The Ability of Bacteriophages to Reduce Biofilms Produced by Pseudomonas aeruginosa Isolated from Corneal Infections
Abstract
1. Introduction
2. Results
2.1. P. aeruginosa Biofilm Formation and Phage Sensitivity Test
2.2. Bacteriophage Propagation and Phage Titration
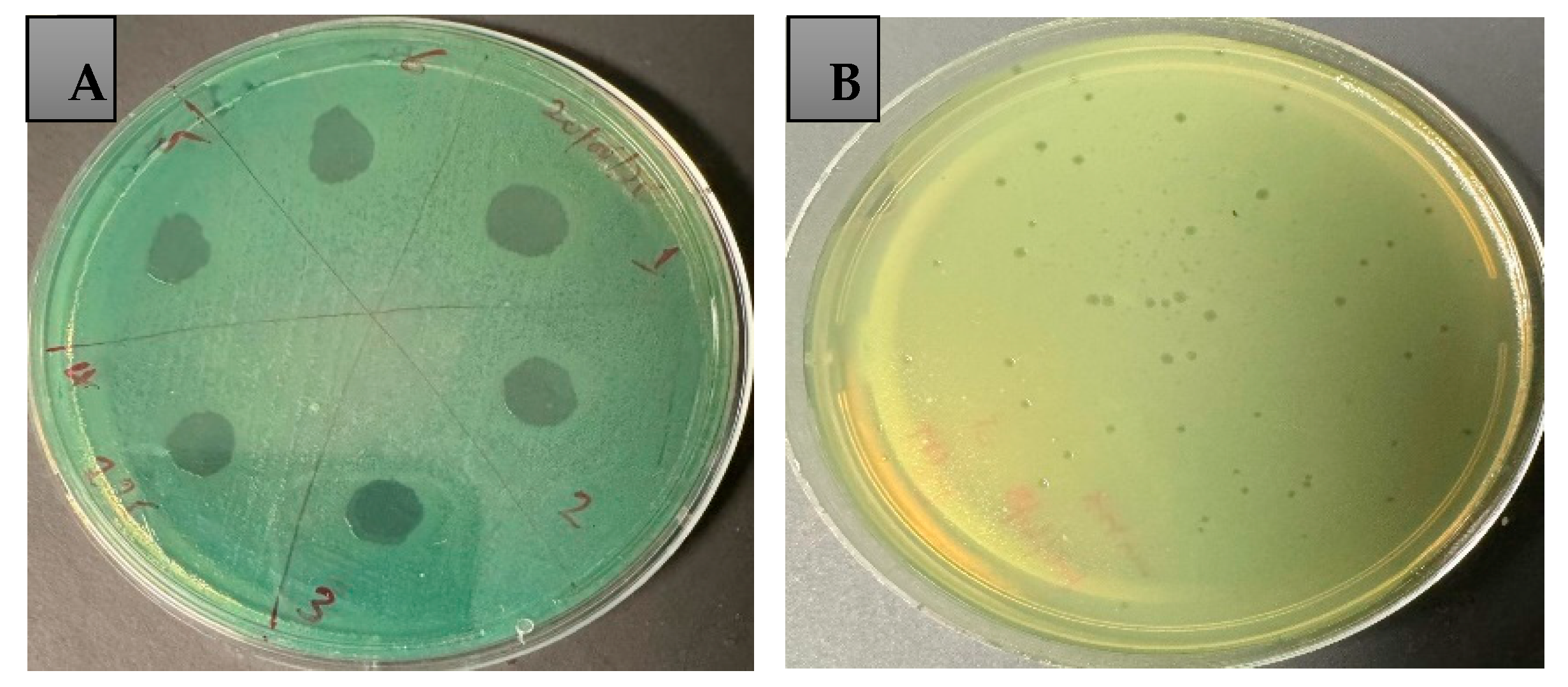
2.3. Host Range and Efficiency of Plating (EOP)
2.4. Time–Kill Assay
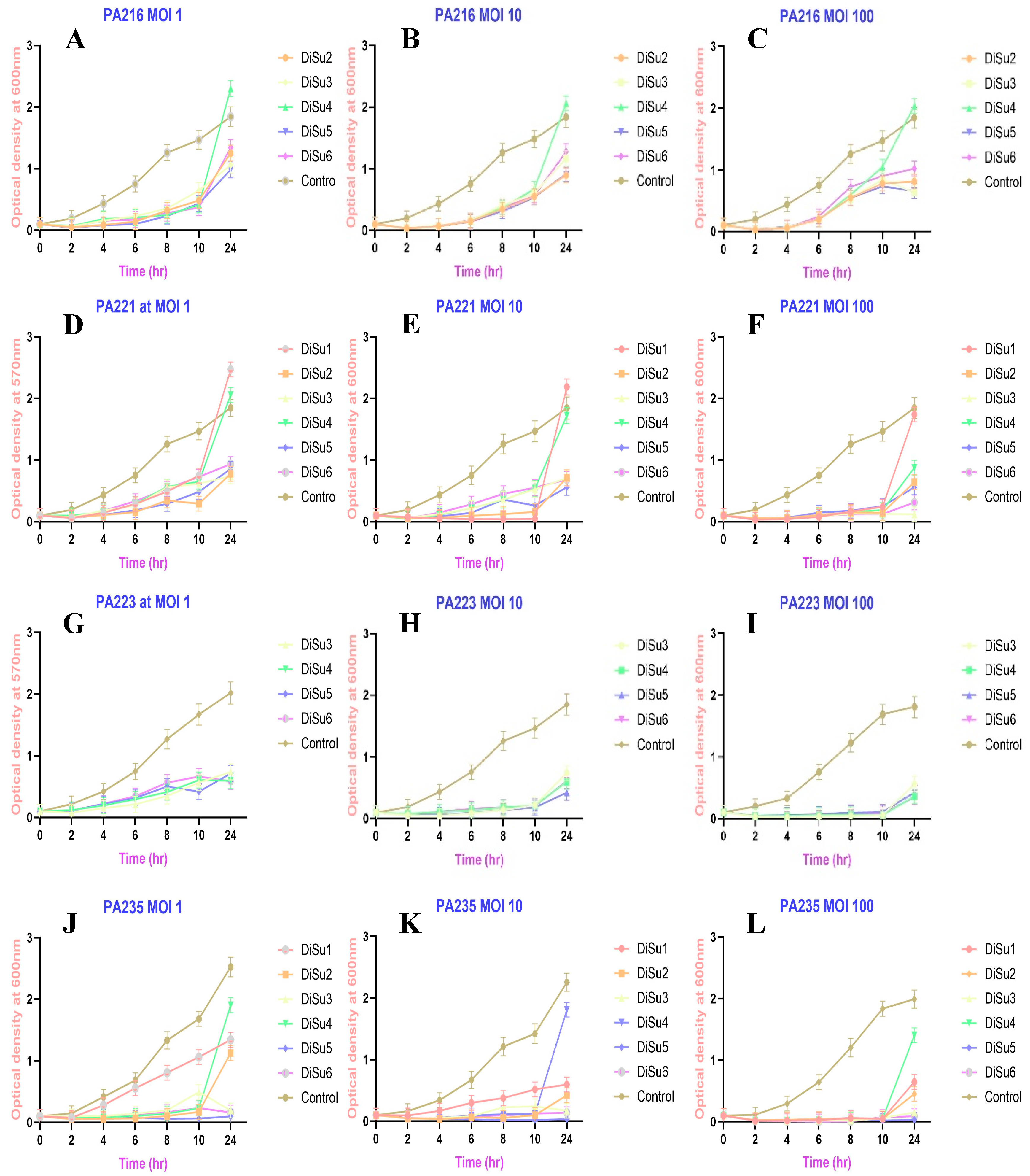
2.5. Biofilm Inhibition with Phages
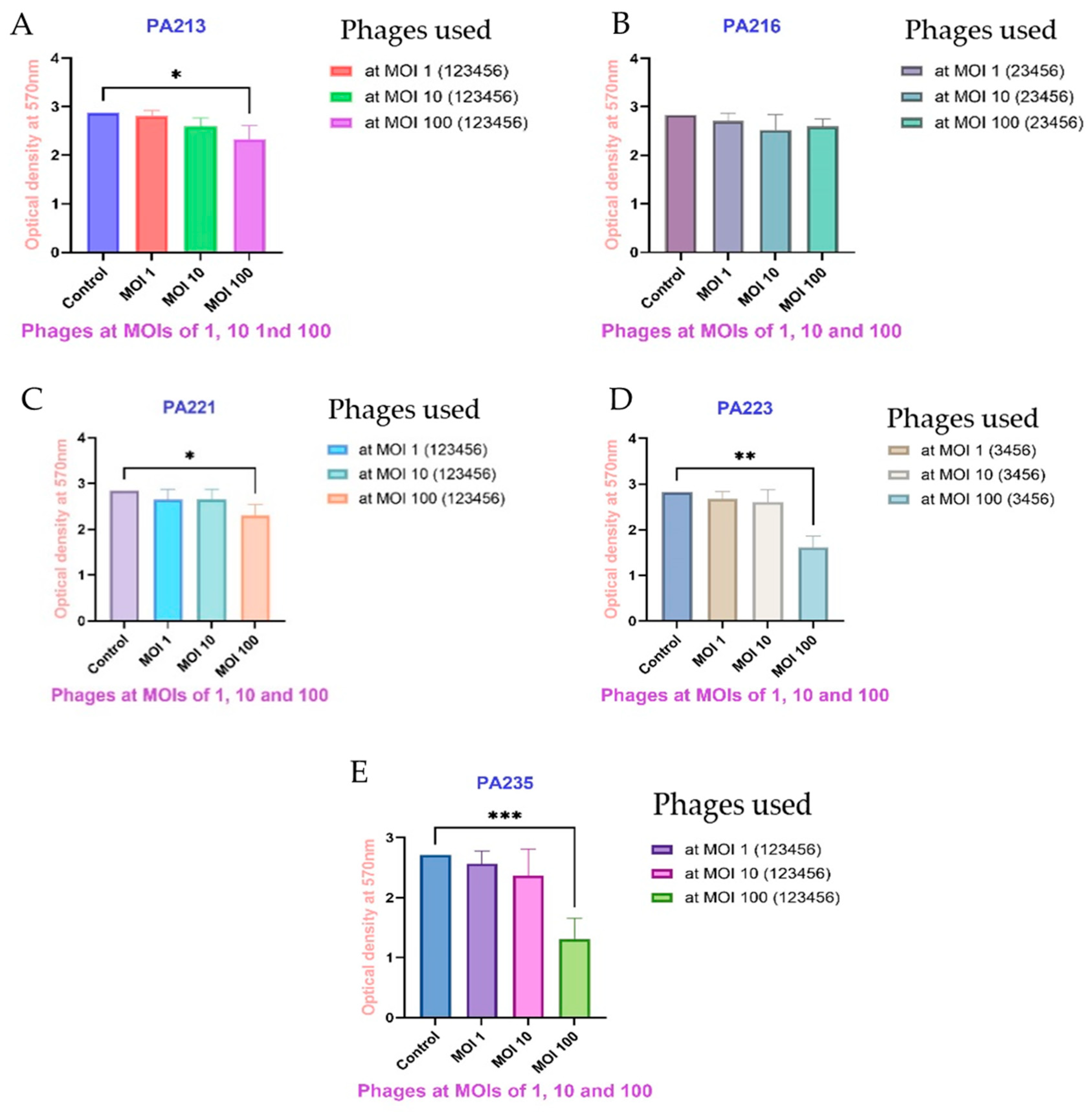
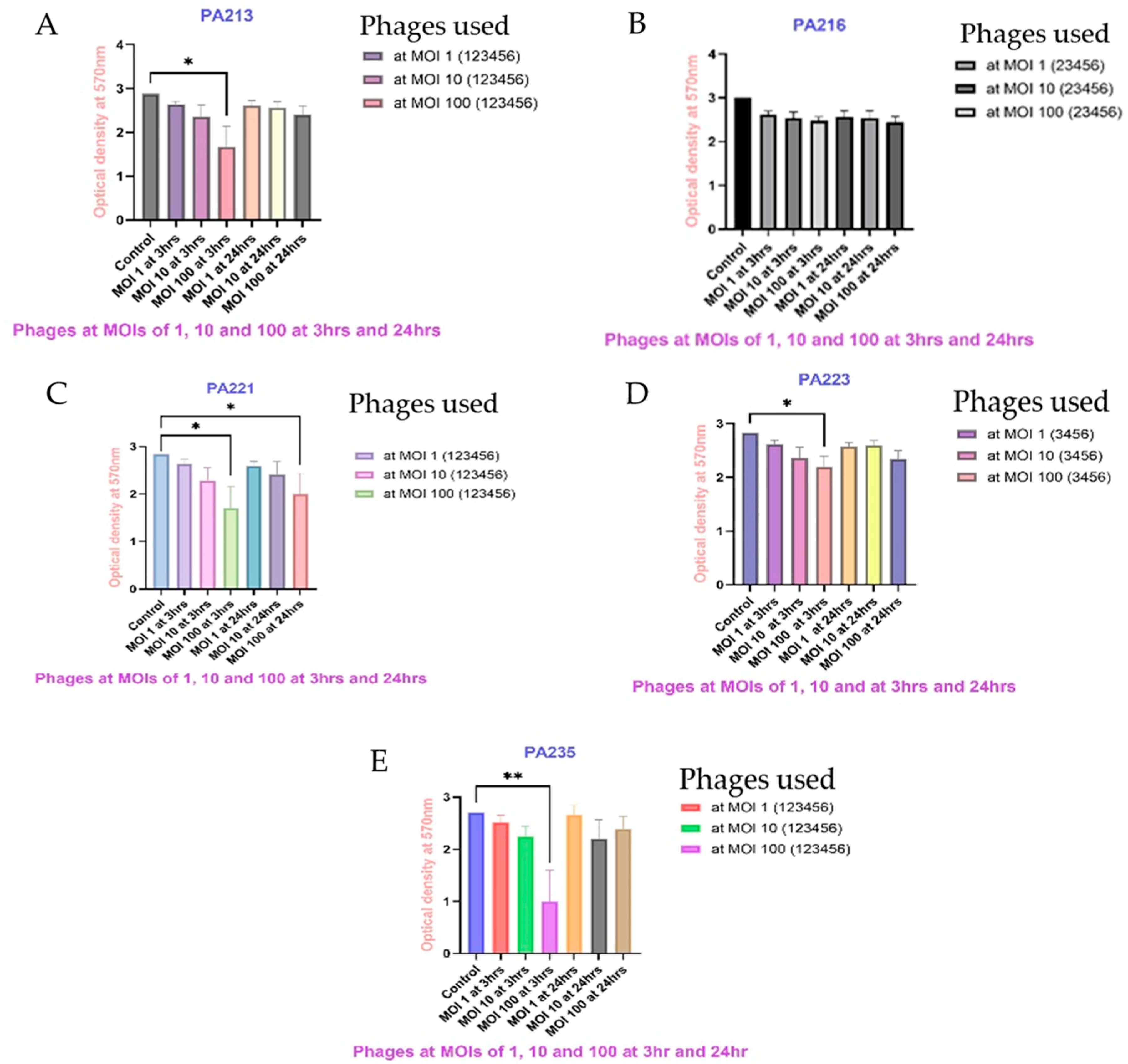
2.6. Biofilm Eradication Assay
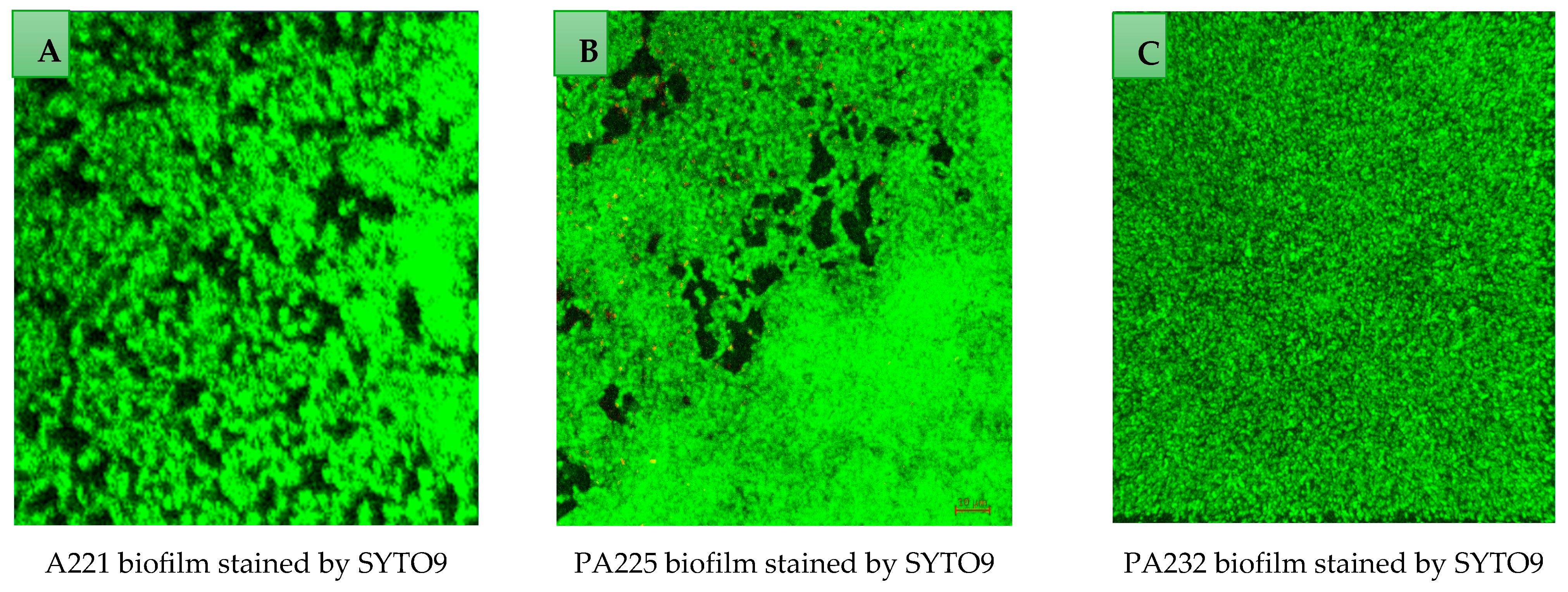
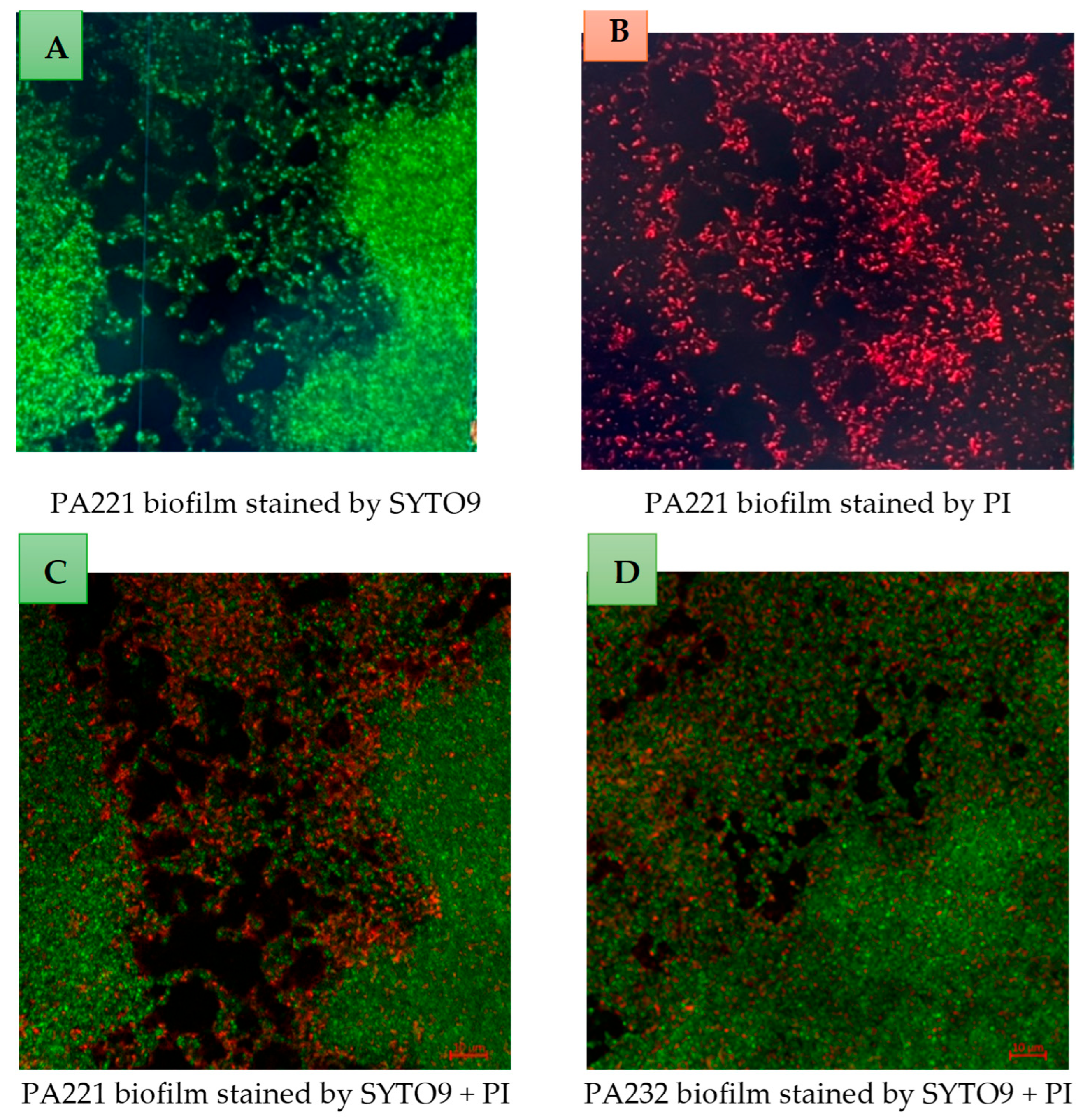
2.7. Examination of Biofilms Using Confocal Scanning Laser Microscope
3. Discussion
4. Materials and Methods
4.1. Assessment of P. aeruginosa Biofilm Formation
4.2. Bacteriophage Isolation
4.3. Bacteriophage Propagation Using Reference Bacteria
4.4. Determination of Host Range and Efficiency of Plating (EOP)
4.5. P. aeruginosa Growth Inhibition in Liquid Medium
4.6. Inhibition of Biofilm Formation by Bacteriophages
4.7. P. aeruginosa Biofilm Eradication Assay
4.8. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Horcajada, J.P.; Montero, M.; Oliver, A.; Sorlí, L.; Luque, S.; Gómez-Zorrilla, S.; Benito, N.; Grau, S. Epidemiology and treatment of multidrug-resistant and extensively drug-resistant Pseudomonas aeruginosa infections. Clin. Microbiol. Rev. 2019, 32, e00031-19. [Google Scholar] [CrossRef]
- World Health Organization. Global Antimicrobial Resistance Surveillance System (GLASS) Report: Early Implementation 2020; World Health Organization: Geneva, Switzerland, 2020. [Google Scholar]
- Munita, J.M.; Arias, C.A. Mechanisms of antibiotic resistance. In Virulence Mechanisms of Bacterial Pathogens; Wiley: Hoboken, NJ, USA, 2016; pp. 481–511. [Google Scholar]
- Blanchette, C.M.; Noone, J.M.; Stone, G.; Zacherle, E.; Patel, R.P.; Howden, R.; Mapel, D. Healthcare cost and utilization before and after diagnosis of Pseudomonas aeruginosa among patients with non-cystic fibrosis bronchiectasis in the US. Med. Sci. 2017, 5, 20. [Google Scholar] [CrossRef] [PubMed]
- Morales, E.; Cots, F.; Sala, M.; Comas, M.; Belvis, F.; Riu, M.; Salvadó, M.; Grau, S.; Horcajada, J.P.; Montero, M.M.; et al. Hospital costs of nosocomial multi-drug resistant Pseudomonas aeruginosa acquisition. BMC Health Serv. Res. 2012, 12, 122. [Google Scholar] [CrossRef] [PubMed]
- Lyczak, J.B.; Cannon, C.L.; Pier, G.B. Establishment of Pseudomonas aeruginosa infection: Lessons from a versatile opportunist. Microbes Infect. 2000, 2, 1051–1060. [Google Scholar] [CrossRef] [PubMed]
- Bassetti, M.; Merelli, M.; Temperoni, C.; Astilean, A. New antibiotics for bad bugs: Where are we? Ann. Clin. Microbiol. Antimicrob. 2013, 12, 22. [Google Scholar] [CrossRef]
- Abjani, F.; Khan, N.A.; Jung, S.Y.; Siddiqui, R. Status of the effectiveness of contact lens disinfectants in Malaysia against keratitis-causing pathogens. Exp. Parasitol. 2017, 183, 187–193. [Google Scholar] [CrossRef]
- Srinivasan, M.; Mascarenhas, J.; Rajaraman, R.; Ravindran, M.; Lalitha, P.; Glidden, D.V.; Ray, K.J.; Hong, K.C.; Oldenburg, C.E.; Lee, S.M.; et al. Corticosteroids for bacterial keratitis: The Steroids for Corneal Ulcers Trial (SCUT). Arch. Ophthalmol. 2012, 130, 143–150. [Google Scholar] [CrossRef]
- Green, M.; Apel, A.; Stapleton, F. Risk factors and causative organisms in microbial keratitis. Cornea 2008, 27, 22–27. [Google Scholar] [CrossRef]
- Shrivastava, S.R.; Shrivastava, P.S.; Ramasamy, J. World health organization releases global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. J. Med. Soc. 2018, 32, 76–77. [Google Scholar] [CrossRef]
- Poole, K. Outer membranes and efflux: The path to multidrug resistance in Gram-negative bacteria. Curr. Pharm. Biotechnol. 2002, 3, 77–98. [Google Scholar] [CrossRef]
- Fajardo, A.; Martínez-Martín, N.; Mercadillo, M.; Galán, J.C.; Ghysels, B.; Matthijs, S.; Cornelis, P.; Wiehlmann, L.; Tümmler, B.; Baquero, F.; et al. The neglected intrinsic resistome of bacterial pathogens. PLoS ONE 2008, 3, e1619. [Google Scholar] [CrossRef] [PubMed]
- Breidenstein, E.B.; de la Fuente-Núñez, C.; Hancock, R.E. Pseudomonas aeruginosa: All roads lead to resistance. Trends Microbiol. 2011, 19, 419–426. [Google Scholar] [CrossRef]
- Abd El-Aziz, A.M.; Elgaml, A.; Ali, Y.M. Bacteriophage therapy increases complement-mediated lysis of bacteria and enhances bacterial clearance after acute lung infection with multidrug-resistant Pseudomonas aeruginosa. J. Infect. Dis. 2019, 219, 1439–1447. [Google Scholar] [CrossRef]
- Bassetti, M.; Vena, A.; Croxatto, A.; Righi, E.; Guery, B. How to manage Pseudomonas aeruginosa infections. Drugs Context 2018, 7, 212527. [Google Scholar] [CrossRef] [PubMed]
- Tolker-Nielsen, T. Pseudomonas aeruginosa biofilm infections: From molecular biofilm biology to new treatment possibilities. Apmis 2014, 122, 1–51. [Google Scholar] [CrossRef]
- Thi, M.T.T.; Wibowo, D.; Rehm, B.H. Pseudomonas aeruginosa biofilms. Int. J. Mol. Sci. 2020, 21, 8671. [Google Scholar] [CrossRef]
- Esmael, A.; Abo-Elmaaty, S.A.; Khafaga, E.M.; Abdelrahman, S.; Hassan, M.G. Efficacy of three lytic bacteriophages for eradicating biofilms of multidrug-resistant Proteus mirabilis. Arch. Virol. 2021, 166, 3311–3322. [Google Scholar] [CrossRef] [PubMed]
- Tian, F.; Li, J.; Nazir, A.; Tong, Y. Bacteriophage—A promising alternative measure for bacterial biofilm control. Infect. Drug Resist. 2021, 14, 205–217. [Google Scholar] [CrossRef]
- Esmael, A.; Azab, E.; Gobouri, A.A.; Nasr-Eldin, M.A.; Moustafa, M.M.A.; Mohamed, S.A.; Badr, O.A.M.; Abdelatty, A.M. Isolation and characterization of two lytic bacteriophages infecting a multi-drug resistant Salmonella Typhimurium and their efficacy to combat salmonellosis in ready-to-use foods. Microorganisms 2021, 9, 423. [Google Scholar] [CrossRef]
- Vahdani, M.; Azimi, L.; Asghari, B.; Bazmi, F.; Lari, A.R. Phenotypic screening of extended-spectrum ß-lactamase and metallo-ß-lactamase in multidrug-resistant Pseudomonas aeruginosa from infected burns. Ann. Burn. Fire Disasters 2012, 25, 78–81. [Google Scholar]
- Ma, L.; Conover, M.; Lu, H.; Parsek, M.R.; Bayles, K.; Wozniak, D.J. Assembly and development of the Pseudomonas aeruginosa biofilm matrix. PLoS Pathog. 2009, 5, e1000354. [Google Scholar] [CrossRef]
- Thomas, P.A.; Geraldine, P. Infectious keratitis. Curr. Opin. Infect. Dis. 2007, 20, 129–141. [Google Scholar] [CrossRef]
- Kaye, S.; Tuft, S.; Neal, T.; Tole, D.; Leeming, J.; Figueiredo, F.; Armstrong, M.; McDonnell, P.; Tullo, A.; Parry, C. Bacterial susceptibility to topical antimicrobials and clinical outcome in bacterial keratitis. Investig. Ophthalmol. Vis. Sci. 2010, 51, 362–368. [Google Scholar] [CrossRef]
- Stapleton, F.; Dart, J. Pseudomonas keratitis associated with biofilm formation on a disposable soft contact lens. Br. J. Ophthalmol. 1995, 79, 864–865. [Google Scholar] [CrossRef] [PubMed]
- McLaughlin-Borlace, L.; Stapleton, F.; Matheson, M.; Dart, J.K. Bacterial biofilm on contact lenses and lens storage cases in wearers with microbial keratitis. J. Appl. Microbiol. 1998, 84, 827–838. [Google Scholar] [CrossRef]
- Tam, C.; Mun, J.J.; Evans, D.J.; Fleiszig, S.M. The impact of inoculation parameters on the pathogenesis of contact lens-related infectious keratitis. Invest. Ophthalmol. Vis. Sci. 2010, 51, 3100–3106. [Google Scholar] [CrossRef] [PubMed]
- McLeod, S.D.; LaBree, L.D.; Tayyanipour, R.; Flowers, C.W.; Lee, P.P.; McDonnell, P.J. The importance of initial management in the treatment of severe infectious corneal ulcers. Ophthalmology 1995, 102, 1943–1948. [Google Scholar] [CrossRef] [PubMed]
- Lalitha, P.; Srinivasan, M.; Manikandan, P.; Bharathi, M.J.; Rajaraman, R.; Ravindran, M.; Cevallos, V.; Oldenburg, C.E.; Ray, K.J.; Toutain-Kidd, C.M.; et al. Relationship of in vitro susceptibility to moxifloxacin and in vivo clinical outcome in bacterial keratitis. Clin. Infect. Dis. 2012, 54, 1381–1387. [Google Scholar] [CrossRef]
- Grossman, M.K.; Rankin, D.A.; Maloney, M.; Stanton, R.A.; Gable, P.; Stevens, V.A.; Ewing, T.; Saunders, K.; Kogut, S.; Nazarian, E.; et al. Extensively drug-resistant Pseudomonas aeruginosa outbreak associated with artificial tears. Clin. Infect. Dis. 2024, 79, 6–14. [Google Scholar] [CrossRef]
- Sundermann, A.J.; Srinivasa, V.R.; Mills, E.G.; Griffith, M.P.; Waggle, K.D.; Ayres, A.M.; Pless, L.; Snyder, G.M.; Harrison, L.H.; Van Tyne, D. Two Artificial Tears Outbreak-Associated Cases of Extensively Drug-Resistant Pseudomonas aeruginosa Detected Through Whole Genome Sequencing-Based Surveillance. J. Infect. Dis. 2024, 229, 517–521. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention. Outbreak of Extensively Drug-Resistant Pseudomonas aeruginosa Associated with Artificial Tears: Centers for Disease Control and Prevention. 2024. Available online: https://archive.cdc.gov/#/details?url=https://www.cdc.gov/hai/outbreaks/crpa-artificial-tears.html (accessed on 13 May 2025).
- Adesanya, O.; Oduselu, T.; Akin-Ajani, O.; Adewumi, O.M.; Ademowo, O.G. An exegesis of bacteriophage therapy: An emerging player in the fight against anti-microbial resistance. AIMS Microbiol. 2020, 6, 204–230. [Google Scholar] [CrossRef] [PubMed]
- Suttle, C.A. Marine viruses—Major players in the global ecosystem. Nat. Rev. Microbiol. 2007, 5, 801–812. [Google Scholar] [CrossRef] [PubMed]
- Fei, B.; Li, D.; Liu, X.; You, X.; Guo, M.; Ren, Y.; Liu, Y.; Wang, C.; Zhu, R.; Li, Y. Characterization and genomic analysis of a broad-spectrum lytic phage HZ2201 and its antibiofilm efficacy against Pseudomonas aeruginosa. Virus Res. 2023, 335, 199184. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Gallardo, M.J.; Villicaña, C.; Yocupicio-Monroy, M.; Alcaraz-Estrada, S.L.; León-Félix, J. Current knowledge in the use of bacteriophages to combat infections caused by Pseudomonas aeruginosa in cystic fibrosis. Folia Microbiol. 2023, 68, 1–16. [Google Scholar] [CrossRef]
- Rai, P.; Shetty, S.S.; Prabell, S.; Kuntar, A.; Pinto, D.; Kumar, B.K.; Divyashree, M.; Raj, J.R.M.; Premanath, R.; Deekshit, V.K.; et al. Characterisation of broad-spectrum phiKZ like jumbo phage and its utilisation in controlling multidrug-resistant Pseudomonas aeruginosa isolates. Microb. Pathog. 2022, 172, 105767. [Google Scholar] [CrossRef]
- Xuan, G.; Lin, H.; Kong, J.; Wang, J.; Yang, X. Phage resistance evolution induces the sensitivity of specific antibiotics in Pseudomonas aeruginosa PAO1. Microbiol. Spectr. 2022, 10, e0135622. [Google Scholar] [CrossRef]
- Yang, X.; Haque, A.; Matsuzaki, S.; Matsumoto, T.; Nakamura, S. The efficacy of phage therapy in a murine model of Pseudomonas aeruginosa pneumonia and sepsis. Front. Microbiol. 2021, 12, 682255. [Google Scholar] [CrossRef]
- Uyttebroek, S.; Chen, B.; Onsea, J.; Ruythooren, F.; Debaveye, Y.; Devolder, D.; Spriet, I.; Depypere, M.; Wagemans, J.; Lavigne, R.; et al. Safety and efficacy of phage therapy in difficult-to-treat infections: A systematic review. Lancet Infect. Dis. 2022, 22, e208–e220. [Google Scholar] [CrossRef]
- Pirnay, J.-P.; Djebara, S.; Steurs, G.; Griselain, J.; Cochez, C.; De Soir, S.; Glonti, T.; Spiessens, A.; Berghe, E.V.; Green, S.; et al. Retrospective, observational analysis of the first one hundred consecutive cases of personalized bacteriophage therapy of difficult-to-treat infections facilitated by a Belgian consortium. MedRxiv 2023. [Google Scholar] [CrossRef]
- Chegini, Z.; Khoshbayan, A.; Taati Moghadam, M.; Farahani, I.; Jazireian, P.; Shariati, A. Bacteriophage therapy against Pseudomonas aeruginosa biofilms: A review. Ann. Clin. Microbiol. Antimicrob. 2020, 19, 45. [Google Scholar] [CrossRef]
- Ding, Y.; Huang, C.; Zhu, W.; Li, Z.; Zhang, Y.; Wang, J.; Pan, H.; Li, H.; Wang, X. Characterization of a novel Jerseyvirus phage T102 and its inhibition effect on biofilms of multidrug-resistant Salmonella. Virus Res. 2023, 326, 199054. [Google Scholar] [CrossRef] [PubMed]
- Henriksen, K.; Rørbo, N.; Rybtke, M.L.; Martinet, M.G.; Tolker-Nielsen, T.; Høiby, N.; Middelboe, M.; Ciofu, O. P. aeruginosa flow-cell biofilms are enhanced by repeated phage treatments but can be eradicated by phage–ciprofloxacin combination:—Monitoring the phage–P. aeruginosa biofilms interactions. Pathog. Dis. 2019, 77, ftz011. [Google Scholar] [CrossRef] [PubMed]
- Danis-Wlodarczyk, K.M.; Wozniak, D.J.; Abedon, S.T. Treating bacterial infections with bacteriophage-based enzybiotics: In vitro, in vivo and clinical application. Antibiotics 2021, 10, 1497. [Google Scholar] [CrossRef]
- Topka-Bielecka, G.; Dydecka, A.; Necel, A.; Bloch, S.; Nejman-Faleńczyk, B.; Węgrzyn, G.; Węgrzyn, A. Bacteriophage-derived depolymerases against bacterial biofilm. Antibiotics 2021, 10, 175. [Google Scholar] [CrossRef] [PubMed]
- Rahimzadeh, G.; Saeedi, M.; Nokhodchi, A.; Moosazadeh, M.; Ghasemi, M.; Rostamkalaei, S.S.; Mortazavi, P.; Eghbali, M.; Pourbakhshian, R.; Rezai, M.S.; et al. Evaluation of in-situ gel-forming eye drop containing bacteriophage against Pseudomonas aeruginosa keratoconjunctivitis in vivo. Bioimpacts 2021, 11, 281–287. [Google Scholar] [CrossRef]
- Sotnikova, L.F.; Goncharova, A.V.; Pimenov, N.V. Use of bacteriophage cocktails for ulcerative keratitis in horses, clinical and ophthalmological justification. IOP Conf. Ser. Earth Environ. Sci. 2021, 677, 022017. [Google Scholar] [CrossRef]
- Fukuda, K.; Ishida, W.; Uchiyama, J.; Rashel, M.; Kato, S.-I.; Morita, T.; Muraoka, A.; Sumi, T.; Matsuzaki, S.; Daibata, M.; et al. Pseudomonas aeruginosa keratitis in mice: Effects of topical bacteriophage KPP12 administration. PLoS ONE 2012, 7, e47742. [Google Scholar] [CrossRef]
- Furusawa, T.; Iwano, H.; Hiyashimizu, Y.; Matsubara, K.; Higuchi, H.; Nagahata, H.; Niwa, H.; Katayama, Y.; Kinoshita, Y.; Hagiwara, K.; et al. Phage therapy is effective in a mouse model of bacterial equine keratitis. Appl. Environ. Microbiol. 2016, 82, 5332–5339. [Google Scholar] [CrossRef]
- Fadlallah, A.; Chelala, E.; Legeais, J.M. Corneal infection therapy with topical bacteriophage administration. Open Ophthalmol. J. 2015, 9, 167–168. [Google Scholar] [CrossRef]
- Khan, M.; Stapleton, F.; Summers, S.; Rice, S.A.; Willcox, M.D.P. Antibiotic Resistance Characteristics of Pseudomonas aeruginosa Isolated from Keratitis in Australia and India. Antibiotics 2020, 9, 600. [Google Scholar] [CrossRef]
- Khan, M.; Ma, K.; Wan, I.; Willcox, M.D. Ciprofloxacin resistance and tolerance of Pseudomonas aeruginosa ocular isolates. Cont. Lens Anterior Eye 2023, 46, 101819. [Google Scholar] [CrossRef]
- Loc-Carrillo, C.; Abedon, S.T. Pros and cons of phage therapy. Bacteriophage 2011, 1, 111–114. [Google Scholar] [CrossRef] [PubMed]
- Sutherland, I.W.; Hughes, K.A.; Skillman, L.C.; Tait, K. The interaction of phage and biofilms. FEMS Microbiol. Lett. 2004, 232, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Glonti, T.; Chanishvili, N.; Taylor, P. Bacteriophage-derived enzyme that depolymerizes the alginic acid capsule associated with cystic fibrosis isolates of Pseudomonas aeruginosa. J. Appl. Microbiol. 2010, 108, 695–702. [Google Scholar] [CrossRef] [PubMed]
- Olszak, T.; Shneider, M.M.; Latka, A.; Maciejewska, B.; Browning, C.; Sycheva, L.V.; Cornelissen, A.; Danis-Wlodarczyk, K.; Senchenkova, S.N.; Shashkov, A.S.; et al. The O-specific polysaccharide lyase from the phage LKA1 tailspike reduces Pseudomonas virulence. Sci. Rep. 2017, 7, 16302. [Google Scholar] [CrossRef]
- Mi, L.; Liu, Y.; Wang, C.; He, T.; Gao, S.; Xing, S.; Huang, Y.; Fan, H.; Zhang, X.; Yu, W.; et al. Identification of a lytic Pseudomonas aeruginosa phage depolymerase and its anti-biofilm effect and bactericidal contribution to serum. Virus Genes. 2019, 55, 394–405. [Google Scholar] [CrossRef]
- Mason, G.; Footer, M.J.; Rojas, E.R. Mechanosensation induces persistent bacterial growth during bacteriophage predation. mBio 2023, 14, e0276622. [Google Scholar] [CrossRef]
- Naureen, Z.; Dautaj, A.; Anpilogov, K.; Camilleri, G.; Dhuli, K.; Tanzi, B.; Maltese, P.E.; Cristofoli, F.; De Antoni, L.; Beccari, T.; et al. Bacteriophages presence in nature and their role in the natural selection of bacterial populations. Acta Biomed. 2020, 91, e2020024. [Google Scholar]
- Rodriguez-Valera, F.; Martin-Cuadrado, A.-B.; Rodriguez-Brito, B.; Pasic, L.; Thingstad, T.F.; Rohwer, F.; Mira, A. Explaining microbial population genomics through phage predation. Nat. Rev. Microbiol. 2009, 7, 828–836. [Google Scholar] [CrossRef]
- Labrie, S.J.; Samson, J.E.; Moineau, S. Bacteriophage resistance mechanisms. Nat. Rev. Microbiol. 2010, 8, 317–327. [Google Scholar] [CrossRef]
- Doron, S.; Melamed, S.; Ofir, G.; Leavitt, A.; Lopatina, A.; Keren, M.; Amitai, G.; Sorek, R. Systematic discovery of antiphage defense systems in the microbial pangenome. Science 2018, 359, eaar4120. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Altae-Tran, H.; Böhning, F.; Makarova, K.S.; Segel, M.; Schmid-Burgk, J.L.; Koob, J.; Wolf, Y.I.; Koonin, E.V.; Zhang, F. Diverse enzymatic activities mediate antiviral immunity in prokaryotes. Science 2020, 369, 1077–1084. [Google Scholar] [CrossRef]
- Betts, A.; Vasse, M.; Kaltz, O.; Hochberg, M.E. Back to the future: Evolving bacteriophages to increase their effectiveness against the pathogen Pseudomonas seudomonas aeruginosa PAO 1. Evol. Appl. 2013, 6, 1054–1063. [Google Scholar] [CrossRef]
- Danis-Wlodarczyk, K.; Vandenheuvel, D.; Bin Jang, H.; Briers, Y.; Olszak, T.; Arabski, M.; Wasik, S.; Drabik, M.; Higgins, G.; Tyrrell, J.; et al. A proposed integrated approach for the preclinical evaluation of phage therapy in Pseudomonas infections. Sci. Rep. 2016, 6, 28115. [Google Scholar] [CrossRef]
- Fiscarelli, E.V.; Rossitto, M.; Rosati, P.; Essa, N.; Crocetta, V.; Di Giulio, A.; Lupetti, V.; Di Bonaventura, G.; Pompilio, A. In vitro newly isolated environmental phage activity against biofilms preformed by Pseudomonas aeruginosa from patients with cystic fibrosis. Microorganisms 2021, 9, 478. [Google Scholar] [CrossRef] [PubMed]
- Hyman, P. Phages for phage therapy: Isolation, characterization, and host range breadth. Pharmaceuticals 2019, 12, 35. [Google Scholar] [CrossRef] [PubMed]
- Leung, C.Y.J.; Weitz, J.S. Modeling the synergistic elimination of bacteria by phage and the innate immune system. J. Theor. Biol. 2017, 429, 241–252. [Google Scholar] [CrossRef]
- Harmsen, M.; Yang, L.; Pamp, S.J.; Tolker-Nielsen, T. An update on Pseudomonas aeruginosa biofilm formation, tolerance, and dispersal. FEMS Immunol. Med. Microbiol. 2010, 59, 253–268. [Google Scholar] [CrossRef]
- Vaitekenas, A.; Tai, A.S.; Ramsay, J.P.; Stick, S.M.; Kicic, A. Pseudomonas aeruginosa Resistance to Bacteriophages and Its Prevention by Strategic Therapeutic Cocktail Formulation. Antibiotics 2021, 10, 145. [Google Scholar] [CrossRef]
- Alshalchi, S.A.; Anderson, G.G. Expression of the lipopolysaccharide biosynthesis gene lpxD affects biofilm formation of Pseudomonas aeruginosa. Arch. Microbiol. 2015, 197, 135–145. [Google Scholar] [CrossRef]
- Guttenplan, S.B.; Kearns, D.B. Regulation of flagellar motility during biofilm formation. FEMS Microbiol. Rev. 2013, 37, 849–871. [Google Scholar] [CrossRef]
- Pires, D.P.; Oliveira, H.; Melo, L.D.; Sillankorva, S.; Azeredo, J. Bacteriophage-encoded depolymerases: Their diversity and biotechnological applications. Appl. Microbiol. Biotechnol. 2016, 100, 2141–2151. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.; Karaynir, A.; Salih, H.; Öncü, S.; Bozdoğan, B. Characterization, genome analysis and antibiofilm efficacy of lytic Proteus phages RP6 and RP7 isolated from university hospital sewage. Virus Res. 2023, 326, 199049. [Google Scholar] [CrossRef]
- Alves, D.R.; Perez-Esteban, P.; Kot, W.; Bean, J.; Arnot, T.; Hansen, L.; Enright, M.C.; Jenkins, A.T.A. A novel bacteriophage cocktail reduces and disperses P seudomonas aeruginosa biofilms under static and flow conditions. Microb. Biotechnol. 2016, 9, 61–74. [Google Scholar] [CrossRef]
- Abedon, S. Phage, bacteria, and food. Appendix: Rate of adsorption is function of phage density. In Bacteriophage Ecology; Cambridge University Press: Cambridge, UK, 2008; pp. 321–324. [Google Scholar]
- Namonyo, S.; Weynberg, K.D.; Guo, J.; Carvalho, G. The effectiveness and role of phages in the disruption and inactivation of clinical P. aeruginosa biofilms. Environ. Res. 2023, 234, 116586. [Google Scholar] [CrossRef] [PubMed]
- Abedon, S.T. Phage therapy dosing: The problem(s) with multiplicity of infection (MOI). Bacteriophage 2016, 6, e1220348. [Google Scholar] [CrossRef] [PubMed]
- Pires, D.; Sillankorva, S.; Faustino, A.; Azeredo, J. Use of newly isolated phages for control of Pseudomonas aeruginosa PAO1 and ATCC 10145 biofilms. Res. Microbiol. 2011, 162, 798–806. [Google Scholar] [CrossRef]
- Alemayehu, D.; Casey, P.G.; McAuliffe, O.; Guinane, C.M.; Martin, J.G.; Shanahan, F.; Coffey, A.; Ross, R.P.; Hill, C. Bacteriophages ϕMR299-2 and ϕNH-4 can eliminate Pseudomonas aeruginosa in the murine lung and on cystic fibrosis lung airway cells. MBio 2012, 3, e00029-12. [Google Scholar] [CrossRef]
- Hanlon, G.W.; Denyer, S.P.; Olliff, C.J.; Ibrahim, L.J. Reduction in exopolysaccharide viscosity as an aid to bacteriophage penetration through Pseudomonas aeruginosa biofilms. Appl. Environ. Microbiol. 2001, 67, 2746–2753. [Google Scholar] [CrossRef]
- Quan, K.; Hou, J.; Zhang, Z.; Ren, Y.; Peterson, B.W.; Flemming, H.-C.; Mayer, C.; Busscher, H.J.; van der Mei, H.C. Water in bacterial biofilms: Pores and channels, storage and transport functions. Crit. Rev. Microbiol. 2022, 48, 283–302. [Google Scholar] [CrossRef]
- Simmons, E.L.; Bond, M.C.; Koskella, B.; Drescher, K.; Bucci, V.; Nadell, C.D. Biofilm structure promotes coexistence of phage-resistant and phage-susceptible bacteria. mSystems 2020, 5, e00877-19. [Google Scholar] [CrossRef] [PubMed]
- Winans, J.B.; Wucher, B.R.; Nadell, C.D. Multispecies biofilm architecture determines bacterial exposure to phages. PLoS Biol. 2022, 20, e3001913. [Google Scholar] [CrossRef]
- Stepanović, S.; Vuković, D.; Hola, V.; Bonaventura, G.D.; Djukić, S.; Ćirković, I.; Ruzicka, F. Quantification of biofilm in microtiter plates: Overview of testing conditions and practical recommendations for assessment of biofilm production by staphylococci. Apmis 2007, 115, 891–899. [Google Scholar] [CrossRef] [PubMed]
- Haney, E.F.; Trimble, M.J.; Cheng, J.T.; Vallé, Q.; Hancock, R.E. Critical assessment of methods to quantify biofilm growth and evaluate antibiofilm activity of host defence peptides. Biomolecules 2018, 8, 29. [Google Scholar] [CrossRef] [PubMed]
- Bonilla, N.; Barr, J.J. Phage on tap: A quick and efficient protocol for the preparation of bacteriophage laboratory stocks. In The Human Virome: Methods and Protocols; Springer: Berlin/Heidelberg, Germany, 2018; pp. 37–46. [Google Scholar]
- Khan Mirzaei, M.; Nilsson, A.S. Isolation of phages for phage therapy: A comparison of spot tests and efficiency of plating analyses for determination of host range and efficacy. PLoS ONE 2015, 10, e0118557. [Google Scholar] [CrossRef]
- González-Villalobos, E.; Ribas-Aparicio, R.M.; Montealegre, G.E.R.; Belmont-Monroy, L.; Ortega-García, Y.; Aparicio-Ozores, G.; Balcázar, J.L.; Eslava-Campos, C.A.; Hernández-Chiñas, U.; Molina-López, J. Isolation and characterization of novel bacteriophages as a potential therapeutic option for Escherichia coli urinary tract infections. Appl. Microbiol. Biotechnol. 2021, 105, 5617–5629. [Google Scholar] [CrossRef]
- Haney, E.F.; Trimble, M.J.; Hancock, R.E. Microtiter plate assays to assess antibiofilm activity against bacteria. Nat. Protoc. 2021, 16, 2615–2632. [Google Scholar] [CrossRef]
- Tsukatani, T.; Sakata, F.; Kuroda, R. A rapid and simple measurement method for biofilm formation inhibitory activity using 96-pin microtiter plate lids. World J. Microbiol. Biotechnol. 2020, 36, 189. [Google Scholar] [CrossRef]
| P. aeruginosa Strains | Antibiotic Resistance [53,54] | Biofilm Formation | DiSu1 | DiSu2 | DiSu3 | DiSu4 | DiSu5 | DiSu6 |
|---|---|---|---|---|---|---|---|---|
| PA209 | Pip, Imi | Moderate | S | S | S | S | S | S |
| PA213 | Cip, Imi, Cef | Strong | S | S | S | S | S | S |
| PA216 | Cip, Lev, Pip, Imi, Cef, PmB | Strong | R | S | S | S | S | S |
| PA217 | Cip, Lev, Pip, Imi, Cef | Strong | R | S | S | S | S | R |
| PA221 | Cip, Lev, Gen, Tob, Pip, Imi, Cef | Strong | S | S | S | S | S | S |
| PA223 | Cip, Pip, Cef | Strong | R | R | S | S | S | S |
| PA225 | Cip, Lev, Imi | Strong | S | S | S | S | S | S |
| PA227 | Cip, Lev, Imi, Cef | Strong | S | S | S | S | R | R |
| PA232 | ND | Strong | S | S | S | S | S | S |
| PA235 | Cip, Pip, Imi, Cef | Moderate | S | S | S | S | S | S |
| Bacterial Strain | PA209 | PA213 | PA216 | PA217 | PA221 | PA223 | PA225 | PA227 | PA232 | PA235 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Phage—MOI = 1 | DiSu1 | −24.4 | −13.9 | R | R | −18.7 | R | −30.1 | −3.0 | −16.7 | 46.8 |
| DiSu2 | 68.9 | 50.7 | 32.1 | 18.2 | 57.8 | R | −23.8 | −1.4 | 28.7 | 55.4 | |
| DiSu3 | 67.2 | 61.7 | 42.6 | −22.8 | 60.0 | 63.3 | −3.0 | 24.6 | 69.6 | 92.3 | |
| DiSu4 | −43.8 | −86.2 | −24.9 | 20.0 | −11.6 | 70.5 | −2.1 | −10.6 | −1.1 | 24.5 | |
| DiSu5 | 79.8 | 69.4 | 46.8 | −15.1 | 53.6 | 64.7 | −4.4 | R | 66.8 | 96.2 | |
| DiSu6 | 70.3 | 57.0 | 27.2 | R | 49.3 | 70.9 | −5.8 | R | 76.3 | 93.6 | |
| Phage—MOI = 10 | DiSu1 | 70.4 | 30.2 | R | R | 5.7 | R | −9.3 | −4.8 | −16.7 | 73.4 |
| DiSu2 | 87.2 | 73.7 | 51.5 | 31.2 | 61.4 | R | −12.3 | −0.3 | 28.7 | 81.3 | |
| DiSu3 | 79.4 | 80.3 | 36.8 | −6.5 | 61.5 | 59.9 | −2.5 | 71.5 | 69.6 | 92.4 | |
| DiSu4 | 35.6 | 51.1 | −11.9 | 20.2 | 6.6 | 67.7 | 24.2 | 0.2 | −1.1 | 19.2 | |
| DiSu5 | 81.1 | 80.0 | 50.6 | −6.8 | 69.7 | 77.1 | 23.6 | R | 66.8 | 98.4 | |
| DiSu6 | 82.3 | 79.7 | 30.6 | R | 62.9 | 67.1 | 33.4 | R | 76.3 | 93.0 | |
| Phage—MOI = 100 | DiSu1 | 72.8 | 58.4 | R | R | −34.1 | R | −1.5 | 23.6 | −0.6 | 67.3 |
| DiSu2 | 88.7 | 84.1 | 55.9 | 40.2 | 65.2 | R | 0.0 | 22.8 | 75.2 | 77.2 | |
| DiSu3 | 92.3 | 92.4 | 65.8 | −4.0 | 93.6 | 69.1 | 36.2 | 92.5 | 94.6 | 92.4 | |
| DiSu4 | 56.3 | 68.2 | −10.6 | 32.5 | 52.6 | 80.3 | 75.7 | 16.1 | 39.6 | 29.0 | |
| DiSu5 | 89.3 | 95.7 | 64.6 | 2.3 | 69.8 | 76.6 | 95.1 | R | 84.8 | 98.5 | |
| DiSu6 | 94.2 | 89.0 | 44.6 | R | 83.1 | 81.1 | 73.0 | R | 96.3 | 95.5 |
| Bacterial Strain | MOI | Percentage of Bacterial Biofilm Inhibition After Coculturing for 24 h | |||||
|---|---|---|---|---|---|---|---|
| DiSu1 | DiSu2 | DiSu3 | DiSu4 | DiSu5 | DiSu6 | ||
| PA209 | 1 | 1.4 | 2.5 | −4.8 | −2.3 | −6.0 | −2.7 |
| 10 | 7.5 | −0.8 | −0.8 | −0.8 | −0.8 | 0.6 | |
| 100 | 27.3 | 21.8 | 21.4 | 12.6 | 12.6 | 5.3 | |
| PA213 | 1 | 0.0 | 2.2 | 10.3 | 2.9 | 0.0 | 0.0 |
| 10 | 4.9 | 11.0 | 11.7 | 4.9 | 20.2 | 6.0 | |
| 100 | 25.1 | 26.8 | 27.4 | 7.2 | 22.0 | 6.0 | |
| PA216 | 1 | R | 13.5 | 5.3 | 3.3 | −1.8 | 1.7 |
| 10 | R | 3.1 | 3.1 | 14.4 | 16.3 | 3.1 | |
| 100 | R | 16.7 | 5.3 | 10.1 | 2.5 | 7.5 | |
| PA217 | 1 | R | 3.3 | 6.4 | 4.9 | −3.7 | R |
| 10 | R | 1.3 | 8.5 | 1.3 | 12.8 | R | |
| 100 | R | 7.0 | 15.2 | 3.5 | 8.4 | R | |
| PA221 | 1 | −1.3 | 14.5 | 17.1 | 8.1 | −1.3 | 4.9 |
| 10 | 3.6 | 3.6 | 3.6 | 3.6 | 22.3 | 3.6 | |
| 100 | 16.8 | 30.6 | 26.5 | 18.4 | 16.5 | 6.1 | |
| PA223 | 1 | R | R | 10.0 | −1.9 | 3.5 | 10.6 |
| 10 | R | R | 3.1 | 3.1 | 21.8 | 3.1 | |
| 100 | R | R | 55.7 | 38.4 | 42.0 | 35.6 | |
| PA225 | 1 | −0.5 | 6.9 | 2.4 | −1.9 | 7.3 | 5.6 |
| 10 | 3.1 | 3.1 | 3.1 | 3.1 | 18.7 | 3.1 | |
| 100 | 18.5 | 28.4 | 16.0 | 16.0 | 52.2 | 26.7 | |
| PA227 | 1 | 9.4 | 3.0 | 17.7 | 3.0 | R | R |
| 10 | 7.7 | 7.7 | 7.7 | 7.7 | R | R | |
| 100 | 33.7 | 14.3 | 20.0 | 26.7 | R | R | |
| PA232 | 1 | 1.5 | −1.9 | 7.5 | −1.9 | −1.9 | −1.9 |
| 10 | 3.1 | 3.1 | 10.0 | 4.0 | 3.1 | 3.1 | |
| 100 | 27.8 | 6.3 | 16.0 | 29.8 | 25.9 | 35.7 | |
| PA235 | 1 | 12.6 | −0.4 | 12.4 | −6.2 | 2.6 | 11.4 |
| 10 | −1.1 | −1.1 | 41.3 | 5.5 | 9.9 | 20.6 | |
| 100 | 49.5 | 41.3 | 83.4 | 20.6 | 73.9 | 32.8 | |
| Bacterial Strain | MOI | Percentage of Bacterial Biofilm Reduction After Exposure to Phages for 3 h | Percentage of Bacterial Biofilm Reduction After Exposure to Phages for 24 h | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DiSu1 | DiSu2 | DiSu3 | DiSu4 | DiSu5 | DiSu6 | DiSu1 | DiSu2 | DiSu3 | DiSu4 | DiSu5 | DiSu6 | ||
| PA209 | 1 | 1.9 | 4.2 | 8.3 | 1.9 | 1.9 | 1.9 | 5.4 | −2.4 | 6.6 | 4.3 | 8.4 | 10.7 |
| 10 | 24.3 | 20.3 | 14.0 | 22.9 | 15.6 | 5.4 | 10.7 | 13.5 | 0.3 | 9.1 | 9.7 | 0.3 | |
| 100 | 25.4 | 25.6 | 41.5 | 27.7 | 16.6 | 13.5 | 17.2 | 5.5 | 12.7 | 11.8 | 5.5 | 0.3 | |
| PA213 | 1 | 7.4 | 7.4 | 13.4 | 7.4 | 7.4 | 7.4 | 11.8 | 3.3 | 8.9 | 6.5 | 12.0 | 14.4 |
| 10 | 19.4 | 23.5 | 23.7 | 7.4 | 27.3 | 7.4 | 12.3 | 17.8 | 5.9 | 11.2 | 11.3 | 5.9 | |
| 100 | 27.3 | 44.6 | 56.7 | 50.0 | 54.0 | 29.3 | 19.0 | 15.7 | 26.7 | 20.4 | 10.8 | 7.4 | |
| PA216 | 1 | R | 8.0 | 11.9 | 5.8 | 5.8 | 5.8 | R | 1.6 | 10.2 | 8.1 | 12.0 | 14.2 |
| 10 | R | 17.3 | 5.8 | 6.1 | 10.5 | 12.1 | R | 16.9 | 4.2 | 12.7 | 13.3 | 4.2 | |
| 100 | R | 16.2 | 15.0 | 13.9 | 9.2 | 9.2 | R | 9.2 | 16.1 | 15.2 | 9.2 | 18.7 | |
| PA217 | 1 | R | 0 | 8.9 | 0 | 0 | R | R | −0.3 | −0.3 | 13.7 | 8.5 | R |
| 10 | R | 15.8 | 4.0 | 4.3 | 8.9 | R | R | 6.2 | 7.1 | 16.0 | 11.2 | R | |
| 100 | R | −0.3 | 8.6 | 6.3 | 10.4 | R | R | 9.6 | 7.5 | 16.3 | 14.0 | R | |
| PA221 | 1 | 6.2 | 6.2 | 14.9 | 6.2 | 6.2 | 6.2 | 11.5 | 2.1 | 8.8 | 10.1 | 11.0 | 10.4 |
| 10 | 24.5 | 6.2 | 16.9 | 29.4 | 11.5 | 28.4 | 15.9 | 16.5 | 4.7 | 29.4 | 18.6 | 4.7 | |
| 100 | 60.3 | 29.1 | 61.3 | 36.4 | 25.3 | 29.4 | 24.5 | 18.6 | 57.7 | 36.4 | 20.0 | 20.5 | |
| PA223 | 1 | R | R | 11.0 | 5.7 | 6.5 | 5.7 | R | R | 6.6 | 9.3 | 12.2 | 7.7 |
| 10 | R | R | 16.4 | 22.4 | 21.3 | 6.6 | R | R | 6.6 | 9.3 | 12.2 | 7.7 | |
| 100 | R | R | 25.2 | 25.3 | 25.6 | 12.2 | R | R | 21.3 | 18.8 | 18.5 | 9.1 | |
| PA225 | 1 | 5.7 | 11.0 | 7.5 | 5.7 | 5.7 | 5.7 | 14.7 | 1.5 | 15.5 | 4.8 | 8.2 | 10.7 |
| 10 | 15.6 | 19.0 | 19.4 | 26.9 | 13.4 | 7.5 | 12.9 | 5.7 | 22.1 | 4.8 | 8.7 | 11.8 | |
| 100 | 39.6 | 22.2 | 56.7 | 48.6 | 13.6 | 22.9 | 36.3 | 5.7 | 21.1 | 18.6 | 9.1 | 9.1 | |
| PA227 | 1 | 10.2 | 20.0 | 13.5 | 10.2 | R | R | 6.2 | 6.2 | 15.5 | 6.3 | R | R |
| 10 | 20.3 | 13.7 | 26.6 | 34.2 | R | R | 15.7 | 8.8 | 16.3 | 23.6 | R | R | |
| 100 | 19.9 | 34.5 | 29.8 | 45.7 | R | R | 13.5 | 8.8 | 19.8 | 25.5 | R | R | |
| PA232 | 1 | 9.5 | 10.6 | 5.7 | 5.7 | 8.5 | 5.7 | 1.5 | 1.5 | 6.6 | 1.5 | 8.6 | 5.3 |
| 10 | 13.2 | 15.8 | 13.3 | 19.2 | 19.5 | 5.7 | 7.9 | 7.7 | 20.4 | 15.2 | 10.5 | 7.7 | |
| 100 | 38.2 | 40.0 | 22.6 | 25.2 | 14.7 | 15.2 | 7.9 | 9.1 | 20.4 | 24.8 | 9.1 | 9.1 | |
| PA235 | 1 | 10.1 | 13.7 | 1.6 | 2.0 | 6.6 | 8.2 | −2.7 | −2.7 | −2.7 | −2.7 | 12.0 | 10.4 |
| 10 | 14.8 | 9.4 | 21.5 | 23.6 | 23.8 | 9.0 | 44.2 | 3.9 | 18.4 | 14.8 | 5.3 | 5.3 | |
| 100 | 88.2 | 70.2 | 90.0 | 41.6 | 44.2 | 46.4 | 9.6 | 12.5 | 29.7 | 10.1 | 14.5 | 16.8 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Urgeya, K.D.; Subedi, D.; Kumar, N.; Willcox, M. The Ability of Bacteriophages to Reduce Biofilms Produced by Pseudomonas aeruginosa Isolated from Corneal Infections. Antibiotics 2025, 14, 629. https://doi.org/10.3390/antibiotics14070629
Urgeya KD, Subedi D, Kumar N, Willcox M. The Ability of Bacteriophages to Reduce Biofilms Produced by Pseudomonas aeruginosa Isolated from Corneal Infections. Antibiotics. 2025; 14(7):629. https://doi.org/10.3390/antibiotics14070629
Chicago/Turabian StyleUrgeya, Kuma Diriba, Dinesh Subedi, Naresh Kumar, and Mark Willcox. 2025. "The Ability of Bacteriophages to Reduce Biofilms Produced by Pseudomonas aeruginosa Isolated from Corneal Infections" Antibiotics 14, no. 7: 629. https://doi.org/10.3390/antibiotics14070629
APA StyleUrgeya, K. D., Subedi, D., Kumar, N., & Willcox, M. (2025). The Ability of Bacteriophages to Reduce Biofilms Produced by Pseudomonas aeruginosa Isolated from Corneal Infections. Antibiotics, 14(7), 629. https://doi.org/10.3390/antibiotics14070629









