The Genetic Background and Culture Medium Only Marginally Affect the In Vitro Evolution of Pseudomonas aeruginosa Toward Colistin Resistance
Abstract
1. Introduction
2. Results
2.1. Experimental Evolution of High-Level Colistin Resistance in Different Media
2.2. Genes Mutated in Colistin-Resistant Clones
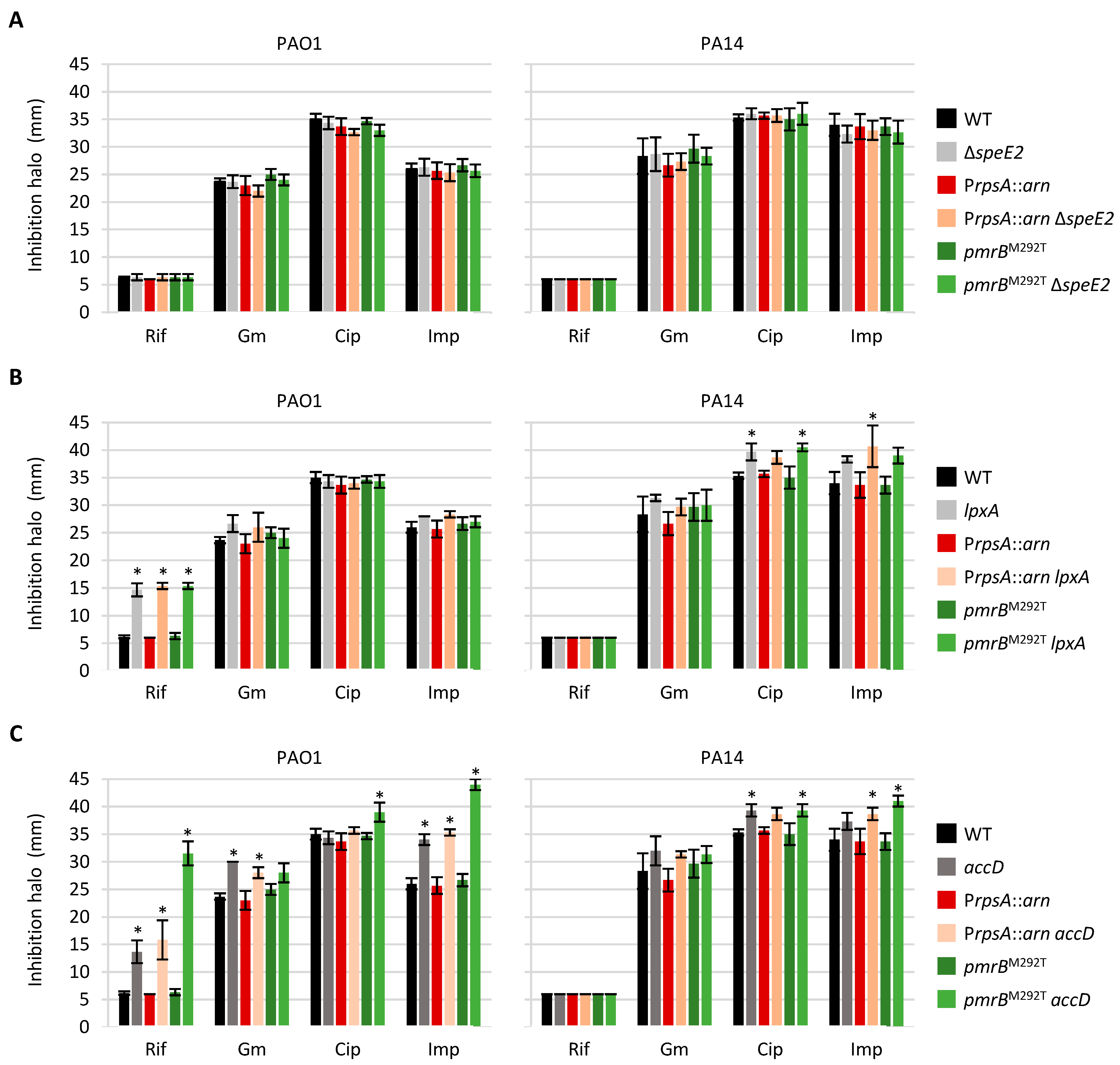
2.3. Norspermidine Has a Negative Impact on Colistin Resistance in P. aeruginosa
2.4. Impaired Expression of an LPS Biosynthetic Gene Has No Impact on Colistin Resistance
2.5. Compromised Fatty Acid Biosynthesis Can Contribute to Colistin Resistance
2.6. Mutations Associated with Colistin Resistance Can Influence Sensitivity to Other Antibiotic Classes
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains, Plasmids, and Growth Conditions
4.2. In Vitro Evolution Assays
4.3. Genome Sequencing and Mutation Analysis
4.4. Generation of Plasmids
4.5. Generation of Deletion, Allele Replacement, and Conditional Mutants
4.6. Growth and Minimum Inhibitory Concentration Assays
4.7. Kirby–Bauer Disk Diffusion Assay
4.8. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ASM | Artificial sputum medium |
| arn | arnBCADTEF-ugd |
| HS | Heat-inactivated human serum |
| L-Ara4N | 4-amino-4-deoxy-L-arabinose |
| LB | Lysogeny broth |
| LPS | Lipopolysaccharide |
| MH | Mueller–Hinton |
| NGS | Next Generation Sequencing |
| PEtN | Phosphoethanolamine |
| TCS | Two-component system |
References
- Theuretzbacher, U.; Bush, K.; Harbarth, S.; Paul, M.; Rex, J.H.; Tacconelli, E.; Thwaites, G.E. Critical analysis of antibacterial agents in clinical development. Nat. Rev. Microbiol. 2020, 18, 286–298. [Google Scholar] [CrossRef]
- Binsker, U.; Käsbohrer, A.; Hammerl, J.A. Global colistin use: A review of the emergence of resistant Enterobacterales and the impact on their genetic basis. FEMS Microbiol. Rev. 2022, 46, fuab049. [Google Scholar] [CrossRef] [PubMed]
- El-Sayed Ahmed, M.A.E.; Zhong, L.L.; Shen, C.; Yang, Y.; Doi, Y.; Tian, G.B. Colistin and its role in the Era of antibiotic resistance: An extended review (2000–2019). Emerg. Microbes Infect. 2020, 9, 868–885. [Google Scholar] [CrossRef] [PubMed]
- Narimisa, N.; Keshtkar, A.; Dadgar-Zankbar, L.; Bostanghadiri, N.; Far, Y.R.; Shahroodian, S.; Zahedi Bialvaei, A.; Razavi, S. Prevalence of colistin resistance in clinical isolates of Pseudomonas. aeruginosa: A systematic review and meta-analysis. Front. Microbiol. 2024, 15, 1477836. [Google Scholar] [CrossRef] [PubMed]
- Poirel, L.; Jayol, A.; Nordmann, P. Polymyxins: Antibacterial Activity, Susceptibility Testing, and Resistance Mechanisms Encoded by Plasmids or Chromosomes. Clin. Microbiol. Rev. 2017, 30, 557–596. [Google Scholar] [CrossRef]
- Sabnis, A.; Hagart, K.L.; Klöckner, A.; Becce, M.; Evans, L.E.; Furniss, R.C.D.; Mavridou, D.A.; Murphy, R.; Stevens, M.M.; Davies, J.C.; et al. Colistin kills bacteria by targeting lipopolysaccharide in the cytoplasmic membrane. Elife 2021, 10, e65836. [Google Scholar] [CrossRef]
- Jeannot, K.; Bolard, A.; Plésiat, P. Resistance to polymyxins in Gram-negative organisms. Int. J. Antimicrob. Agents 2017, 49, 526–535. [Google Scholar] [CrossRef]
- Hamel, M.; Rolain, J.M.; Baron, S.A. The History of Colistin Resistance Mechanisms in Bacteria: Progress and Challenges. Microorganisms 2021, 9, 442. [Google Scholar] [CrossRef]
- Reynolds, D.; Kollef, M. The Epidemiology and Pathogenesis and Treatment of Pseudomonas aeruginosa Infections: An Update. Drugs. 2021, 81, 2117–2131. [Google Scholar] [CrossRef]
- Cervoni, M.; Sposato, D.; Lo Sciuto, A.; Imperi, F. Regulatory Landscape of the Pseudomonas aeruginosa Phosphoethanolamine Transferase Gene eptA in the Context of Colistin Resistance. Antibiotics 2023, 12, 200. [Google Scholar] [CrossRef]
- McPhee, J.B.; Bains, M.; Winsor, G.; Lewenza, S.; Kwasnicka, A.; Brazas, M.D.; Brinkman, F.S.; Hancock, R.E. Contribution of the PhoP-PhoQ and PmrA-PmrB two-component regulatory systems to Mg2+-induced gene regulation in Pseudomonas aeruginosa. J. Bacteriol. 2006, 188, 3995–4006. [Google Scholar] [CrossRef] [PubMed]
- Barrow, K.; Kwon, D.H. Alterations in two-component regulatory systems of phoPQ and pmrAB are associated with polymyxin B resistance in clinical isolates of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2009, 53, 5150–5154. [Google Scholar] [CrossRef] [PubMed]
- Schurek, K.N.; Sampaio, J.L.; Kiffer, C.R.; Sinto, S.; Mendes, C.M.; Hancock, R.E. Involvement of pmrAB and phoPQ in polymyxin B adaptation and inducible resistance in non-cystic fibrosis clinical isolates of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2009, 53, 4345–4351. [Google Scholar] [CrossRef] [PubMed]
- Moskowitz, S.M.; Brannon, M.K.; Dasgupta, N.; Pier, M.; Sgambati, N.; Miller, A.K.; Selgrade, S.E.; Miller, S.I.; Denton, M.; Conway, S.P.; et al. PmrB mutations promote polymyxin resistance of Pseudomonas aeruginosa isolated from colistin-treated cystic fibrosis patients. Antimicrob. Agents Chemother. 2012, 56, 1019–1030. [Google Scholar] [CrossRef]
- Jochumsen, N.; Marvig, R.L.; Damkiær, S.; Jensen, R.L.; Paulander, W.; Molin, S.; Jelsbak, L.; Folkesson, A. The evolution of antimicrobial peptide resistance in Pseudomonas aeruginosa is shaped by strong epistatic interactions. Nat. Commun. 2016, 7, 13002. [Google Scholar] [CrossRef]
- Lo Sciuto, A.; Imperi, F. Aminoarabinosylation of Lipid A Is Critical for the Development of Colistin Resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2018, 62, e01820-17. [Google Scholar] [CrossRef]
- Lee, J.Y.; Park, Y.K.; Chung, E.S.; Na, I.Y.; Ko, K.S. Evolved resistance to colistin and its loss due to genetic reversion in Pseudomonas aeruginosa. Sci. Rep. 2016, 6, 25543. [Google Scholar] [CrossRef]
- Dößelmann, B.; Willmann, M.; Steglich, M.; Bunk, B.; Nübel, U.; Peter, S.; Neher, R.A. Rapid and Consistent Evolution of Colistin Resistance in Extensively Drug-Resistant Pseudomonas. aeruginosa during Morbidostat Culture. Antimicrob. Agents Chemother. 2017, 61, e00043-17. [Google Scholar] [CrossRef]
- Erdmann, M.B.; Gardner, P.P.; Lamont, I.L. The PitA protein contributes to colistin susceptibility in Pseudomonas aeruginosa. PLoS ONE 2023, 18, e0292818. [Google Scholar] [CrossRef]
- Lo Sciuto, A.; Cervoni, M.; Stefanelli, R.; Mancone, C.; Imperi, F. Effect of lipid A aminoarabinosylation on Pseudomonas aeruginosa colistin resistance and fitness. Int. J. Antimicrob. Agents 2020, 55, 105957. [Google Scholar] [CrossRef]
- Gutu, A.D.; Rodgers, N.S.; Park, J.; Moskowitz, S.M. Pseudomonas aeruginosa high-level resistance to polymyxins and other antimicrobial peptides requires cprA, a gene that is disrupted in the PAO1 strain. Antimicrob. Agents Chemother. 2015, 59, 5377–5387. [Google Scholar] [CrossRef] [PubMed]
- Chung, E.S.; Lee, J.Y.; Rhee, J.Y.; Ko, K.S. Colistin resistance in Pseudomonas aeruginosa that is not linked to arnB. J. Med. Microbiol. 2017, 66, 833–841. [Google Scholar] [CrossRef] [PubMed]
- Freschi, L.; Jeukens, J.; Kukavica-Ibrulj, I.; Boyle, B.; Dupont, M.J.; Laroche, J.; Larose, S.; Maaroufi, H.; Fothergill, J.L.; Moore, M.; et al. Clinical utilization of genomics data produced by the international Pseudomonas. aeruginosa consortium. Front. Microbiol. 2015, 6, 1036. [Google Scholar] [CrossRef] [PubMed]
- Han, M.L.; Zhu, Y.; Creek, D.J.; Lin, Y.W.; Gutu, A.D.; Hertzog, P.; Purcell, T.; Shen, H.H.; Moskowitz, S.M.; Velkov, T.; et al. Comparative Metabolomics and Transcriptomics Reveal Multiple Pathways Associated with Polymyxin Killing in Pseudomonas aeruginosa. Msystems. 2019, 4, e00149-18. [Google Scholar] [CrossRef]
- Han, M.L.; Zhu, Y.; Creek, D.J.; Lin, Y.W.; Anderson, D.; Shen, H.H.; Tsuji, B.; Gutu, A.D.; Moskowitz, S.M.; Velkov, T.; et al. Alterations of Metabolic and Lipid Profiles in Polymyxin-Resistant Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2018, 62, e02656-17. [Google Scholar] [CrossRef]
- Macfarlane, E.L.; Kwasnicka, A.; Ochs, M.M.; Hancock, R.E. PhoP-PhoQ homologues in Pseudomonas. aeruginosa regulate expression of the outer-membrane protein OprH and polymyxin B resistance. Mol. Microbiol. 1999, 34, 305–316. [Google Scholar] [CrossRef]
- Gooderham, W.J.; Gellatly, S.L.; Sanschagrin, F.; McPhee, J.B.; Bains, M.; Cosseau, C.; Levesque, R.C.; Hancock, R.E.W. The sensor kinase PhoQ mediates virulence in Pseudomonas aeruginosa. Microbiology 2009, 155, 699–711. [Google Scholar] [CrossRef]
- Miller, A.K.; Brannon, M.K.; Stevens, L.; Johansen, H.K.; Selgrade, S.E.; Miller, S.I.; Høiby, N.; Moskowitz, S.M. PhoQ mutations promote lipid A modification and polymyxin resistance of Pseudomonas. aeruginosa found in colistin-treated cystic fibrosis patients. Antimicrob. Agents Chemother. 2011, 55, 5761–5769. [Google Scholar] [CrossRef]
- Gutu, A.D.; Sgambati, N.; Strasbourger, P.; Brannon, M.K.; Jacobs, M.A.; Haugen, E.; Kaul, R.K.; Johansen, H.K.; Høiby, N.; Moskowitz, S.M. Polymyxin resistance of Pseudomonas aeruginosa. phoQ mutants is dependent on additional two-component regulatory systems. Antimicrob. Agents Chemother. 2013, 57, 2204–2215. [Google Scholar] [CrossRef]
- Abraham, N.; Kwon, D.H. A single amino acid substitution in PmrB is associated with polymyxin B resistance in clinical isolate of Pseudomonas aeruginosa. FEMS Microbiol. Lett. 2009, 298, 249–254. [Google Scholar] [CrossRef]
- Bolard, A.; Schniederjans, M.; Haüssler, S.; Triponney, P.; Valot, B.; Plésiat, P.; Jeannot, K. Production of Norspermidine Contributes to Aminoglycoside Resistance in pmrAB Mutants of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2019, 63, e01044-19. [Google Scholar] [CrossRef] [PubMed]
- Raetz, C.R.; Guan, Z.; Ingram, B.O.; Six, D.A.; Song, F.; Wang, X.; Zhao, J. Discovery of new biosynthetic pathways: The lipid A story. J. Lipid Res. 2009, 50, S103–S108. [Google Scholar] [CrossRef] [PubMed]
- Mdluli, K.E.; Witte, P.R.; Kline, T.; Barb, A.W.; Erwin, A.L.; Mansfield, B.E.; McClerren, A.L.; Pirrung, M.C.; Tumey, L.N.; Warrener, P.; et al. Molecular validation of LpxC as an antibacterial drug target in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2006, 50, 2178–2184. [Google Scholar] [CrossRef] [PubMed]
- Turner, K.H.; Wessel, A.K.; Palmer, G.C.; Murray, J.L.; Whiteley, M. Essential genome of Pseudomonas aeruginosa in cystic fibrosis sputum. Proc. Natl. Acad. Sci. USA 2015, 112, 4110–4115. [Google Scholar] [CrossRef]
- Lee, S.A.; Gallagher, L.A.; Thongdee, M.; Staudinger, B.J.; Lippman, S.; Singh, P.K.; Manoil, C. General and condition-specific essential functions of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 2015, 112, 5189–5194. [Google Scholar] [CrossRef]
- Lo Sciuto, A.; Cervoni, M.; Stefanelli, R.; Spinnato, M.C.; Di Giamberardino, A.; Mancone, C.; Imperi, F. Genetic Basis and Physiological Effects of Lipid A Hydroxylation in Pseudomonas. aeruginosa PAO1. Pathogens 2019, 8, 291. [Google Scholar] [CrossRef]
- Nowicki, E.M.; O’Brien, J.P.; Brodbelt, J.S.; Trent, M.S. Characterization of Pseudomonas. aeruginosa LpxT reveals dual positional lipid A kinase activity and co-ordinated control of outer membrane modification. Mol. Microbiol. 2014, 94, 728–741. [Google Scholar] [CrossRef]
- Hofstaedter, C.E.; Chandler, C.E.; Met, C.M.; Gillespie, J.J.; Harro, J.M.; Goodlett, D.R.; Rasko, D.A.; Ernst, R.K. Divergent Pseudomonas. aeruginosa LpxO enzymes perform site-specific lipid A 2-hydroxylation. mBio 2024, 15, e0282323. [Google Scholar] [CrossRef]
- Boechat, A.L.; Kaihami, G.H.; Politi, M.J.; Lépine, F.; Baldini, R.L. A novel role for an ECF sigma factor in fatty acid biosynthesis and membrane fluidity in Pseudomonas aeruginosa. PLoS ONE 2013, 8, e84775. [Google Scholar] [CrossRef]
- Lo Sciuto, A.; Fernández-Piñar, R.; Bertuccini, L.; Iosi, F.; Superti, F.; Imperi, F. The periplasmic protein TolB as a potential drug target in Pseudomonas aeruginosa. PLoS ONE 2014, 9, e103784. [Google Scholar] [CrossRef][Green Version]
- Fernández-Piñar, R.; Lo Sciuto, A.; Rossi, A.; Ranucci, S.; Bragonzi, A.; Imperi, F. In vitro and in vivo screening for novel essential cell-envelope proteins in Pseudomonas aeruginosa. Sci. Rep. 2015, 5, 17593. [Google Scholar] [CrossRef] [PubMed]
- Pasqua, M.; Visaggio, D.; Lo Sciuto, A.; Genah, S.; Banin, E.; Visca, P.; Imperi, F. Ferric Uptake Regulator Fur Is Conditionally Essential in Pseudomonas aeruginosa. J. Bacteriol. 2017, 199, e00472-17. [Google Scholar] [CrossRef] [PubMed]
- Lo Sciuto, A.; Martorana, A.M.; Fernández-Piñar, R.; Mancone, C.; Polissi, A.; Imperi, F. Pseudomonas. aeruginosa LptE is crucial for LptD assembly, cell envelope integrity, antibiotic resistance and virulence. Virulence 2018, 9, 1718–1733. [Google Scholar] [CrossRef] [PubMed]
- Ozer, E.A.; Allen, J.P.; Hauser, A.R. Characterization of the core and accessory genomes of Pseudomonas. aeruginosa using bioinformatic tools Spine and AGEnt. BMC Genom. 2014, 15, 737. [Google Scholar] [CrossRef]
- Qu, L.; She, P.; Wang, Y.; Liu, F.; Zhang, D.; Chen, L.; Luo, Z.; Xu, H.; Qi, Y.; Wu, Y. Effects of norspermidine on Pseudomonas aeruginosa biofilm formation and eradication. Microbiologyopen 2016, 5, 402–412. [Google Scholar] [CrossRef]
- Anjum, M.F.; Duggett, N.A.; AbuOun, M.; Randall, L.; Nunez-Garcia, J.; Ellis, R.J.; Rogers, J.; Horton, R.; Brena, C.; Williamson, S.; et al. Colistin resistance in Salmonella and Escherichia coli isolates from a pig farm in Great Britain. J. Antimicrob. Chemother. 2016, 71, 2306–2313. [Google Scholar] [CrossRef]
- López-Rojas, R.; Jiménez-Mejías, M.E.; Lepe, J.A.; Pachón, J. Acinetobacter. baumannii resistant to colistin alters its antibiotic resistance profile: A case report from Spain. J. Infect. Dis. 2011, 204, 1147–1148. [Google Scholar] [CrossRef]
- Carfrae, L.A.; Rachwalski, K.; French, S.; Gordzevich, R.; Seidel, L.; Tsai, C.N.; Tu, M.M.; MacNair, C.R.; Ovchinnikova, O.G.; Clarke, B.R.; et al. Inhibiting fatty acid synthesis overcomes colistin resistance. Nat. Microbiol. 2023, 8, 1026–1038. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed on 16 May 2025).
- Krueger, F. Trim Galore: A Wrapper Tool Around Cutadapt and FastQC to Consistently Apply Quality and Adapter Trimming to FastQ Files, with Some Extra Functionality for MspI-Digested RRBS-Type (Reduced Representation Bisufite-Seq) Libraries. 2012. Available online: https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/ (accessed on 16 May 2025).
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- R Core Team. R Language Definition; R Foundation for Statistical Computing: Vienna, Austria, 2020. [Google Scholar]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual, 2nd ed.; Cold Spring Harbor Laboratory: Cold Spring Harbor, NY, USA, 1989. [Google Scholar]
- Milton, D.L.; O’Toole, R.; Horstedt, P.; Wolf-Watz, H. Flagellin A is essential for the virulence of Vibrio anguillarum. J. Bacteriol. 1996, 178, 1310–1319. [Google Scholar] [CrossRef]
- Lo Sciuto, A.; Spinnato, M.C.; Pasqua, M.; Imperi, F. Generation of Stable and Unmarked Conditional Mutants in Pseudomonas aeruginosa. Methods Mol. Biol. 2022, 2548, 21–35. [Google Scholar] [CrossRef] [PubMed]
- Hoang, T.T.; Kutchma, A.J.; Becher, A.; Schweizer, H.P. Integration-proficient plasmids for Pseudomonas. aeruginosa: Site-specific integration and use for engineering of reporter and expression strains. Plasmid 2000, 43, 59–72. [Google Scholar] [CrossRef] [PubMed]
- Simon, R.; Priefer, U.; Pühler, A. A Broad Host Range Mobilization System for In Vivo Genetic Engineering: Transposon Mutagenesis in Gram Negative Bacteria. Nat. Biotechnol. 1983, 1, 784–791. [Google Scholar] [CrossRef]
- de Lorenzo, V.; Timmis, K.N. Analysis and construction of stable phenotypes in gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol. 1994, 235, 386–405. [Google Scholar] [CrossRef]
- Rahme, L.G.; Stevens, E.J.; Wolfort, S.F.; Shao, J.; Tompkins, R.G.; Ausubel, F.M. Common virulence factors for bacterial pathogenicity in plants and animals. Science 1995, 268, 1899–1902. [Google Scholar] [CrossRef]
- Hoang, T.T.; Karkhoff-Schweizer, R.R.; Kutchma, A.J.; Schweizer, H.P. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: Application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 1998, 212, 77–86. [Google Scholar] [CrossRef]
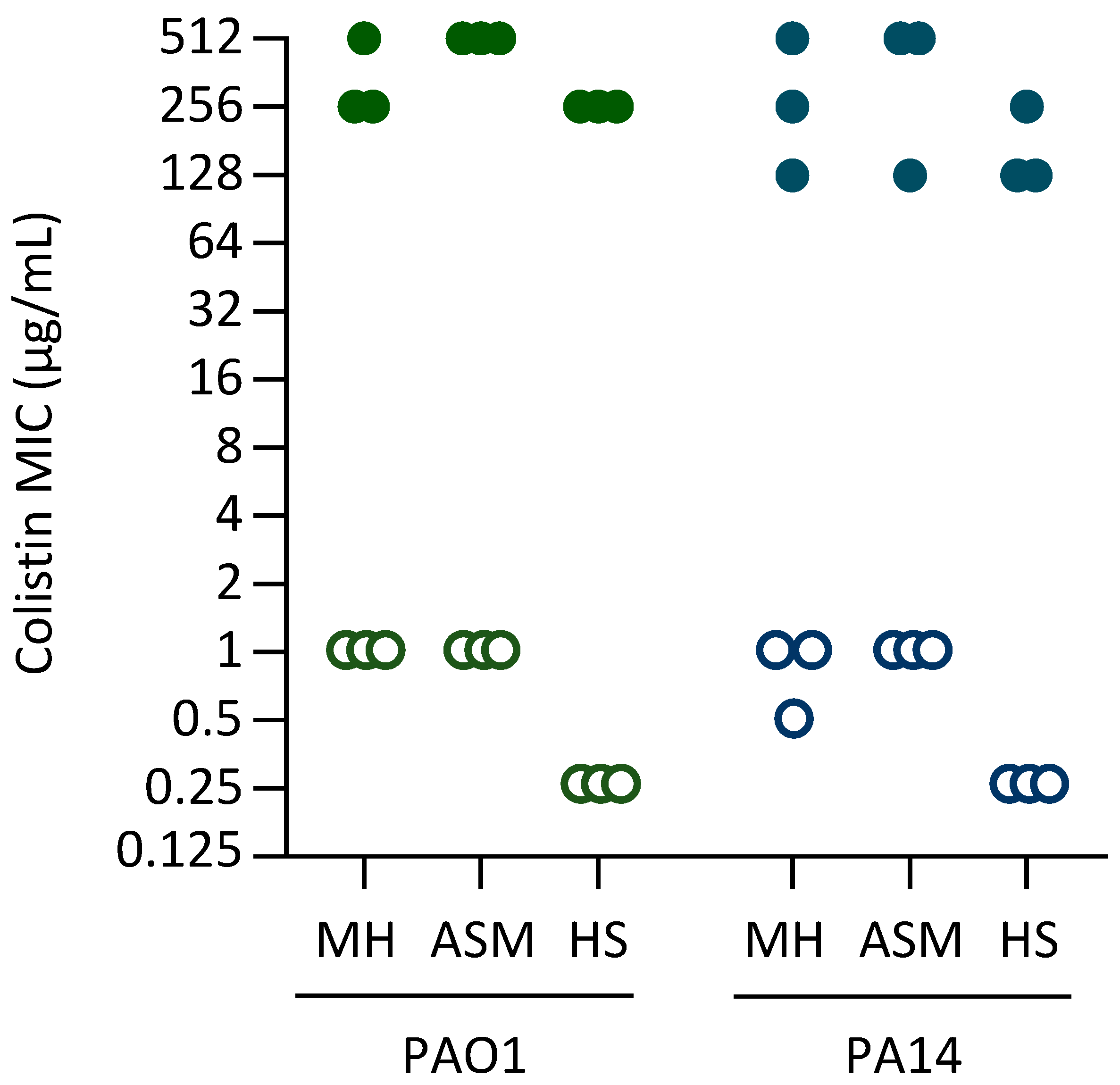
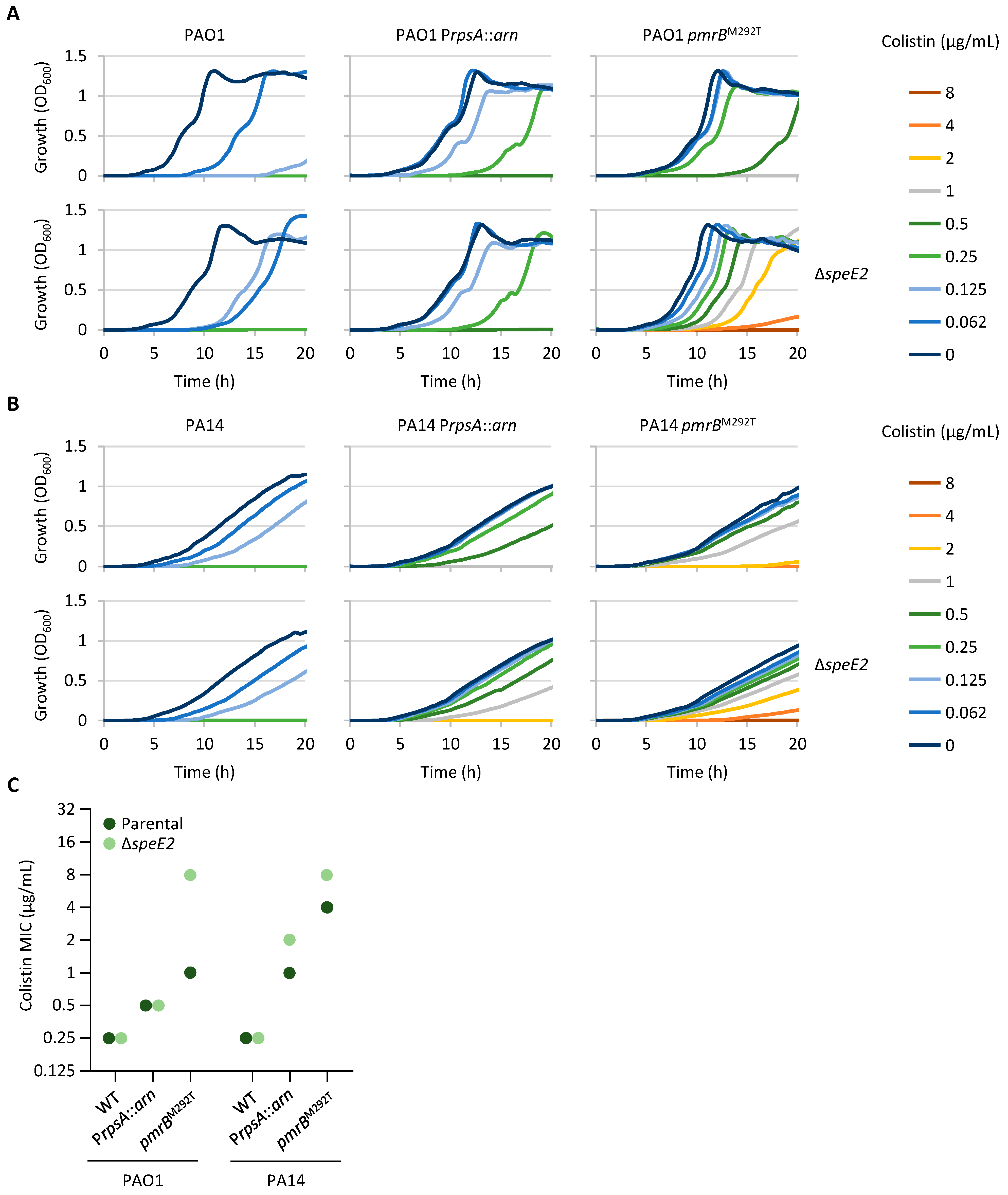
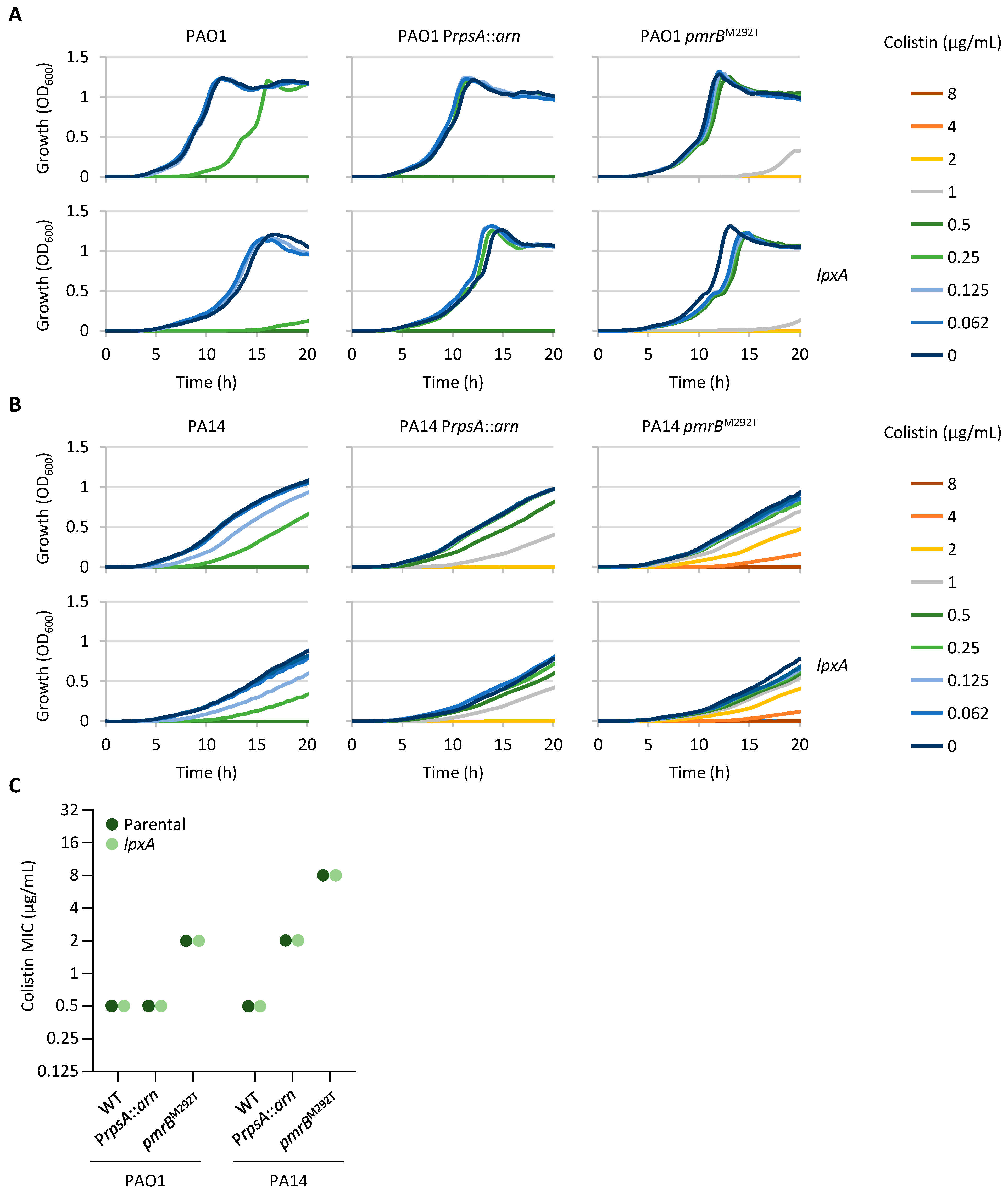
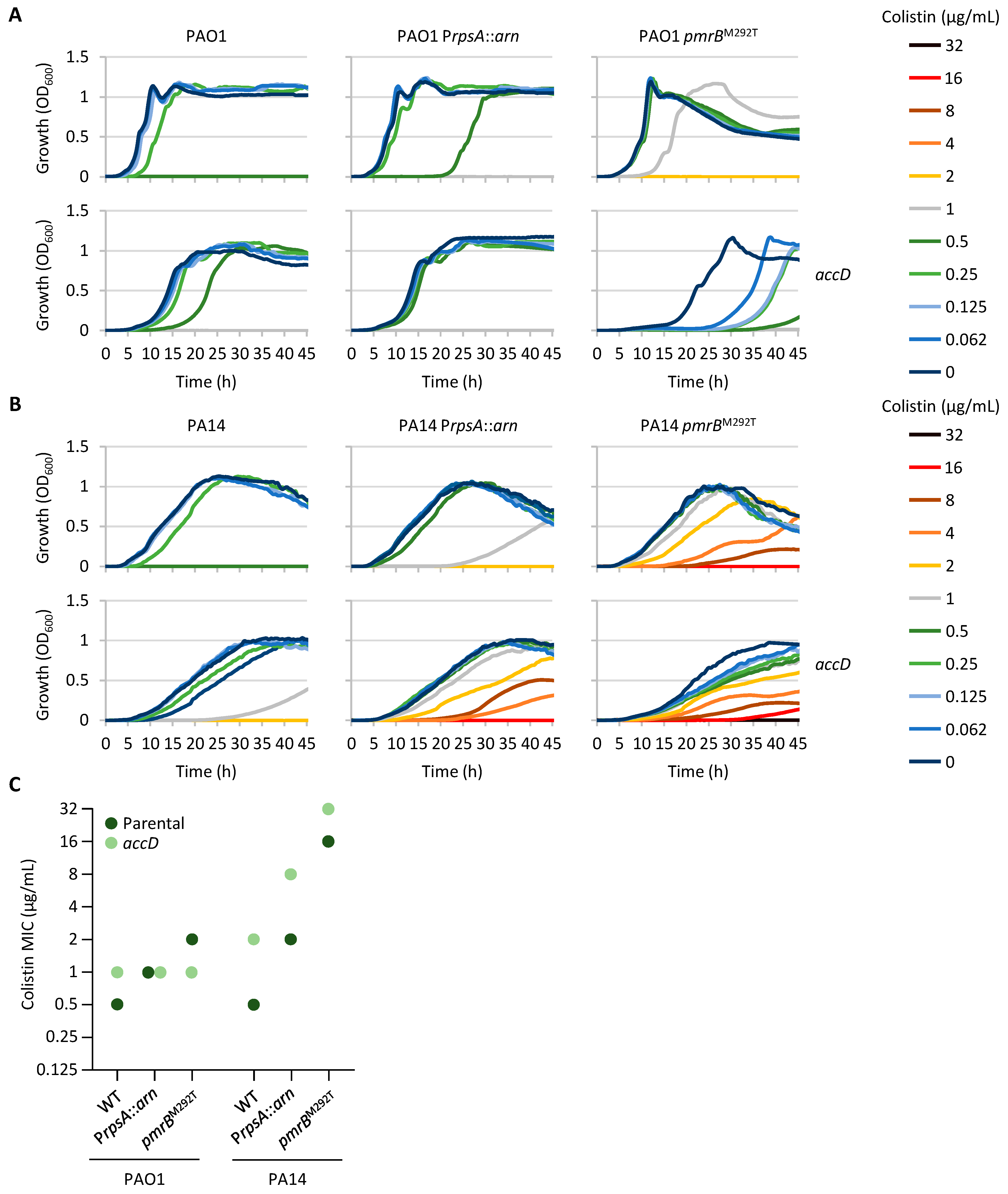
| Strain | Medium | TCSs Controlling Lipid A Aminoarabinosylation | LPS Synthesis and Modification | Norspermidine Synthesis | Fatty Acid Synthesis | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| pmrA | pmrB | phoP | phoQ | lpxA | lpxC | lpxD | lpxT | lpxO2 | galU | speD2 | speE2 | accD | acpP | fabB | fabY | ||
| PAO1 | MH | 3 | 2 | ||||||||||||||
| ASM | 1 | 2 | 1 | 1 | 1 | 3 | |||||||||||
| HS | 2 | 1 | 1 | 2 | 1 | ||||||||||||
| PA14 | MH | 1 | 1 | 2 | 1 | 1 | |||||||||||
| ASM | 3 | 1 | 2 | 1 | 1 | ||||||||||||
| HS | 1 | 2 | 1 | 2 | 1 | 1 | 1 | 1 | |||||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cervoni, M.; Ferriero, A.M.; Lo Sciuto, A.; Guidi, F.; Babić Jordamović, N.; Piazza, S.; Jousson, O.; Esposito, A.; Imperi, F. The Genetic Background and Culture Medium Only Marginally Affect the In Vitro Evolution of Pseudomonas aeruginosa Toward Colistin Resistance. Antibiotics 2025, 14, 601. https://doi.org/10.3390/antibiotics14060601
Cervoni M, Ferriero AM, Lo Sciuto A, Guidi F, Babić Jordamović N, Piazza S, Jousson O, Esposito A, Imperi F. The Genetic Background and Culture Medium Only Marginally Affect the In Vitro Evolution of Pseudomonas aeruginosa Toward Colistin Resistance. Antibiotics. 2025; 14(6):601. https://doi.org/10.3390/antibiotics14060601
Chicago/Turabian StyleCervoni, Matteo, Antonio Maria Ferriero, Alessandra Lo Sciuto, Francesca Guidi, Naida Babić Jordamović, Silvano Piazza, Olivier Jousson, Alfonso Esposito, and Francesco Imperi. 2025. "The Genetic Background and Culture Medium Only Marginally Affect the In Vitro Evolution of Pseudomonas aeruginosa Toward Colistin Resistance" Antibiotics 14, no. 6: 601. https://doi.org/10.3390/antibiotics14060601
APA StyleCervoni, M., Ferriero, A. M., Lo Sciuto, A., Guidi, F., Babić Jordamović, N., Piazza, S., Jousson, O., Esposito, A., & Imperi, F. (2025). The Genetic Background and Culture Medium Only Marginally Affect the In Vitro Evolution of Pseudomonas aeruginosa Toward Colistin Resistance. Antibiotics, 14(6), 601. https://doi.org/10.3390/antibiotics14060601






