Multidrug Efflux Systems in Microaerobic and Anaerobic Bacteria
Abstract
:1. Introduction
2. Classification of Efflux Pumps and Their Regulation
| Species | Major Efflux Pumps | Regulators | Antimicrobial Agents Being Pumped | Reference |
|---|---|---|---|---|
| Microaerobic niches | ||||
| Campylobacter spp. | CmeABC (RND) | CmeR (TetR) | Ciprofloxacin, Norfloxacin, Cefotaxime, Fusidic Acid, Erythromycin | [14,15,16] |
| CosR (OmpR) | ||||
| CmeDEF (RND) | Ampicillin, Polymyxin B, Ethidium Bromide | [14] | ||
| CmeG (MFS) | Ciprofloxacin, Erythromycin, Gentamicin, Tetracycline, Rifampicin, Ethidium Bromide, Cholic Acid, Hydrogen Peroxide | [17] | ||
| NhaA1/NhaA2 (cation/proton antiporters) | Trisodium Phosphate | [18] | ||
| Helicobacter pylori | HefABC (RND) | Metronidazole, Tetracycline, Erythromycin, Penicillin G, Ciprofloxacin | [19,20] | |
| HefDEF (RND) | [19] | |||
| HefGHI (RND) | [19] | |||
| Staphylococcus aureus | NorB | MgrA (MarR) | Moxifloxacin, Sparfloxacin | [21] |
| Anaerobic niches | ||||
| Bacteroides fragilis | BmeABC1-16 (RND) | BmeR (TetR) | Cephems, Polypeptide Antibiotics, Fusidic Acid, Novobiocin, Puromycin Ampicillin, Cefoxitin, Cefoperazone, Ciprofloxacin, Metronidazole, Imipenem, Ethidium Bromide, Sodium Dodecyl Sulfate. | [22,23] |
| BexA | Fluoroquinolone | [24] | ||
| Clostridium perfringens | bcrABD | bcrR | Phenotypic Bacitracin | [25] |
| Escherichia coli | MdtEF (RND) | ArcAB | Indole Nitrosative Derivatives erythromycin | [26,27] |
| (TCS) | ||||
| MnmE | ||||
| H-NS | ||||
| CusCBA (RND) | CusSR (TCS) | Cu(I) | [28] | |
| Porphyromonas gingivalis | XepCAB (RND) | Rifampin, Puromycin, Ethidium Bromide | [29] | |
| Salmonella enterica | AcrAB (RND) | [30] | ||
| TolC | [31] | |||
| TetA | Etracycline, Ethidium Bromide | [31] | ||
| Staphylococcus aureus | MnhF | Bile Salts | [32] |
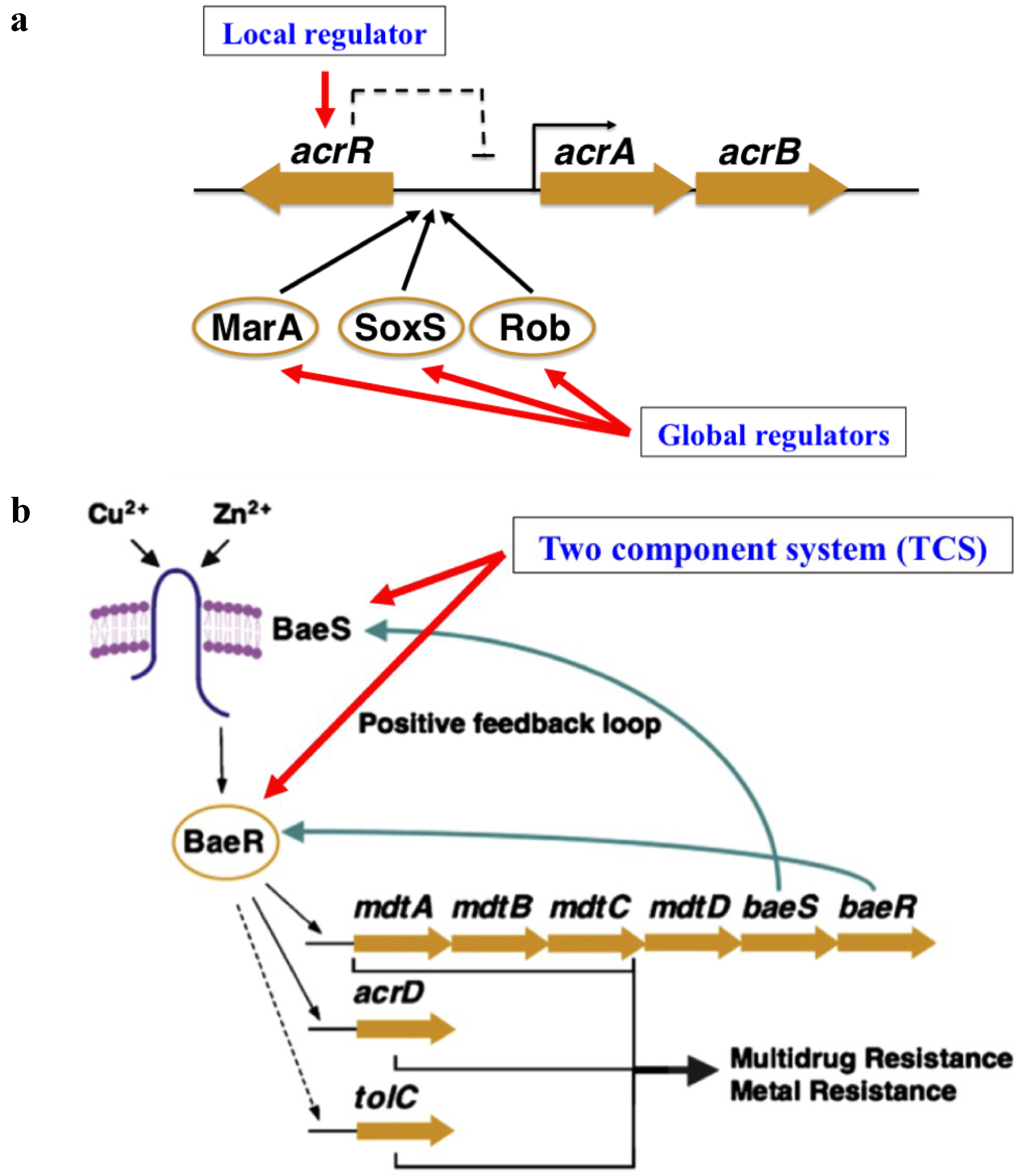
3. Drug Efflux Pumps in Microaerobic Niches
3.1. Campylobacter spp. Efflux Pumps
3.2. Helicobacter pylori Efflux Pumps
3.3. Staphylococcus aureus Efflux Pumps
4. Drug Efflux Pumps in Anaerobic Niches
5. Physiological Roles of Efflux Pumps during the Anaerobic Adaptation of Facultative Bacteria
5.1. Escherichia coli Efflux Pumps
5.2. Salmonella enterica Efflux Pumps
5.3. Role of Efflux Pumps in the Anaerobic Niches of Bacterial Communities
6. Other Efflux Systems
7. Inhibition of Drug Efflux in Microaerobic and Anaerobic Niches
8. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| ABC | the ATP (adenosine triphosphate)-binding cassette superfamily |
| CCCP | carbonyl cyanide m-chlorophenylhydrazone |
| CLR | clarithromycin |
| EB | ethidium bromide |
| EPI | efflux pump inhibitor |
| MATE | the multidrug and toxic compound extrusion family |
| MFS | the major facilitator superfamily |
| MIC | minimum inhibitory concentration |
| Mtz | metronidazole |
| PaβN | Phe-Arg β-naphthylamide dihydrochloride |
| RND | the resistance-nodulation-division family |
| SDS | sodium dodecyl sulfate |
| SMR | the small multidrug resistance family |
| TCS | two component system |
| TSP | trisodium phosphate |
References
- Poole, K. Efflux pumps as antimicrobial resistance mechanisms. Ann. Med. 2007, 39, 162–176. [Google Scholar] [CrossRef] [PubMed]
- Hirakawa, H.; Inazumi, Y.; Senda, Y.; Kobayashi, A.; Hirata, T.; Nishino, K.; Yamaguchi, A. N-Acetyl-d-Glucosamine induces the expression of multidrug exporter genes, mdtEF, via catabolite activation in Escherichia coli. J. Bacteriol. 2006, 188, 5851–5858. [Google Scholar] [CrossRef] [PubMed]
- Piddock, L.J. Multidrug-resistance efflux pumps—Not just for resistance. Nat. Rev. Microbiol. 2004, 4, 629–636. [Google Scholar] [CrossRef] [PubMed]
- Jantsch, J.; Schodel, J. Hypoxia and hypoxia-inducible factors in myeloid cell-driven host defense and tissue homeostasis. Immunobiology 2015, 220, 305–314. [Google Scholar] [CrossRef] [PubMed]
- Marteyn, B.; Scorza, F.B.; Sansonetti, P.J.; Tang, C. Breathing life into pathogens: The influence of oxygen on bacterial virulence and host responses in the gastrointestinal tract. Cell Microbiol. 2011, 13, 171–176. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Deng, Z.; Yan, A. Bacterial multidrug efflux pumps: Mechanisms, physiology and pharmacological exploitations. Biochem. Biophys. Res. Commun. 2014, 453, 254–267. [Google Scholar] [CrossRef] [PubMed]
- Putman, M.; Veen, H.W.; Konings, W. Molecular properties of bacterial multidrug transporters. Microbiol. Mol. Biol. Rev. 2000, 64, 672–693. [Google Scholar] [CrossRef] [PubMed]
- Perera, I.C.; Grove, A. Molecular mechanisms of ligand-mediated attenuation of DNA binding by MarR family transcriptional regulators. J. Mol. Cell Biol. 2010, 2, 243–254. [Google Scholar] [CrossRef] [PubMed]
- Brown, N.L.; Stoyanov, J.V.; Kidd, S.P.; Hobman, J.L. The MerR family of transcriptional regulators. FEMS Microbiol. Rev. 2003, 27, 145–163. [Google Scholar] [CrossRef]
- Ramos, J.L.; Martinez-Bueno, M.; Molina-Henares, A.J.; Teran, W.; Watanabe, K.; Zhang, X.; Gallegos, M.T.; Brennan, R.; Tobes, R. The TetR family of transcriptional repressors. Microbiol. Mol. Biol. Rev. 2005, 69, 326–356. [Google Scholar] [CrossRef] [PubMed]
- Nikaido, E.; Shirosaka, I.; Yamaguchi, A.; Nishino, K. Regulation of the AcrAB multidrug efflux pump in Salmonella enterica serovar Typhimurium in response to indole and paraquat. Microbiology 2011, 157, 648–655. [Google Scholar] [CrossRef] [PubMed]
- Hoch, J.A. Two-component and phosphorelay signal transduction. Curr. Opin. Microbiol. 2000, 3, 165–170. [Google Scholar] [CrossRef]
- Baranova, N.; Nikaido, H. The BaeSR two-component regulatory system activates transcription of the yegMNOB (mdtABCD) transporter gene cluster in Escherichia coli and increases its resistance to novobiocin and deoxycholate. J. Bacteriol. 2002, 184, 4168–4176. [Google Scholar] [CrossRef] [PubMed]
- Akiba, M.; Lin, J.; Barton, Y.W.; Zhang, Q. Interaction of CmeABC and CmeDEF in conferring antimicrobial resistance and maintaining cell viability in Campylobacter jejuni. J. Antimicrob. Chemoth. 2006, 57, 52–60. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Akiba, M.; Sahin, O.; Zhang, Q. CmeR functions as a transcriptional repressor for the multidrug efflux pump CmeABC in Campylobacter jejuni. Antimicrob. Agents Chemother. 2005, 49, 1067–1075. [Google Scholar] [CrossRef] [PubMed]
- Hwang, S.; Zhang, Q.; Ryu, S.; Jeon, B. Transcriptional regulation of the CmeABC multidrug efflux pump and the KatA catalase by CosR in Campylobacter jejuni. J. Bacteriol. 2012, 194, 6883–6891. [Google Scholar] [CrossRef] [PubMed]
- Jeon, B.; Wang, Y.; Hao, H.; Barton, Y.W.; Zhang, Q. Contribution of CmeG to antibiotic and oxidative stress resistance in Campylobacter jejuni. J. Antimicrob. Chemother. 2011, 66, 79–85. [Google Scholar] [CrossRef] [PubMed]
- Riedel, C.T.; Cohn, M.T.; Stabler, R.A.; Wren, B.; Brondsted, L. Cellular Response of Campylobacter jejuni to Trisodium Phosphate. Appl. Environ. Microbiol. 2011, 78, 1411–1415. [Google Scholar] [CrossRef] [PubMed]
- Bina, J.E.; Alm, R.A.; Uria-nichelsen, M.; Thomas, S.R.; Trust, T.J.; Hancock, R.E.W. Helicobacter pylori uptake and efflux: Basis for intrinsic susceptibility to antibiotics in vitro. Antimicrob. Agents Chemother. 2000, 44, 248–254. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.Q. Efflux pump gene hefA of Helicobacter pylori plays an important role in multidrug resistance. World J. Gastroenterol. 2008, 14, 5217–5222. [Google Scholar] [CrossRef] [PubMed]
- Truong-Bolduc, Q.C.; Hsing, L.C.; Villet, R.; Bolduc, G.R.; Estabrooks, Z.; Taguezem, G.F.; Hooper, D.C. Reduced aeration affects the expression of the NorB efflux pump of Staphylococcus aureus by posttranslational modification of MgrA. J. Bacteriol. 2012, 194, 1823–1834. [Google Scholar] [CrossRef] [PubMed]
- Ueda, O.; Wexler, H.M.; Hirai, K.; Shibata, Y.; Yoshimura, F.; Fujimura, S. Sixteen homologs of the mex-type multidrug resistance efflux pump in Bacteroides fragilis. Antimicrob. Agents Chemother. 2005, 49, 2807–2815. [Google Scholar] [CrossRef] [PubMed]
- Pumbwe, L.; Chang, A.; Smith, R.L.; Wexler, H.M. BmeRABC5 is a multidrug efflux system that can confer metronidazole resistance in Bacteroides fragilis. Microbiol. Drug Resist. 2007, 13, 96–101. [Google Scholar] [CrossRef] [PubMed]
- Eitel, Z.; Soki, J.; Urban, E.; Nagy, E. Infection ESGoA. The prevalence of antibiotic resistance genes in Bacteroides fragilis group strains isolated in different European countries. Anaerobe 2013, 21, 43–49. [Google Scholar] [CrossRef] [PubMed]
- Charlebois, A.; Jalbert, L.A.; Harel, J.; Masson, L.; Archambault, M. Characterization of genes encoding for acquired bacitracin resistance in Clostridium perfringens. PLoS ONE 2012, 7, e44449. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Xiao, M.; Horiyama, T.; Zhang, Y.; Li, X.; Nishino, K.; Yan, A. The multidrug efflux pump MdtEF protects against nitrosative damage during the anaerobic respiration in Escherichia coli. J. Biol. Chem. 2011, 286, 26576–26584. [Google Scholar] [CrossRef] [PubMed]
- Deng, Z.; Shan, Y.; Pan, Q.; Gao, X.; Yan, A. Anaerobic expression of the gadE-mdtEF multidrug efflux operon is primarily regulated by the two-component system ArcBA through antagonizing the H-NS mediated repression. Front. Microbiol. 2013, 4, e194. [Google Scholar] [CrossRef] [PubMed]
- Fung, D.K.C.; Lau, W.Y.; Chan, W.T.; Yan, A. Copper efflux is induced during anaerobic amino acid limitation in Escherichia coli to protect iron-sulfur cluster enzymes and biogenesis. J. Bacteriol. 2013, 195, 4556–4568. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, T.; Yoshimura, F. A resistance-nodulation-cell division family xenobiotic efflux pump in an obligate anaerobe, Porphyromonas gingivalis. Antimicrob. Agents Chemother. 2002, 46, 3257–3260. [Google Scholar] [CrossRef] [PubMed]
- Webber, M.A.; Bailey, A.M.; Blair, J.M.; Morgan, E.; Stevens, M.P.; Hinton, J.C.; Ivens, A.; Wain, J.; Piddock, L.J. The global consequence of disruption of the AcrAB-TolC efflux pump in Salmonella enterica includes reduced expression of SPI-1 and other attributes required to infect the host. J. Bacteriol. 2009, 191, 4276–4285. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Ahn, J. Assessment of efflux-mediated antibiotic-resistant Salmonella enterica serovar Typhimurium under simulated gastrointestinal conditions. Ann. Microbiol. 2013, 64, 581–587. [Google Scholar] [CrossRef]
- Sannasiddappa, T.H.; Hood, G.A.; Hanson, K.J.; Costabile, A.; Gibson, G.R.; Clarke, S.R. Staphylococcus aureus MnhF mediates cholate efflux and facilitates survival under human colonic conditions. Infect Immun. 2015, 83, 2350–2357. [Google Scholar] [CrossRef] [PubMed]
- Nishino, K.; Nikaido, E.; Yamaguchi, A. Regulation and physiological function of multidrug efflux pumps in Escherichia coli and Salmonella. Biochim. Biophys. Acta 2009, 1794, 834–843. [Google Scholar] [CrossRef] [PubMed]
- Blair, J.M.; Webber, M.A.; Baylay, A.J.; Ogbolu, D.O.; Piddock, L.J. Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol. 2015, 13, 42–51. [Google Scholar] [CrossRef] [PubMed]
- Blaser, M.J. Epidemiologic and clinical features of Campylobacter jejuni infections. J. Infect. Dis. 1997, 176, S103–S105. [Google Scholar] [CrossRef] [PubMed]
- Bolton, D.; Patriarchi, A.; Fox, Á.; Fanning, S. A study of the molecular basis of quinolone and macrolide resistance in a selection of Campylobacter isolates from intensive poultry flocks. Food Control. 2013, 30, 222–226. [Google Scholar] [CrossRef]
- Li, X.; Nikaido, H. Efflux-mediated drug resistance in bacteria: An update. Drugs 2009, 69, 1555–1623. [Google Scholar] [CrossRef] [PubMed]
- Jeon, B.; Zhang, Q. Sensitization of Campylobacter jejuni to fluoroquinolone and macrolide antibiotics by antisense inhibition of the CmeABC multidrug efflux transporter. J. Antimicrob. Chemoth. 2009, 63, 946–948. [Google Scholar] [CrossRef] [PubMed]
- Gibreel, A.; Wetsch, N.M.; Taylor, D.E. Contribution of the CmeABC efflux pump to macrolide and tetracycline resistance in Campylobacter jejuni. Antimicrob. Agents Chemother. 2007, 51, 3212–3216. [Google Scholar] [CrossRef] [PubMed]
- Hwang, S.; Kim, M.; Ryu, S.; Jeon, B. Regulation of oxidative stress response by CosR, an essential response regulator in Campylobacter jejuni. PLoS ONE 2011, 6, e22300. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Cagliero, C.; Guo, B.; Barton, Y.W.; Maurel, M.C.; Payot, S.; Zhang, Q. Bile salts modulate expression of the CmeABC multidrug efflux pump in Campylobacter jejuni. J. Bacteriol. 2005, 187, 7417–7424. [Google Scholar] [CrossRef] [PubMed]
- Thanassi, D.G.; Cheng, L.W.; Nikaido, H. Active efflux of bile salts by Escherichia coli. J. Bacteriol. 1997, 179, 2512–2518. [Google Scholar]
- Hartog, E.; Menashe, O.; Kler, E.; Yaron, S. Salicylate reduces the antimicrobial activity of ciprofloxacin against extracellular Salmonella enterica serovar Typhimurium, but not against Salmonella in macrophages. J. Antimicrob. Chemother. 2010, 65, 888–896. [Google Scholar] [CrossRef] [PubMed]
- Shen, Z.; Pu, X.Y.; Zhang, Q. Salicylate functions as an efflux pump inducer and promotes the emergence of fluoroquinolone-resistant Campylobacter jejuni mutants. Appl. Environ. Microbiol. 2011, 77, 7128–7133. [Google Scholar] [CrossRef] [PubMed]
- Oyarzabal, O.A. Reduction of Campylobacter spp. by commercial antimicrobials applied during the processing of broiler chickens: A review from the United States Perspective. J. Food Protect. 2005, 68, 1752–1760. [Google Scholar]
- Sampathkumar, B.; Khachatourians, G.G.; Korber, D.R. High pH during trisodium phosphate treatment causes membrane damage and destruction of Salmonella enterica Serovar Enteritidis. Appl. Environ. Microbiol. 2003, 69, 122–129. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, H.; Hibi, T.; Marshall, B.J. Helicobacter pylori: Present status and future prospects in Japan. J. Gastroenterol. 2007, 42, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Qureshi, W.A.; Graham, D.Y. Antibiotic-resistant H. pylori infection and its treatment. Curr. Pharm. Design. 2000, 6, 1537–1544. [Google Scholar] [CrossRef]
- Hirata, K.; Suzuki, H.; Nishizawa, T.; Tsugawa, H.; Muraoka, H.; Saito, Y.; Matsuzaki, J.; Hibi, T. Contribution of efflux pumps to clarithromycin resistance in Helicobacter pylori. J. Gastroen. Hepatol. 2010, 25, S75–S79. [Google Scholar] [CrossRef] [PubMed]
- Belzer, C.; Stoof, J.; Breijer, S.; Kusters, J.G.; Kuipers, E.J.; van Vliet, A.H.M. The Helicobacter hepaticus hefA gene is involved in resistance to amoxicillin. Helicobacter 2009, 14, 72–79. [Google Scholar] [CrossRef] [PubMed]
- Tsugawa, H.; Suzuki, H.; Muraoka, H.; Ikeda, F.; Hirata, K.; Matsuzaki, J.; Saito, Y.; Hibi, T. Enhanced bacterial efflux system is the first step to the development of metronidazole resistance in Helicobacter pylori. Biochem. Biophys. Res. Commun. 2011, 404, 656–660. [Google Scholar] [CrossRef] [PubMed]
- Yonezawa, H.; Osaki, T.; Hanawa, T.; Kurata, S.; Ochiai, K.; Kamiya, S. Impact of Helicobacter pylori biofilm formation on clarithromycin susceptibility and generation of resistance mutations. PLoS ONE 2013, 8, e73301. [Google Scholar] [CrossRef] [PubMed]
- Carron, M.A.; Tran, V.R.; Sugawa, C.; Coticchia, J.M. Identification of Helicobacter pylori biofilms in human gastric mucosa. J. Gastrointest. Surg. 2006, 10, 712–717. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, S.; Pane-Farre, J.; Kohler, C.; Hecker, M.; Engelmann, S. Anaerobic gene expression in Staphylococcus aureus. J. Bacteriol. 2007, 189, 4275–4289. [Google Scholar] [CrossRef] [PubMed]
- Kosmidis, C.; Schindler, B.D.; Jacinto, P.L.; Patel, D.; Bains, K.; Seo, S.M.; Kaatz, G.W. Expression of multidrug resistance efflux pump genes in clinical and environmental isolates of Staphylococcus aureus. Int. J. Antimicrob. Agents 2012, 40, 204–209. [Google Scholar] [CrossRef] [PubMed]
- Truong-Bolduc, Q.C.; Strahilevitz, J.; Hooper, D.C. NorC, a new efflux pump regulated by MgrA of Staphylococcus aureus. Antimicrob. Agents Chemother. 2006, 50, 1104–1107. [Google Scholar] [CrossRef] [PubMed]
- Trotonda, M.P.; Tamber, S.; Memmi, G.; Cheung, A.L. MgrA Represses biofilm formation in Staphylococcus aureus. Infect Immun. 2008, 76, 5645–5654. [Google Scholar] [CrossRef] [PubMed]
- Sannasiddappa, T.H.; Costabile, A.; Gibson, G.R.; Clarke, S.R. The influence of Staphylococcus aureus on gut microbial ecology in an in vitro continuous culture human colonic model system. PLoS ONE 2011, 6, e23227. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, F.L.; Silva, D.N.; Pauer, H.; Ferreira, L.Q.; Ferreira Ede, O.; Domingues, R.M.; Lobo, L.A. The role of BmoR, a MarR family regulator, in the survival of Bacteroides fragilis during oxidative stress. Int. J. Med. Microbiol. 2013, 303, 443–448. [Google Scholar] [CrossRef] [PubMed]
- Wexler, H.M. Bacteroides: The good, the bad, and the nitty-gritty. Clin. Microbiol. Rev. 2007, 20, 593–621. [Google Scholar] [CrossRef] [PubMed]
- Urban, E.; Horvath, Z.; Soki, J.; Lazar, G. First Hungarian case of an infection caused by multidrug-resistant Bacteroides fragilis strain. Anaerobe 2015, 31, 55–58. [Google Scholar] [CrossRef] [PubMed]
- Herin, O.; Hedberg, M.; Edlund, C. Efflux-mediated fluoroquinolone resistance in the Bacteroides fragilis Group. Anaerobe 2002, 8, 277–282. [Google Scholar] [CrossRef]
- Pumbwe, L.; Ueda, O.; Yoshimura, F.; Chang, A.; Smith, R.L.; Wexler, H.M. Bacteroides fragilis BmeABC efflux systems additively confer intrinsic antimicrobial resistance. J. Antimicrob. Chemother. 2006, 58, 37–46. [Google Scholar] [CrossRef] [PubMed]
- Nishino, K.; Yamaguchi, A. Analysis of a complete library of putative drug transporter genes in Escherichia coli. J. Bacteriol. 2001, 183, 5803–5812. [Google Scholar] [CrossRef] [PubMed]
- Braymer, J.J.; Giedroc, D.P. Recent developments in copper and zinc homeostasis in bacterial pathogens. Curr. Opin. Chem. Biol. 2014, 19, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Hood, M.I.; Skaar, E.P. Nutritional immunity: Transition metals at the pathogen-host interface. Nat. Rev. Microbiol. 2012, 10, 525–537. [Google Scholar] [CrossRef] [PubMed]
- Bird, L.J.; Coleman, M.L.; Newman, D.K. Iron and copper act synergistically to delay anaerobic growth of bacteria. Appl. Environ. Microbiol. 2013, 79, 3619–3627. [Google Scholar] [CrossRef] [PubMed]
- White, C.; Lee, J.; Kambe, T.; Fritsche, K.; Petris, M.J. A role for the ATP7A copper-transporting ATPase in macrophage bactericidal activity. J. Biol. Chem. 2009, 284, 33949–33956. [Google Scholar] [CrossRef] [PubMed]
- Macomber, L.; Imlay, J.A. The iron-sulfur clusters of dehydratases are primary intracellular targets of copper toxicity. Proc. Natl. Acad. Sci. USA 2009, 106, 8344–8349. [Google Scholar] [CrossRef] [PubMed]
- Nishino, K.; Latifi, T.; Groisman, E.A. Virulence and drug resistance roles of multidrug efflux systems of Salmonella enterica serovar Typhimurium. Mol. Microbiol. 2006, 59, 126–141. [Google Scholar] [CrossRef] [PubMed]
- Eaves, D.J.; Ricci, V.; Piddock, L.J.V. Expression of acrB, acrF, acrD, marA, and soxS in Salmonella enterica Serovar Typhimurium: Role in multiple antibiotic resistance. Antimicrob. Agents Chemother. 2004, 48, 1145–1150. [Google Scholar] [CrossRef] [PubMed]
- Ricci, V.; Tzakas, P.; Buckley, A.; Piddock, L.J. Ciprofloxacin-resistant Salmonella enterica serovar Typhimurium strains are difficult to select in the absence of AcrB and TolC. Antimicrob. Agents Chemother. 2006, 50, 38–42. [Google Scholar] [CrossRef] [PubMed]
- Buckley, A.M.; Webber, M.A.; Cooles, S.; Randall, L.P.; la Ragione, R.M.; Woodward, M.J.; Piddock, L.J. The AcrAB-TolC efflux system of Salmonella enterica serovar Typhimurium plays a role in pathogenesis. Cell Microbiol. 2006, 8, 847–856. [Google Scholar] [CrossRef] [PubMed]
- Nikaido, E.; Yamaguchi, A.; Nishino, K. AcrAB multidrug efflux pump regulation in Salmonella enterica serovar Typhimurium by RamA in response to environmental signals. J. Biol. Chem. 2008, 283, 24245–24253. [Google Scholar] [CrossRef] [PubMed]
- Balleste-Delpierre, C.; Sole, M.; Domenech, O.; Borrell, J.; Vila, J.; Fabrega, A. Molecular study of quinolone resistance mechanisms and clonal relationship of Salmonella enterica clinical isolates. Int. J. Antimicrob. Agents 2014, 43, 121–125. [Google Scholar] [CrossRef] [PubMed]
- Olliver, A.; Valle, M.; Chaslus-Dancla, E.; Cloeckaert, A. Role of an acrR mutation in multidrug resistance of in vitro-selected fluoroquinolone-resistant mutants of Salmonella enterica serovar Typhimurium. FEMS Microbiol. Lett. 2004, 238, 267–272. [Google Scholar] [PubMed]
- Costerton, J.W.; Lewandowski, Z.; Caldwell, D.E.; Korber, D.R.; Lappin-Scott, H.M. Microbial biofilms. Annu. Rev. Microbiol. 1995, 49, 711–745. [Google Scholar] [CrossRef] [PubMed]
- Fox, E.P.; Cowley, E.S.; Nobile, C.J.; Hartooni, N.; Newman, D.K.; Johnson, A.D. Anaerobic bacteria grow within Candida albicans biofilms and induce biofilm formation in suspension cultures. Curr. Biol. 2014, 24, 2411–2416. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Kern, S.E.; Newman, D.K. Endogenous phenazine antibiotics promote anaerobic survival of Pseudomonas aeruginosa via extracellular electron transfer. J. Bacteriol. 2010, 192, 365–369. [Google Scholar] [CrossRef] [PubMed]
- Price-Whelan, A.; Dietrich, L.E.P.; Newman, D.K. Rethinking “secondary” metabolism: Physiological roles for phenazine antibiotics. Nat. Chem. Biol. 2006, 2, 71–78. [Google Scholar] [CrossRef] [PubMed]
- Dietrich, L.E.P.; Teal, T.K.; Price-Whelan, A.; Newman, D.K. Redox-active antibiotics control gene expression and community behavior in divergent bacteria. Science 2008, 321, 1203–1206. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wilks, J.C.; Danhorn, T.; Ramos, I.; Croal, L.; Newman, D.K. Phenazine-1-carboxylic acid promotes bacterial biofilm development via ferrous iron acquisition. J. Bacteriol. 2011, 193, 3606–3617. [Google Scholar] [CrossRef] [PubMed]
- Dietrich, L.E.P.; Price-Whelan, A.; Petersen, A.; Whiteley, M.; Newman, D.K. The phenazine pyocyanin is a terminal signalling factor in the quorum sensing network of Pseudomonas aeruginosa. Mol. Microbiol. 2006, 61, 1308–1321. [Google Scholar] [CrossRef] [PubMed]
- King, P.; Citron, D.M.; Griffith, D.C.; Lomovskaya, O.; Dudley, M.N. Effect of oxygen limitation on the in vitro activity of levofloxacin and other antibiotics administered by the aerosol route against Pseudomonas aeruginosa from cystic fibrosis patients. Diagn. Micr. Infec. Dis. 2010, 66, 181–186. [Google Scholar] [CrossRef] [PubMed]
- Schaible, B.; Taylor, C.T.; Schaffer, K. Hypoxia increases antibiotic resistance in Pseudomonas aeruginosa through altering the composition of multidrug efflux pumps. Antimicrob. Agents Chemother. 2012, 56, 2114–2118. [Google Scholar] [CrossRef] [PubMed]
- Arioli, S.; Guglielmetti, S.; Amalfitano, S.; Viti, C.; Marchi, E.; Decorosi, F.; Giovannetti, L.; Mora, D. Characterization of tetA-like gene encoding for a major facilitator superfamily efflux pump in Streptococcus thermophilus. FEMS Microbiol. Lett. 2014, 355, 61–70. [Google Scholar] [CrossRef] [PubMed]
- Sherrard, L.J.; Schaible, B.; Graham, K.A.; McGrath, S.J.; McIlreavey, L.; Hatch, J.; Wolfgang, M.C.; Muhlebach, M.S.; Gilpin, D.F.; Schneiders, T.; et al. Mechanisms of reduced susceptibility and genotypic prediction of antibiotic resistance in Prevotella isolated from cystic fibrosis (CF) and non-CF patients. J. Antimicrob. Chemother. 2014, 69, 2690–2698. [Google Scholar] [CrossRef] [PubMed]
- Bhardwaj, A.K.; Mohanty, P. Bacterial efflux pumps involved in multidrug resistance and their inhibitors: rejuvinating the antimicrobial chemotherapy. Recent Pat. Antiinfect. Drug Discov. 2012, 7, 73–89. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Martinez, A. Effect of efflux pump inhibitors on bile resistance and in vivo colonization of Campylobacter jejuni. J. Antimicrob. Chemother. 2006, 58, 966–972. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z. Influence of efflux pump inhibitors on the multidrug resistance of Helicobacter pylori. World J. Gastroenterol. 2010, 16, 1279–1284. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.Q.; Huang, G.R.; Wu, M.H.; Tang, H.Y.; Huang, Z.S.; Zhou, X.H.; Yu, W.Q.; Su, J.W.; Mo, X.Q.; Chen, B.P.; et al. Inhibitory effects of emodin, baicalin, schizandrin and berberine on hefA gene: Treatment of Helicobacter pylori-induced multidrug resistance. World J. Gastroenterol. 2015, 21, 4225–4231. [Google Scholar] [CrossRef] [PubMed]
- Yim, G.; McClure, J.; Surette, M.G.; Davies, J.E. Modulation of Salmonella gene expression by subinhibitory concentrations of quinolones. J. Antibiot. 2011, 64, 73–78. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.H.; Molla, M.N.; Cantor, C.R.; Collins, J.J. Bacterial charity work leads to poplation-wide resistance. Nature 2010, 467, 82–85. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, R.; Sakurai, K.; Yamasaki, S.; Hayashi, K.; Nagata, C.; Hoshino, K.; Onodera, Y.; Nishino, K.; Yamaguchi, A. Structural basis for the inhibition of bacterial multidrug exporters. Nature 2013, 500, 102–106. [Google Scholar] [CrossRef] [PubMed]
- Boyanova, L.; Davidkov, L.; Gergova, G.; Kandilarov, N.; Evstatiev, I.; Panteleeva, E.; Mitov, I. Helicobacter pylori susceptibility to fosfomycin, rifampin, and 5 usual antibiotics for H. pylori eradication. Diagn. Micr. Infec. Dis. 2014, 79, 358–361. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, Z.; Yan, A. Multidrug Efflux Systems in Microaerobic and Anaerobic Bacteria. Antibiotics 2015, 4, 379-396. https://doi.org/10.3390/antibiotics4030379
Xu Z, Yan A. Multidrug Efflux Systems in Microaerobic and Anaerobic Bacteria. Antibiotics. 2015; 4(3):379-396. https://doi.org/10.3390/antibiotics4030379
Chicago/Turabian StyleXu, Zeling, and Aixin Yan. 2015. "Multidrug Efflux Systems in Microaerobic and Anaerobic Bacteria" Antibiotics 4, no. 3: 379-396. https://doi.org/10.3390/antibiotics4030379
APA StyleXu, Z., & Yan, A. (2015). Multidrug Efflux Systems in Microaerobic and Anaerobic Bacteria. Antibiotics, 4(3), 379-396. https://doi.org/10.3390/antibiotics4030379





