Abstract
Bacteriophages and phage enzymes are considered as possible alternatives to antibiotics in the treatment of infections caused by antibiotic-resistant bacteria. Due to the ability to cleave the capsular polysaccharides (CPS), one of the main virulence factors of Klebsiella pneumoniae, phage depolymerases, has potential in the treatment of K. pneumoniae infections. Here, we characterized in vivo two novel phage-encoded polysaccharide depolymerases as therapeutics against clinical isolates of K. pneumoniae. The depolymerases Dep_kpv79 and Dep_kpv767 encoded by Klebsiella phages KpV79 (Myoviridae; Jedunavirus) and KpV767 (Autographiviridae, Studiervirinae, Przondovirus), respectively, were identified as specific β-galactosidases that cleave the K. pneumoniae K57 type CPS by the hydrolytic mechanism. They were found to be highly effective at combating sepsis and hip infection caused by K. pneumoniae in lethal mouse models. Here, 80–100% of animals were protected against death by a single dose (e.g., 50 μg/mouse) of the enzyme injected 0.5 h after infection by K. pneumoniae strains of the K57 capsular type. The therapeutic effect of the depolymerases is because they strip the capsule and expose the underlying bacterium to the immune attack such as complement-mediated killing. These data provide one more confirmation that phage polysaccharide depolymerases represent a promising tool for antimicrobial therapy.
1. Introduction
Antibiotic resistance of pathogenic bacteria has become an increasingly pressing clinical issue worldwide. Klebsiella pneumoniae is a well-known Gram-negative bacterium notorious for contributing to multidrug resistance. So far, K. pneumoniae has been considered as an opportunistic pathogen, primarily causing different types of healthcare-associated infections in immunocompromised patients, including pneumonia, urinary and intestinal tract infections, and wound or surgical site infections [1,2]. However, in recent years, a new type of hypervirulent K. pneumoniae (hvKp) which causes community-acquired invasive life-threatening infections characterized by pyogenic liver abscesses complicated with meningitis and endophthalmitis, has emerged worldwide, especially in Southeast Asia [2,3,4].
K. pneumoniae cells produce a number of virulence factors including fimbrial adhesins, lipopolysaccharides (O antigen), siderophore iron acquisition systems, and a polysaccharide capsule (K antigen) [2]. An essential virulence factor and a defense barrier of K. pneumoniae are a polysaccharide capsule (CPS) that allows a bacterial cell to survive and spread inside the host overcoming the protective mechanisms of the immune system. The capsule impairs phagocytosis and opsonophagocytosis of the K. pneumoniae cells by immune cells hinders the bactericidal action of antimicrobial peptides and blocks complement components, such as C3, thus preventing complement-mediated killing [2,5,6]. There is a tremendous amount of structural variability among K. pneumoniae capsules, and at least 134 capsular types have been identified, including 77 K-types recognized by serological typing [7] and new K-types characterized by molecular genotyping and sequencing of the capsule synthesis locus (K-locus, KL) [8,9]. Of all CPS-types K. pneumoniae, K1, K2, K5, K20, K54, and K57 strains overproducing capsular polysaccharides are the most virulent (hvKp) human pathogens [10]. In addition, the CPS is recognized as a primary receptor for bacteriophages that possess capsule depolymerization activities [11]. These enzymatic activities allow phage to get access to the cell surface and bind to the secondary receptor. Bacteriophage-encoded polysaccharide-degrading enzymes, known as polysaccharide (PS) depolymerases, are highly specific enzymes, typically associated with the tail structure of the virions such as tail fibers, tailspikes, base plate or tail tubular proteins [12,13,14]. Since depolymerases target and destroy cell structures, which are important for bacteria survival and virulence, they have been suggested as potential anti-virulence agents to control and prevent bacterial infections. Recently, there have been several reports on the successful application of PS-depolymerases in the treatment of infections caused by Escherichia coli [15], Pasteurella multocida [16], Pseudomonas aeruginosa [17], Acinetobacter baumannii [18]. It was shown that depolymerases successfully rescued mice and Galleria mellonella larvae infected by K. pneumoniae, including hvKp strains [19,20,21,22].
As previously reported, a number of PS-depolymerases specific for K. pneumoniae capsular type K1 [19,23], K2 [24], K3, K21 [22], K5, K8, K30/K69 [21], K11, K25, K35, K64, KN5 [23], K63 [20], KN1, KN3, KN4, and K56 [25] have been isolated and characterized. Pan et al. [26] identified nine PS-depolymerases in a broad host range bacteriophage infecting K. pneumoniae and correlated the activities of the gene products to all host strains that belonged to capsule types K1, K11, K21, K25, K30/K69, K35, K64, KN4, and KN5. However, depolymerases, specific for K.pneumoniae of K57 capsule type, have not been characterized yet.
Previously, we reported on the isolation and genome characterization of bacteriophage KpV767 that can produce polysaccharide depolymerase and infect hypermucoviscous K.pneumoniae of K57 capsular type [24]. In this study, we describe a new lytic phage KpV79 against K57 capsule type K. pneumoniae and characterize two K57 specific polysaccharide depolymerases from phages KpV79 and KpV767.
2. Results and Discussion
2.1. Bacteriophages KpV79 and KpV767 Are Specific for Klebsiella pneumoniae of Capsular Type K57
Bacteriophages KpV767 and KpV79 were originally isolated from sewage in Russia in November 2015 and February 2016, respectively. We evaluated the lytic spectrum of KpV767 and KpV79 on a collection of K. pneumoniae clinical isolates (N = 250) including 21 strains of capsular type K57 (Table 1). Both phages multiplied and lysed K. pneumoniae capsular type K57 to form plaques with a halo expanding during incubation, which we suspected is related to a putative phage-derived depolymerase (Figure 1A). The phages did not multiply with plaque formation on some K57-type strains. However, in these cases, a translucent spot resembling the plaque halo was observed in the spot test by using undiluted phage suspension (Table 2). It should be noted that both phages were not active against others (non-K57 type) strains of the test collection (n = 229), as indicated by the absence of plaques of lysis or translucent spots on a bacterial lawn. These results indicate that phages KpV767 and KpV79 exhibit specificity to K57 capsular polysaccharides.

Table 1.
General features of bacteriophages KpV79 and KpV767.
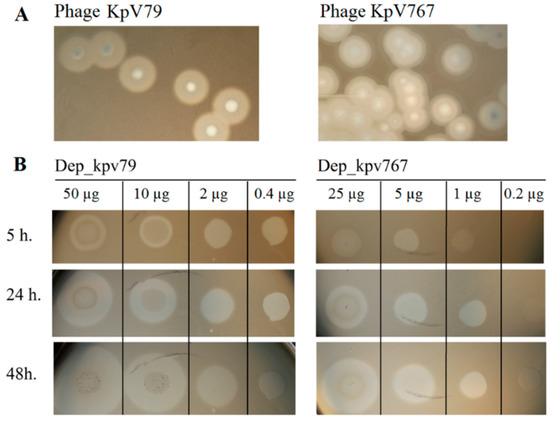
Figure 1.
The activity of depolymerases Dep_kpv79 and Dep_kpv767 against Klebsiella pneumoniae strains. (A) Haloes surrounding phage KpV79 and KpV767 plaques, suggesting the presence of phage depolymerases. (B) Spot tests of purified depolymerases Dep_kpv79 and Dep_kpv767 on K. pneumoniae KPi8289 lawn. Aliquots (10 µL) of serial five-fold dilutions of Dep_79 and Dep_767 were spotted onto a plate containing the K. pneumoniae strain. The plates were observed for 5 h, 24 h, and 48 h of incubation at 37 °C.

Table 2.
The activity of bacteriophages KpV79 and KpV767 and their PS-depolymerases Dep79 and Dep767 against the K. pneumoniae strains of capsular type K57.
2.2. Genomes of Bacteriophages KpV79 and KpV767, Representing Distinct Taxonomic Groups, Contain Genes Encoding Polysaccharide Depolymerases with a High Level of Similarity at the Amino Acid Level
In a previous study [24], we presented the sequence of the bacteriophage KpV767 genome, consisting of 40,395 bp with a G + C content of 52.28%. The KpV79 phage genome has also been sequenced and analyzed. This phage has a linear double-stranded DNA genome of 47,760 bp. Its G + C content is 48.96%, and 75 open reading frames (ORF) are identified and annotated. In both phage genomes, putative functions were assigned to ORF products involved in phage DNA replication, transcription, packaging DNA into the capsid, host lysis, head, and tail morphogenesis (Figure 2).
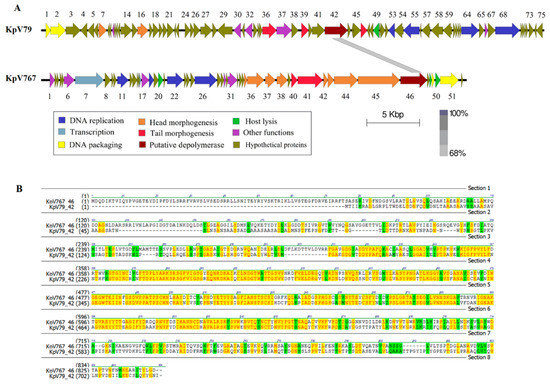
Figure 2.
Genomic analysis of bacteriophages KpV79 and KpV767. (A) Open reading frame organization and comparison of phages genomes. (B) Comparison of amino acid sequences encoded by the PS-depolymerase genes kpvV79_42 and kpv767_46 from bacteriophages KpV79 and KpV767, respectively.
In a number of bacteriophages, the proteins containing the depolymerase domain are usually located in tail spikes and tail fibers attached to the base plate, though they may also be located in other components of the virion, such as internal head or viral membrane [13]. Such proteins identified in genomes of the studied phages, as well as some hypothetical proteins with unknown functions, were analyzed for the presence of amino-acid and structural homology with carbohydrate-hydrolyzing enzymes in protein databases using PSI-BLAST and HHpred tools. To all appearances, the products of genes kpv79_42 and kpv767_46 of phages KpV79 and KpV767, respectively, are structural proteins with predicted PS depolymerase activity involved in the phage interaction with host bacterium. The proteins have significant homology within about 360 aa in the regions 309–671 aa of kpv767_46 protein and 177–539 aa of kpv79_42 protein, respectively (Figure 2).
A high level of similarity at the amino acid level in the central domain of proteins may indicate that these domains determine phage specificity and are responsible for the catalytic cleavage of polysaccharides. In contrast, differences in their N-terminal domains that are responsible for binding to phage particles reflect the fact that phages KpV79 and KpV767 are representatives of different families.
To confirm whether the predicted proteins have polysaccharide-degrading activity, we cloned genes kpv767_46 (phage KpV767) and kpv79_42 (phage KpV79) into the pET22b+ expression vector. His-tag fusion proteins Dep_kpv767 and Dep_kpv79 with a predicted molecular mass of 64 kDa and 77 kDa, respectively, were expressed and purified.
The activities of Dep_kpv767 and Dep_kpv79 were tested on 50 K. pneumoniae strains including 21 strains of K57 type. On spotting to a plate inoculated with K. pneumoniae K57 strains, the recombinant proteins generated a translucent spot resembling the plaque halo (Table 2; Figure 1B). At the same time, translucent spots were not formed on any bacterial lawn of non-K57 type K. pneumoniae strains. The presented results show that depolymerases Dep_kpv79 and Dep_kpv767, like parental phages KpV79 and KpV767, exhibit specificity to K57-capsulated K. pneumoniae.
2.3. Depolymerases Dep_767 and Dep_79 Inhibit the Adsorption of Corresponding Bacteriophages on a Capsule of Bacterial Cells
To study the interaction of bacterial viruses with surface receptors of a bacterial cell, quite simple methods are often used, such as inhibiting the adsorption of bacteriophages on the surface of bacteria by proteins of the phage receptor apparatus, including PS depolymerase, as well as phage inactivation by primary bacterial receptors such as capsule polysaccharides. We used a competition assay to assess the role of Dep_kpv79 and Dep_kpv767 in the KpV79 phage-cell interaction at the initial stages of the infection process. According to our preliminary results, at least 90% of the phage KpV79 are adsorbed to the K. pneumoniae cells in 5 min, and the phage latent period is about 30 min. The phage was added to the K. pneumoniae cells pre-treated with PS-depolymerases, and after 5 min of incubation at 37 °C, the titer of the phage that remained free in solution was measured. In one result, both K57 specific depolymerases Dep_kpv79 and Dep_kpv767 inhibited the phage-host binding. In the same conditions, K1-specific depolymerase Dep_kpv71 [24] did not affect phage adsorption (Figure 3A).
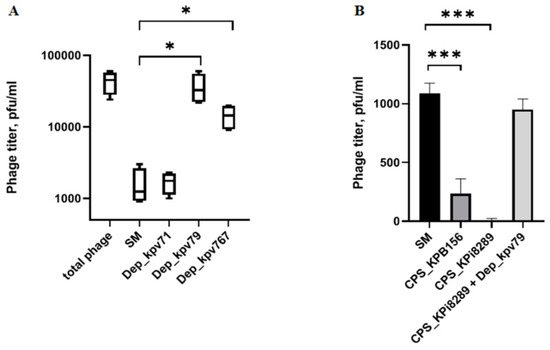
Figure 3.
Phage KpV79 adsorption inhibition assay and phage inactivation by CPS. (A) From left to right, a titer of the total phage sample and titers of the free phage particles after the 5 min incubation with KpV79-sensitive K. pneumoniae KPi8289 cells in SM buffer and in the presence of Dep_kpv71, Dep_kpv79, and Dep_kpv767. (B) Purified CPS’s from K. pneumoniae KPB156 and KPi8289, as well as CPS KPi8289 pre-treated with PS-depolymerase Dep_kpv79, were added to identical phage samples and, after 10 min of incubation, aliquots were taken and titrated. Pure SM buffer was used as a negative control. Average titers from three independent experiments are shown and compared using Student’s t-test analysis. * p < 0.05, *** p < 0.001.
Primary bacterial receptors usually competitively inhibit phage adsorption on a bacterial cell. To evaluate the inhibitory effect of capsular polysaccharides on the bacteriophage KpV79, we conducted the phage inactivation by CPS. As a result, it turned out that the CPS of both strains can inactivate the bacteriophages. Furthermore, phage KpV79 inactivation did not occur if the CPS is pre-treated with Dep_kpv79 depolymerase (Figure 3B).
The data on specificity of the bacteriophages KpV79 and KpV767 to K. pneumoniae of the capsular type K57, as well as the inhibition of the phages by the K57-CPS, suggest that the capsule is the primary receptor for these phages. Both Dep_767 and Dep_79 inhibit the phage adsorption to K. pneumoniae cells, indicating that the phage and depolymerases compete for the same set of receptors on the host cell surface, that are capsular polysaccharides. These results are consistent with that Dep_kpv79 and Dep_kpv767 proteins were bioinformatics predicted as putative tail fibers/spikes that are responsible for host cell recognition. Based on adsorption experiments, we suppose that Dep_kpv79 and Dep_kpv767 proteins are responsible for the reversible binding and subsequent degradation of capsular polysaccharides of K. pneumoniae cells. This mechanism, followed by a stage of irreversible adsorption to the main receptors of the bacterial outer membrane, has been described for other capsule-specific phages [11].
2.4. Depolymerases Dep_kpv79 and Dep_kpv767 Specifically Cleave the β-Galactosidic Linkages in the K57 Capsular Polysaccharide of K. pneumoniae
Depending on the way of degradation of bacterial polysaccharides, phage-associated enzymes fall into the two main groups—glycoside hydrolases (EC 3.2.1.) and lyases (EC 4.2.2.)—which are divided into different subclasses [13]. Usually, the mechanism of the enzymatic action of phage enzymes is predicted according to amino-acid and structural homology with carbohydrate- degrading enzymes in protein databases. At the same time, only a few experimental data that relate to the enzymatic mechanisms of phage depolymerase action are available [17,27,28].
In this work, we elucidated the mechanism and characterized for the first time the product of cleavage of the K. pneumoniae K57 CPS by phage depolymerases Dep_kpv79 and Dep_kpv767.
The composition and structure of the K57 CPS of K. pneumoniae have been known [29]. CPS structure of the strain KPi8289 was confirmed by the assignment of the 1H and 13C NMR spectra of the CPS using two-dimensional 1H,1H COSY, 1H,1H TOCSY, and 1H,13C HSQC experiments (Table 3). The CPS is built up of branched tetrasaccharide repeats (K units) containing two residues of d-mannose (units B and D) and one residue each of d-galactose (unit A) and d-galacturonic acid (unit C) (Figure 4).

Table 3.
1H and 13C NMR chemical shifts (δ, ppm) of the K57 CPS from K. pneumoniae KPi8289 and oligosaccharide 1 (OS) derived by cleavage of the CPS with depolymerase Dep_kpv79.
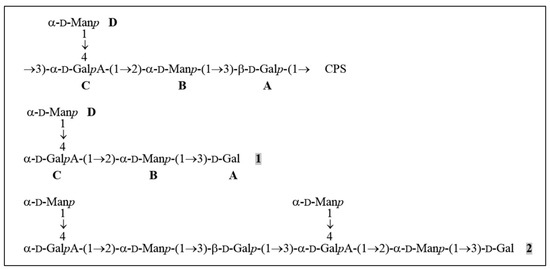
Figure 4.
Structures of the K57 CPS of K. pneumoniae KPi8289 and oligosaccharides derived by cleavage of the CPS with depolymerase Dep_kpv79. CPS, tetrasaccharide repeat, containing two residues of D-mannose (units B and D) and one residue each of D-galactose (unit A) and D-galacturonic acid (unit C). 1 and 2, oligosaccharides obtained by cleavage of the CPS_KPi8289 with Dep_kpv79 (the same oligosaccharides were obtained by CPS splitting with Dep_kpv767).
The CPS was cleaved with depolymerase Dep_kpv79 and the products were fractionated by gel permeation chromatography on Fractogel TSK HW-40S to give two oligosaccharides 1 and 2 (Figure 4). The same oligosaccharides were obtained by cleavage of the CPS_KPi8289 with Dep_kpv767 depolymerase (data not shown).
Analysis by negative ion mode high-resolution electrospray ionization mass spectrometry (HR ESI MS) showed that a lower oligosaccharide 1 corresponded to the K unit of the CPS and oligosaccharide 2 to a dimer of the K unit (the spectra showed [M-H]− ion peaks for Hex3HexA1 and Hex6HexA2 at m/z 679.1841 and 1341.3834; calculated values m/z 679.1938 and 1341.3844, respectively).
The 1H and 13C NMR spectra of oligosaccharide 1 were assigned as described above for the CPS (Table 3). As in the K unit of the CPS, four monosaccharide residues (units A-D) were identified. Whereas the 13C NMR chemical shifts of B and D were essentially the same in the CPS and 1, those of units A and C were different. The monosaccharide residue at the reducing end of 1 that was identified as the Gal residue (unit A) occurred in two anomeric pyranose forms (δH 5.28 and 4.61, δC 93.6 and 97.3 for the α- and β-anomers; compare published data [30] δH 5.22 and 4.53, δC 93.18 and 97.37 for α-Galp and β-Galp, respectively). This residue is β-linked in the CPS. The GalA residue (unit C) is 3,4-disubstituted in the CPS and 4-substituted in 1, as followed from a marked displacement of the signal for C-3 of unit C from δ 78.4 in the CPS to δ 70.3 in 1.
Therefore, taking into account the CPS structure, oligosaccharide 1 has the structure shown in Figure 4. The same oligosaccharide has been obtained by mild acid hydrolysis of the K68 CPS of K. pneumoniae, which differs from the K57 CPS in the presence of a pyruvic acid acetal linked to the terminal Man residue [31].
The 1H and 13C NMR spectra of oligosaccharide 2 showed two series of signals, one corresponding to oligosaccharide 1 and the other to the K unit within the CPS. Particularly, there were signals for two units A, one β-linked as in the CPS and the other at the reducing end as in 1, and two units C, one 3,4-disubstituted and the other 4-substituted. Therefore, 2 represented a dimer of the K unit having the structure shown in Figure 4.
Thus, the data obtained indicated that depolymerases Dep_kpv79 and Dep_kpv767 are β-glycosidases that cleave specifically the β-d-Galp-(1→3)-d-GalpA (A→C) linkages in the CPS of K. pneumoniae KPi8289 by the hydrolytic mechanism.
2.5. Capsule Depolymerases Are Effective Therapeutics in Mouse Infection Models
Clinical manifestations of diseases caused by K. pneumoniae are extremely diverse [1,2]. Therefore, it is advisable to evaluate the therapeutic efficacy of bacteriophages on different laboratory models that differ in the route of infection and the infectious process development. We used two previously described mouse models of K. pneumoniae primary sepsis [32] and thigh soft tissue infection [33]. Two strains KPB156 and KPB550, differing in the degree of lysis by bacteriophages KpV79 and KpV767, were used to modeling K. pneumoniae infections in mice. As noted in Table 2, strain KPB550 was sensitive only to phage KpV79, whereas strain KPB156 was lysed by both phages with high efficiency of plating. Three groups of infected mice with 10 animals in each were used in each experiment: control (depolymerase untreated mouse) and two experimental (30 min after infection, the depolymerase Dep_kpv79 or Dep_kpv767 at a dose of 50 μg/mouse was injected intraperitoneally to mice of these groups).
As a result, nine of 10 (90%) and six of 10 (60%) mice infected with an intraperitoneal injection of K. pneumoniae KPB550 and KPB156 strains, respectively (primary sepsis model), and eight of 10 (80%) mice infected with an intramuscular injection of K. pneumoniae KPB550 (soft tissue infection model) died in the control groups (without PS-depolymerase treatment). In these cases, a culture of the infecting strain was isolated from all mice, those that died and those that survived. In contrast, 80% to 100% of the animals survived in the experimental groups treated with Dep_kpv79 and Dep_kpv767 depolymerase (Figure 5). It should be noted that no pathology was detected in the organs and tissues of the animals that survived the challenge with K. pneumoniae. In total, a culture of K. pneumoniae was isolated from five of the 53 surviving animals that were treated with Dep_kpv79 or Dep_kpv767 depolymerase.
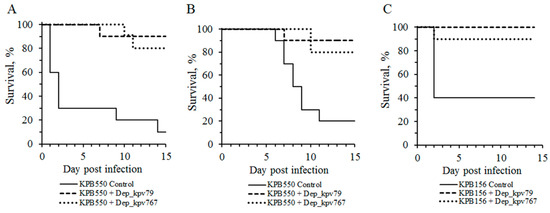
Figure 5.
Protective effects of K57-specific depolymerases Dep_kpv79 and Dep_kpv767 of bacteriophages KpV79 and KpV767, respectively, in mice challenged with K. pneumoniae strains. Depolymerases (50 μg per mouse) were administered intraperitoneally 30 min after infection. (A) Primary sepsis caused by K. pneumoniae KPB550 (intraperitoneal injection). (B) Thigh soft tissue infection caused by K. pneumoniae KPB550 (intramuscular injection). (C) Primary sepsis caused by K. pneumoniae KPB156 (intraperitoneal injection).
The bacteria of strain KPB156 are less virulent compared to KPB550. Therefore, when mice were infected in the thigh muscles with a culture of strain KPB156 no death of animals was observed. However, seven of ten infected mice showed soft tissue edema at the injection site. After depolymerase treatment, edema was observed on day 3 in three of 10 mice, followed by its disappearance on day 7. In the control group, 30% of mice remained edematous for up to 10 days after infection.
Therefore, Dep_kpv79 and Dep_kpv767 depolymerases that cleave the K57 type CPS, have a therapeutic effect against K. pneumoniae–induced infections in mice.
Previously, a few works have been published on the in vivo therapeutic efficacy of CPS specific depolymerases against K. pneumoniae-induced infections [19,20,22]. Lin et al. demonstrated that treatment with the recombinant depolymerase provided increased survival in mice infected with K.pneumoniae of capsular type K1 [19]. Experiments by Majkowska-Skrobek et al. have shown that depolymerizing enzymes were effective against K. pneumoniae strains, representing K3, K21, and K63 capsular types, in a G. mellonella infecting model. In particular, the application of K3 and K21 specific depolymerases considerably increased the lifespan of G. mellonella larvae in a time- and K. pneumoniae strain–dependent manner [22]. The other depolymerase was effective against a native capsule of K. pneumoniae strains, representing the K63 type, and significantly inhibited Klebsiella-induced mortality of G. mellonella larvae in a time-dependent manner [20].
Our results on the therapeutic efficacy of two K57 specific depolymerases against K. pneumoniae-induced infection in mice provide one more confirmation that phage polysaccharide depolymerases represent a promising tool for antimicrobial therapy.
2.6. Depolymerases Increase Sensitivity to Serum-Complement-Mediated Killing
It is believed that the therapeutic efficacy of depolymerases is due to their antivirulence potential [20]. Since the polysaccharide capsule is one of the major virulence factors of K. pneumoniae, the CPS destruction makes bacteria less virulent and more susceptible to innate host defense including pathogen phagocytosis and complement-mediated killing [19,22,23,34].
In our work, the influence of the depolymerases on the K. pneumoniae serum resistance was studied. As shown in Figure 6, the depolymerases did not affect bacterial survival in the absence of serum. Also, serum alone did not affect the killing of K. pneumoniae strains KPB550 and KPi8289. However, treatment of K. pneumoniae cells with depolymerase Dep_kpv79 or Dep_kpv767 resulted in a decrease of the bacteria survival by four orders of magnitude after 3-h incubation. The pre-heated serum did not affect the viability of the K. pneumoniae cells. Therefore, increased bactericidal serum activity is associated with complement-mediated killing.
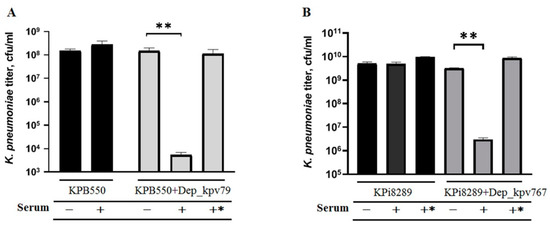
Figure 6.
Serum sensitivity assay. Bacteria of serum-resistant K. pneumoniae strains KPB550 (A) and Kpi8289 (B) were pre-incubated with or without depolymerase Dep_kpv79 and Dep_kpv767, respectively (100 µg/mL, overnight at 4 °C). Mixtures were diluted 10 times, incubated for 3 h at 37 °C with 50% sheep serum (+), heat-inactivated serum (+*) or PBS (-), and then plated to determine colony-forming units. Average titers from three independent experiments are shown and compared by Student’s t-test analysis. ** p < 0.01.
These results show that the depolymerases increase K. pneumoniae sensitivity to serum complement-mediated killing, consistent with other findings [19,22,34]. In addition, our data confirmed that the CPS is responsible for the complement resistance since other virulence factors of K. pneumoniae were unchanged after depolymerase treatment.
3. Materials and Methods
3.1. Bacterial Strains and Bacteriophages
Bacterial strains used in the experiments were obtained from the State Collection of Pathogenic Microorganisms and Cell Cultures (SCPM, Obolensk, Russia). All bacteria were routinely cultured in the Nutrient Medium No.1 (SRCAMB, Obolensk, Russia), or on LB broth agar (Difco Laboratories, Detroit, MI, USA) plates at 37 °C stationary or with shaking.
Bacteriophages were propagated using liquid cultures of K. pneumoniae sensitive strains as previously described [24]. Phage suspensions were titrated by a standard soft agar overlay method and stored at 4 °C until use.
3.2. Determination of the Host Range of Phage and Polysaccharide Depolymerase
Bacteriophages were screened against K. pneumoniae strains using the efficiency of the plating micro method [24]. PS depolymerase activity was assessed using the spot assay on K. pneumoniae strains of different capsular types and varying the enzyme concentrations. For K. pneumoniae lawn formation, 0.3 mL of bacterial suspension (~3 × 108 CFU/mL) was mixed with 4 mL of soft agar (LB broth supplemented with 0.6% agarose) at 45 °C and poured onto the surface of the Nutrient Medium No.1 plate. The plate was incubated at 37 °C overnight. These bacterial lawns as well as lawns inactivated by chloroform vapor were used for depolymerase testing.
3.3. Analysis of the Phage Genomes
Phage genomes were sequenced using the Ion Torrent PGM (Life Technologies, Carlsbad, CA, USA), according to the manufacturer’s instructions. De novo assembly of sequence reads was performed using Newbler v.2.9. The correctness of assembly was checked using the SeqMan NGen software (DNAStar, Madison, WI, USA).
Protein-encoding genes and their putative functions were automatically predicted by RAST [35] and compared with known gene and protein sequences using the BLASTx and BLASTp algorithms [36] at the NCBI website. Linear genome comparisons of the phages and visualization of the coding regions were performed with Easyfig [37].
To identify putative polysaccharide depolymerase, a tail fiber, tail spike, and hypothetical proteins were analyzed by PSI-BLAST at the NCBI website and were screened through the HHpred web server [38].
The annotated genome sequences of bacteriophages KpV79 and KpV767 were deposited in the GenBank database and are available under accession numbers MF663761, and KX712070, respectively.
3.4. Cloning of CPS Depolymerase Genes and Preparation of Recombinant Proteins
Coding sequences for genes kpv79_42 (GenBank: ATI16495.1) and kpv767_46 (GenBank: AOZ65519.1) containing the putative depolymerase motifs were amplified from DNA of phages KpV79 and KpV767, respectively, by PCR using the following oligonucleotide primer pairs 79_XhoI: 5′-CTTACTCGAGGTTACCATAATACTGCAGAGAA-3′ and 79_NcoI: 5′-TATACCATGGACATTATCAAACGCGCAGAC-3′ (kpv79_42 gene cloning); 767_XhoI; 5′-TTAGTCCTCGAGATACGTGATGCGAGCCTC-3′ and 767_NcoI: 5′-CTTCACCATGGCAATGACCACAGAGTCTAG-3′ (kpv767_46 gene cloning).
The amplified genes were cloned into pET22b expression vector (Novagen, Madison, WI, USA) via the NcoI and XhoI sites by using Escherichia coli strain DH5α. The kpv79_42 gene was cloned entirely; the kpv767_46 gene was cloned without N-terminus fragment encoding the first 252 amino acid residues. Recombinant plasmids were verified by Sanger sequencing and then transformed into E. coli BL21 (DE3) for protein expression.
The production of recombinant proteins in LB broth (Difco Laboratories, Detroit, MI, USA) supplemented with 100 µg/mL ampicillin was induced by the addition of isopropyl-β-D-thiogalactopyranoside (IPTG) to the final concentration of 0.5 mM for 20 h at 22 °C. The recombinant proteins were separated from contaminating proteins by affinity chromatography using a Ni-iminodiacetic acid-Sepharose 5 mL column. The CPS-degrading activity of the recombinant proteins was assayed in the spot-test using K. pneumoniae strains of different capsular types [24].
3.5. Determination of the Structures of the Oligosaccharides Derived by the Treatment of K. pneumoniae CPS with Phage Depolymerases
Polysaccharides were isolated from K. pneumoniae KPi8289 cells by the modified water—phenol extraction method described previously [39].
Gel-permeation chromatography was carried out on a column (85 × 2.5 cm) of Fractogel TSK HW-40S (Merck, Germany). Elution was performed with 0.1% HOAc at a flow rate of 0.5 mL/min and monitored with a differential refractometer (Knauer, Germany).
1H and 13C NMR spectra were recorded on a Bruker Avance II 600 MHz spectrometer (Germany). Before measurement, samples were freeze-dried from 99.9% D2O and then examined in 99.95% D2O at 60 °C using sodium 3-(trimethylsilyl)propanoate-2,2,3,3-d4 (δH 0, δC −1.6) as the internal reference for calibration. Two-dimensional NMR spectra were obtained using standard Bruker software, and the TopSpin 2.1 program (Bruker, Germany) was employed to acquire and to process the data. A MLEV-17 spin-lock time of 60 ms was used in the TOCSY experiment.
HR ESI MS was performed in the negative ion mode using a micrOTOF II instrument (Bruker Daltonics). Oligosaccharide samples (~50 ng/μL) were dissolved in a 1:1 (v/v) water/acetonitrile mixture and injected with a syringe at a flow rate of 3 μL/min. The capillary entrance voltage was set at 3200 V, and the interface temperature at 180 °C. Nitrogen was used as the drying gas. The mass range was from m/z 50 to 3500 Da. Internal calibration was done with ESI Calibrant Solution (Agilent).
3.6. Phage Adsorption Inhibition Test and Phage Inactivation by CPS
The effect of PS-depolymerases Dep_767 and Dep_79 on the bacteriophage adsorption to K. pneumoniae cells was evaluated as described previously [40] with some modifications. In brief, a bacterial cell suspension (~2 × 108 cfu/mL) in SM buffer (8 mM MgCl2, 100 mM NaCl, 50 mMTris-HCl pH 7.5) with added depolymerase (400 μg/mL) and a phage sample at 105 pfu/mL were first brought to 37 °C. Then equal volumes of the phage and cell suspension were mixed. After 5 min incubation at 37 °C, bacterial cells with adsorbed phage were precipitated by centrifugation at 14,000 g for 1 min, and the titer of non-adsorbed phages was determined in the supernatant.
The phage inactivation by CPS was performed as follows. Phage samples (103 pfu/mL) in SM buffer with added CPS at a final concentration of 1 μg/mL were incubated at 37 °C for 10 min. After incubation, the phage titer was assessed by the double agar layer technique.
3.7. In vivo K. pneumoniae Infection Models
Female white outbred mice (age 6–8 weeks) were used in experiments. Mice were housed under standard conditions of light, temperature, and water and food availability.
Two previously described mouse models of K. pneumoniae primary sepsis [32] and thigh soft tissue infection [33] were used to evaluate the therapeutic efficacy of PS-depolymerases. In the first case, K. pneumoniae strain KPB550 (LD50 = 6 × 106 cfu) or KPB156 (LD50 = 1 × 108 cfu) was used for intraperitoneal infection of mice (2 × 108 cfu in a volume of 0.5 mL). In the second model, mice were injected intramuscularly (in the left thigh) with a culture of K. pneumoniae strains and at the same dose in a volume of 0.1 mL. In each case, control and experimental groups of mice were used (10 animals each). PS-depolymerases were injected into the mice of the experimental group half an hour after infection (once, intraperitoneal, 50 µg per mouse). Mice of the control groups were not treated with depolymerases. All animals were sacrificed two weeks after infection, and blood and parenchymal organs were examined for the presence of a K. pneumoniae culture. Mice that died during the experiment were also autopsied and examined for bacterial dissemination.
Mouse experiments were performed following the Directive 2010/63/EU of the European Parliament and of the Council of 22 September 2010 on the protection of animals used for scientific purposes. The experiments were approved by the Animal Ethics Committee of State Research Center for Applied Microbiology and Biotechnology.
3.8. Serum Complement Activity
A log-phase bacterial culture (~108 cfu/mL) were preincubated with or without depolymerase (at the final concentration of 100 µg/mL) overnight at 4 °C. Mixtures were diluted 10 times, incubated for 3 h at 37 °C with 50% sheep serum, heat-inactivated serum (56 °C, 30 min) or phosphate-buffered saline (PBS), and then plated to determine colony-forming units. Each test was performed at least three independent experiments.
3.9. Statistical Analysis
Computation of data and statistical tests were performed using GraphPad Prism 8.0 software (GraphPad Software, Inc., La Jolla, CA, USA). Statistical significance was determined using a t-test. p < 0.05 was considered statistically significant.
4. Conclusions
In this work, we characterized two new PS-depolymerases Dep_kpv79 and Dep_kpv767 encoded by myovirus KpV79 and phage KpV767 of the Autographiviridae family both specific to K. pneumoniae K57. The depolymerases have a high level of similarity at the amino acid level in the central domain, which is responsible for the catalytic cleavage of polysaccharides. To the best of our knowledge, this is the first report of two identical depolymerases with the same CPS specificity derived from phages of a different family.
The phages recognize the K57 CPS as the primary receptor for adsorption, which is the first step in the phage infection process. These results were confirmed by a phage inactivation assay performed with the CPS isolated from K. pneumoniae strains of the K57 capsular type.
For the first time, we elucidated the mechanism and characterized the products of cleavage of the K. pneumoniae K57 CPS by depolymerases Dep_kpv79 and Dep_kpv767 encoded by phages with different tail structures. Both depolymerases were identified as specific glycosidases that cleave the CPS of K. pneumoniae strains by the hydrolytic mechanism.
Due to the ability to cleave the CPS, one of the main virulence factors of K. pneumoniae, depolymerases show a therapeutic effect in the treatment of K. pneumoniae–induced infections. Previously it has been reported that CPS-specific depolymerases successfully rescued mice and G. mellonella larvae infected by K. pneumoniae strains of capsular type K1 [19], K3, K21 [22], K63 [20], and K64 [34]. The Dep_kpv79 and Dep_kpv767 depolymerases were highly effective at combating sepsis and hip infection caused by K. pneumoniae in lethal mouse models. Both depolymerases exerted a therapeutic effect in mice infected with strains that differ in virulence and phage sensitivity. Indeed, 80–100% of animals were protected against death by a single dose (i.p., 50 μg/mouse) of the enzyme injected 0.5 h after infection by K. pneumoniae strains of K57 capsular type. The therapeutic effect of depolymerases is because they strip capsules and expose the underlying bacterium to immune attack such as complement-mediated killing.
Our study provides another confirmation that phage-borne depolymerases are able to target with high specificity such virulence factors as a bacterial capsule. Therefore, they represent a promising tool for typing and treatment of infections caused by bacterial pathogens.
Author Contributions
Conceptualization, N.V.V., and Y.A.K.; methodology, N.V.V., A.I.B., V.V.V., and A.S.S.; formal analysis, A.M.S., E.V.K. and V.M.K., investigation, A.M.S., A.I.B., V.M.K., E.V.K., A.S.S. and V.V.V.; writing—original draft preparation, review, and editing, N.V.V. and Y.A.K.; supervision and funding acquisition, N.V.V. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Russian Science Foundation (grant number 15-15-00058) and the Ministry of Science and Higher Education of the Russian Federation (grant number 075-15-2019-1671).
Acknowledgments
We are grateful to Olga Korobova, Tatyana Kombarova, and Irina Ufimtseva for assistance in mouse experiments. We also thank Angelina Kislichkina for help with DNA sequencing and Alexander Chizhov for measuring high-resolution mass spectra.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Podschun, R.; Ullmann, U. Klebsiella spp. as nosocomial pathogens: Epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin. Microbiol. Rev. 1998, 11, 589–603. [Google Scholar] [CrossRef] [PubMed]
- Paczosa, M.K.; Mecsas, J. Klebsiella pneumoniae: Going on the Offense with a Strong Defense. Microbiol. Mol. Biol. Rev. 2016, 80, 629–661. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.H.; Liu, Y.C.; Lee, S.S.; Yen, M.Y.; Chen, Y.S.; Wang, J.H.; Wann, S.R.; Lin, H.H. Primary liver abscess due to Klebsiella pneumoniae in Taiwan. Clin. Infect. Dis. 1998, 26, 1434–1438. [Google Scholar] [CrossRef]
- Turton, J.F.; Englender, H.; Gabriel, S.N.; Turton, S.E.; Kaufmann, M.E.; Pitt, T.L. Genetically similar isolates of Klebsiella pneumoniae serotype K1 causing liver abscesses in three continents. J. Med. Microbiol. 2007, 56, 593–597. [Google Scholar] [CrossRef] [PubMed]
- Cortes, G.; Borrell, N.; de Astorza, B.; Gómez, C.; Sauleda, J.; Albertí, S. Molecular analysis of the contribution of the capsular polysaccharide and the lipopolysaccharide O side chain to the virulence of Klebsiella pneumoniae in a murine model of pneumonia. Infect. Immun. 2002, 70, 2583–2590. [Google Scholar] [CrossRef] [PubMed]
- Domenico, P.; Salo, R.J.; Cross, A.S.; Cunha, B.A. Polysaccharide capsule-mediated resistance to opsonophagocytosis in Klebsiella pneumoniae. Infect. Immun. 1994, 62, 4495–4499. [Google Scholar] [CrossRef] [PubMed]
- Ørskov, I.; Fife-Asbury, M.A. New Klebsiella capsular antigen K82 and the deletion of five of those previously assigned. Int. J. Syst. Bacteriol. 1977, 27, 386–387. [Google Scholar] [CrossRef]
- Pan, Y.J.; Lin, T.L.; Chen, C.T.; Chen, Y.Y.; Hsieh, P.F.; Hsu, C.R.; Wu, M.C.; Wang, J.T. Genetic analysis of capsular polysaccharide synthesis gene clusters in 79 capsular types of Klebsiella spp. Sci. Rep. 2015, 5, 15573. [Google Scholar] [CrossRef]
- Wyres, K.L.; Wick, R.R.; Gorrie, C.; Jenney, A.; Follador, R.; Thomson, N.R.; Holt, K.E. Identification of Klebsiella capsule synthesis loci from whole genome data. Microb. Genom. 2016, 2, e000102. [Google Scholar] [CrossRef]
- Shon, A.S.; Bajwa, R.P.; Russo, T.A. Hypervirulent (hypermucoviscous) Klebsiella pneumoniae: A new and dangerous breed. Virulence 2013, 4, 107–118. [Google Scholar] [CrossRef]
- Rakhuba, D.V.; Kolomiets, E.I.; Dey, E.S.; Novik, G.I. Bacteriophage receptors, mechanisms of phage adsorption and penetration into host cell. Pol. J. Microbiol. 2010, 59, 145–155. [Google Scholar] [CrossRef]
- Latka, A.; Maciejewska, B.; Majkowska-Skrobek, G.; Briers, Y.; Drulis-Kawa, Z. Bacteriophage-encoded virion-associated enzymes to overcome the carbohydrate barriers during the infection process. Appl. Microbiol. Biotechnol. 2017, 101, 3103–3119. [Google Scholar] [CrossRef] [PubMed]
- Pires, D.P.; Oliveira, H.; Melo, L.D.R.; Sillankorva, S.; Azeredo, J. Bacteriophage-encoded depolymerases: Their diversity and biotechnological applications. Appl. Microbiol. Biotechnol. 2016, 100, 2141–2151. [Google Scholar] [CrossRef] [PubMed]
- Brzozowska, E.; Pyra, A.; Pawlik, K.; Janik, M.; Górska, S.; Urbańska, N.; Drulis-Kawa, Z.; Gamian, A. Hydrolytic activity determination of Tail Tubular Protein A of Klebsiella pneumoniae bacteriophages towards saccharide substrates. Sci. Rep. 2017, 7, 18048. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; Paff, M.L.; Molineux, I.J.; Bull, J.J. Therapeutic application of phage capsule depolymerases against K1, K5, and K30 capsulated E. coli in mice. Front. Microbiol. 2017, 8, 2257. [Google Scholar] [CrossRef]
- Chen, Y.; Sun, E.; Yang, L.; Song, J.; Wu, B. Therapeutic Application of Bacteriophage PHB02 and its Putative Depolymerase Against Pasteurella multocida Capsular Type A in Mice. Front. Microbiol. 2018, 9, 1678. [Google Scholar] [CrossRef]
- Olszak, T.; Shneider, M.M.; Latka, A.; Maciejewska, B.; Browning, C.; Sycheva, L.V.; Cornelissen, A.; Danis-Wlodarczyk, K.; Senchenkova, S.N.; Shashkov, A.S.; et al. The O-specific polysaccharide lyase from the phage LKA1 tailsike reduces Pseudomonas virulence. Sci. Rep. 2017, 7, 16302. [Google Scholar] [CrossRef]
- Hernandez-Morales, A.C.; Lessor, L.L.; Wood, T.L.; Migl, D.; Mijalis, E.M.; Cahill, J.; Russell, W.K.; Young, R.F.; Gill, J.J. Genomic and Biochemical Characterization of Acinetobacter Podophage Petty Reveals a Novel Lysis Mechanism and Tail-Associated Depolymerase Activity. J. Virol. 2018, 92, e01064-17. [Google Scholar] [CrossRef]
- Lin, T.L.; Hsieh, P.F.; Huang, Y.T.; Lee, W.C.; Tsai, Y.T.; Su, P.A.; Pan, Y.J.; Hsu, C.R.; Wu, M.C.; Wang, J.T. Isolation of a bacteriophage and its depolymerase specific for K1 capsule of Klebsiella pneumoniae: Implication in typing and treatment. J. Infect. Dis. 2014, 210, 1734–1744. [Google Scholar] [CrossRef]
- Majkowska-Skrobek, G.; Łątka, A.; Berisio, R.; Maciejewska, B.; Squeglia, F.; Romano, M.; Lavigne, R.; Struve, C.; Drulis-Kawa, Z. Capsule-Targeting Depolymerase, Derived from Klebsiella KP36 Phage, as a Tool for the Development of Anti-Virulent Strategy. Viruses 2016, 8, 324. [Google Scholar] [CrossRef]
- Hsieh, P.F.; Lin, H.H.; Lin, T.L.; Chen, Y.Y.; Wang, J.T. Two T7-like Bacteriophages, K5-2 and K5-4, Each Encodes Two Capsule Depolymerases: Isolation and Functional Characterization. Sci. Rep. 2017, 7, 4624. [Google Scholar] [CrossRef]
- Majkowska-Skrobek, G.; Latka, A.; Berisio, R.; Squeglia, F.; Maciejewska, B.; Briers, Y.; Drulis-Kawa, Z. Phage-Borne depolymerases decrease Klebsiella pneumoniae resistance to innate defense mechanisms. Front. Microbiol. 2018, 9, 2517. [Google Scholar] [CrossRef]
- Pan, Y.J.; Lin, T.L.; Lin, Y.T.; Su, P.A.; Chen, C.T.; Hsieh, P.F.; Hsu, C.R.; Chen, C.C.; Hsieh, Y.C.; Wang, J.T. Identification of capsular types in carbapenem-resistant Klebsiella pneumoniae strains by wzc sequencing and implications for capsule depolymerase treatment. Antimicrob. Agents Chemother. 2015, 59, 1038–1047. [Google Scholar] [CrossRef]
- Solovieva, E.V.; Myakinina, V.P.; Kislichkina, A.A.; Krasilnikova, V.M.; Verevkin, V.V.; Mochalov, V.V.; Lev, A.I.; Fursova, N.K.; Volozhantsev, N.V. Comparative genome analysis of novel Podoviruses lytic for hypermucoviscous Klebsiella pneumoniae of K1, K2, and K57 capsular types. Virus Res. 2018, 243, 10–18. [Google Scholar] [CrossRef]
- Pan, Y.J.; Lin, T.L.; Chen, Y.Y.; Lai, P.H.; Tsai, Y.T.; Hsu, C.R.; Hsieh, P.F.; Lin, Y.T.; Wang, J.T. Identification of three podoviruses infecting Klebsiella encoding capsule depolymerases that digest specific capsular types. Microb. Biotechnol. 2019, 12, 472–486. [Google Scholar] [CrossRef]
- Pan, Y.J.; Lin, T.L.; Chen, C.C.; Tsai, Y.T.; Cheng, Y.H.; Chen, Y.Y.; Hsieh, P.F.; Lin, Y.T.; Wang, J.T. Klebsiella Phage ΦK64-1 Encodes Multiple Depolymerases for Multiple Host Capsular Types. J. Virol. 2017, 91, e02457-16. [Google Scholar] [CrossRef]
- Knirel, Y.A.; Shneider, M.M.; Popova, A.V.; Kasimova, A.A.; Senchenkova, S.N.; Shashkov, A.S.; Chizhov, A.O. Mechanisms of Acinetobacter baumannii Capsular Polysaccharide Cleavage by Phage Depolymerases. Biochemistry 2020, 85, 567–574. [Google Scholar] [CrossRef]
- Lee, I.M.; Tu, I.F.; Yang, F.L.; Ko, T.P.; Liao, J.H.; Lin, N.T.; Wu, C.Y.; Ren, C.T.; Wang, A.H.; Chang, C.M.; et al. Structural basis for fragmenting the exopolysaccharide of Acinetobacter baumannii by bacteriophage ΦAB6 tailspike protein. Sci. Rep. 2017, 17, 42711. [Google Scholar] [CrossRef]
- Kamerling, J.P.; Lindberg, B.; Lönngren, J.; Nimmich, W. Structural studies of the Klebsiella type 57 capsular polysaccharide. Acta Chem. Scand. 1975, 29, 593–598. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Jansson, P.E.; Kenne, L.; Widmalm, G. Computer-assisted structural analysis of polysaccharides with an extended version of CASPER using 1H- and 13C-n.m.r. data. Carbohydr. Res. 1989, 188, 169–191. [Google Scholar] [CrossRef]
- Dutton, G.G.S.; Parolis, H.; Parolis, L.A.S. The structural elucidation of the capsular polysaccharide of Klebsiella K68. Carbohydr. Res. 1986, 152, 249–259. [Google Scholar] [CrossRef]
- Borzilov, A.I.; Myakinina, V.P.; Korobova, O.V.; Kombarova, T.I.; Krasilnikova, V.M.; Verevkin, V.V.; Volozhantsev, N.V. Evaluation of preventive and therapeutic efficacy of Klebsiella pneumoniae bacteriophage vB_KpnP_KpV289 on the model of acute sepsis in mice. Bacteriology 2017, 2, 73–77. [Google Scholar] [CrossRef]
- Borzilov, A.I.; Volozhantsev, N.V.; Korobova, O.V.; Kombarova, T.I.; Myakinina, V.P.; Krasilnikova, V.M.; Verevkin, V.V.; Svetoch, E.A.; Dyatlov, I.A. The effectiveness of bacteriophage KpV289 in treatment of acute pneumonia and a hip infection caused by Klebsiella pneumoniae in mice. Infekc. Bolezni (Infect. Dis.) 2017, 15, 48–56. [Google Scholar] [CrossRef]
- Liu, Y.; Leung, S.; Huang, Y.; Guo, Y.; Jiang, N.; Li, P.; Chen, J.; Wang, R.; Bai, C.; Mi, Z.; et al. Identification of Two Depolymerases From Phage IME205 and Their Antivirulent Functions on K47 Capsule of Klebsiella pneumoniae. Front. Microbiol. 2020, 11, 218. [Google Scholar] [CrossRef] [PubMed]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST server: Rapid annotations using subsystems technology. BMC Genomics 2008, 9, 75. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Sullivan, M.J.; Petty, N.K.; Beatson, S.A. Easyfig: A genome comparison visualizer. Bioinformatics 2011, 27, 1009–1010. [Google Scholar] [CrossRef]
- Söding, J.; Biegert, A.; Lupas, A.N. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005, 33, 244–248. [Google Scholar] [CrossRef]
- Volozhantsev, N.V.; Shpirt, A.M.; Kislichkina, A.A.; Shashkov, A.S.; Verevkin, V.V.; Fursova, N.K.; Knirel, Y.A. Structure and gene cluster of the capsular polysaccharide of multidrug resistant carbapenemase OXA-48-producing Klebsiella pneumoniae strain KPB536 of the genetic line ST147. Res. Microbiol. 2020, 171, 74–79. [Google Scholar] [CrossRef]
- Prokhorov, N.S.; Riccio, C.; Zdorovenko, E.L.; Shneider, M.M.; Browning, C.; Knirel, Y.A.; Leiman, P.G.; Letarov, A.V. Function of bacteriophage G7C esterase tailspike in host cell adsorption. Mol. Microbiol. 2017, 105, 385–398. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).