Rapid decline of bacterial drug-resistance in an antibiotic-free environment through phenotypic reversion. Elife 2019, 8, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Word Health Organization. Critically Important Antimicrobials for Human Medicine, 6th ed.; Word Health Organization: Geneva, Switzerland, 2019; ISBN 9789241515528. [Google Scholar]
- Presumptive Use of Azithromycin. Available online: https://www.who.int/maternal_child_adolescent/guidelines/development/provision-of-azithromycin-to-infants/en/ (accessed on 7 December 2019).
- Bergman, M.; Huikko, S.; Huovinen, P.; Paakkari, P.; Seppälä, H.; Thompson, A.K.; Rautio, M.; Renkonen, R.; Muotiala, A.; Vaara, M.; et al. Macrolide and azithromycin use are linked to increased macrolide resistance in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 2006, 50, 3646–3650. [Google Scholar] [CrossRef]
- Lin, L.; Nonejuie, P.; Munguia, J.; Hollands, A.; Olson, J.; Dam, Q.; Kumaraswamy, M.; Rivera, H.; Corriden, R.; Rohde, M.; et al. Azithromycin Synergizes with Cationic Antimicrobial Peptides to Exert Bactericidal and Therapeutic Activity Against Highly Multidrug-Resistant Gram-Negative Bacterial Pathogens. EBioMedicine 2015, 2, 690–698. [Google Scholar] [CrossRef]
- Gomes, C.; Ruiz-Roldán, L.; Mateu, J.; Ochoa, T.J.; Ruiz, J. Azithromycin resistance levels and mechanisms in Escherichia coli. Sci. Rep. 2019, 9, 1–10. [Google Scholar] [CrossRef]
- Horner, P.J. Azithromycin antimicrobial resistance and genital Chlamydia trachomatis infection: Duration of therapy may be the key to improving efficacy. Sex. Transm. Infect. 2012, 88, 154–156. [Google Scholar] [CrossRef]
- Gomes, C.; Martínez-Puchol, S.; Palma, N.; Horna, G.; Ruiz-Roldán, L.; Pons, M.J.; Ruiz, J. Macrolide resistance mechanisms in Enterobacteriaceae: Focus on azithromycin. Crit. Rev. Microbiol. 2017, 43, 1–30. [Google Scholar] [CrossRef]
- Levin, B.R.; McCall, I.C.; Perrot, V.; Weiss, H.; Ovesepian, A.; Baquero, F. A Numbers Game: Ribosome Densities, Bacterial Growth, and Antibiotic- Mediated Stasis and Death. MBio 2017, 8, e02253-16. [Google Scholar] [CrossRef] [PubMed]
- Tadmor, A.D.; Tlusty, T. A coarse-grained biophysical model of E. coli and its application to perturbation of the rRNA operon copy number. PLoS Comput. Biol. 2008, 4, e1000038. [Google Scholar] [CrossRef] [PubMed]
- Greulich, P.; Scott, M.; Evans, M.R.; Allen, R.J. Growth-dependent bacterial susceptibility to ribosome-targeting antibiotics. Mol. Syst. Biol. 2015, 11, 796. [Google Scholar] [CrossRef] [PubMed]
- Garneau-Tsodikova, S.; Labby, K.J. Mechanisms of resistance to aminoglycoside antibiotics: Overview and perspectives. Medchemcomm 2016, 7, 11–27. [Google Scholar] [CrossRef] [PubMed]
- Toan, N.D.; Darton, T.C.; Boinett, C.J.; Campbell, J.I.; Karkey, A.; Kestelyn, E.; Thinh, L.Q.; Mau, N.K.; Thanh Tam, P.T.; Nhan, L.N.T.; et al. Clinical features, antimicrobial susceptibility patterns and genomics of bacteria causing neonatal sepsis in a children’s hospital in Vietnam: Protocol for a prospective observational study. BMJ Open 2018, 8, e019611. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2018. [Google Scholar]
- Inouye, M.; Dashnow, H.; Raven, L.A.; Schultz, M.B.; Pope, B.J.; Tomita, T.; Zobel, J.; Holt, K.E. SRST2: Rapid genomic surveillance for public health and hospital microbiology labs. Genome Med. 2014, 6, 1–16. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Page, A.J.; Cummins, C.A.; Hunt, M.; Wong, V.K.; Reuter, S.; Holden, M.T.G.; Fookes, M.; Falush, D.; Keane, J.A.; Parkhill, J. Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics 2015, 31, 3691–3693. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
| Isolate ID | Minimum Inhibition Concentrations (mg/L) | ||||
|---|---|---|---|---|---|
| CIP 1 | AZI 1 | CRO 1 | TE 1 | CN 1 | |
| E3585 | 32 (R) | 48 (R) | 256 (R) | 256 (R) | 256 (R) |
| E3823 | 32 (R) | 48 (R) | 256 (R) | 256 (R) | 256 (R) |
| E4569 | 0.064 (S) | 12 (S) | 32 (R) | 4 (S) | 1 (S) |
| E6348 | 0.38 (S) | 8 (S) | 256 (R) | 4 (S) | 256 (R) |
| E8964 | 0.38 (S) | 64 (R) | 256 (R) | 256 (R) | 64 (R) |
| E9833 | 0.38 (S) | 16 (S) | 256 (R) | 256 (R) | 1 (S) |
| E10085 | 32 (R) | 48 (R) | 256 (R) | 16 (R) | 256 (R) |
| E10487 | 32 (R) | 256 (R) | 256 (R) | 256 (R) | 96 (R) |
| E10996 | 0.094 (S) | 16 (S) | 256 (R) | 256 (R) | 1 (S) |
| E11030 | 32 (R) | 4 (S) | 0.094 (S) | 256 (R) | 64 (R) |
| E12236 | 0.25 (R) | 256 (R) | 1.5 (I) | 256 (R) | 1 (S) |
| E12241 | 0.094 (S) | 8 (S) | 0.064 (S) | 128 (R) | 1 (S) |
| E12674 | 32 (R) | 6 (S) | 64 (R) | 128 (R) | 1.5 (S) |
| E14252 | 12 (R) | 16 (S) | 256 (R) | 256 (R) | 32 (R) |
| E14488 | 32 (R) | 256 (R) | 256 (R) | 4 (S) | 16 (R) |
| E15475 | 32 (R) | 256 (R) | 256 (R) | 256 (R) | 1 (S) |
| E15476 | 32 (R) | 256 (R) | 256 (R) | 256 (R) | 1.5 (S) |
| E15583 | 32 (R) | 256 (R) | 256 (R) | 256 (R) | 1 (S) |
| E1456 | 0.25 (S) | 48 (R) | 256 (R) | 64 (R) | 256 (R) |
| E1965 | 32 (R) | 256 (R) | 256 (R) | 4 (S) | 256 (R) |
| E2408 | 32 (R) | 256 (R) | 256 (R) | 4 (S) | 64 (R) |
| E2542 | 32 (R) | 256 (R) | 256 (R) | 4 (S) | 256 (R) |
| E4751 | 32 (R) | 256 (R) | 256 (R) | 256 (R) | 256 (R) |
| E5306 | 0.38 (S) | 48 (R) | 256 (R) | 256 (R) | 256 (R) |
| E5610 | 32 (R) | 256 (R) | 256 (R) | 4 (S) | 256 (R) |
| E5896 | 0.38 (S) | 256 (R) | 128 (R) | 256 (R) | 256 (R) |
| E6227 | 32 (R) | 256 (R) | 256 (R) | 256 (R) | 1.5 (S) |
| ATCC25922 | 0.094 (S) | 4 (S) | 0.125 (S) | 4 (S) | 1.5 (S) |
| Susceptible | 8/27 (29.6%) | 8/27 (29.6%) | 2/27(7.4%) | 7/27 (25.9%) | 10/27 (37.0%) |
| Intermediate | 0/27 (0.0%) | 0/27 (0.0%) | 1/27 (3.7%) | 0/27 (0.0%) | 0/27 (0.0%) |
| Resistant | 19/27 (70.4%) | 19/27 (70.4%) | 24/27 (88.9%) | 20/27 (74.1%) | 17/27 (63.0%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tuan-Anh, T.; Tuyen, H.T.; Minh Chau, N.N.; Toan, N.D.; Triet, T.H.; Triet, L.M.; Trang, N.H.T.; To, N.T.N.; Bartholdson Scott, J.; The, H.C.; et al. Pathogenic Escherichia coli Possess Elevated Growth Rates under Exposure to Sub-Inhibitory Concentrations of Azithromycin. Antibiotics 2020, 9, 735. https://doi.org/10.3390/antibiotics9110735
Tuan-Anh T, Tuyen HT, Minh Chau NN, Toan ND, Triet TH, Triet LM, Trang NHT, To NTN, Bartholdson Scott J, The HC, et al. Pathogenic Escherichia coli Possess Elevated Growth Rates under Exposure to Sub-Inhibitory Concentrations of Azithromycin. Antibiotics. 2020; 9(11):735. https://doi.org/10.3390/antibiotics9110735
Chicago/Turabian StyleTuan-Anh, Tran, Ha Thanh Tuyen, Nguyen Ngoc Minh Chau, Nguyen Duc Toan, Tran Hanh Triet, Le Minh Triet, Nguyen Hoang Thu Trang, Nguyen Thi Nguyen To, Josefin Bartholdson Scott, Hao Chung The, and et al. 2020. "Pathogenic Escherichia coli Possess Elevated Growth Rates under Exposure to Sub-Inhibitory Concentrations of Azithromycin" Antibiotics 9, no. 11: 735. https://doi.org/10.3390/antibiotics9110735
APA StyleTuan-Anh, T., Tuyen, H. T., Minh Chau, N. N., Toan, N. D., Triet, T. H., Triet, L. M., Trang, N. H. T., To, N. T. N., Bartholdson Scott, J., The, H. C., Thanh, D. P., Clapham, H., & Baker, S. (2020). Pathogenic Escherichia coli Possess Elevated Growth Rates under Exposure to Sub-Inhibitory Concentrations of Azithromycin. Antibiotics, 9(11), 735. https://doi.org/10.3390/antibiotics9110735





