P3MA: A Promising Mycobacteriophage Infecting Mycobacterium abscessus
Abstract
1. Introduction
2. Results
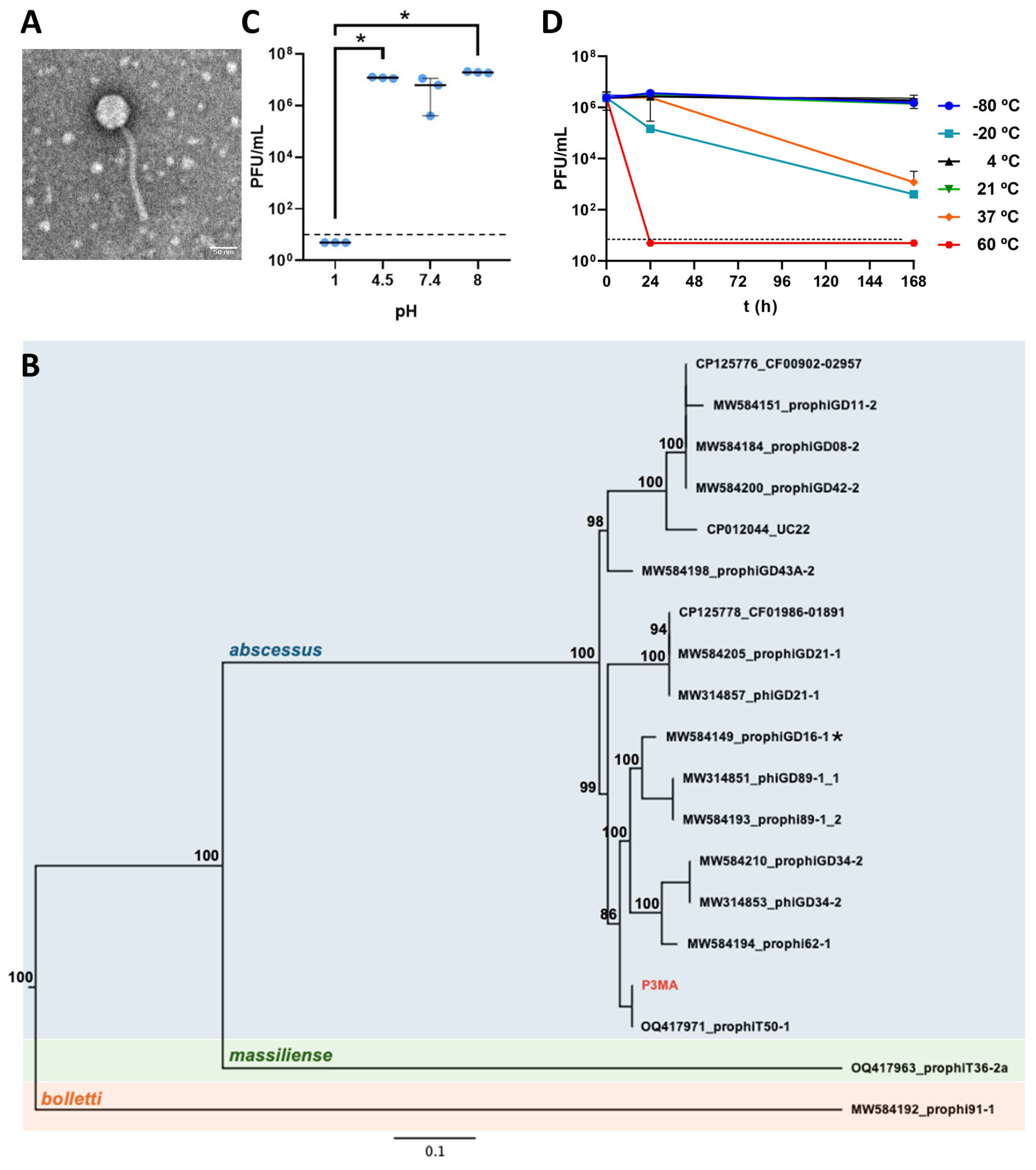
2.1. P3MA Is a Siphovirus with 17 Sequences Genetically Near and Stable at Physiological Conditions
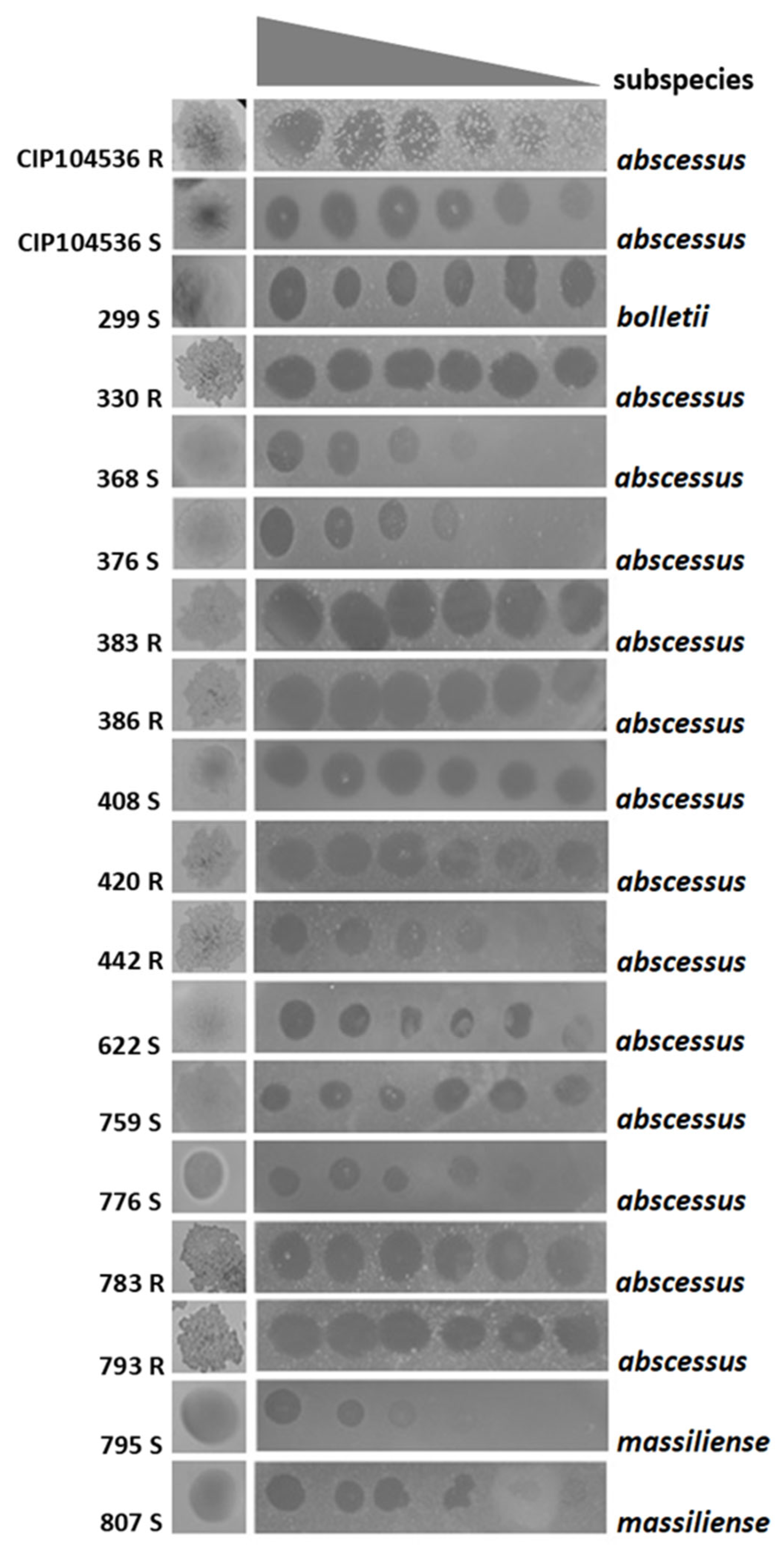
2.2. P3MA Infects Different Clinical M. abscessus Strains
2.3. Synergistic Effect of P3MA with Imipenem and Antagonism with Clarithromycin
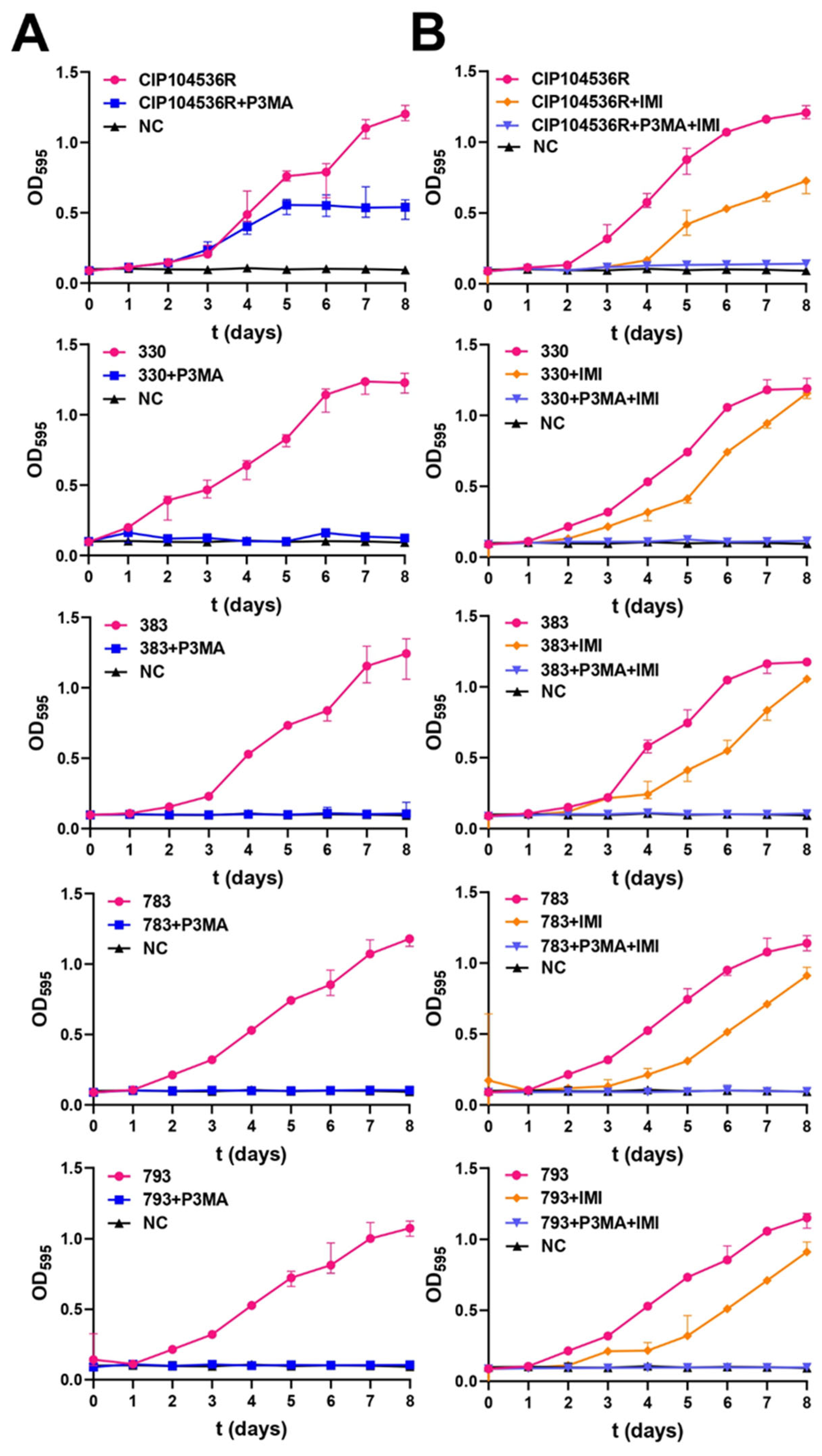
2.4. P3MA Inhibits M. abscessus Growth Alone and Combined with Imipenem
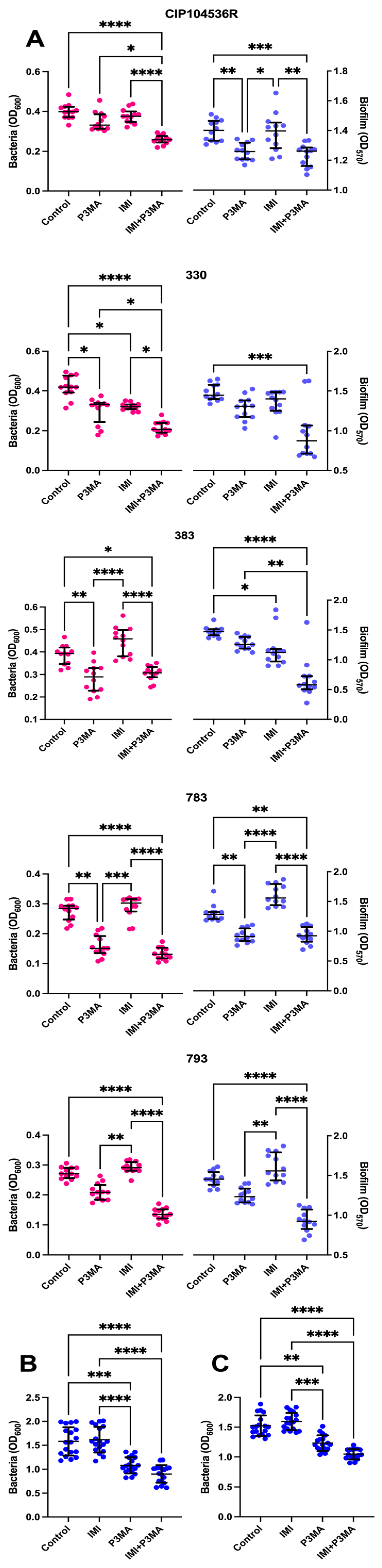
2.5. Mycobacteriophage Effect on M. abscessus Biofilms and a Granuloma-like Model
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains
4.2. Bacteriophage Isolation
4.3. Comparative Genomics
4.4. Bacteriophage Enrichment
4.5. Bacteriophage Stability Testing
4.6. Bacteriophage Host Range Analysis
4.7. Electron Microscopy
4.8. Checkerboard
4.9. Bacteriophage Inhibition Assays
4.10. Mycobacteriophage Effect on M. abscessus Biofilms
4.11. Granuloma Model
4.12. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tortoli, E. Microbiological features and clinical relevance of new species of the genus Mycobacterium. Clin. Microbiol. Rev. 2014, 27, 727–752. [Google Scholar] [CrossRef]
- Cho, Y.-J.; Yi, H.; Chun, J.; Cho, S.-N.; Daley, C.L.; Koh, W.-J.; Jae Shin, S. The genome sequence of Mycobacterium massiliense strain CIP 108297 suggests the independent taxonomic status of the Mycobacterium abscessus complex at the subspecies level. PLoS ONE 2013, 8, e81560. [Google Scholar] [CrossRef] [PubMed]
- Sassi, M.; Drancourt, M. Genome analysis reveals three genomospecies in Mycobacterium abscessus. BMC Genom. 2014, 15, 359. [Google Scholar] [CrossRef]
- Johansen, M.D.; Herrmann, J.-L.; Kremer, L. Non-tuberculous mycobacteria and the rise of Mycobacterium abscessus. Nat. Rev. Microbiol. 2020, 18, 392–407. [Google Scholar] [CrossRef]
- Bernut, A.; Herrmann, J.-L.; Kissa, K.; Dubremetz, J.-F.; Gaillard, J.-L.; Lutfalla, G.; Kremer, L. Mycobacterium abscessus cording prevents phagocytosis and promotes abscess formation. Proc. Natl. Acad. Sci. USA 2014, 111, E943–E952. [Google Scholar] [CrossRef] [PubMed]
- Schenkel, A.R.; Mitchell, J.D.; Cool, C.D.; Bai, X.; Groshong, S.; Koelsch, T.; Verma, D.; Ordway, D.; Chan, E.D. Characterization of immune cells from the lungs of patients with chronic non-tuberculous mycobacteria or Pseudomonas aeruginosa infection. Immune Netw. 2022, 22, e27. [Google Scholar] [CrossRef]
- Dorhoi, A.; Reece, S.T.; Kaufmann, S.H.E. For better or for worse: The immune response against Mycobacterium tuberculosis balances pathology and protection. Immunol. Rev. 2011, 240, 235–251. [Google Scholar] [CrossRef]
- To, K.; Cao, R.; Yegiazaryan, A.; Owens, J.; Venketaraman, V. General overview of nontuberculous mycobacteria opportunistic pathogens: Mycobacterium avium and Mycobacterium abscessus. J. Clin. Med. 2020, 9, 2541. [Google Scholar] [CrossRef]
- Catherinot, E.; Roux, A.-L.; Macheras, E.; Hubert, D.; Matmar, M.; Dannhoffer, L.; Chinet, T.; Morand, P.; Poyart, C.; Heym, B.; et al. Acute respiratory failure involving an R variant of Mycobacterium abscessus. J. Clin. Microbiol. 2009, 47, 271–274. [Google Scholar] [CrossRef]
- Hedin, W.; Fröberg, G.; Fredman, K.; Chryssanthou, E.; Selmeryd, I.; Gillman, A.; Orsini, L.; Runold, M.; Jönsson, B.; Schön, T.; et al. A rough colony morphology of Mycobacterium abscessus is associated with cavitary pulmonary disease and poor clinical outcome. J. Infect. Dis. 2023, 227, 820–827. [Google Scholar] [CrossRef] [PubMed]
- Richards, J.P.; Ojha, A.K. Mycobacterial biofilms. Microbiol. Spectr. 2014, 2. [Google Scholar] [CrossRef]
- Strnad, L.; Winthrop, K. Treatment of Mycobacterium abscessus complex. Semin. Respir. Crit. Care Med. 2018, 39, 362–376. [Google Scholar] [CrossRef]
- Haworth, C.S.; Banks, J.; Capstick, T.; Fisher, A.J.; Gorsuch, T.; Laurenson, I.F.; Leitch, A.; Loebinger, M.R.; Milburn, H.J.; Nightingale, M.; et al. British thoracic society guideline for the management of non-tuberculous mycobacterial pulmonary disease (NTM-PD). BMJ Open Respir. Res. 2017, 4, e000242. [Google Scholar] [CrossRef]
- Daley, C.L.; Iaccarino, J.M.; Lange, C.; Cambau, E.; Wallace, R.J.; Andrejak, C.; Böttger, E.C.; Brozek, J.; Griffith, D.E.; Guglielmetti, L.; et al. Treatment of nontuberculous mycobacterial pulmonary disease: An official ATS/ERS/ESCMID/IDSA clinical practice guideline. Clin. Infect. Dis. 2020, 71, 905–913. [Google Scholar] [CrossRef]
- Broncano-Lavado, A.; Senhaji-Kacha, A.; Santamaría-Corral, G.; Esteban, J.; García-Quintanilla, M. Alternatives to antibiotics against Mycobacterium abscessus. Antibiotics 2022, 11, 1322. [Google Scholar] [CrossRef]
- List of sequenced Actinobacteriophages. Available online: https://phagesdb.org/phages/ (accessed on 24 June 2025).
- Hatfull, G.F. Mycobacteriophages. Microbiol. Spectr. 2018, 6. [Google Scholar] [CrossRef]
- Dedrick, R.M.; Smith, B.E.; Garlena, R.A.; Russell, D.A.; Aull, H.G.; Mahalingam, V.; Divens, A.M.; Guerrero-Bustamante, C.A.; Zack, K.M.; Abad, L.; et al. Mycobacterium abscessus strain morphotype determines phage susceptibility, the repertoire of therapeutically useful phages, and phage resistance. mBio 2021, 12, e03431-20. [Google Scholar] [CrossRef]
- Bonacorsi, A.; Ferretti, C.; Di Luca, M.; Rindi, L. Mycobacteriophages and their applications. Antibiotics 2024, 13, 926. [Google Scholar] [CrossRef] [PubMed]
- Cristinziano, M.; Shashkina, E.; Chen, L.; Xiao, J.; Miller, M.B.; Doligalski, C.; Coakley, R.; Lobo, L.J.; Footer, B.; Bartelt, L.; et al. Use of epigenetically modified bacteriophage and dual beta-lactams to treat a Mycobacterium abscessus sternal wound infection. Nat. Commun. 2024, 15, 10360. [Google Scholar] [CrossRef] [PubMed]
- Dedrick, R.M.; Smith, B.E.; Cristinziano, M.; Freeman, K.G.; Jacobs-Sera, D.; Belessis, Y.; Whitney Brown, A.; Cohen, K.A.; Davidson, R.M.; Van Duin, D.; et al. Phage therapy of Mycobacterium infections: Compassionate use of phages in 20 patients with drug-resistant mycobacterial disease. Clin. Infect. Dis. 2023, 76, 103–112. [Google Scholar] [CrossRef] [PubMed]
- Gorzynski, M.; De Ville, K.; Week, T.; Jaramillo, T.; Danelishvili, L. Understanding the phage-host interaction mechanism toward improving the efficacy of current antibiotics in Mycobacterium abscessus. Biomedicines 2023, 11, 1379. [Google Scholar] [CrossRef]
- Johansen, M.D.; Alcaraz, M.; Dedrick, R.M.; Roquet-Banères, F.; Hamela, C.; Hatfull, G.F.; Kremer, L. Mycobacteriophage–antibiotic therapy promotes enhanced clearance of drug-resistant Mycobacterium abscessus. Dis. Models Mech. 2021, 14, dmm049159. [Google Scholar] [CrossRef]
- Broncano-Lavado, A.; Roquet-Banères, F.; Kremer, L.; Russell, D.A.; Jacobs-Sera, D.; Reidy, T.G.; Aguilera-Correa, J.J.; Esteban, J.; Hatfull, G.F.; García-Quintanilla, M. Genome sequence of Mycobacterium abscessus phage P3MA. Microbiol. Resour. Announc. 2025, 14, e0016925. [Google Scholar] [CrossRef] [PubMed]
- Amarh, E.D.; Dedrick, R.M.; Garlena, R.A.; Russell, D.A.; Gauthier, C.H.; Aull, H.G.; Abad, L.; Jacobs-Sera, D.; Akusobi, C.; Rubin, E.J.; et al. Unusual prophages in Mycobacterium abscessus genomes and strain variations in phage susceptibilities. PLoS ONE 2023, 18, e0281769. [Google Scholar] [CrossRef] [PubMed]
- Woods, G.L.; Brown-Elliott, B.A.; Conville, P.S.; Desmond, E.P.; Hall, G.S.; Lin, G.; Pfyffer, G.E.; Ridderhof, J.C.; Siddiqi, S.H.; Wallace, R.J.; et al. Susceptibility Testing of Mycobacteria, Nocardiae, and Other Aerobic Actinomycetes. In CLSI Standards: Guidelines for Health Care Excellence, 2nd ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2011; ISBN 978-1-56238-746-4. [Google Scholar]
- Sornsuvit, C.; Wientong, P.; Uitrakul, S.; Okonogi, S.; Katip, W. Influence of concentration and temperature on stability of imipenem focused on solutions for extended infusion. Dose Response 2021, 19, 15593258211059325. [Google Scholar] [CrossRef]
- Degiacomi, G.; Sammartino, J.C.; Chiarelli, L.R.; Riabova, O.; Makarov, V.; Pasca, M.R. Mycobacterium abscessus, an emerging and worrisome pathogen among cystic fibrosis patients. Int. J. Mol. Sci. 2019, 20, 5868. [Google Scholar] [CrossRef]
- Santamaría-Corral, G.; Pagán, I.; Aguilera-Correa, J.J.; Esteban, J.; García-Quintanilla, M. A novel bacteriophage infecting multi-drug- and extended-drug-resistant Pseudomonas aeruginosa strains. Antibiotics 2024, 13, 523. [Google Scholar] [CrossRef]
- Chang, R.Y.K.; Wallin, M.; Kutter, E.; Morales, S.; Britton, W.; Li, J.; Chan, H.-K. Storage stability of inhalable phage powders containing lactose at ambient conditions. Int. J. Pharm. 2019, 560, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Malik, D.J. Bacteriophage encapsulation using spray drying for phage therapy. Curr. Issues Mol. Biol. 2021, 40, 303–316. [Google Scholar] [CrossRef]
- Cao Yao, J.C.; Garcia Cehic, D.; Quer, J.; Méndez, J.N.; Gorrín, A.D.; Hevia, L.G.; Fernández, M.T.T. Complete genome sequences of four mycobacteriophages involved in directed evolution against undisputed Mycobacterium abscessus clinical strains. Microorganisms 2024, 12, 374. [Google Scholar] [CrossRef]
- Ross, A.; Ward, S.; Hyman, P. More is better: Selecting for broad host range bacteriophages. Front. Microbiol. 2016, 7, 1352. [Google Scholar] [CrossRef] [PubMed]
- Holtappels, D.; Alfenas-Zerbini, P.; Koskella, B. Drivers and consequences of bacteriophage host range. FEMS Microbiol. Rev. 2023, 47, fuad038. [Google Scholar] [CrossRef] [PubMed]
- Knezevic, P.; Curcin, S.; Aleksic, V.; Petrusic, M.; Vlaski, L. Phage-antibiotic synergism: A possible approach to combatting Pseudomonas aeruginosa. Res. Microbiol. 2013, 164, 55–60. [Google Scholar] [CrossRef]
- Kortright, K.E.; Chan, B.K.; Koff, J.L.; Turner, P.E. Phage therapy: A renewed approach to combat antibiotic-resistant bacteria. Cell Host Microbe 2019, 25, 219–232. [Google Scholar] [CrossRef] [PubMed]
- Vázquez, R.; Díez-Martínez, R.; Domingo-Calap, P.; García, P.; Gutiérrez, D.; Muniesa, M.; Ruiz-Ruigómez, M.; Sanjuán, R.; Tomás, M.; Tormo-Mas, M.Á.; et al. Essential topics for the regulatory consideration of phages as clinically valuable therapeutic agents: A perspective from Spain. Microorganisms 2022, 10, 717. [Google Scholar] [CrossRef]
- Comeau, A.M.; Tétart, F.; Trojet, S.N.; Prère, M.-F.; Krisch, H.M. Phage-Antibiotic Synergy (PAS): Beta-lactam and quinolone antibiotics stimulate virulent phage growth. PLoS ONE 2007, 2, e799. [Google Scholar] [CrossRef]
- Gu Liu, C.; Green, S.I.; Min, L.; Clark, J.R.; Salazar, K.C.; Terwilliger, A.L.; Kaplan, H.B.; Trautner, B.W.; Ramig, R.F.; Maresso, A.W. Phage-Antibiotic Synergy is driven by a unique combination of antibacterial mechanism of action and stoichiometry. mBio 2020, 11, e01462-20. [Google Scholar] [CrossRef]
- Supina, B.S.I.; Dennis, J.J. The current landscape of Phage–Antibiotic Synergistic (PAS) interactions. Antibiotics 2025, 14, 545. [Google Scholar] [CrossRef]
- Labrie, S.J. Bacteriophage resistance mechanisms. Nat. Rev. Microbiol. 2010, 8, 317–327. [Google Scholar] [CrossRef]
- Simmons, E.L.; Bond, M.C.; Koskella, B.; Drescher, K.; Bucci, V.; Nadell, C.D. Biofilm structure promotes coexistence of phage-resistant and phage-susceptible bacteria. mSystems 2020, 5, e00877-19. [Google Scholar] [CrossRef]
- Abedon, S.T. Ecology and evolutionary biology of hindering phage therapy: The phage tolerance vs. phage resistance of bacterial biofilms. Antibiotics 2023, 12, 245. [Google Scholar] [CrossRef] [PubMed]
- Fajardo-Lubian, A.; Venturini, C. Use of bacteriophages to target intracellular pathogens. Clin. Infect. Dis. 2023, 77, S423–S432. [Google Scholar] [CrossRef] [PubMed]
- Bichet, M.C.; Chin, W.H.; Richards, W.; Lin, Y.-W.; Avellaneda-Franco, L.; Hernandez, C.A.; Oddo, A.; Chernyavskiy, O.; Hilsenstein, V.; Neild, A.; et al. Bacteriophage uptake by mammalian cell layers represents a potential sink that may impact phage therapy. iScience 2021, 24, 102287. [Google Scholar] [CrossRef] [PubMed]
- Lehti, T.A.; Pajunen, M.I.; Skog, M.S.; Finne, J. Internalization of a polysialic acid-binding Escherichia coli bacteriophage into eukaryotic neuroblastoma cells. Nat. Commun. 2017, 8, 1915. [Google Scholar] [CrossRef]
- Schmalstig, A.A.; Wiggins, A.; Badillo, D.; Wetzel, K.S.; Hatfull, G.F.; Braunstein, M. Bacteriophage infection and killing of intracellular Mycobacterium abscessus. mBio 2024, 15, e02924-23. [Google Scholar] [CrossRef]
- Twest, R.; Kropinski, A.M. Bacteriophage enrichment from water and soil. In Bacteriophages; Clokie, M.R.J., Kropinski, A.M., Eds.; Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2009; Volume 501, pp. 15–21. ISBN 978-1-58829-682-5. [Google Scholar]
- Minh, B.Q.; Nguyen, M.A.T.; Von Haeseler, A. Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 2013, 30, 1188–1195. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; Von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Nguyen, L.-T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Gill, J.; Hyman, P. Phage choice, isolation, and preparation for phage therapy. CPB 2010, 11, 2–14. [Google Scholar] [CrossRef]
- Dedrick, R.M.; Guerrero-Bustamante, C.A.; Garlena, R.A.; Russell, D.A.; Ford, K.; Harris, K.; Gilmour, K.C.; Soothill, J.; Jacobs-Sera, D.; Schooley, R.T.; et al. Engineered bacteriophages for treatment of a patient with a disseminated drug resistant Mycobacterium abscessus. Nat. Med. 2019, 25, 730–733. [Google Scholar] [CrossRef]
- Ackermann, H.-W. Bacteriophage electron microscopy. In Advances in Virus Research; Elsevier: Amsterdam, The Netherlands, 2012; Volume 82, pp. 1–32. ISBN 978-0-12-394621-8. [Google Scholar]
- Odds, F.C. Synergy, antagonism, and what the chequerboard puts between them. J. Antimicrob. Chemother. 2003, 52, 1. [Google Scholar] [CrossRef]
- Bellio, P.; Fagnani, L.; Nazzicone, L.; Celenza, G. New and simplified method for drug combination studies by checkerboard assay. MethodsX 2021, 8, 101543. [Google Scholar] [CrossRef]
- Wetzel, K.S.; Illouz, M.; Abad, L.; Aull, H.G.; Russell, D.A.; Garlena, R.A.; Cristinziano, M.; Malmsheimer, S.; Chalut, C.; Hatfull, G.F.; et al. Therapeutically useful mycobacteriophages BPs and Muddy require trehalose polyphleates. Nat. Microbiol. 2023, 8, 1717–1731. [Google Scholar] [CrossRef]
- Rodloff, A.C.; Goldstein, E.J.C.; Torres, A. Two decades of imipenem therapy. J. Antimicrob. Chemother. 2006, 58, 916–929. [Google Scholar] [CrossRef]
- Esteban, J.; Martín-de-Hijas, N.Z.; Kinnari, T.J.; Ayala, G.; Fernández-Roblas, R.; Gadea, I. Biofilm development by potentially pathogenic non-pigmented rapidly growing mycobacteria. BMC Microbiol. 2008, 8, 184. [Google Scholar] [CrossRef]
- Pettit, R.K.; Weber, C.A.; Pettit, G.R. Application of a high throughput Alamar blue biofilm susceptibility assay to Staphylococcus aureus biofilms. Ann. Clin. Microbiol. Antimicrob. 2009, 8, 28. [Google Scholar] [CrossRef] [PubMed]
- Ackart, D.F.; Hascall-Dove, L.; Caceres, S.M.; Kirk, N.M.; Podell, B.K.; Melander, C.; Orme, I.M.; Leid, J.G.; Nick, J.A.; Basaraba, R.J. Expression of antimicrobial drug tolerance by attached communities of Mycobacterium tuberculosis. Pathog. Dis. 2014, 70, 359–369. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, P.; Kumar, A. The extracellular matrix of mycobacterial biofilms: Could we shorten the treatment of mycobacterial infections? Microb. Cell 2019, 6, 105–122. [Google Scholar] [CrossRef] [PubMed]
| Antibiotic | Strain | ATB (µg/mL) (Phenotype) * | ATB-P3MA (µg/mL) | P3MA (PFU/mL) | P3MA-ATB (PFU/mL) | FICI | FICI Interpretation |
|---|---|---|---|---|---|---|---|
| Amikacin | CIP 104536 R | 32 (I) | 32 | 1011 | 1011 | 2 | Indifference |
| 330 | 16 (S) | 16 | 1011 | 1011 | 2 | Indifference | |
| 383 | 32 (I) | 32 | 1011 | 1011 | 2 | Indifference | |
| 783 | 16 (S) | 32 | 1011 | 1011 | 2 | Indifference | |
| 793 | 16 (S) | 32 | 1011 | 1011 | 2 | Indifference | |
| Imipenem | CIP 104536 R | 32 (I) | 2 | 1011 | 1010 | 0.1625 | Synergy |
| 330 | 64 (R) | 2 | 1011 | 1010 | 0.13125 | Synergy | |
| 383 | 64 (R) | 4 | 1011 | 1010 | 0.1625 | Synergy | |
| 783 | 64 (R) | 8 | 1011 | 1010 | 0.225 | Synergy | |
| 793 | 64 (R) | 8 | 1011 | 1010 | 0.225 | Synergy | |
| Clarithromycin | CIP 104536 R | 2 (S) | 8 | 1011 | 1011 | 5 | Antagonism |
| 330 | 4 (I) | 32 | 1011 | 1011 | 9 | Antagonism | |
| 383 | 2 (S) | 16 | 1011 | 1011 | 9 | Antagonism | |
| 783 | 4 (I) | 16 | 1011 | 1011 | 5 | Antagonism | |
| 793 | 4 (S) | 64 | 1011 | 1011 | 17 | Antagonism |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Broncano-Lavado, A.; Aguilera-Correa, J.J.; Roquet-Banères, F.; Kremer, L.; Mediero, A.; Seoane-Blanco, M.; van Raaij, M.J.; Pagán, I.; Esteban, J.; García-Quintanilla, M. P3MA: A Promising Mycobacteriophage Infecting Mycobacterium abscessus. Antibiotics 2025, 14, 801. https://doi.org/10.3390/antibiotics14080801
Broncano-Lavado A, Aguilera-Correa JJ, Roquet-Banères F, Kremer L, Mediero A, Seoane-Blanco M, van Raaij MJ, Pagán I, Esteban J, García-Quintanilla M. P3MA: A Promising Mycobacteriophage Infecting Mycobacterium abscessus. Antibiotics. 2025; 14(8):801. https://doi.org/10.3390/antibiotics14080801
Chicago/Turabian StyleBroncano-Lavado, Antonio, John Jairo Aguilera-Correa, Françoise Roquet-Banères, Laurent Kremer, Aránzazu Mediero, Mateo Seoane-Blanco, Mark J. van Raaij, Israel Pagán, Jaime Esteban, and Meritxell García-Quintanilla. 2025. "P3MA: A Promising Mycobacteriophage Infecting Mycobacterium abscessus" Antibiotics 14, no. 8: 801. https://doi.org/10.3390/antibiotics14080801
APA StyleBroncano-Lavado, A., Aguilera-Correa, J. J., Roquet-Banères, F., Kremer, L., Mediero, A., Seoane-Blanco, M., van Raaij, M. J., Pagán, I., Esteban, J., & García-Quintanilla, M. (2025). P3MA: A Promising Mycobacteriophage Infecting Mycobacterium abscessus. Antibiotics, 14(8), 801. https://doi.org/10.3390/antibiotics14080801








