Beyond Culture: Real-Time PCR Performance in Detecting Causative Pathogens and Key Antibiotic Resistance Genes in Hospital-Acquired Pneumonia
Abstract
1. Introduction
2. Results
2.1. Demographic and Clinical Features
2.2. Clinical Data and Outcomes
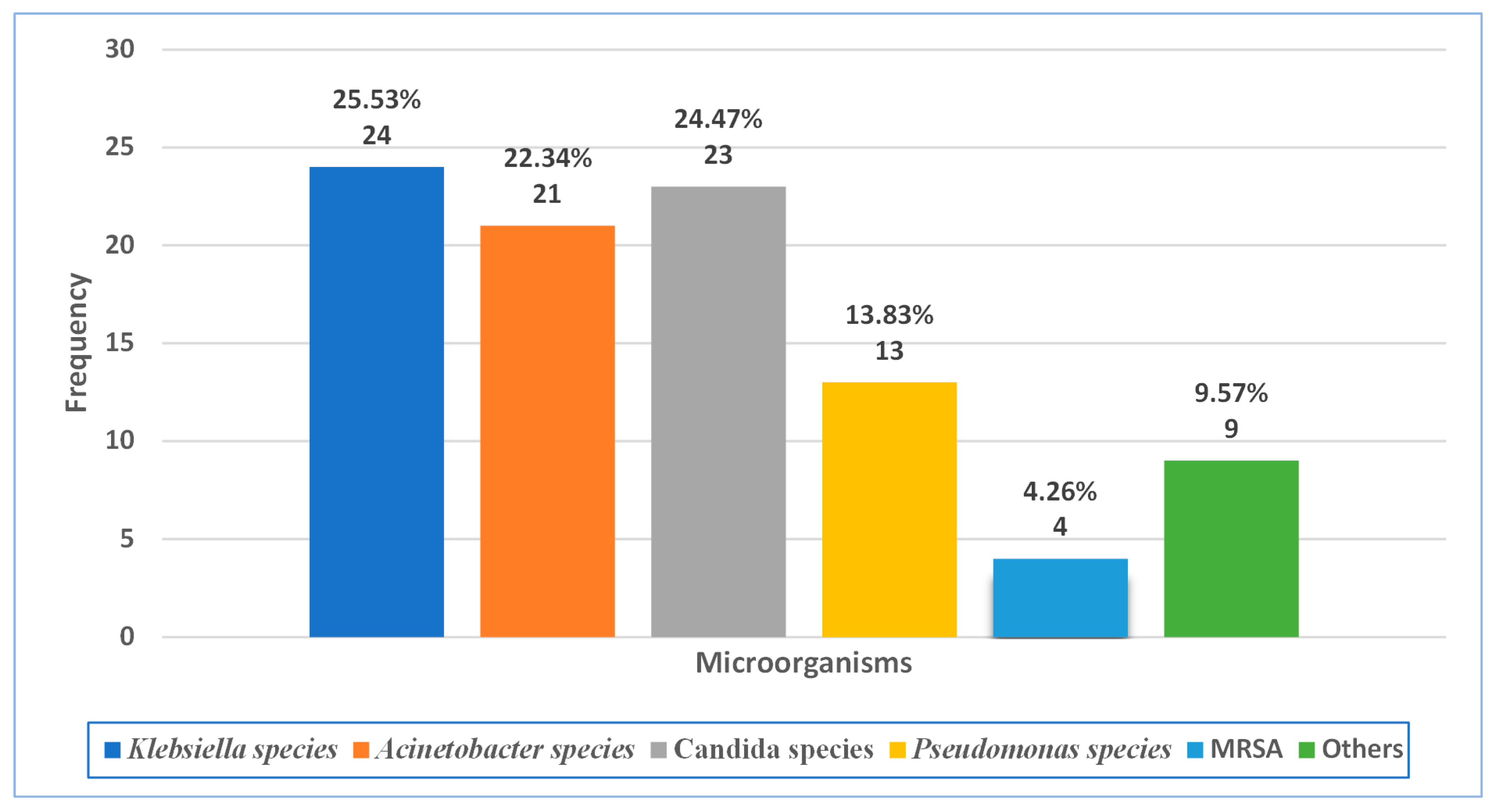
2.3. Microbiological Data
2.4. Antimicrobial Use
2.5. Bacterial Sensitivity Toward Antibiotics
2.6. Culture and qPCR in Microbial Detection
2.7. Antibiotic Resistance Gene Detection
2.7.1. Antibiotic Resistance Gene Prevalence
2.7.2. Culture and qPCR Antibiotic Resistance Detection
2.7.3. Mortality Risk
3. Discussion
- Carbapenem resistance genes: CRE genes (KPC, NDM, VIM, IMP) and OXA genes (OXA-51, OXA-23/OXA-58, OXA-48);
- Quinolone resistance gene: qnr;
- Methicillin resistance genes: mecA and mecC;
- ESBL resistance gene: CTX-M;
- Vancomycin resistance genes: VanA/VanB.
Future Work
- Optimization of new kits that include a larger spectrum of target pathogens to be detected using real-time PCR and involving other, less common causative pathogens such as fungi (e.g., Candida albicans, Candida tropicalis);
- Validate the efficacy, selectivity, and sensitivity of real-time PCR in detecting lower respiratory tract microorganisms in a larger-sized sample and via a multicenter study of HAP patients;
- Consider other important emerging resistance genes in the University of Jordan hospital toward effective antibiotics such as colistin;
- Study the most prevalent resistance genes in The University of Jordan hospital as mortality risk predictors.
4. Materials and Methods
4.1. Study Design
4.2. Study Sample Selection
4.2.1. Sample Types
4.2.2. Handling and Storage
4.3. Clinical Data and Outcomes
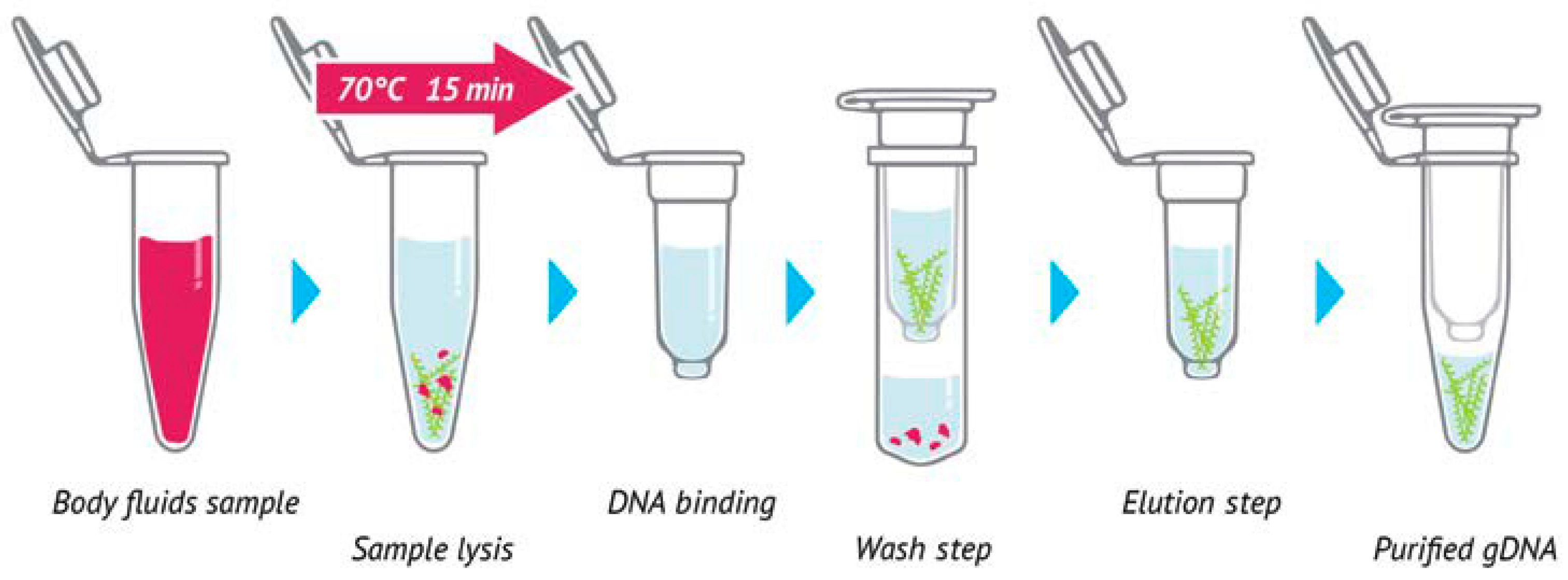
4.4. DNA Extraction
4.4.1. Materials and Equipment
- Proteinase K solution (Qiagen, Hilden, German)
- 2.
- Lysis buffer
- 3.
- Ethanol
- 4.
- Washing buffer
- 5.
- RNase-free water
- 6.
- NZYspin Bacterial/Viral column
- 7.
- Collection tubes (2 mL)
- 8.
- ml Microcentrifuge tubes (Labselect, Beijing, China);
- 9.
- Micropipettes (Acura 825, Socorex, Ecublens, Switzerland);
- 10.
- Sterile disposable tips (Tarsons, Kolkata, India);
- 11.
- Centrifuge (MPW-260R, MPW, Warsaw, Poland);
- 12.
- Vortex mixer (KMC-1300V, Vision Scientific Co., Ltd., Daejeon, Republic of Korea);
- 13.
- Thermostatic Water Bath (Gemmy Industrial Corp., Taiwan, China).
4.4.2. Sample Preparation
4.4.3. Cell Lysis
4.4.4. DNA Binding
4.4.5. Washing
4.4.6. Dry Silica Membrane
4.4.7. DNA Elution
4.5. Real-Time PCR for Lower Respiratory Bacteria Detection
4.5.1. Materials and Equipment
- Negative Control: A no-template control (NTC) allowing for contamination control during qPCR;
- Positive Control (PC): We used a PC containing an artificial DNA sequence that corresponds to lower respiratory bacteria oligonucleotide (LRB Oligo) mix targets and the primer set that detects it, working as a reagent stability control (six PCs: PC-LRB 1/PC-LRB 2/PC-LRB 3/PC-LRB 4/PC-LRB 5/PC-LRB 6);
- qPCR Mix: We used an optimized ready-to-use mix for the qPCR assay. The mixture contains all components necessary for PCR, including a green intercalating dye, dNTPs, stabilizers, and enhancers;
- LRB Oligo Mix: This mix contained specific nucleic acid amplification and detection targets. LRB Oligo Mixes and their specific nucleic acid detection targets are summarized in Table 11;
- Micropipettes (Biosan, Riga, Lativa);
- Compatible filtered pipette tips (nuclease-free): Suitable for transferring 1–10, 10–100, and 100–1000 μL of liquid (Stent, Beijing, China);
- Eppendorf tubes (Extra gene, Taiwan, China);
- PCR plate (Genetics company, Shenzhen, China);
- PCR plate seal (Genetics company, China);
- Vortex for PCR plates (CVP-2, Biosan, Lativa);
- Centrifuge (Combi-Spin, Biosan, Lativa);
- Real-Time PCR Instrument (Quant Gene 9600, Bioer, Tokyo, Japan).
4.5.2. Procedure
- The PCR kit was taken out of the −20 °C freezer.
- LRB Oligo Mixes and 2X qPCR Mix were spun via mini spin.
- (Sample Count + 3) × 5 µL of LRB Oligo Mix 1 was pipetted into an empty Eppendorf tube.
- (Sample Count + 3) × 10 µL of 2X qPCR Mix was added into the Eppendorf tube prepared in step 2.
- The master mix was spun, using mini spin, to become homogenous.
- Steps 3, 4, and 5 were repeated for all master mixes (6 master mixes in total).
- Next, 15 μL of each master mix was pipetted into the relative PCR wells.
- Then, 5 μL of each extracted sample was mixed by pipetting up and down into the relative PCR wells.
- Then, 5 μL of NTC was mixed by pipetting up and down into negative control PCR wells.
- Next, 5 μL of PC-LRB 1 was mixed by pipetting up and down into the first well of the PC plate, repeated for all PCs.
- The PCR plate was sealed firmly and spun via vortex for each PCR plate for 30 s.
- The qPCR device was programed according to the manufacturer’s qPCR program protocol. qPCR program protocol details are summarized in Table 12.
- The lid of the instrument was opened to place the PCR plate and then closed so that the instrument could be used for 40 min.
4.6. Real-Time PCR for Resistance Gene Detection
4.6.1. Materials and Equipment
- Urinary Tract Antibiotic Resistance qPCR Panel kit (qPCR Mix, Urinary tract antibiotic resistance (UTABR) Oligo Mix 2/3, PC-UTABR 2/3, NTC);
- Carbapenem Resistance qPCR Kit (qPCR Mix, Carbapenem resistance Enterobacteriaceae (CRE) Oligo Mix, OXA Oligo Mix, PC-CRE, PC-OXA, NTC).
- 3.
- Micropipettes (Biosan, Lativa);
- 4.
- Compatible filtered pipette tips (nuclease-free) (Stent, China);
- 5.
- Eppendorf tubes (Extra gene, Taichung, Taiwan);
- 6.
- PCR plate (Genetics company, China);
- 7.
- PCR plate seal (Genetics company, China);
- 8.
- Vortex for PCR plates (CVP-2, Biosan, Lativa);
- 9.
- Centrifuge (Combi-Spin, Biosan, Lativa);
- 10.
- Real-time PCR instrument (Quant Gene 9600, Bioer, Japan).
4.6.2. Procedure
- The PCR kit was taken out of the −20 °C freezer.
- UTABR Oligo Mixes, CRE Oligo Mixes, and VRE Oligo Mix with 2X qPCR Mix for each kit were spun using mini spin.
- (Sample Count + 3) × 5 µL of UTABR Oligo Mix 2, CRE Oligo Mix, and VRE Oligo Mix was pipetted into an empty Eppendorf tube for each one.
- (Sample Count + 3) × 10 µL of 2X qPCR Mix for each kit was added into the related Eppendorf tube prepared in step 2.
- The master mix was spun, using mini spin, to become homogenous.
- Steps 3, 4, and 5 were repeated for all master mixes (5 master mixes in total).
- Then, 15 μL of each master mix was pipetted into the relative PCR wells.
- Next, 5 μL of each extracted sample was mixed by pipetting up and down into the relative PCR wells.
- Then, 5 μL of each NTC was mixed by pipetting up and down into related Negative Control PCR wells.
- Next, 5 μL of PC- UTABR 2/3, PC-CRE/OXA, and PC-VRE were mixed by pipetting up and down into the first relative well of the PC plate, and this was repeated for all PCs.
- The PCR plate was sealed firmly and spun via vortex for all PCR plates for 30 s.
- The qPCR device was programed according to the same manufacturer’s qPCR program protocol used during the lower respiratory bacteria detection procedure.
- The lid of the instrument was opened to place the PCR plate and then closed to use the instrument for 40 min.
4.7. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ALI | Acute Lung Injury |
| AMP | Antimicrobial Peptides |
| AP | Aspiration Pneumonia |
| ARDS | Acute Respiratory distress |
| BALF | Bronchoalveolar Lavage Fluid |
| CAP | Community-Acquired Pneumonia |
| CDC | Centers for Disease Control and Prevention |
| CRE | Carbapenem Resistance Enterobacteriaceae |
| CY5 | Indodicarbocyanine |
| dNTPs | Deoxynucleotide Triphosphates |
| DNA | Deoxyribonucleic Acid |
| ESBL | Extended-Spectrum β-lactamases |
| ESR | Erythrocyte Sedimentation Rate |
| FAM | 6-Carboxyfluorescein |
| GERD | Gastroesophageal Reflux Disease |
| GNB | Gram-Negative Bacteria |
| HAP | Hospital-Acquired Pneumonia |
| HEX | Hexachloro-6-Carboxyfluorescein |
| ICU | Intensive Care Unit |
| JASP | Jeffreys’s Amazing Statistics Program |
| LRB | Lower Respiratory Bacteria |
| MDR | Multi-Drug Resistance |
| MDR-GNB | Multi-drug-Resistant Gram-Negative Bacteria |
| MRSA | Methicillin-Resistant Staphylococcus aureus |
| NTC | No-Template Control |
| Oligo | Oligonucleotide |
| PBP2a | Penicillin-Binding Protein 2a |
| PDR | Pan-Drug-Resistant |
| PCR | Polymerase Chain Reaction |
| qPCR | Quantitative Polymerase Chain Reaction |
| RNA | Ribonucleic Acid |
| ROX | 6-Carboxy-X-Rhodamine |
| SOB | Shortness Of Breath |
| SPSS | Statistical Package for the Social Sciences |
| UTABR | Urinary Tract Antibiotic Resistance |
| VAP | Ventilator-Associated Pneumonia |
| VRE | Vancomycin-Resistant Enterococci |
References
- Koenig, S.M.; Truwit, J.D. Ventilator-Associated Pneumonia: Diagnosis, Treatment, and Prevention. Clin. Microbiol. Rev. 2006, 19, 637–657. [Google Scholar] [CrossRef]
- Torres, A.; Cilloniz, C.; Niederman, M.S.; Menéndez, R.; Chalmers, J.D.; Wunderink, R.G.; Van Der Poll, T. Pneumonia. Nat. Rev. Dis. Primers. 2021, 7, 25. [Google Scholar] [CrossRef]
- Plata-Menchaca, E.P.; Ferrer, R. Current Treatment of Nosocomial Pneumonia and Ventilator-Associated Pneumonia. Rev. Esp. Quimioter. 2022, 35, 25–29. [Google Scholar] [CrossRef]
- Rodrigues, D.S.; De Souza, P.T.D.R.; Orsi, J.S.R.; Souza, P.H.C.; Azevedo-Alanis, L.R. Oral Care to Reduce Costs and Increase Clinical Effectiveness in Preventing Nosocomial Pneumonia: A Systematic Review. J. Evid.-Based Dent. Pract. 2023, 23, 101834. [Google Scholar] [CrossRef]
- Fagon, J.; Chastre, J. Diagnosis and Treatment of Nosocomial Pneumonia in ALI/ARDS Patients. Eur. Respir. J. 2003, 22, 77s–83s. [Google Scholar] [CrossRef]
- Urban-Chmiel, R.; Marek, A.; Stępień-Pyśniak, D.; Wieczorek, K.; Dec, M.; Nowaczek, A.; Osek, J. Antibiotic Resistance in Bacteria—A Review. Antibiotics 2022, 11, 1079. [Google Scholar] [CrossRef] [PubMed]
- Adams, J.; Ferguson, K.; Hirschy, R.; Konopka, E.; Meckel, J.; Benanti, G.; Kuhrau, S.; Albarillo, F.; Chang, K.; Santarossa, M.; et al. Antimicrobial Stewardship Techniques for Critically Ill Patients with Pneumonia. Antibiotics 2023, 12, 295. [Google Scholar] [CrossRef]
- Murphy, C.N.; Fowler, R.; Balada-Llasat, J.M.; Carroll, A.; Stone, H.; Akerele, O.; Buchan, B.; Windham, S.; Hopp, A.; Ronen, S.; et al. Multicenter Evaluation of the BioFire FilmArray Pneumonia/Pneumonia Plus Panel for Detection and Quantification of Agents of Lower Respiratory Tract Infection. J. Clin. Microbiol. 2020, 58, e00128-20. [Google Scholar] [CrossRef]
- Alsayed, A.R.; Abed, A.; Khader, H.A.; Al-Shdifat, L.M.; Hasoun, L.; Al-Rshaidat, M.M.; Alkhatib, M.; Zihlif, M. Molecular Accounting and Profiling of Human Respiratory Microbial Communities: Toward Precision Medicine by Targeting the Respiratory Microbiome for Disease Diagnosis and Treatment. Int. J. Mol. Sci. 2023, 24, 4086. [Google Scholar] [CrossRef] [PubMed]
- Candel, F.J.; Salavert, M.; Estella, A.; Ferrer, M.; Ferrer, R.; Gamazo, J.J.; García-Vidal, C.; Del Castillo, J.G.; González-Ramallo, V.J.; Gordo, F.; et al. Ten Issues to Update in Nosocomial or Hospital-Acquired Pneumonia: An Expert Review. J. Clin. Med. 2023, 12, 6526. [Google Scholar] [CrossRef] [PubMed]
- Artika, I.M.; Dewi, Y.P.; Nainggolan, I.M.; Siregar, J.E.; Antonjaya, U. Real-Time Polymerase Chain Reaction: Current Techniques, Applications, and Role in COVID-19 Diagnosis. Genes 2022, 13, 2387. [Google Scholar] [CrossRef] [PubMed]
- Alsayed, A.R.; Abed, A.; Khader, H.A.; Hasoun, L.; Al Maqbali, M.; Al Shawabkeh, M.J. The Role of Human Rhinovirus in COPD Exacerbations in Abu Dhabi: Molecular Epidemiology and Clinical Significance. Libyan J. Med. 2024, 19, 2307679. [Google Scholar] [CrossRef] [PubMed]
- Alsayed, A.R.; Talib, W.; Al-Dulaimi, A.; Daoud, S.; Al Maqbali, M. The First Detection of Pneumocystis Jirovecii in Asthmatic Patients Post-COVID-19 in Jordan. Bosn. J. Basic Med. Sci. 2022, 22, 784. [Google Scholar] [CrossRef]
- Al-Dulaimi, A.; Alsayed, A.R.; Al Maqbali, M.; Zihlif, M. Investigating the Human Rhinovirus Co-Infection in Patients with Asthma Exacerbations and COVID-19. Pharm. Pract. 2022, 20, 2665. [Google Scholar] [CrossRef]
- Al-Shaibari, K.S.A.; Mousa, H.A.-L.; Alqumber, M.A.A.; Alqfail, K.A.; Mohammed, A.; Bzeizi, K. The Diagnostic Performance of Various Clinical Specimens for the Detection of COVID-19: A Meta-Analysis of RT-PCR Studies. Diagnostics 2023, 13, 3057. [Google Scholar] [CrossRef]
- Yoshii, Y.; Shimizu, K.; Morozumi, M.; Chiba, N.; Ubukata, K.; Uruga, H.; Hanada, S.; Wakui, H.; Ito, S.; Takasaka, N.; et al. Identification of Pathogens by Comprehensive Real-Time PCR versus Conventional Methods in Community-Acquired Pneumonia in Japanese Adults. Infect. Dis. 2016, 48, 782–788. [Google Scholar] [CrossRef] [PubMed]
- Gadsby, N.J.; Russell, C.D.; McHugh, M.P.; Mark, H.; Conway Morris, A.; Laurenson, I.F.; Hill, A.T.; Templeton, K.E. Comprehensive Molecular Testing for Respiratory Pathogens in Community-Acquired Pneumonia. Clin. Infect. Dis. 2016, 62, 817–823. [Google Scholar] [CrossRef]
- Pintea-Simon, I.-A.; Bancu, L.; Mare, A.D.; Ciurea, C.N.; Toma, F.; Man, A. Rapid Molecular Diagnostics of Pneumonia Caused by Gram-Negative Bacteria: A Clinician’s Review. Antibiotics 2024, 13, 805. [Google Scholar] [CrossRef]
- Varela, M.F.; Stephen, J.; Lekshmi, M.; Ojha, M.; Wenzel, N.; Sanford, L.M.; Hernandez, A.J.; Parvathi, A.; Kumar, S.H. Bacterial Resistance to Antimicrobial Agents. Antibiotics 2021, 10, 593. [Google Scholar] [CrossRef]
- Gajic, I.; Kabic, J.; Kekic, D.; Jovicevic, M.; Milenkovic, M.; Mitic Culafic, D.; Trudic, A.; Ranin, L.; Opavski, N. Antimicrobial Susceptibility Testing: A Comprehensive Review of Currently Used Methods. Antibiotics 2022, 11, 427. [Google Scholar] [CrossRef]
- Al-Tamimi, M.; Albalawi, H.; Alkhawaldeh, M.; Alazzam, A.; Ramadan, H.; Altalalwah, M.; Alma’aitah, A.; Al Balawi, D.; Shalabi, S.; Abu-Raideh, J.; et al. Multidrug-Resistant Acinetobacter Baumannii in Jordan. Microorganisms 2022, 10, 849. [Google Scholar] [CrossRef] [PubMed]
- Swedan, S.; Alabdallah, E.A.; Ababneh, Q. Resistance to Aminoglycoside and Quinolone Drugs among Klebsiella Pneumoniae Clinical Isolates from Northern Jordan. Heliyon 2024, 10, e23368. [Google Scholar] [CrossRef]
- Borg, M.A.; De Kraker, M.; Scicluna, E.; Van De Sande-Bruinsma, N.; Tiemersma, E.; Monen, J.; Grundmann, H.; on behalf of the ARMed Project members and collaborators. Prevalence of Methicillin-Resistant Staphylococcus Aureus (MRSA) in Invasive Isolates from Southern and Eastern Mediterranean Countries. J. Antimicrob. Chemother. 2007, 60, 1310–1315. [Google Scholar] [CrossRef]
- van Hoek, A.H.A.M.; Mevius, D.; Guerra, B.; Mullany, P.; Roberts, A.P.; Aarts, H.J.M. Acquired Antibiotic Resistance Genes: An Overview. Front. Microbiol. 2011, 2, 203. [Google Scholar] [CrossRef]
- Boyd, S.E.; Holmes, A.; Peck, R.; Livermore, D.M.; Hope, W. OXA-48-Like β-Lactamases: Global Epidemiology, Treatment Options, and Development Pipeline. Antimicrob. Agents Chemother. 2022, 66, e00216-22. [Google Scholar] [CrossRef]
- Mulvey, M.R.; Grant, J.M.; Plewes, K.; Roscoe, D.; Boyd, D.A. New Delhi Metallo-β-Lactamase in Klebsiella Pneumoniae and Escherichia Coli, Canada. Emerg. Infect. Dis. 2011, 17, 103–106. [Google Scholar] [CrossRef] [PubMed]
- Dung, T.T.N.; Phat, V.V.; Vinh, C.; Lan, N.P.H.; Phuong, N.L.N.; Ngan, L.T.Q.; Thwaites, G.; Thwaites, L.; Rabaa, M.; Nguyen, A.T.K.; et al. Development and Validation of Multiplex Real-Time PCR for Simultaneous Detection of Six Bacterial Pathogens Causing Lower Respiratory Tract Infections and Antimicrobial Resistance Genes. BMC Infect. Dis. 2024, 24, 164. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Shen, Z.; Ding, S.; Wang, S. A TaqMan-Based Multiplex Real-Time PCR Assay for the Rapid Detection of Tigecycline Resistance Genes from Bacteria, Faeces and Environmental Samples. BMC Microbiol. 2020, 20, 174. [Google Scholar] [CrossRef]

| Variables | Mdn (IQR) | |
|---|---|---|
| Age | 63 (36.5) | |
| Variables | N | % |
| Gender | ||
| Male | 51 | 61.45% |
| Female | 32 | 38.55% |
| Comorbidities | ||
| Cardiovascular diseases | 48 | 57.83% |
| Diabetes Mellitus | 32 | 38.55% |
| Malignancy | 20 | 24.10% |
| CKD | 19 | 22.89% |
| Another Lung disease * | 19 | 22.89% |
| Hepatic Impairment | 6 | 7.23% |
| Others | 15 | 18.07% |
| Risk Factors for HAP &/or VAP | ||
| Stroke or other neurologic disorders, seizures | 30 | 36.14% |
| Use of antibiotics in the last 3 months | 28 | 33.73% |
| Intubation and mechanical ventilation | 27 | 32.53% |
| Tube feeding | 26 | 31.33% |
| Supine positioning of the patient | 24 | 28.92% |
| Hyperglycemia | 7 | 8.43% |
| Aspiration | 7 | 8.43% |
| Others | 5 | 6.02% |
| Initial Findings | ||
| Chest radiography (segmental infiltrate) | 60 | 72.29% |
| Tachycardia (HR > 100 beats/min) | 25 | 30.12% |
| Altered breath sounds or localized rales | 22 | 26.51% |
| Cough and sputum production | 21 | 25.30% |
| SOB | 20 | 24.10% |
| Tachypnoea | 18 | 21.69% |
| Hypotension (BP < 90 mmHg) | 12 | 14.46% |
| Vomiting | 6 | 7.23% |
| Loss of appetite | 6 | 7.23% |
| Others | 9 | 10.84% |
| Variables | Mdn (IQR) | N (%) * |
|---|---|---|
| Number of Previous Hospitalization | 5 (6) | |
| Hospitalization Location | ||
| Ward | 31 (37.35%) | |
| ICU | 52 (62.65%) | |
| Hospital Length of stay (LOS) | 25 (28) | |
| ICU LOS | 19.5 (22.75) | |
| Pneumonia Classification | ||
| HAP (excluding VAP) | 60 (72.29%) | |
| VAP | 23 (27.71%) | |
| Number of Previous Pneumonia | 2 (1) | |
| Sample Type | ||
| Sputum | 80 (96.39%) | |
| BALF | 3 (3.61%) | |
| C-Reactive Protein (CRP) | ||
| At diagnosis | 89 (106.5) | |
| At discharge | 80.65 (111.8) | |
| Total duration (days) of ventilation | ||
| Invasive mechanical ventilation | 3 (18) | |
| O2 therapy | 5.5 (19) | |
| Outcome | ||
| Survived | 42 (51.85%) | |
| Not Survived | 39 (48.15%) |
| Antibiotic | Antibiotic Selection Strategy (N %) | ||
|---|---|---|---|
| Empiric * (n = 241) | Initial ** (n = 65) | Next *** (n = 51) | |
| Carbapenem | |||
| Imipenem–cilastatin | 26 (10.79%) | 3 (4.62%) | 2 (3.92%) |
| Meropenem | 40 (16.60%) | 7 (10.77%) | 3 (5.88%) |
| Ertapenem | 1 (0.41%) | - | 1 (1.96%) |
| Penicillin | |||
| Piperacillin–tazobactam | 26 (10.79%) | - | 2 (3.92%) |
| Cephalosporin | |||
| Ceftriaxone | 4 (1.66%) | - | - |
| Ceftazidime–avibactam | 2 (0.83%) | 3 (4.62%) | - |
| Others | 1 (0.41%) | 1 (1.54%) | 2 (3.92%) |
| Macrolide | |||
| Azithromycin | 3 (1.24%) | - | - |
| Erythromycin | - | 1 (1.54%) | - |
| Glycopeptide | |||
| Vancomycin | 49 (20.33%) | 7 (10.77%) | 4 (7.84%) |
| Teicoplanin | - | - | 1 (1.96%) |
| Quinolone | |||
| Levofloxacin | 28 (11.62%) | - | 5 (9.80%) |
| Aminoglycoside | |||
| Amikacin | 14 (5.81%) | 6 (9.23%) | 3 (5.88%) |
| Gentamicin | 11 (4.56%) | 5 (7.69%) | 3 (5.88%) |
| Antifungal | |||
| Anidulafungin | 10 (4.15%) | 3 (4.62%) | 5 (9.80%) |
| Voriconazole | 1 (0.41%) | 1 (1.54%) | 3 (3.614%) |
| Others | 4 (1.66%) | - | 2 (3.92%) |
| Other antimicrobial agents | |||
| Colistin | 13 (5.39%) | 14 (21.54%) | 7 (13.73%) |
| Tigecycline | 2 (0.83%) | 6 (9.23%) | 5 (9.80%) |
| Trimethoprim–sulfamethoxazole | 2 (0.83%) | 3 (4.62%) | 2 (3.92%) |
| Others | 4 (1.66%) | 5 (7.69%) | 1 (1.96%) |
| Bacteria (N) * | Sensitivity N (%) ** | ||
|---|---|---|---|
| Sensitive | Intermediate | Resistance | |
| Acinetobacter Baumannii (16) | Amikacin | ||
| - | - | 15 (100%) | |
| Aztreonam | |||
| - | - | 5 (100%) | |
| Cefazolin | |||
| - | - | 5 (100%) | |
| Cefepime | |||
| - | 1 (6.25%) | 15 (93.75%) | |
| Cefotaxime | |||
| - | 1 (7.14%) | 13 (92.86%) | |
| Cefoxitin | |||
| - | - | 7 (100%) | |
| Ceftazidime | |||
| 1 (7.14%) | - | 13 (92.86%) | |
| Ceftriaxone | |||
| - | - | 14 (100%) | |
| Ciprofloxacin | |||
| 1 (7.69%) | - | 12 (92.31%) | |
| Colistin | |||
| 12 (92.31%) | - | 1 (7.69%) | |
| Doxycycline | |||
| 2 (28.57%) | - | 5 (71.43%) | |
| Ertapenem | |||
| - | - | 9 (100%) | |
| Gentamicin | |||
| 2 (15.39%) | - | 11 (84.62%) | |
| Imipenem | |||
| - | - | 16 (100%) | |
| Levofloxacin | |||
| - | - | 7 (100%) | |
| Meropenem | |||
| - | - | 12 (100%) | |
| Minocycline | |||
| - | 1 (14.29%) | 6 (85.71%) | |
| Piperacillin–tazobactam | |||
| - | - | 15 (100%) | |
| Tetracycline | |||
| - | - | 7 (100%) | |
| Tobramycin | |||
| - | - | 6 (100%) | |
| Trimethoprim–sulfamethoxazole | |||
| 1 (20%) | - | 4 (80%) | |
| Klebsiella pneumoniae (19) | Amikacin | ||
| 2 (11.11%) | 11 (61.11%) | 5 (27.78%) | |
| Amoxicillin–clavulanic acid | |||
| - | - | 11 (100%) | |
| Cefazolin | |||
| - | - | 11 (100%) | |
| Cefepime | |||
| 1 (5.56%) | - | 17 (94.44%) | |
| Cefotaxime | |||
| 1 (9.09%) | - | 10 (90.91%) | |
| Cefoxitin | |||
| - | - | 6 (100%) | |
| Ceftazidime | |||
| 1 (5.88%) | - | 16 (94.12%) | |
| Ceftalozane–sulbactam | |||
| - | - | 5 (100%) | |
| Ceftriaxone | |||
| - | - | 16 (100%) | |
| Cefuroxime | |||
| - | - | 8 (100%) | |
| Ciprofloxacin | |||
| 1 (5.56%) | - | 17 (94.44%) | |
| Colistin | |||
| 4 (44.44%) | - | 5 (55.56%) | |
| Ertapenem | |||
| 1 (5.88%) | - | 16 (94.12%) | |
| Gentamicin | |||
| 2 (11.11%) | 1 (5.56%) | 15 (83.33%) | |
| Imipenem | |||
| 1 (5.56%) | - | 17 (94.44%) | |
| Meropenem | |||
| - | - | 14 (100%) | |
| Piperacillin–tazobactam | |||
| - | - | 15 (100%) | |
| Tigecycline | |||
| 6 (85.71%) | - | 1 (14.29%) | |
| Trimethoprim–sulfamethoxazole | |||
| 10 (83.33%) | - | 2 (16.67%) | |
| Pseudomonas aeruginosa (12) | Amikacin | ||
| 4 (44.44%) | - | 5 (55.56%) | |
| Aztreonam | |||
| 3 (37.5%) | - | 5 (62.5%) | |
| Cefepime | |||
| 4 (36.36%) | - | 7 (63.64%) | |
| Ceftazidime | |||
| 3 (30%) | - | 7 (70%) | |
| Ciprofloxacin | |||
| 4 (40%) | - | 6 (60%) | |
| Colistin | |||
| 3 (100%) | - | - | |
| Gentamicin | |||
| 5 (62.5%) | - | 3 (37.5%) | |
| Imipenem | |||
| 9 (90%) | - | 1 (10%) | |
| Meropenem | |||
| 5 (50%) | - | 5 (50%) | |
| Piperacillin–tazobactam | |||
| 7 (63.64%) | 1 (9.09%) | 3 (27.27%) | |
| MRSA (4) | Clindamycin | ||
| 2 (50%) | - | 2 (50%) | |
| Erythromycin | |||
| 1 (25%) | - | 3 (75%) | |
| Gentamicin | |||
| 3 (75%) | 1 (25%) | - | |
| Levofloxacin | |||
| 2 (50%) | - | 2 (50%) | |
| Oxacillin | |||
| - | - | 4 (100%) | |
| Vancomycin | |||
| 4 (100%) | - | - | |
| Microorganism | Culture | qPCR | p-Value (Pearson Chi-Square Test) | ||
|---|---|---|---|---|---|
| N | % | N | % | ||
| Proteus species | 1/83 | 1.21% | 2/82 | 2.44% | <0.001 |
| Streptococcus pneumoniae | - | - | 4/82 | 4.88% | - |
| Streptococcus pyogen | 1/83 | 1.21% | 1/82 | 1.22% | - |
| Streptococcus agalactiae | - | - | 1/82 | 1.22% | - |
| Mycoplasma pneumoniae | - | - | - | - | - |
| Haemophilus influenzae | - | - | 4/82 | 4.88% | - |
| Moraxella Catarrhalis | - | - | 1/82 | 1.22% | - |
| Klebsiella aerogenes | - | - | - | - | - |
| Klebsiella oxytoca | 1/83 | 1.21% | 4/82 | 4.88% | <0.001 |
| Enterobacteriaceae | - | - | 39/82 | 47.56% | - |
| Escherichia coli | - | - | 10/82 | 12.16% | - |
| Enterobacter cloacae | 1/83 | 1.21% | 9/82 | 10.98% | 0.004 |
| Serratia marcescens | - | - | 5/82 | 6.10% | - |
| Legionella pneumophila | - | - | - | - | - |
| Pseudomonas aeruginosa | 12/83 | 14.46% | 11/82 | 13.42% | <0.001 |
| Acinetobacter baumannii | 16/83 | 19.28% | 51/82 | 62.20% | 0.020 |
| Klebsiella pneumoniae | 19/83 | 22.89% | 37/82 | 45.12% | <0.001 |
| Staphylococcus aureus | - | - | 5/82 | 6.10% | - |
| Pandorea species | 1/83 | 1.21% | - | - | - |
| Burkholderia cepacia | 3/83 | 3.61% | - | - | - |
| Sphingomonas paucimobilis | 1/83 | 1.21% | - | - | - |
| Gram-negative-Staphylococcus species | 1/83 | 1.21% | - | - | - |
| Acinetobacter species | 5/83 | 6.02% | - | - | - |
| Klebsiella species | 4/83 | 4.82% | - | - | - |
| Pseudomonas species | 1/83 | 1.21% | - | - | - |
| MRSA | 4/83 | 4.82% | - | - | - |
| Candida albicans | 15/83 | 18.07% | - | - | - |
| Candida tropicalis | 2/83 | 2.41% | - | - | - |
| Candida krusei | 1/83 | 1.22% | - | - | - |
| Candida glabrata | 3/83 | 3.61% | - | - | - |
| Candida species | 1/83 | 1.21% | - | - | - |
| Total Number of Detections per Patient | M (±SD) | Pearson Correlation | p-Value | |
|---|---|---|---|---|
| Culture | 0–2 | 0.579 (±0.587) | 0.287 | 0.001 |
| qPCR | 0–8 | 2.244 (±1.796) |
| Antibiotic | Mechanism of Resistance | Resistance Gene | N (%) * |
|---|---|---|---|
| Carbapenem | Carbapenemase-hydrolyzing enzymes | oxa-23 | 38/65 (58.46%) |
| oxa-58 | 38/65 (58.46%) | ||
| oxa-48 | 38/65 (58.46%) | ||
| oxa-51 | 22/65 (33.85%) | ||
| Imp | 2/65 (3.08%) | ||
| kpc | - | ||
| ndm | 39/65 (60%) | ||
| vim | 4/65 (6.15%) | ||
| Vancomycin | Synthesis of modified peptidoglycan precursors with reduced binding affinity | vanA/B | 1/5 (20%) |
| Quinolone | Synthesis of barrier protein for DNA gyrase | qnr | 31/64 (48.44%) |
| Aminoglycoside β-lactam Cephalosporin Quinolone Trimethoprim–sulfamethoxazole | ESBL-hydrolyzing enzymes | CTX-M | 26/64 (40.63%) |
| Methicillin | Encoding penicillin-binding protein 2a (PBP2a), which has a low affinity for beta-lactams | mecA/mecC | 32/64 (50%) |
| Antibiotic (N) * | Culture | Resistance Genes | qPCR | p-Value (Pearson Chi-Square Test) | ||
|---|---|---|---|---|---|---|
| Resistance N (%) | Sensitive N (%) | Detected N (%) ** | Not detected N (%) ** | |||
| Imipenem (47) | 36/47 (76.60%) | 12/47 (23.40%) | oxa-23 | 27/36 (75%) | 9/36 (25%) | 0.530 |
| oxa-58 | 27/36 (75%) | 9/36 (25%) | 0.530 | |||
| oxa-48 | 32/36 (88.89%) | 4/36 (11.11%) | 0.004 | |||
| oxa-51 | 19/36 (52.78%) | 17/34 (47.22%) | 0.029 | |||
| OXA mix *** | 35/36 (97.22%) | 1/36 (2.78%) | 0.023 | |||
| imp | 1/36 (2.78%) | 35/36 (97.22%) | 0.855 | |||
| kpc | - | 36/36 (100%) | - | |||
| ndm | 29/36 (80.56%) | 7/36 (19.44%) | 0.671 | |||
| vim | 3/36 (8.33%) | 33/36 (91.67%) | 0.613 | |||
| CRE mix *** | 30/36 (83.33%) | 6/36 (16.67%) | 0.565 | |||
| Overall *** | 36/36 (100%) | - | 0.003 | |||
| Meropenem (37) | 31/37 (83.78%) | 6/37 (16.22%) | oxa-23 | 24/31 (77.42%) | 7/31 (22.58%) | 0.747 |
| oxa-58 | 24/31 (77.42%) | 7/31 (22.58%) | 0.747 | |||
| oxa-48 | 26/31 (83.87%) | 5/31 (16.13%) | 0.065 | |||
| oxa-51 | 16/31 (51.61%) | 15/31 (48.39%) | 0.116 | |||
| OXA mix *** | 30/31 (96.77%) | 1/31 (3.23%) | 0.183 | |||
| imp | - | 32/32 (100%) | - | |||
| kpc | - | 32/32 (100%) | - | |||
| ndm | 24/31 (77.42%) | 7/31 (22.58%) | 0.747 | |||
| vim | 1/31 (3.23%) | 30/31 (96.77%) | 0.656 | |||
| CRE mix *** | 25/31 (80.65%) | 6/31 (19.35%) | 0.878 | |||
| Overall *** | 31/31 (100%) | - | 0.021 | |||
| Ertapenem (29) | 26/29 (89.66%) | 3/29 (10.34%) | oxa-23 | 18/26 (69.23%) | 8/26 (30.77%) | 0.019 |
| oxa-58 | 18/26 (69.23%) | 8/26 (30.77%) | 0.019 | |||
| oxa-48 | 24/26 (92.31%) | 2/26 (7.69%) | 0.005 | |||
| oxa-51 | 13/26 (50%) | 13/26 (50%) | 0.099 | |||
| Oxa mix *** | 26/26 (100%) | - | <0.001 | |||
| imp | 1/26 (3.85%) | 25/26 (96.15%) | 0.730 | |||
| kpc | - | 26/26 (100%) | - | |||
| ndm | 25/26 (96.15%) | 1/26 (3.85%) | <0.001 | |||
| vim | 2/26 (7.69%) | 24/26 (92.31%) | 0.619 | |||
| CRE mix *** | 25/26 (96.15%) | 1/26 (3.85%) | <0.001 | |||
| Overall *** | 26/26 (100%) | - | <0.001 | |||
| Levofloxacin (16) | 14/16 (87.5%) | 2/16 (12.5%) | qnr | 5/14 (35.71%) | 9/14 (64.29%) | 0.696 |
| CTX-M | 5/14 (35.71%) | 9/14 (64.29%) | 0.308 | |||
| Ciprofloxacin (47) | 35/47 (74.47%) | 12/47 (25.53%) | qnr | 24/35 (68.57%) | 11/35 (31.43%) | 0.248 |
| CTX-M | 21/35 (60.00%) | 14/35 (40.00%) | 0.270 | |||
| Ampicillin–sulbactam (13) | 12/13 (92.31%) | 1/13 (7.69%) | CTX-M | 6/12 (50%) | 6/12 (50%) | 0.335 |
| Amoxicillin–clavulanic acid (12) | 11/12 (91.67%) | 1/12 (8.31%) | CTX-M | 8/11 (72.73%) | 3/11 (27.27%) | 0.546 |
| Piperacillin–tazobactam (39) | 30/39 (76.92%) | 9/39 (23.08%) | CTX-M | 17/30 (56.67%) | 13/30 (43.33%) | 0.404 |
| Aztreonam (14) | 8/14 (57.14%) | 6/14 (42.86%) | CTX-M | 3/8 (37.5%) | 5/8 (62.5%) | 0.872 |
| Cefoxitin (19) | 17/19 (89.47%) | 2/19 (10.53%) | CTX-M | 10/17 (58.82%) | 7/17 (41.18%) | 0.811 |
| Cefotaxime (31) | 27/31 (87.10) | 4/31 (12.90%) | CTX-M | 15/27 (55.56%) | 12/27 (44.44%) | 0.441 |
| Ceftazidime (41) | 33/41 (80.49%) | 8/41 (19.51%) | CTX-M | 18/33 (54.55%) | 15/33 (45.45%) | 0.817 |
| Ceftazidime–avibactam (10) | 7/10 (70%) | 3/10 (30%) | CTX-M | 5/7 (71.43%) | 2/7 (28.57%) | 0.665 |
| Ceftriaxone (33) | 31/33 (93.94%) | 2/33 (6.06%) | CTX-M | 19/31 (61.29%) | 12/31 (38.71%) | 0.751 |
| Cefepime (48) | 39/48 (81.25%) | 9/48 (18.75%) | CTX-M | 23/39 (58.97%) | 16/39 (41.03%) | 0.465 |
| Trimethoprim–sulfamethoxazole (20) | 8/20 (40%) | 12/20 (60%) | CTX-M | 3/8 (37.5%) | 5/8 (62.5%) | 0.199 |
| Gentamicin (48) | 30/48 (62.50%) | 18/48 (37.50%) | CTX-M | 18/30 (60%) | 12/30 (40%) | 0.570 |
| Amikacin (46) | 28/46 (60.87%) | 18/46 (39.13%) | CTX-M | 14/28 (50%) | 14/28 (50%) | 0.239 |
| Inclusion criteria |
| Gender: Men and Women |
| Diagnosis: Patients with a clinical diagnosis of hospital-acquired pneumonia (HAP), including ventilator-associated pneumonia (VAP), as confirmed by medical records |
| Exclusion criteria |
| Diagnosis: Patients with clinical backgrounds that do not match with the HAP diagnosis criteria |
| Samples: Samples that were insufficient or contaminated were excluded from the analysis |
| Demographics |
| Gender |
| Age |
| Weight |
| Body mass index |
| Comorbidities |
| Location (ICU or Ward) |
| Laboratory investigations |
| Temperature |
| Blood pressure, heart rate, and respiratory rate |
| Biochemistry parameters including potassium, sodium, and creatinine |
| Hematology parameters including hemoglobin, hematocrit, and white blood cell count |
| Acid and base balance parameters including PH, PCO2, PO2, HCO3, and O2 saturation. |
| Inflammation parameters including CRP and ESR |
| Risk factors for HAP and/or VAP |
| Intubation and mechanical ventilation (including duration, in days, of oxygen therapy or mechanical ventilation) |
| Aspiration |
| Tube feedings |
| Oral/dental disease, poor oral hygiene, use of antibiotics in the last three months, use of oral antiseptics, poor infection control measures |
| Hyperglycemia |
| Supine positioning of the patient |
| Stroke or other neurologic disorders, seizures |
| Alcoholism |
| Gastroesophageal reflux disease (GERD) |
| Initial Findings |
| Symptoms onset (date and time) |
| Pleuritic chest pain |
| Shortness of breath (SOB), dyspnea, tachypnoea |
| Cough |
| Sputum production |
| Hemoptysis or rust-colored sputum |
| Muscular or joint pain, headache |
| Nausea and vomiting |
| Loss of appetite |
| Hypotension (BP < 90 mmHg) |
| Tachycardia (HR > 100 beats/min) |
| Altered breath sounds or localized rales |
| Chest radiography (dense lobar or segmental infiltrate) |
| Microbiological Diagnosis |
| Microbiology sputum culture and susceptibility results |
| Antibiotic used |
| Antibiotic treatment (empirical, initial, and next) with detailed dosage regimen and duration. |
| Outcome |
| Outcome of the patient, categorized as improved, partially improved, did not improve, worsening, or death |
| Time to clinical improvement, if applicable. |
| LRB Oligo Mix | Specific Nucleic Acid Amplification and Detection |
|---|---|
| LRB Oligo Mix 1 | 6-Carboxyfluorescein (FAM): Streptococcus pyogenes Hexachloro-6-carboxyfluorescein (HEX): Human Internal Control (IC) 6-Carboxy-X-rhodamine (ROX): Enterobacteriaceae |
| LRB Oligo Mix 2 | FAM: Haemophilus influenzae ROX: Mycoplasma pneumoniae |
| LRB Oligo Mix 3 | FAM: Pseudomonas aeruginosa ROX: Streptococcus agalactiae Indodicarbocyanine (CY5): Escherichia coli |
| LRB Oligo Mix 4 | FAM: Proteus spp. HEX: Serratia marcescens ROX: Klebsiella pneumoniae CY5: Acinetobacter calcoaceticus-baumannii complex |
| LRB Oligo Mix 5 | FAM: Legionella pneumophila HEX: Klebsiella aerogenes ROX: Enterobacter cloacae complex CY5: Streptococcus pneumoniae |
| LRB Oligo Mix 6 | FAM: Staphylococcus aureus ROX: Klebsiella oxytoca CY5: Moraxella catarrhalis |
| Step | Cycle | Temperature | Duration |
|---|---|---|---|
| Enzyme Activation | 1 cycle | 52 °C | 3 min |
| Pre-Incubation | 1 cycle | 95 °C | 10 s |
| Denaturation | 95 °C | 1 s | |
| Annealing and Extension | 40 cycles | 95 °C | 1 s |
| 55 °C | 15 s | ||
| Detection (Reading) | (FAM-Green) (HEX-Yellow) | (ROX-Orange) (CY5-Red) |
| Urinary Tract Antibiotic Resistance Kit | Carbapenem Resistance Kit | Vancomycin Resistance Kit | |||
|---|---|---|---|---|---|
| Oligo Mix | Intended Use | Oligo Mix | Intended Use | Oligo Mix | Intended Use |
| UTABR Oligo Mix 2 | CRE Oligo Mix | FAM: KPC | |||
| FAM: qnr—Quinolone resistance | HEX: NDM | VRE Oligo Mix | FAM: vanA/vanB | ||
| ROX: vanB—Vancomycin resistance | ROX: VIM | HEX: Human (Internal Control) | |||
| CY5: OXA-48—Carbapenem resistance | CY5: IMP | ||||
| UTABR Oligo Mix 3 | OXA Oligo Mix | ||||
| FAM: mecA/mecC—Methicillin resistance | FAM: OXA-51 | ||||
| ROX: CTX-M ESBL | HEX: Human (Internal Control) | ||||
| ROX: OXA-23/OXA-58 | |||||
| CY5: OXA-48 | |||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abu Khadija, L.H.; Alomari, S.M.; Alsayed, A.R.; Khader, H.A.; Permana, A.D.; Hasoun, L.Z.; Zraikat, M.S.; Ashran, W.; Zihlif, M. Beyond Culture: Real-Time PCR Performance in Detecting Causative Pathogens and Key Antibiotic Resistance Genes in Hospital-Acquired Pneumonia. Antibiotics 2025, 14, 937. https://doi.org/10.3390/antibiotics14090937
Abu Khadija LH, Alomari SM, Alsayed AR, Khader HA, Permana AD, Hasoun LZ, Zraikat MS, Ashran W, Zihlif M. Beyond Culture: Real-Time PCR Performance in Detecting Causative Pathogens and Key Antibiotic Resistance Genes in Hospital-Acquired Pneumonia. Antibiotics. 2025; 14(9):937. https://doi.org/10.3390/antibiotics14090937
Chicago/Turabian StyleAbu Khadija, Lana Hani, Shatha M. Alomari, Ahmad R. Alsayed, Heba A. Khader, Andi Dian Permana, Luai Z. Hasoun, Manar Saleh Zraikat, Walaa Ashran, and Malek Zihlif. 2025. "Beyond Culture: Real-Time PCR Performance in Detecting Causative Pathogens and Key Antibiotic Resistance Genes in Hospital-Acquired Pneumonia" Antibiotics 14, no. 9: 937. https://doi.org/10.3390/antibiotics14090937
APA StyleAbu Khadija, L. H., Alomari, S. M., Alsayed, A. R., Khader, H. A., Permana, A. D., Hasoun, L. Z., Zraikat, M. S., Ashran, W., & Zihlif, M. (2025). Beyond Culture: Real-Time PCR Performance in Detecting Causative Pathogens and Key Antibiotic Resistance Genes in Hospital-Acquired Pneumonia. Antibiotics, 14(9), 937. https://doi.org/10.3390/antibiotics14090937






