Abstract
A series of new urea derivatives, containing aryl moieties as potential antimicrobial agents, were designed, synthesized, and characterized by 1H NMR, 13C NMR, FT-IR, and LCMS spectral techniques. All newly synthesized compounds were screened in vitro against five bacterial strains (Escherichia coli, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Staphylococcus aureus) and two fungal strains (Candida albicans and Cryptococcus neoformans). Variable levels of interaction were observed for these urea derivatives. However, and of major importance, many of these molecules exhibited promising growth inhibition against Acinetobacter baumannii. In particular, to our delight, the adamantyl urea adduct 3l demonstrated outstanding growth inhibition (94.5%) towards Acinetobacter baumannii. In light of this discovery, molecular docking studies were performed in order to elucidate the binding interaction mechanisms of the most active compounds, as reported herein.
1. Introduction
Bacterial and fungal diseases have become increasingly more prominent and complex in recent years, particularly when compared to the last half of the twentieth century [1]. As a result, the need for new antibacterial and antifungal agents has become a goal of extreme importance, especially in light of the documented emergence of multi-drug-resistant (MDR) strains in recent years [2,3,4,5,6,7]. These MDR strains already pose a well-recognized health threat to the world population and are frequently associated with increased healthcare cost and prolonged hospital stays. Despite recent advances to understanding the pathogenesis of infection, research laboratories have become increasingly focused on the discovery of new and more effective drug candidates as the MDR strains continue to increase. In particular, various, novel, synthetic small molecules have been synthesized and screened as potential candidates to combat these resistant strains of bacteria and fungi [8,9,10,11,12]. We have been actively involved in the design and discovery of such new bioactive molecules to tackle MDR strains [13,14,15,16,17,18,19,20,21,22] and have reported our most recent studies herein, which include some very promising discoveries.
Since urea functionality is a core moiety in many drug candidates, we have recently commenced a research program focused on the development of potential urea functionalized antimicrobial agents [23]. Our initial research efforts have identified the novel urea derivatives I and II as the initial hits for bacterial strain Staphylococcus aureus and fungal strain Cryptococcus neoformans, respectively, (Figure 1A) [23]. We have also studied the antimicrobial effects of N,N-disubstituted urea derivatives, where one of the urea nitrogen was part of a piperazine ring system (e.g., III which showed moderate inhibition against Candida albicans [24]. Encouraged by these results, we embarked on further exploring novel new urea derivatives for antimicrobial activities, as shown in Figure 1B.
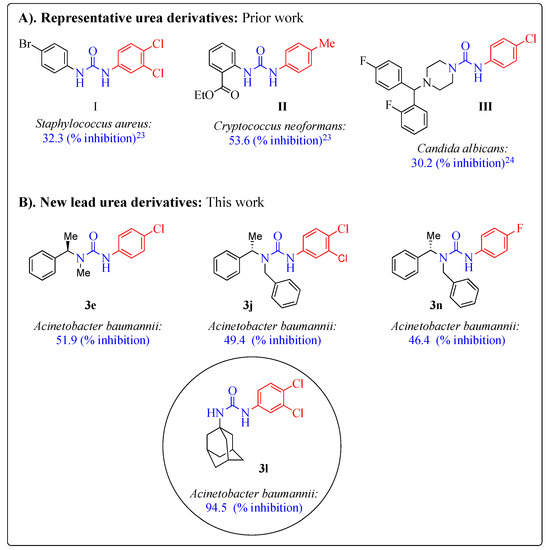
Figure 1.
Antimicrobial urea derivatives.
In this work, we explored the effects of diverse substitutions present on the nitrogen atoms of the urea backbone. For instance, aryl or aliphatic substitutions were studied (Scheme 1). Very interestingly, N,N-disubstituted compounds; (R)-3-(4-chlorophenyl)-1-methyl-1-(1-phenylethyl)urea (3e), (S)-1-benzyl-3-(3,4-dichlorophenyl)-1-(1-phenylethyl)urea (3j), and (S)-1-benzyl-3-(4-fluorophenyl)-1-(1-phenylethyl)urea (3n) demonstrated a good inhibition towards Acinetobacter baumannii. Remarkably, this study identified a new potential lead drug candidate, 1-((3S,5S,7S)-adamantan-1-yl)-3-(3,4-dichlorophenyl)urea (3l) for Acinetobacter baumannii. This compound (3l) exhibited selective and outstanding inhibition (94.5%) towards Acinetobacter baumannii. A unique feature of this compound was that it included the lipophilic adamantane moiety (Figure 1). Notably, this adamantane functionality is extensively used in drug design and is part of several drug candidates [25,26,27,28].
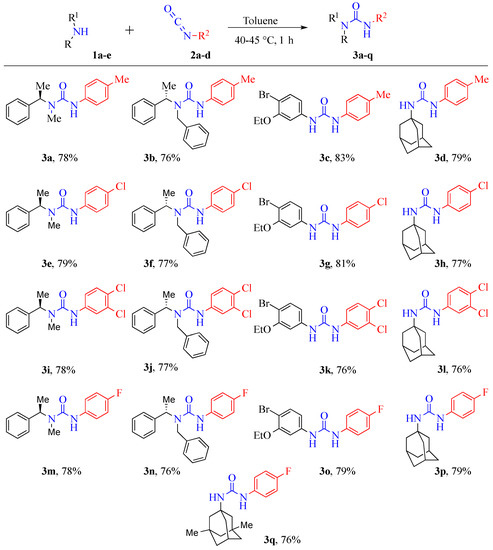
Scheme 1.
Synthesis of urea derivatives (3a–q).
2. Results and Discussion
2.1. Chemistry
The basic synthetic approach for preparation of the seventeen new urea derivatives, containing aryl moieties, is illustrated in Scheme 1. All compounds (3a–q) were successfully synthesized in a simple, one-step method, via the reaction of amines with commercially available isocyanates, at 40–45 °C, in toluene. The attractive features of this method were, (1) simple one-pot procedure, (2) mild reaction conditions, (3) short reaction time, (4) easier work-up, and (5) high yields (76% to 83%).
2.2. Spectroscopic Characterization
The chemical structures of the reported urea derivatives (3a–q) were confirmed by 1H NMR, 13C NMR, FT-IR, and LCMS spectral techniques. The spectral data of the compounds (3a–q) are presented in the experimental section; these were in accordance with assigned structures for urea derivatives containing aryl moieties (Scheme 1). The FT–IR spectra, for the title urea compounds, were recorded within the region of 400 to 4000 cm−1. The IR spectra of all urea derivatives exhibited bands at 3264–3374 and 1648–1626 cm−1 assigned to the ν(NH) and ν(C=O) groups, respectively. IR observed signals at 3028–2905 and 1595–1504 cm−1 might be assigned to the C-H and C=C stretches of the aromatic rings. The chemical structure of all molecules was further confirmed by 1H NMR spectroscopy and further confirmed by the 13C NMR spectra. The 13C NMR spectra of all urea derivatives (3a–q) exhibited a characteristic signal of the urea carbonyl functional group (C=O) between 157.5 and 153.5 ppm. Further confirmation of the molecular structures of the reported compounds was assessed by mass spectra. The molecular ion peak (M + H)+ and the base peak for all urea compounds were clearly found in the mass spectrometry study. Thus, the molecular ion peaks agreed with the molecular weight of the respective compounds.
2.3. Antimicrobial Activity
The in vitro antimicrobial activity of the freshly produced urea derivatives (3a–q) was evaluated against five bacteria [Escherichia coli, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Staphylococcus aureus] and two fungal [Candida albicans and Cryptococcus neoformans] species. Colistin was used as a positive inhibitor standard for Gram-negative bacteria; Vancomycin was used for the Gram-positive bacteria; and Fluconazole was used as a positive fungal inhibitor standard for both fungi. The antimicrobial potential/results of these urea derivatives are summarized in Table 1.

Table 1.
Antimicrobial activity of compounds (3a–q) with the concentration set at 32 μg/mL in DMSO.
The antimicrobial screening disclosed that some of the tested compounds demonstrated excellent to moderate growth inhibition towards various tested microbial strains. The result indicated that among the tested compounds, compounds 3c, 3e, 3f, 3i, 3j, 3l, and 3n showed moderate to excellent growth inhibition against A. baumannii. Further, it was found that compounds (3c and 3g) showed moderate growth inhibition towards K. pneumonia, whereas compound (3k) demonstrated moderate growth inhibition against S. aureus. In contrast, all urea derivatives screened (3a–q) against E. coli, P. aeruginosa, and C. albicans exhibited moderate to poor growth inhibition. Compound (3a) showed moderate growth inhibition against the C. neoformans fungi, whereas the remaining compounds (3b–q) revealed a poor growth inhibition. Overall, the results indicated that the adamantyl urea derivative (3l) showed the highest growth inhibition towards the A. baumannii bacteria (94.5%) (Table 1), a very exciting observation, which suggested development and biological evaluation of other adamantyl urea derivatives.
2.4. Molecular Docking Studies
As noted above, antimicrobial screening revealed that our urea derivatives were superior inhibitors against A. baumannii. Therefore, to investigate the mechanism of this antibacterial action, and explore the intermolecular interactions between the synthesized compounds with the receptor, molecular docking studies were performed on the crystal structure of the A. baumannii PBP1a, in complex with penicillin G (PDB ID 3UDI, 2.6 Å X-ray resolution), using the surflex-dock program of the sybyl-X 2.0 software. All inhibitors, along with the ligand, were docked into the active site of enoyl-(acyl-carrier-protein) reductase (ENR), as shown in Figure 2. The predicted binding energies of the compounds are listed in Table 2. The docking study revealed that all the compounds exhibited very good docking scores against the enzyme.
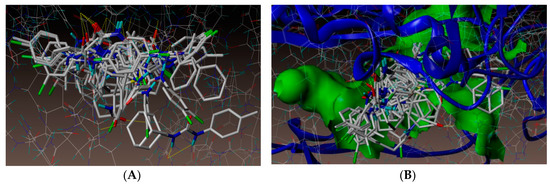
Figure 2.
(A) All compounds docked into the active site of enoyl-(acyl-carrier-protein) reductase (ENR). (B) Docked view of all the compounds at the active site of enzyme PDB ID: 3UDI.

Table 2.
Surflex Docking Score (kcal/mol) of the urea derivatives.
As depicted in Figure 3, compound (3n) makes a hydrogen bonding interaction at the active site of the enzyme (PDB ID: 3UDI), the oxygen atom of carbonyl group interacts with the hydrogen atom of SER487 (C=O------H-SER487, 1.99 Å). As depicted in Figure 4, compound (3j) makes a hydrogen bonding interaction at the active site of the enzyme (PDB ID: 3UDI), oxygen atom of the carbonyl group interacts with the hydrogen atom of THR670 (C=O------H-THR670, 2.14 Å). The binding interaction of 3UDI_ligand with enzyme active sites shows six bonding interactions and the docked view of the same; as depicted in Figure 5. Figure 6A,B represents the hydrophobic and hydrophilic amino acids surrounded to the studied compound (3n and 3j).
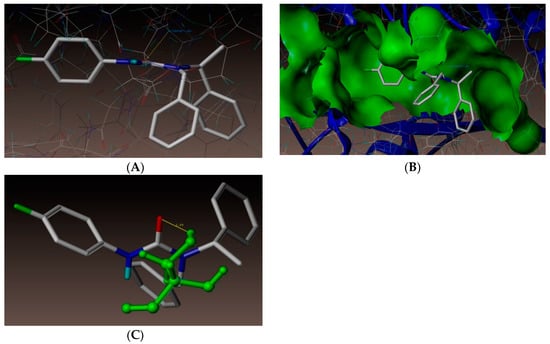
Figure 3.
(A) Docked view of compound 3n. (B) Docked view of compound 3n at the active site of enzyme PDB ID: 3UDI. (C) Hydrogen-bond interaction of compound 3n at the active site.
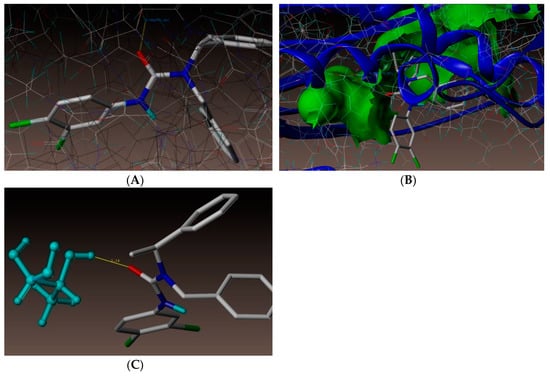
Figure 4.
(A) Docked view of compound 3j. (B) Docked view of compound 3j at the active site of enzyme PDB ID: 3UDI. (C) Hydrogen-bond interaction of compound 3j at the active site.
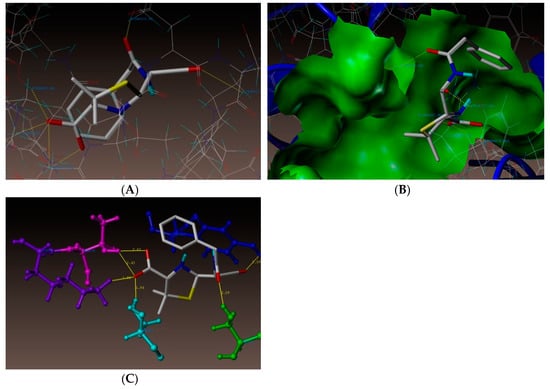
Figure 5.
(A) Docked views of 3UDI_ligand. (B) Docked view of 3UDI_ligand at the active site of enzyme PDB ID: 3UDI. (C) Neighboring interactions of 3UDI_ligand.
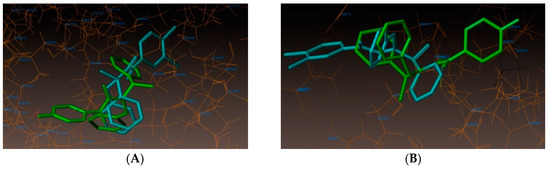
Figure 6.
(A) Hydrophobic amino acids surrounded to compounds 3n (green color) and 3j (cyan color). (B) Hydrophilic amino acids surrounded by compounds 3n and 3j.
All compounds showed consensus scores in the range 5.19–2.91, indicating the summary of all forces of interaction between ligands and the enzyme. Moreover, it was observed that the studied compounds displayed the same type of interactions with amino acid residues SER487 and THR670, as that of the reference 3UDI_ligand. This indicated that molecules preferentially bound to the enzyme in comparison to the reference 3UDI_ligand (Table 2).
As documented above, new urea derivative (3l) showed highest growth inhibition against A. baumannii. Hence, a molecular docking study was also performed for 3l. As represented in Figure 7, compound 3l showed hydrogen bonding interaction at the active site of the enzyme (PDB ID: 3UDI). The adamantane ring was surrounded by hydrophobic amino acids ALA537, ILE566, GLY675, ILE555, PHE554, ALA682, and VAL565 (Figure 8), which might have resulted in the increased activity of compound (3l). Superimposition of 3l (magenta color) with benzyl penicillin (purple color) has been depicted in Figure 9. As shown in the figure, the adamantane moiety was superimposed with β-lactam and thiazolidine rings.
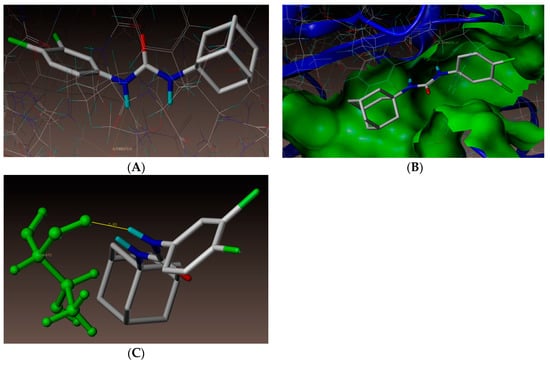
Figure 7.
(A) Docked view of compound 3l. (B) Docked view of compound 3l at the active site of enzyme PDB ID: 3UDI. (C) Hydrogen-bond interaction of compound 3l at the active site.
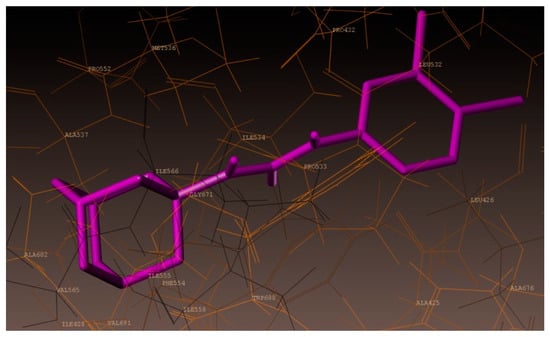
Figure 8.
Hydrophobic amino acids surrounding compound 3l (magenta color).
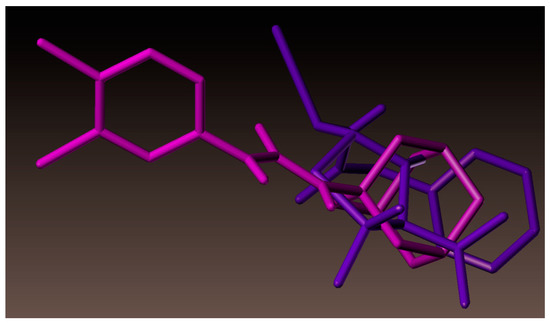
Figure 9.
Superimposition of compound 3l (magenta color) with benzyl penicillin (purple color).
3. Materials and Methods
3.1. General Consideration
All chemicals, including isocyanates, were purchased from Sigma-Aldrich chemical company and were used without further purification. All solvents were of analytical grade and were used without further purification. All reactions were carried out under aerobic conditions, in oven-dried glassware with magnetic stirring. Heating was accomplished by either a heating mantle or silicone oil bath. Reactions were monitored by thin-layer chromatography (TLC) performed on 0.25 mm Merck TLC silica gel plates, using UV light as a visualizing agent. Purification of the reaction products was carried out by flash column chromatography using silica gel 60 (230–400 mesh). Yields refer to the chromatographically pure material. Concentration in vacuo refers to the removal of volatile solvent using a rotary evaporator attached to a dry diaphragm pump (10–15 mm Hg), followed by pumping to a constant weight with an oil pump (<300 mTorr). 1H spectra were recorded on JEOL Eclipse Plus 500 (500 MHz) and were reported relative to CDCl3 (δ 7.26) or DMSO-d6 (δ 2.50). 1H NMR coupling constants (J) were reported in Hertz (Hz) and multiplicities were indicated as follows—s (singlet), d (doublet), t (triplet), quint (quintet), m (multiplet). Proton-decoupled 13C NMR spectra were recorded on the JEOL Eclipse Plus 500 (125 MHz) and were reported relative to CDCl3 (δ 77.00) or DMSO-d6 (δ 39.52). IR spectra were recorded on an Alpha-P Bruker FT/IR spectrometer. Liquid chromatography mass spectra (LC-MS) were recorded on Agilent technologies quadrupole LC–MS system.
3.2. Syntheses
3.2.1. General Experimental Procedure for the Synthesis of Urea Derivatives
To a solution of isocyanate (1.877 mmol) in toluene (2.5 mL) a solution of amine (1.877 mmol) in toluene (1.0 mL) was added. The reaction mixture was heated at 40–45 °C for 1 h. Then, the reaction mixture was cooled down to the room temperature. The resulting solids were filtered and washed with toluene (2.0 mL). Additional toluene (2.5 mL) was added to the solids and stirred at room temperature for about 30 minutes, filtered, and washed with more toluene (2.0 mL) to obtain the crude urea derivatives. Finally, the crude urea derivatives were purified by silica gel flash column chromatography, using hexane/ethyl acetate (9:1) as the eluents, to afford pure adducts. 1H and 13C NMR data of all the compounds (3a–q) in Supplementary Materials.
Synthesis of (R)-1-methyl-1-(1-phenylethyl)-3-(p-tolyl)urea (3a)
Compound (3a) was synthesized from 4-methylphenyl isocyanate (0.25 g, 1.87 mmol) and (R)-(+)-N, α-dimethylbenzylamine (0.25 g, 1.87 mmol), according to the general procedure. It was a white solid; yield— 78% (0.39 g). 1H NMR (CDCl3, 500 MHz): δ 7.39–7.36 (m, 4H), 7.29 (d, J = 8.0 Hz, 3H), 7.09 (d, J = 8.0 Hz, 2H), 6.53 (br, 1H), 5.70 (q, J = 6.9 Hz, 1H), 2.73 (s, 3H), 2.30 (s, 3H), 1.56 (d, J = 6.9 Hz, 3H). 13C NMR (CDCl3, 125 MHz): δ 155.5, 140.9, 136.3, 132.6, 129.3, 128.5, 127.3, 127.1, 120.0, 52.6, 29.4, 20.7, 16.6. IR (KBr): ῡ = 3304.2, 2923.4, 1633.9, 1595.6, 1291.7, 811.2.LC-MS for C17H20N2O (268.35): m/z = 269.32 [M + H]+.
Synthesis of (S)-1-benzyl-1-(1-phenylethyl)-3-(p-tolyl)urea (3b)
Compound (3b) was synthesized from 4-methylphenyl isocyanate (0.25 g, 1.87 mmol) and (S)-(-)-N-benzyl-α-methylbenzylamine (0.39 g, 1.87 mmol), according to the general procedure. It was a white solid; yield—76% (0.49 g). 1H NMR (DMSO-d6, 500 MHz): δ 8.26 (s, 1H), 7.38–7.33 (m, 4H), 7.30 (d, J = 8.6 Hz, 2H), 7.28–7.24 (m, 3H), 7.20–7.15 (m, 3H),7.02 (d, J = 8.0 Hz, 2H), 5.72 (q, J = 7.5 Hz, 1H), 4.68 (d, J = 17.8 Hz, 1H), 4.16 (d, J = 17.2 Hz, 1H), 2.21 (s, 3H), 1.44 (d, J = 7.5 Hz, 3H). 13C NMR (DMSO-d6, 125 MHz): δ 155.7, 142.0, 139.9, 137.8, 130.7, 128.7, 128.4, 128.1, 127.1, 127.0, 126.5, 126.4, 120.0, 52.9, 45.4, 20.3, 17.9. IR (KBr): ῡ = 3353.6, 2918.3, 1633.2, 1511.5, 1239.1, 816.3. LC-MS for C23H24N2O (344.45): m/z = 345.43 [M + H]+.
Synthesis of 1-(4-bromo-3-ethoxyphenyl)-3-(p-tolyl)urea (3c)
Compound (3c) was synthesized from 4-methylphenyl isocyanate (0.25 g, 1.87 mmol) and 4-bromo-3-ethoxyaniline (0.40 g, 1.87 mmol), according to the general procedure. It was a white solid; yield—83% (0.54 g). 1H NMR (DMSO-d6, 500 MHz): δ 8.76 (s, 1H), 8.58 (s, 1H), 7.41 (d, J = 8.6 Hz, 1H), 7.39 (d, J = 2.9 Hz, 1H), 7.33 (d, J = 8.6 Hz, 2H), 7.08 (d, J = 8.0 Hz, 2H), 6.87 (dd, J = 8.6, 2.3 Hz, 1H), 4.06 (q, J = 6.9 Hz, 1H), 2.24 (s, 3H), 1.36 (t, J = 7.6 Hz, 1H). 13C NMR (DMSO-d6, 125 MHz): δ 154.8, 152.5, 140.7, 136.9, 132.6, 130.9, 129.2, 118.5, 111.4, 103.6, 102.4, 64.1, 20.4, 14.6. IR (KBr): ῡ = 3340.0, 2983.6, 1648.7, 1321.5, 1045.4, 821.9. LC-MS for C16H17BrN2O2 (349.22): m/z = 350.21 [M + H]+.
Synthesis of 1-((3s,5s,7s)-adamantan-1-yl)-3-(p-tolyl)urea (3d)
Compound (3d) was synthesized from 4-methylphenyl isocyanate (0.25 g, 1.87 mmol) and 1-amino adamantane (0.28 g, 1.87 mmol), according to the general procedure. It was a white solid; yield—79% (0.42 g). 1H NMR (DMSO-d6, 500 MHz): δ 8.11 (s, 1H), 7.21 (d, J = 8.6 Hz, 2H), 6.99 (d, J = 8.0 Hz, 2H), 5.80 (s, 1H), 2.20 (s, 3H), 2.02 (s, 3H), 1.95–1.90 (m, 6H), 1.62 (s, 6H). 13C NMR (DMSO-d6, 125 MHz): δ 154.1, 138.1, 129.4, 129.0, 117.4, 49.8, 41.7, 36.1, 28.9, 20.3. IR (KBr): ῡ = 3324.0, 2908.2, 2884.3, 1642.4, 1556.6, 1235.8, 810.3.LC-MS for C18H24N2O (284.40): m/z = 285.39 [M + H]+.
Synthesis of (R)-3-(4-chlorophenyl)-1-methyl-1-(1-phenylethyl)urea (3e)
Compound (3e) was synthesized from 4-chlorophenyl isocyanate (0.25 g, 1.62 mmol) and (R)-(+)-N, α-dimethylbenzylamine (0.22 g, 1.62 mmol), according to the general procedure. It was a white solid; yield—79% (0.37 g). 1H NMR (DMSO-d6, 500 MHz): δ 8.48 (s, 3H), 7.59–7.56 (m, 2H), 7.36 (t, J = 7.5 Hz, 2H), 7.33–7.25 (m, 5H), 5.64 (q, J = 6.9 Hz, 1H), 2.67 (s, 3H), 1.48 (d, J = 6.9 Hz, 3H). 13C NMR (DMSO-d6, 125 MHz): δ 155.5, 141.5, 139.7, 128.4, 128.1, 126.9, 126.8, 125.3, 121.3, 51.3, 28.7, 16.4. IR (KBr): ῡ = 3306.3, 3028.5, 1638.3, 1518.7, 1241.5, 821.0. LC-MS for C16H17ClN2O (288.77): m/z = 289.65 [M + H]+.
Synthesis of (S)-1-benzyl-3-(4-chlorophenyl)-1-(1-phenylethyl)urea (3f)
Compound (3f) was synthesized from 4-chlorophenyl isocyanate (0.25 g, 1.62 mmol) and (S)-(-)-N-benzyl-α-methylbenzylamine (0.34 g, 1.62 mmol), according to the general procedure. It was a white solid; yield—77% (0.45 g). 1H NMR (DMSO-d6, 500 MHz): δ 8.55 (s, 1H), 7.48 (d, J = 9.2 Hz, 2H), 7.38–7.33 (m, 4H), 7.28–7.33 (m, 5H), 7.18–7.15 (m, 3H), 5.71 (q, J = 7.5 Hz, 1H), 4.68 (d, J = 17.2 Hz, 1H), 4.17 (d, J = 17.2 Hz, 1H), 1.45 (d, J = 6.9 Hz, 3H). 13C NMR (DMSO-d6, 125 MHz): δ 155.5, 141.8, 139.6, 139.4, 128.4, 128.1 (2C), 127.1, 127.0, 126.4 (2C), 125.5, 121.3, 53.1, 45.5, 17.9. IR (KBr): ῡ =3373.7, 2972.8, 2934.0, 1636.8, 1520.7, 1232.3, 829.9. LC-MS for C22H21ClN2O (364.87): m/z = 365.83 [M + H]+.
Synthesis of 1-(4-bromo-3-ethoxyphenyl)-3-(4-chlorophenyl)urea (3g)
Compound (3g) was synthesized from 4-chlorophenyl isocyanate (0.25 g, 1.62 mmol) and 4-bromo-3-ethoxyaniline (0.351g, 1.62 mmol), according to the general procedure. It was a white solid; yield—81% (0.48 g). 1H NMR (DMSO-d6, 500 MHz): δ 8.85 (s, 1H), 8.84 (s, 1H), 7.49 (d, J = 9.2 Hz, 2H), 7.43-7.38 (m, 2H), 7.32 (d, J = 9.2 Hz, 2H), 6.89 (dd, J = 8.6, 2.3 Hz, 1H), 4.06 (q, J = 6.9 Hz, 2H), 1.36 (q, J = 6.9 Hz, 3H). 13C NMR (DMSO-d6, 125 MHz): δ 154.8, 152.3, 140.4, 138.5, 132.6, 128.6, 125.6, 119.9, 111.6, 103.8, 102.8, 64.2, 14.6. IR (KBr): ῡ = 3295.0, 2984.5, 2887.0, 1645.0, 1555.8, 1196.8, 824.2. LC-MS for C15H14BrClN2O2 (369.64): m/z = 370.62 [M + H]+.
Synthesis of 1-((3S,5S,7S)-adamantan-1-yl)-3-(4-chlorophenyl)urea (3h)
Compound (3h) was synthesized from 4-chlorophenyl isocyanate (0.25 g, 1.62 mmol) and 1-amino adamantane (0.24 g, 1.62 mmol), according to the general procedure. It was a white solid; yield—77% (0.38 g). 1H NMR (DMSO-d6, 500 MHz): δ 8.37 (s, 1H), 7.36 (d, J = 8.6 Hz, 2H), 7.22 (d, J = 8.6 Hz, 2H), 5.88 (s, 1H), 2.01 (s, 3H), 1.95–1.90 (m, 6H), 1.62 (s, 6H). 13C NMR (DMSO-d6, 125 MHz): δ 153.8, 139.6, 128.4, 124.1, 118.8, 49.9, 41.6, 36.0, 28.9. IR (KBr): ῡ = 3327.3, 2905.4, 2847.8, 1644.5, 1554.0, 1231.6, 815.2.LC-MS for C17H21ClN2O (304.81): m/z = 305.78 [M + H]+.
Synthesis of (R)-3-(3,4-dichlorophenyl)-1-methyl-1-(1-phenylethyl)urea (3i)
Compound (3i) was synthesized from 3,4-dichlorophenyl isocyanate (0.25 g, 1.32 mmol) and (R)-(+)-N, α-dimethylbenzylamine (0.17 g, 1.32 mmol), according to the general procedure. It was a white solid; yield—78% (0.335 g). 1H NMR (DMSO-d6, 500 MHz): δ 8.64 (s, 1H), 7.93 (d, J = 2.3 Hz, 1H), 7.54 (dd, J = 8.6, 2.3 Hz, 1H), 7.47 (d, J = 9.2 Hz, 1H), 7.36 (t, J = 7.5 Hz, 2H), 7.30 (d, J = 7.5 Hz, 2H), 7.26 (d, J = 6.9 Hz, 1H), 5.63 (q, J = 6.9 Hz, 1H), 2.67 (s, 3H), 1.48 (d, J = 6.9 Hz, 3H). 13C NMR (DMSO-d6, 125 MHz): δ 155.2, 141.3, 141.0, 130.5, 130.1, 128.4, 127.0, 126.9, 122.9, 120.7, 119.5, 51.4, 28.8, 16.3. IR (KBr): ῡ = 3263.3, 2982.5, 1636.0, 1580.8, 1236.1, 817.8. LC-MS for C16H16Cl2N2O (323.22): m/z = 324.15 [M + H]+.
Synthesis of (S)-1-benzyl-3-(3,4-dichlorophenyl)-1-(1-phenylethyl)urea (3j)
Compound (3j) was synthesized from 3,4-dichlorophenyl isocyanate (0.25g, 1.32 mmol) and (S)-(-)-N-benzyl-α-methylbenzylamine (0.28g, 1.32 mmol), according to the general procedure. It was a pale yellow liquid; yYield—77% (0.40 g). 1H NMR (DMSO-d6, 500 MHz): δ 8.75 (s, 1H), 8.32 (s, 1H), 7.86 (s, 1H), 7.45 (d, J = 1.2 Hz, 2H), 7.38–7.32 (m, 4H), 7.29–7.24 (m, 3H), 7.16 (d, J = 8.6 Hz, 3H), 5.69 (q, J = 6.9 Hz, 1H), 4.68 (d, J = 17.2 Hz, 1H), 4.19 (d, J = 17.2 Hz, 1H), 1.45 (d, J = 7.5 Hz, 3H). 13C NMR (DMSO-d6, 125 MHz): δ 155.2, 141.6, 140.7, 139.4, 130.6, 130.1, 128.5, 128.2, 127.2, 127.0, 126.5, 126.4, 123.1, 120.7, 119.6, 53.2, 45.6, 17.8. IR (KBr): ῡ = 3330.0, 2976.4, 2876.1, 1638.8, 1510.2, 1232.4, 750.7. LC-MS for C22H20Cl2N2O (399.31): m/z = 400.27 [M + H]+.
Synthesis of 1-(4-bromo-3-ethoxyphenyl)-3-(3,4-dichlorophenyl)urea (3k)
Compound (3k) was synthesized from 3,4-dichlorophenyl isocyanate (0.25 g, 1.32 mmol) and 4-bromo-3-ethoxyaniline (0.28 g, 1.32 mmol), according to the general procedure. It was a white solid; yield—76% (0.405 g). 1H NMR (DMSO-d6, 500 MHz): δ 9.01 (s, 1H), 8.94 (s, 1H), 7.88 (d, J = 2.3 Hz, 1H), 7.50 (d, J = 9.2 Hz, 1H), 7.42 (d, J = 8.6 Hz, 1H), 7.38 (d, J = 2.3 Hz, 1H), 7.32 (dd, J = 8.9, 2.3 Hz, 1H), 6.89 (dd, J 8.6, 2.3 Hz, 1H), 4.06 (q, J = 6.9 Hz, 2H), 1.36 (t, J = 7.5 Hz, 3H). 13C NMR (DMSO-d6, 125 MHz): δ 154.8, 152.2, 140.2, 139.8, 132.7, 131.1, 130.6, 123.3, 119.4, 118.5, 111.7, 103.9, 103.0, 64.2, 14.6. IR (KBr): ῡ = 3292.3, 2982.6, 2928.9, 1638.4, 1544.2, 1231.2, 800.2. LC-MS for C15H13BrCl2N2O2 (404.09): m/z = 405.10 [M + H]+.
Synthesis of 1-((3S,5S,7S)-adamantan-1-yl)-3-(3,4-dichlorophenyl)urea (3l)
Compound (3l) was synthesized from 3,4-dichlorophenyl isocyanate (0.25 g, 1.32 mmol) and 1-amino adamantane (0.20 g, 1.32 mmol), according to the general procedure. It was a white solid; yield—76% (0.34 g). 1H NMR (DMSO-d6, 500 MHz): δ 8.55 (s, 1H), 7.83 (d, J = 2.3 Hz, 1H), 7.40 (d, J = 8.6 Hz, 1H), 7.11 (dd, J = 8.9, 1.7 Hz, 1H), 5.96 (s, 1H), 2.01 (s, 3H), 1.95–1.88 (m, 6H), 1.62 (s, 6H). 13C NMR (DMSO-d6, 125 MHz): δ 153.5, 140.8, 131.0, 130.4, 121.9, 118.3, 117.4, 50.1, 41.5, 36.0, 28.9. IR (KBr): ῡ = 3331.1, 2905.5, 2850.5, 1646.7, 1548.3, 1226.7, 808.5. LC-MS for C17H20Cl2N2O (339.26): m/z = 340.15 [M + H]+.
Synthesis of (R)-3-(4-fluorophenyl)-1-methyl-1-(1-phenylethyl)urea (3m)
Compound (3m) was synthesized from 4-fluorophenyl isocyanate (0.25 g, 1.82 mmol) and (R)-(+)-N, α-dimethylbenzylamine (0.24 g, 1.82 mmol), according to the general procedure. It was a white solid; yield—78% (0.38 g). 1H NMR (DMSO-d6, 500 MHz): δ 8.39 (s, 1H), 7.56–7.53 (m, 2H), 7.39–7.25 (m, 5H), 7.08 (t, J = 9.2 Hz, 2H), 5.66 (q, J = 6.9 Hz, 1H), 2.67 (s, 3H), 1.48 (d, J = 6.9 Hz, 3H). 13C NMR (DMSO-d6, 125 MHz): δ 157.4 (d, 1JC,F = 238.7 Hz), 155.7, 141.6, 137.0, 128.4, 126.9, 126.8, 121.7 (d, 3JC,F = 8.4 Hz), 114.7 (d, 2JC,F = 22.8 Hz), 51.3, 28.7, 16.4. IR (KBr): ῡ = 3264.8, 3067.4, 1634.8, 1507.9, 1212.5, 827.7. LC-MS for C16H17FN2O (272.32): m/z = 273.30 [M + H]+.
Synthesis of (S)-1-benzyl-3-(4-fluorophenyl)-1-(1-phenylethyl)urea (3n)
Compound (3n) was synthesized from 4-fluorophenyl isocyanate (0.25 g, 1.82 mmol) and (S)-(-)-N-benzyl-α-methylbenzylamine (0.38 g, 1.82 mmol), according to the general procedure. It was a white solid; yield—76% (0.479 g). 1 H NMR (DMSO-d6, 500 MHz): δ 8.46 (s, 1H), 7.45 (dd, J = 8.6, 5.2 Hz, 2H), 7.36 (d, J = 4.0 Hz, 4H), 7.26 (t, J = 7.5 Hz, 3H), 7.22–7.16 (m, 3H), 7.06 (t, J = 9.2 Hz, 2H), 5.73 (q, J = 6.9 Hz, 1H), 4.70 (d, J = 17.2 Hz, 1H), 4.17 (d, J = 17.2 Hz, 1H), 1.45 (d, J = 6.9 Hz, 3H). 13C NMR (DMSO-d6, 125 MHz): δ 157.5 (d, 1JC,F = 237.5 Hz), 155.7, 141.9, 139.8, 136.7, 128.4, 128.1, 127.1, 127.0, 126.4, 126.3, 121.7 (d, 3JC,F = 7.2 Hz), 114.7 (d, 2JC,F = 22.8 Hz), 53.0, 45.5, 17.9. IR (KBr): ῡ = 3306.6, 2929.4, 2875.6, 1633.9, 1507.4, 1208.3, 828.1. LC-MS for C22H21FN2O (348.41): m/z = 349.36 [M + H]+.
Synthesis of 1-(4-bromo-3-ethoxyphenyl)-3-(4-fluorophenyl)urea (3o)
Compound (3o) was synthesized from 4-fluorophenyl isocyanate (0.25 g, 1.82 mmol) and 4-bromo-3-ethoxyaniline (0.39 g, 1.82 mmol), according to the general procedure. It was a white solid; yield—79% (0.64 g). 1H NMR (DMSO-d6, 500 MHz): δ 8.81 (s, 1H), 8.73 (s, 1H), 7.48–7.45 (m, 2H), 7.41 (d, J = 8.6 Hz, 1H), 7.38 (d, J = 2.3 Hz, 1H), 7.14–7.09 (m, 2H), 6.89 (dd, J = 8.6, 2.3 Hz, 1H), 4.06 (q, J = 6.9 Hz, 2H), 1.36 (d, J = 6.9 Hz, 3H). 13C NMR (DMSO-d6, 125 MHz): δ 157.5 (d, 1JC,F = 238.7 Hz), 154.8, 152.5, 140.6, 135.8, 132.6, 120.2 (d, 3JC,F = 7.2 Hz), 115.3 (d, 2JC,F = 22.8 Hz), 111.5, 103.7, 102.6, 64.2, 14.6. IR (KBr): ῡ = 3284.9, 2983.7, 2900.8, 1626.6, 1504.4, 1212.5, 805.5. LC-MS for C15H14BrFN2O2 (353.19): m/z = 354.16 [M + H]+.
Synthesis of 1-((3S,5S,7S)-adamantan-1-yl)-3-(4-fluorophenyl)urea (3p)
Compound (3p) was synthesized from 4-fluorophenyl isocyanate (0.25 g, 1.82 mmol) and 1-amino adamantane (0.27 g, 1.82 mmol), according to the general procedure. It was a white solid; yield—79% (0.41 g). 1H NMR (DMSO-d6, 500 MHz): δ 8.26 (s, 1H), 7.35–7.32 (m, 2H), 7.04–6.99 (m, 2H), 5.82 (s, 1H), 2.01 (s, 3H), 1.96–1.89 (m, 6H), 1.62 (s, 6H). 13C NMR (DMSO-d6, 125 MHz): δ 156.7 (d, 1JC,F = 237.5 Hz), 154.0, 137.0, 118.8 (d, 3JC,F = 7.2 Hz), 115.0 (d, 2JC,F = 22.8 Hz), 49.8, 41.7, 36.1, 28.9. IR (KBr): ῡ = 3342.7, 2906.5, 2851.6, 1646.6, 1508.2, 1208.7, 828.2. LC-MS for C17H21FN2O (288.36): m/z = 289.33 [M + H]+.
Synthesis of 1-((3R,5S,7R)-3,5-dimethyladamantan-1-yl)-3-(4-fluorophenyl)urea (3q)
Compound (3q) was synthesized from 4-fluoro phenyl isocyanate (0.25g, 1.82 mmol) and 1-amino-3,5-dimethyladamantane (0.32 g, 1.82 mmol), according to the general procedure. It was a white solid; yield—76% (0.43 g). 1H NMR (DMSO-d6, 500 MHz): δ 8.25 (s, 1H), 7.36–7.32 (m, 2H), 7.04–6.99 (m, 2H), 5.84 (s, 1H), 2.08–2.06 (m, 1H), 1.75 (d, J = 2.9 Hz, 2H), 1.57 (s, 4H), 1.32 (d, J = 12.6 Hz, 2H), 1.24 (d, J = 12.6 Hz, 2H), 1.10 (s, 2H), 0.81 (s, 6H). 13C NMR (DMSO-d6, 125 MHz): δ 156.7 (d, 1JC,F = 237.5 Hz), 154.1, 137.0, 118.8 (d, 3JC,F = 7.2 Hz), 115.0 (d, 2JC,F = 21.2 Hz), 51.5, 50.3, 47.8, 42.3, 40.2, 31.9, 30.1, 29.6. IR (KBr): ῡ = 3305.8, 2947.6, 2894.7, 1648.7, 1553.8, 1212.9, 830.2. LC-MS for C19H25FN2O (316.41): m/z = 317.35 [M + H]+.
3.3. Antimicrobial Studies
Samples were prepared in DMSO and water to a final testing concentration of 32 μg/mL or 20 μM (unless otherwise indicated in the datasheet), in 384-well, non-binding surface plate (NBS) for each bacterial/fungal strain, in duplicates (n = 2); the final DMSO concentration was kept to a maximum of 1% DMSO [29,30,31,32,33]. All sample preparations for the antimicrobial studies were done using liquid handling robots.
3.3.1. Antimicrobial Assay
Primary antimicrobial screening study, by whole cell growth inhibition assays, were conducted using the compounds (3a–q) at a single concentration, in duplicates (n = 2). The inhibition of growth was measured against five bacteria—Escherichia coli (E. coli) ATCC 25922, Klebsiella pneumonia (K. pneumoniae) ATCC 700603, Acinetobacter baumannii (A. Baumannii) ATCC 19606, Pseudomonas aeruginosa (P. aeruginosa) ATCC 27853) and Staphylococcus aureus (S. aureus) ATCC 43300, and two fungi—Candida albicans ATCC 90028 and Cryptococcus neoformans ATCC 208821 [34].
Procedure
All bacteria were cultured in Cation-adjusted Mueller Hinton broth (CAMHB) at 37 °C, overnight. A sample of each culture was then diluted 40-fold in fresh broth and incubated at 37 °C for 1.5–3 h. The resultant mid-log phase cultures were diluted (CFU/mL measured by OD600) and then added to each well of the compound containing plates, giving a cell density of 5 × 105 CFU/mL and a total volume of 50 μL. All plates were covered and incubated at 37 °C for 18 h without shaking.
Analysis
Inhibition of bacterial growth was determined by measuring absorbance at 600 nm (OD600), using a Tecan M1000 Pro monochromator plate reader. The percentage of growth inhibition was calculated for each well, using negative control (media only) and positive control (bacteria without inhibitors) on the same plate as references. The significance of the inhibition values was determined by modified Z-scores, calculated using the median and median absolute deviation (MAD) of the samples (no controls) on the same plate. Samples with inhibition value above 80% and a Z-Score above 2.5 for either replicate (n = 2 on different plates) were classed as actives. Samples with inhibition values between 50–80% and Z-Score above 2.5 for either replicate (n = 2 on different plates) were classed as partial actives.
3.3.2. Antifungal Assay
Procedure
Fungi strains were cultured for 3 days on Yeast Extract–Peptone–Dextrose (YPD) agar at 30 °C. A yeast suspension of 1 × 106 to 5 × 106 CFU/mL (as determined by OD530) was prepared from five colonies. The suspension was subsequently diluted and added to each well of the compound-containing plates, giving a final cell density of the fungi suspension of 2.5 × 103 CFU/mL and a total volume of 50 μL. All plates were covered and incubated at 35 °C for 24 h without shaking.
Analysis
Growth inhibition of C. albicans was determined by measuring absorbance at 530 nm (OD530), while the growth inhibition of C. neoformans was determined measuring the difference in absorbance between 600 and 570 nm (OD600–570), after the addition of resazurin (0.001% final concentration) and incubation at 35 °C for additional 2 h. The absorbance was measured using a Biotek Synergy HTX plate reader. The percentage of growth inhibition was calculated for each well, using a negative control (media only) and a positive control (fungi without inhibitors) on the same plate. The significance of the inhibition values was determined by the modified Z-scores, calculated using the median and the MAD of the samples (no controls) on the same plate. Samples with inhibition values above 80% and a Z-Score above 2.5 for either replicates (n = 2 on different plates) were classed as actives. Samples with inhibition values between 50–80% and Z-Score above 2.5 for either replicates (n = 2 on different plates) were classed as partial actives.
3.4. Docking Simulations
Molecular docking was used to clarify the binding mode of the compounds to provide straight forward information for further structural optimization. The crystal structure of A. baumannii PBP1a in complex with penicillin G (PDB ID 3UDI, 2.6 Å X-ray resolution) was extracted from the Brookhaven Protein Database (PDB http://www.rcsb.org/pdb). The proteins were prepared for docking by adding polar hydrogen atom with Gasteiger–Huckel charges and the water molecules were removed. The 3D structure of the ligands was generated by the SKETCH module implemented in the SYBYL program (Tripos Inc., St. Louis, USA) and its energy-minimized confirmation was obtained with the help of the Tripos force field using Gasteiger–Huckel [35] charges. Molecular docking was performed with the Surflex-Dock program, which was interfaced with Sybyl-X 2.0 [36], and other miscellaneous parameters were assigned with the default values given by the software.
4. Conclusions
In conclusion, we synthesized seventeen new urea derivatives (3a–q) with a simple one-step method in short reaction time (1 h) and with good yields (76–83%). The structures of all synthesized compounds were confirmed by IR, 1H NMR, 13C NMR, and mass spectroscopic techniques. The newly synthesized compounds were evaluated for their antimicrobial activity. Compounds 3c, 3e, 3f, 3i, 3j, 3l, and 3n exhibited moderate to excellent growth inhibition against the A. baumannii bacterial strain. Among these seven compounds, compound 3l displayed the highest growth inhibition towards the Gram-negative bacteria, A. baumannii (94.5%). Therefore, this 1-adamantyl urea could be considered as a promising antimicrobial lead and could form the structural backbone for further design and synthesis of other urea derivatives to be developed and screened. Furthermore, molecular docking studies of all synthesized compounds were carried out and good docking score towards A. baumannii were observed for all reported urea compounds.
Supplementary Materials
Supplementary data [1H and 13C NMR data of all the compounds (3a–q)]. Supplementary data to this article can be found online at https://www.mdpi.com/2079-6382/8/4/178/s1.
Author Contributions
Conceptualization, S.A.P. (Siddappa A. Patil) and A.B.; methodology, M.P.; software, S.D.J.; validation, S.A.P. (Shivaputra A. Patil), and A.B.; formal analysis, A.N.-P.; investigation, M.P. and A.N.-P.; resources, A.B. and S.A.P. (Siddappa A. Patil); data curation, A.B. and S.A.P. (Siddappa A. Patil); writing—original draft preparation, A.B. and S.A.P. (Siddappa A. Patil); writing—review and editing, A.B.; visualization, S.D.J., A.B., and S.A.P. (Siddappa A. Patil); supervision, A.B. and S.A.P. (Siddappa A. Patil); project administration, A.B. and S.A.P. (Siddappa A. Patil); funding acquisition, A.B. and S.A.P. (Siddappa A. Patil).
Funding
Florida Gulf Coast University partially supported this work. This research work also received funding from the American Chemical Society Petroleum Research Fund (grant # 58269-ND1). Author S.A.P. thanks the DST-Nanomission, India (SR/NM/NS-20/2014), DST-SERB, India [SERB/F/1423/2017–18 (File No. YSS/2015/000010)] and Jain University, India for financial support received. The antimicrobial screening was performed by CO-ADD (The Community for Antimicrobial Drug Discovery), funded by the Welcome Trust (UK) and The University of Queensland (Australia).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Casadevall, A. Fungal Diseases in the 21st century: The near and far horizons. Pathog. Immun. 2018, 3, 183–196. [Google Scholar] [CrossRef]
- Gould, I.M. Coping with antibiotic resistance: The impending crisis. Int. J. Antimicrob. Agents 2010, 36, S1–S2. [Google Scholar] [CrossRef]
- Piddock, L.J. The crisis of no new antibiotics—What is the way forward? Lancet Infect. Dis. 2012, 12, 249–253. [Google Scholar] [CrossRef]
- Padiyara, P.; Inoue, H.; Sprenger, M. Global governance mechanisms to address antimicrobial resistance. Infect. Dis. Res. Treat. 2018, 11, 1178633718767887. [Google Scholar] [CrossRef] [PubMed]
- Chattopadhyay, M.K.; Chakraborty, R.; Grossart, H.P.; Reddy, G.S.; Jagannadham, M.V. Antibiotic resistance of bacteria. Biomed. Res. Int. 2015, 2015, 501658. [Google Scholar] [CrossRef] [PubMed]
- Blair, J.M.; Webber, M.A.; Baylay, A.J.; Ogbolu, D.O.; Piddock, L.J. Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol. 2015, 13, 42–51. [Google Scholar] [CrossRef]
- Schillaci, D.; Spano, V.; Parrino, B.; Carbone, A.; Montalbano, A.; Barraja, P.; Diana, P.; Cirrincione, G.; Cascioferro, S. Pharmaceutical approaches to target antibiotic resistance mechanisms. J. Med. Chem. 2017, 60, 8268–8297. [Google Scholar] [CrossRef] [PubMed]
- Bush, K.; Pucci, M.J. New antimicrobial agents on the horizon. Biochem. Pharm. 2011, 82, 1528–1539. [Google Scholar] [CrossRef]
- Narayana, B.; Ashalatha, B.; Raj, K.; Kumari, N.S. Synthesis of 3-amino-2-methyl/ethyl-5, 6, 7, 8-tetrahydro [1] benzothieno [2, 3-d] pyrimidin-4 (3H)-one and its Schiff bases as possible antimicrobial and non-steroidal antiinflammatory agents. Indian J. Chem. 2006, 45B, 2696–2703. [Google Scholar] [CrossRef]
- Abdel-Rahman, A.E.; Bakhite, E.A.; Al-Taifi, E.A. Synthesis and Antimicrobial Activity of New Pyridothienopyrimidines and Pyridothienotriazines. J. Chin. Chem. Soc. 2002, 49, 223–231. [Google Scholar] [CrossRef]
- Narayana, B.; Vijaya Raj, K.K.; Ashalatha, B.V.; Kumari, N.S.; Sarojini, B.K. Synthesis of some new 5-(2-substituted-1,3-thiazol-5-yl)-2-hydroxy benzamides and their 2-alkoxy derivatives as possible antifungal agents. Eur. J Med. Chem. 2004, 39, 867–872. [Google Scholar] [CrossRef] [PubMed]
- Isloor, A.M.; Kalluraya, B.; Shetty, P. Regioselective reaction: Synthesis, characterization and pharmacological studies of some new Mannich bases derived from 1,2,4-triazoles. Eur. J Med. Chem. 2009, 44, 3784–3787. [Google Scholar] [CrossRef] [PubMed]
- Patil, S.; Claffey, J.; Deally, A.; Hogan, M.; Gleeson, B.; Menéndez Méndez, L.M.; Müller-Bunz, H.; Paradisi, F.; Tacke, M. Synthesis, cytotoxicity and antibacterial studies of p-methoxybenzyl-substituted and benzyl-substituted N-heterocyclic carbene-silver complexes. Eur. J. Inorg. Chem. 2010, 1020–1031. [Google Scholar] [CrossRef]
- Patil, S.; Deally, A.; Gleeson, B.; Muller-Bunz, H.; Paradisi, F.; Tacke, M. Novel benzyl-substituted N-heterocyclic carbene-silver acetate complexes: Synthesis, cytotoxicity and antibacterial studies. Metallomics 2011, 3, 74–88. [Google Scholar] [CrossRef] [PubMed]
- Patil, S.A.; Patil, S.A.; Patil, R. Medicinal applications of (benz)imidazole- and indole-based macrocycles. Chem. Biol. Drug Des. 2017, 89, 639–649. [Google Scholar] [CrossRef] [PubMed]
- Patil, S.A.; Patil, S.A.; Patil, R.; Keri, R.S.; Budagumpi, S.; Balakrishna, G.R.; Tacke, M. N-heterocyclic carbene metal complexes as bio-organometallic antimicrobial and anticancer drugs. Future Med. Chem. 2015, 7, 1305–1333. [Google Scholar] [CrossRef]
- Patil, V.; Barragan, E.; Patil, S.A.; Patil, S.A.; Bugarin, A. Direct synthesis and antimicrobial evaluation of structurally complex chalcones. Chem. Sel. 2016, 1, 3647–3650. [Google Scholar] [CrossRef]
- Shahini, C.R.; Achar, G.; Budagumpi, S.; Müller–Bunz, H.; Tacke, M.; Patil, S.A. Benzoxazole and dioxolane substituted benzimidazole–based N –heterocyclic carbene–silver(I) complexes: Synthesis, structural characterization and in vitro antimicrobial activity. J. Organomet. Chem. 2018, 868, 1–13. [Google Scholar] [CrossRef]
- Shahini, C.R.; Achar, G.; Budagumpi, S.; Tacke, M.; Patil, S.A. Non-symmetrically p-nitrobenzyl-substituted N-heterocyclic carbene-silver(I) complexes as metallopharmaceutical agents. Appl. Organomet. Chem. 2017, 31, e3819. [Google Scholar] [CrossRef]
- Shahini, C.R.; Achar, G.; Budagumpi, S.; Tacke, M.; Patil, S.A. Synthesis, structural investigation and antibacterial studies of non–symmetrically p–nitrobenzyl substituted benzimidazole N–heterocyclic carbene–silver(I) complexes. Inorg. Chim. Acta 2017, 466, 432–441. [Google Scholar] [CrossRef]
- Sharkey, M.; O’Gara, J.; Gordon, S.; Hackenberg, F.; Healy, C.; Paradisi, F.; Patil, S.; Schaible, B.; Tacke, M. Investigations into the antibacterial activity of the silver-based antibiotic drug candidate SBC3. Antibiotics 2012, 1, 25–28. [Google Scholar] [CrossRef]
- Subramanya Prasad, T.V.; Shahini, C.R.; Patil, S.A.; Huang, X.; Bugarin, A.; Patil, S.A. Non-symmetrically p-nitrobenzyl- and p-cyanobenzyl-substituted N-heterocyclic carbene-silver(I) complexes: Synthesis, characterization and antibacterial studies. J. Coord. Chem. 2017, 70, 600–614. [Google Scholar] [CrossRef]
- Patil, M.; Noonikara-Poyil, A.; Joshi, S.D.; Patil, S.A.; Patil, S.A.; Bugarin, A. Synthesis, molecular docking studies, and antimicrobial evaluation of new structurally diverse ureas. Bioorg. Chem. 2019, 87, 302–311. [Google Scholar] [CrossRef] [PubMed]
- Patil, M.; Noonikara-Poyil, A.; Joshi, S.D.; Patil, S.A.; Patil, S.A.; Bugarin, A. Design, synthesis, and molecular docking study of new piperazine derivative as potential antimicrobial agents. Bioorg. Chem. 2019, 92, 103217. [Google Scholar] [CrossRef] [PubMed]
- Davies, W.L.; Grunert, R.R.; Haff, R.F.; McGahen, J.W.; Neumayer, E.M.; Paulshock, M.; Watts, J.C.; Wood, T.R.; Hermann, E.C.; Hoffmann, C.E. Antiviral activity of 1-adamantanamine (amantadie). Science 1964, 144, 862–863. [Google Scholar] [CrossRef] [PubMed]
- Dolin, R.; Reichman, R.C.; Madore, H.P.; Maynard, R.; Linton, P.N.; Webber-Jones, J. A controlled trial of amantadine and rimantadine in the prophylaxis of influenza A infection. N. Eng. J. Med. 1982, 307, 580–584. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Obando, D.; Liao, V.; Lifa, T.; Codd, R. The many faces of the adamantyl group in drug design. Eur. J Med. Chem. 2011, 46, 1949–1963. [Google Scholar] [CrossRef] [PubMed]
- Wanka, L.; Iqbal, K.; Schreiner, P.R. The Lipophilic Bullet Hits the Targets: Medicinal Chemistry of Adamantane Derivatives. Chem. Rev. 2013, 113, 3516–3604. [Google Scholar] [CrossRef]
- Ahameethunisa, A.R.; Hopper, W. Antibacterial activity of Artemisia nilagirica leaf extracts against clinical and phytopathogenic bacteria. BMC Complement. Altern. Med. 2010, 10, 6. [Google Scholar] [CrossRef]
- Balouiri, M.; Sadiki, M.; Ibnsouda, S.K. Methods for in vitro evaluating antimicrobial activity: A review. J. Pharm. Anal. 2016, 6, 71–79. [Google Scholar] [CrossRef]
- Forbes, L.; Ebsworth-Mojica, K.; DiDone, L.; Li, S.G.; Freundlich, J.S.; Connell, N.; Dunman, P.M.; Krysan, D.J. A High throughput screening assay for anti-mycobacterial small molecules based on adenylate kinase release as a reporter of cell lysis. PLoS ONE 2015, 10, e0129234. [Google Scholar] [CrossRef] [PubMed]
- Greis, K.D.; Zhou, S.; Siehnel, R.; Klanke, C.; Curnow, A.; Howard, J.; Layh-Schmitt, G. Development and validation of a whole-cell inhibition assay for bacterial methionine aminopeptidase by surface-enhanced laser desorption ionization-time of flight mass spectrometry. Antimicrob. Agents Chemother. 2005, 49, 3428–3434. [Google Scholar] [CrossRef] [PubMed]
- Nasrin, F.; Bulbul, I.; Begum, Y. In vitro antimicrobial and cytotoxicity screening of n-hexane, chloroform and ethyl acetate extracts of Lablab purpureus (L.) leaves. Agric. Biol. J. North Am. 2012, 3, 43–48. [Google Scholar] [CrossRef]
- Edwards, I.A.; Elliott, A.G.; Kavanagh, A.M.; Zuegg, J.; Blaskovich, M.A.T.; Cooper, M.A. Contribution of amphipathicity and hydrophobicity to the antimicrobial activity and cytotoxicity of β-hairpin peptides. ACS Infect. Dis. 2016, 2, 442–450. [Google Scholar] [CrossRef] [PubMed]
- Gasteiger, J.; Marsili, M. Iterative partial equalization of orbital electronegativity—a rapid access to atomic charges. Tetrahedron 1980, 36, 3219–3228. [Google Scholar] [CrossRef]
- Tripos International. Sybyl-X 2.0, Tripos International: St. Louis, MO, USA, 2012.
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).