Current Insights into Antibiotic Resistance in Uropathogenic Escherichia coli and Interventions Using Selected Bioactive Phytochemicals
Abstract
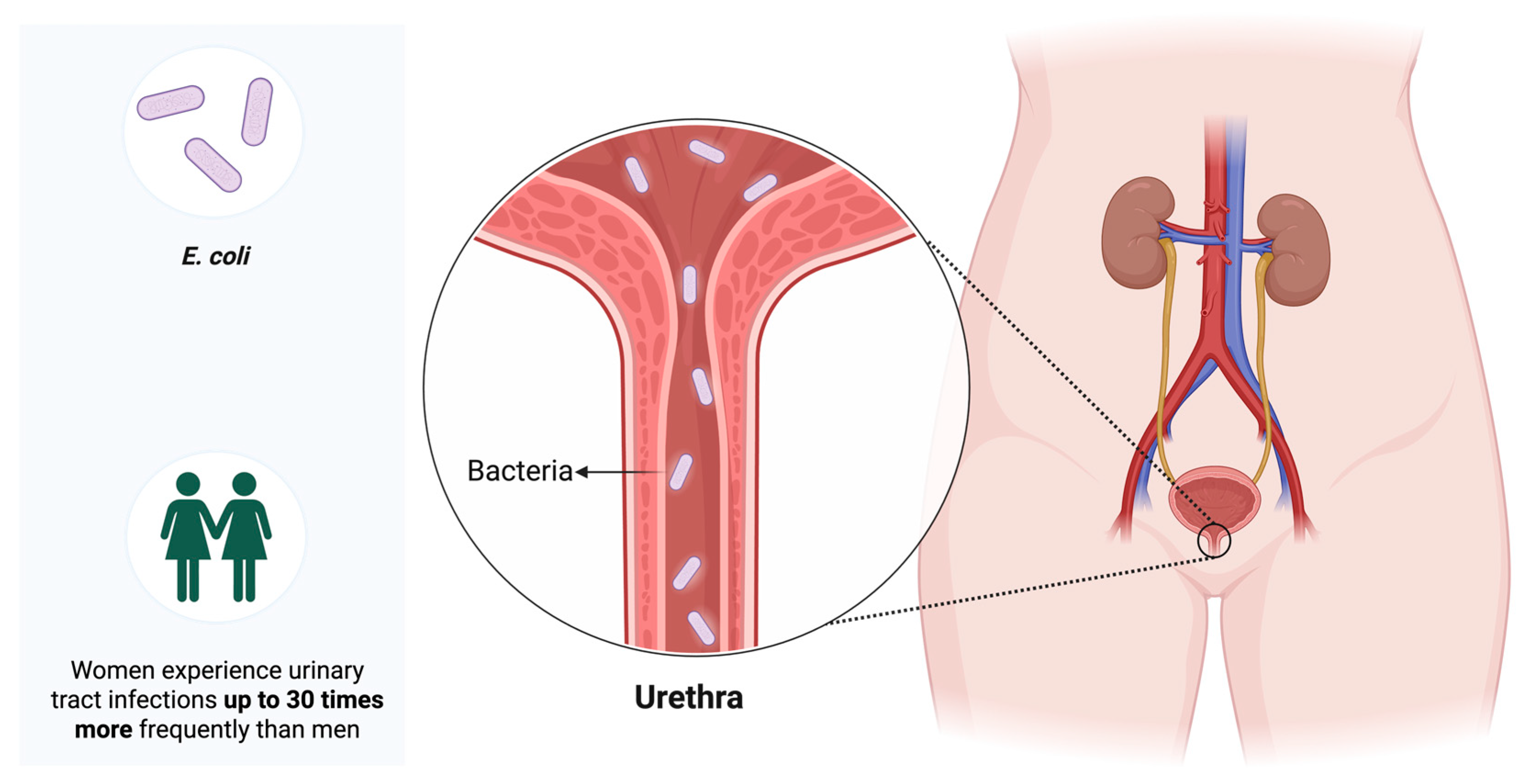
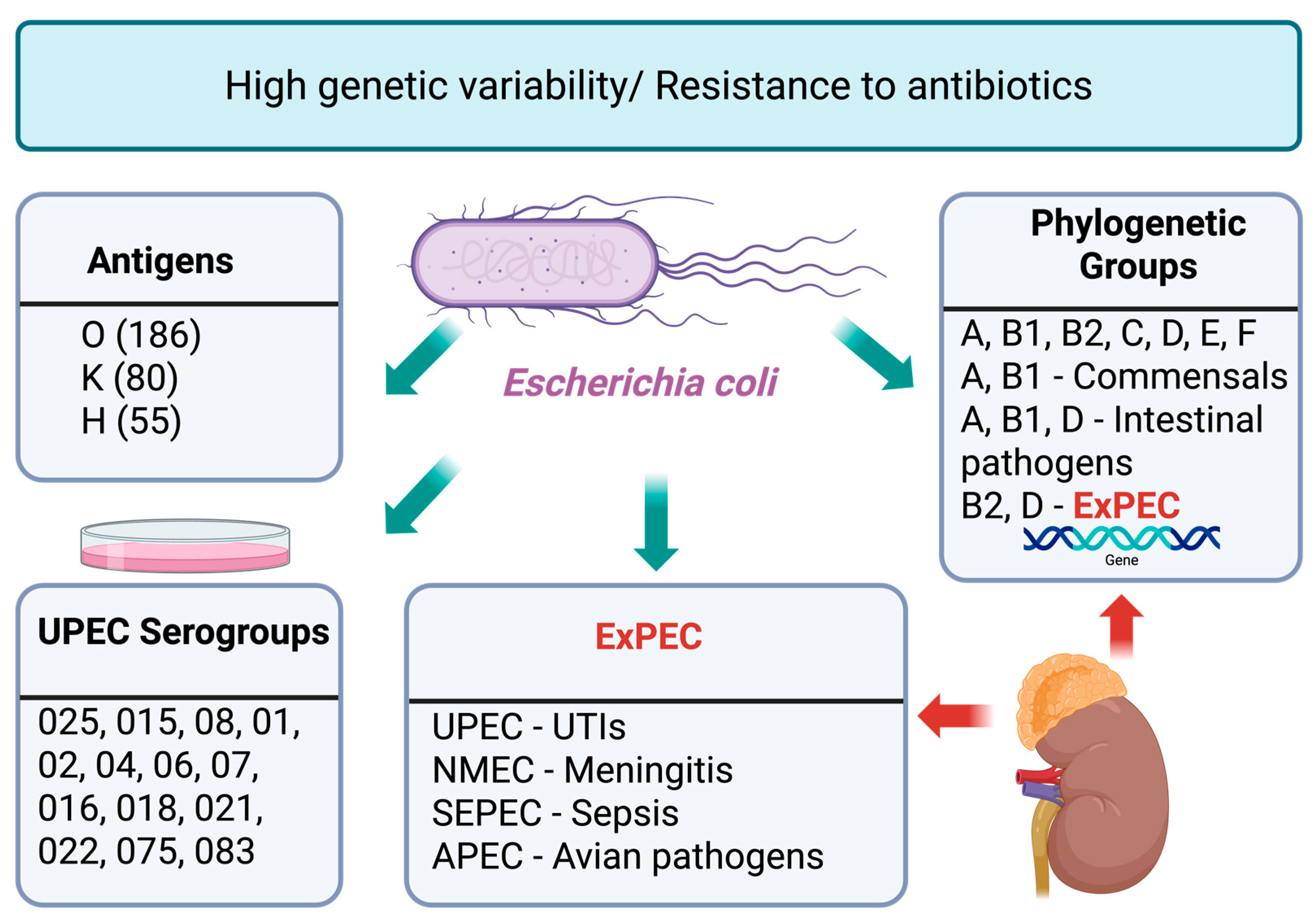
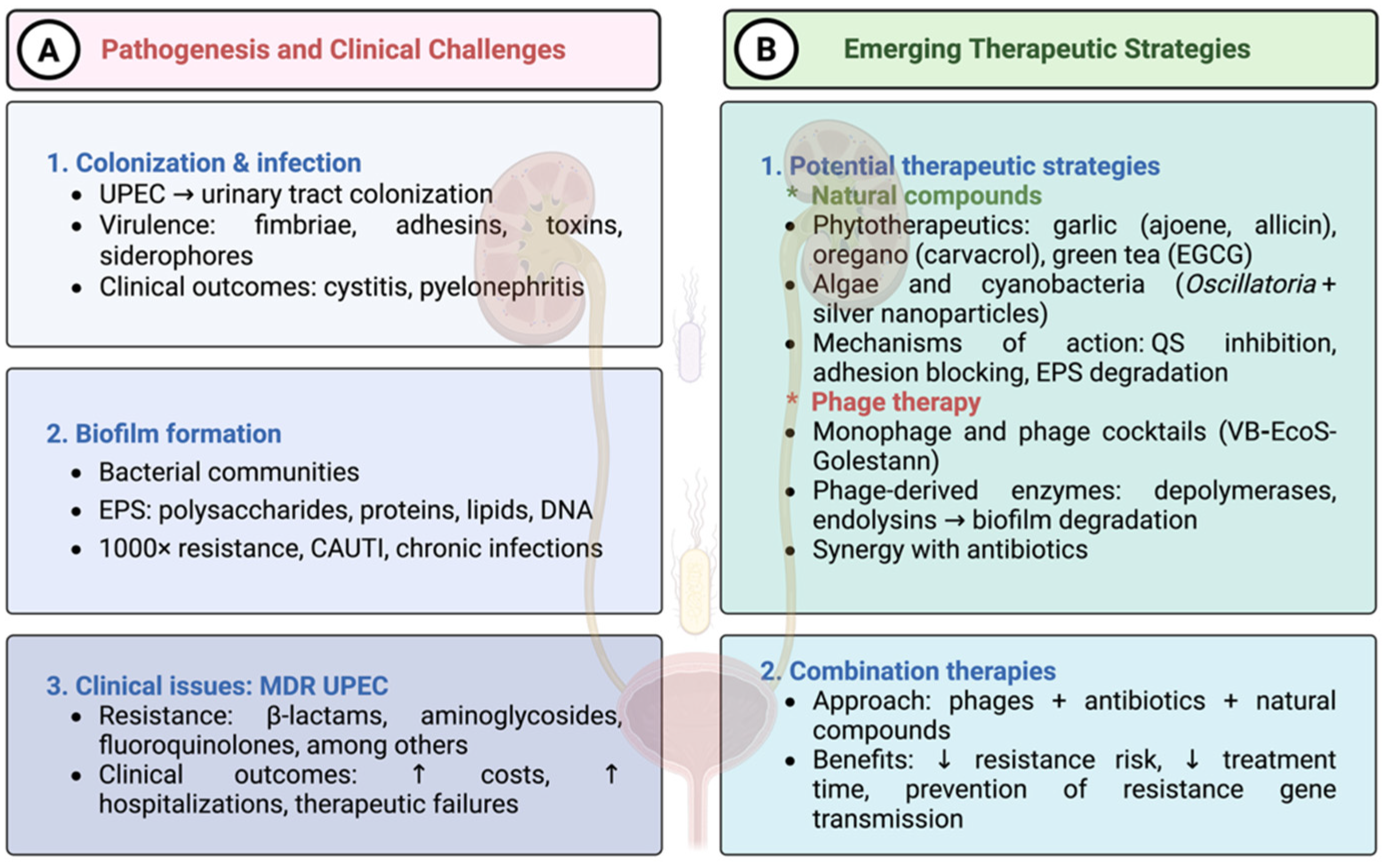
1. Introduction
2. E. coli Strategies for Antibiotic Resistance
3. Virulence and Adaptive Mechanisms of UPEC
3.1. Basis of Two-Component Systems in Bacterial Stress and Drug Resistance
3.2. Role of Adhesins in UPEC Infections
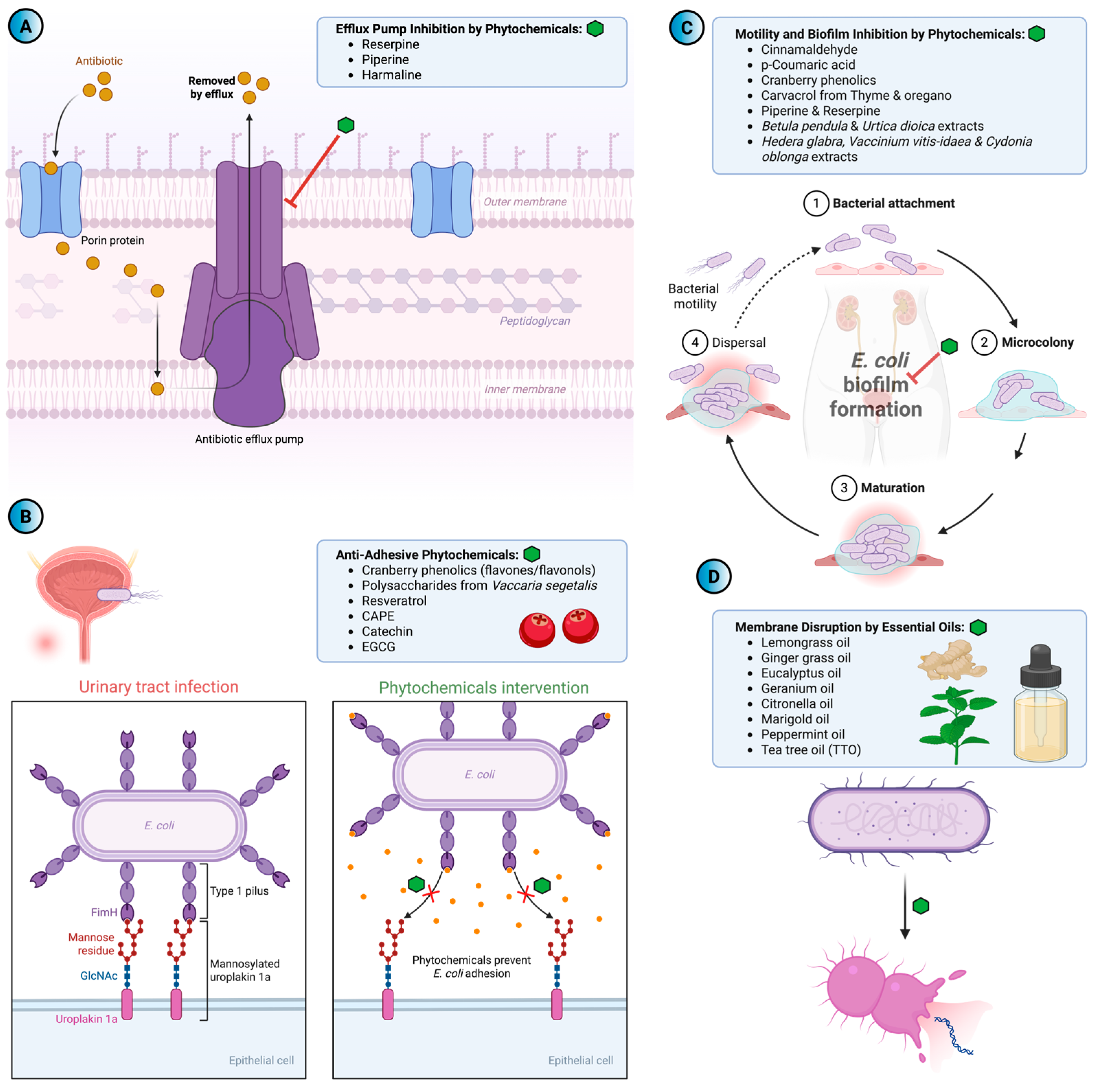
4. Efflux-Mediated Antibiotic Resistance and the Search for Natural Inhibitors
5. Phytochemicals-Based Approaches to Overcome Antibiotic Resistance in UPEC
5.1. Anti-Adhesive Phytochemicals
5.2. Phytochemicals as Inhibitors of Motility and Biofilm Formation
| Phytochemical | Primary Target | Mechanism of Action | Observed Outcome | References |
|---|---|---|---|---|
| Cranberry (Vaccinium macrocarpon) | Adhesion, Motility, Biofilm | Inhibition of FimH-mediated adhesion; downregulation of fliC; reduction of flagellar motility | Decrease in UPEC adhesion and colonization; decrease in biofilm formation | [125,128,129] |
| Propolis + Cranberry extract | Motility, Biofilm | Synergistic inhibition of flagellar motility and EPS-associated biofilm formation | Strong impact on the motility and the biofilm formation of UPEC | [127] |
| Cinnamaldehyde (CNMA) | Adhesion, Biofilm | Inhibits UPEC biofilm formation by suppressing motility, reducing fimbriae, and damaging the bacterial membrane | Strong reduction of UPEC biofilm, fimbriae, motility and growth | [130] |
| p-Coumaric acid | Growth, Membrane integrity | Disruption of bacterial cell membranes and possible binding to bacterial genomic DNA | Loss of membrane integrity, disruption of cellular functions, inhibition of growth | [131,132] |
| Allyl isothiocyanate (AITC) 2-Phenylethyl isothiocyanate (PEITC) | Growth | Disruption of the bacterial cell membrane | Potassium leakage, altered surface properties, growth inhibition, bactericidal activity | [133] |
| Resveratrol | Adhesion, Invasion | Inhibition of FAK (Y576) phosphorylation and block actin-dependent invasion | Strong inhibition of UPEC invasion with minimal effect on adhesion | [135] |
| CAPE (caffeic acid phenethyl ester) | Invasion | Inhibition of FAK (Y576) phosphorylation and block actin-dependent invasion | Strong inhibition of UPEC invasion with minimal effect on adhesion | [135] |
| Catechin, Epigallocatechin gallate—EGCG | Adhesion | More subtle, but still discernable, effects on pFAK (Y576) | Have minimal effects on UPEC adhesion and modest reduction of invasion | [135] |
| Bean pods extract (Phaseoli pericarpium) | Adhesion | Antiadhesive activity against | ↓ E. coli NU14 adhesion to T24 human bladder cells (dose-dependent) | [119] |
| Quince (Cydonia oblonga) extract | Adhesion | — | ↓ Bacterial adhesion; moderate antibacterial activity | [120] |
| Betula pendula, Urtica dioica extracts | Motility | — | ↓ UPEC motility in E. coli clinical isolates | [121] |
| Vaccaria segetalis polysaccharides | Adhesion, Invasion, Motility | Inhibits UPEC adhesion, invasion and motility by downregulating fimbrial adhesins, TLR signalling and uroplakin expression | Significant reduction of UPEC adhesion, invasion, motility, and bladder colonization | [134] |
| Lemongrass essential oil (Cymbopogon flexuosus) | Growth | Possible cell membrane disruption | Antibacterial activity effect against E. coli; no resistance after 30 passages | [138] |
| Tea tree oil (Melaleuca alternifolia) | Growth/ Resistance modulation | Sub-MIC exposure may induce cross-resistance to antibiotics | Potential risk of reduced antibiotic susceptibility in E. coli after repeated exposure | [139] |
| Piperine, Reserpine | Motility, Biofilm | Downregulation of motA, motB; upregulation of fimA, papA, uvrY | ↓ Motility; ↑ adhesion; enhanced antibiotic efficacy | [117] |
6. Summary
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mahshouri, P.; Alikhani, M.Y.; Momtaz, H.E.; Doosti-Irani, A.; Shokoohizadeh, L. Analysis of phylogroups, biofilm formation, virulence factors, antibiotic resistance and molecular typing of uropathogenic Escherichia coli strains isolated from patients with recurrent and non-recurrent urinary tract infections. BMC Infect. Dis. 2025, 25, 267. [Google Scholar] [CrossRef]
- Chegini, Z.; Khoshbayan, A.; Vesal, S.; Moradabadi, A.; Hashemi, A.; Shariati, A. Bacteriophage therapy for inhibition of multi drug-resistant uropathogenic bacteria: A narrative review. Ann. Clin. Microbiol. Antimicrob. 2021, 20, 30. [Google Scholar] [CrossRef] [PubMed]
- Mancuso, G.; Midiri, A.; Gerace, E.; Marra, M.; Zummo, S.; Biondo, C. Urinary tract infections: The current scenario and future prospects. Pathogens 2023, 12, 623. [Google Scholar] [CrossRef]
- Czajkowski, K.; Broś-Konopielko, M.; Teliga-Czajkowska, J. Urinary Tract Infection in Women. Prz Menopauzalny 2021, 20, 40–47. [Google Scholar] [CrossRef]
- Medina, M.; Castillo-Pino, E. An introduction to the epidemiology and burden of urinary tract infections. Ther. Adv. Urol. 2019, 11, 1756287219832172. [Google Scholar] [CrossRef]
- Moreira de Gouveia, M.I.; Bernalier-Donadille, A.; Jubelin, G. Enterobacteriaceae in the human gut: Dynamics and ecological roles in health and disease. Biology 2024, 13, 142. [Google Scholar] [CrossRef]
- Whelan, S.; Lucey, B.; Finn, K. Uropathogenic Escherichia coli (UPEC)-aassociated urinary tract infections: The molecular basis for challenges to effective treatment. Microorganisms 2023, 11, 2169. [Google Scholar] [CrossRef]
- Silva, N.; Igrejas, G.; Gonçalves, A.; Poeta, P. Commensal Gut Bacteria: Distribution of Enterococcus species and prevalence of Escherichia coli phylogenetic groups in animals and humans in Portugal. Ann. Microbiol. 2012, 62, 449–459. [Google Scholar] [CrossRef]
- Clermont, O.; Bonacorsi, S.; Bingen, E. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 2000, 66, 4555–4558. [Google Scholar] [CrossRef] [PubMed]
- Escobar-Páramo, P.; Clermont, O.; Blanc-Potard, A.-B.; Bui, H.; Le Bouguénec, C.; Denamur, E. A specific genetic background is required for acquisition and expression of virulence factors in Escherichia coli. Mol. Biol. Evol. 2004, 21, 1085–1094. [Google Scholar] [CrossRef] [PubMed]
- Sarowska, J.; Futoma-Koloch, B.; Jama-Kmiecik, A.; Frej-Madrzak, M.; Ksiazczyk, M.; Bugla-Ploskonska, G.; Choroszy-Krol, I. Virulence factors, prevalence and potential transmission of extraintestinal pathogenic Escherichia coli isolated from different sources: Recent Reports. Gut Pathog. 2019, 11, 10. [Google Scholar] [CrossRef] [PubMed]
- Lüthje, P.; Brauner, A. Virulence factors of uropathogenic E. coli and their interaction with the host. Adv. Microb. Physiol. 2014, 65, 337–372. [Google Scholar]
- Zhou, Y.; Zhou, Z.; Zheng, L.; Gong, Z.; Li, Y.; Jin, Y.; Huang, Y.; Chi, M. Urinary tract iinfections caused by uropathogenic Escherichia coli: Mechanisms of infection and treatment options. Int. J. Mol. Sci. 2023, 24, 10537. [Google Scholar] [CrossRef]
- Kaper, J.B.; Nataro, J.P.; Mobley, H.L.T. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2004, 2, 123–140. [Google Scholar] [CrossRef]
- Tajbakhsh, E.; Ahmadi, P.; Abedpour-Dehkordi, E.; Arbab-Soleimani, N.; Khamesipour, F. Biofilm formation, antimicrobial susceptibility, serogroups and virulence genes of uropathogenic E. coli isolated from clinical samples in Iran. Antimicrob. Resist. Infect. Control. 2016, 5, 11. [Google Scholar] [CrossRef]
- Rahme, D.; Nakkash Chmaisse, H.; Salameh, P. Unraveling the length of hospital stay for patients with urinary tract infections: Contributing factors and microbial susceptibility. Antibiotics 2025, 14, 421. [Google Scholar] [CrossRef]
- Ahmed, S.K.; Hussein, S.; Qurbani, K.; Ibrahim, R.H.; Fareeq, A.; Mahmood, K.A.; Mohamed, M.G. Antimicrobial resistance: Impacts, challenges, and future prospects. J. Med. Surg. Public Health 2024, 2, 100081. [Google Scholar] [CrossRef]
- World Health Organization. Bacterial Priority Pathogens List, 2024: Bacterial Pathogens of Public Health Importance to Guide Research, Development and Strategies to Prevent and Control Antimicrobial Resistance; World Health Organization: Geneva, Switzerland, 2024. [Google Scholar]
- Yazdi, M.; Bouzari, M.; Ghaemi, E.A.; Shahin, K. Isolation, Characterization and genomic analysis of a novel bacteriophage VVB_EcoS-Golestan infecting multidrug-resistant Escherichia coli isolated from urinary tract infection. Sci. Rep. 2020, 10, 7690. [Google Scholar] [CrossRef]
- González-Villalobos, E.; Ribas-Aparicio, R.M.; Montealegre, G.E.R.; Belmont-Monroy, L.; Ortega-García, Y.; Aparicio-Ozores, G.; Balcázar, J.L.; Eslava-Campos, C.A.; Hernández-Chiñas, U.; Molina-López, J. Isolation and characterization of novel bacteriophages as a potential therapeutic option for Escherichia coli urinary tract infections. Appl. Microbiol. Biotechnol. 2021, 105, 5617–5629. [Google Scholar] [CrossRef] [PubMed]
- Zulk, J.J.; Clark, J.R.; Ottinger, S.; Ballard, M.B.; Mejia, M.E.; Mercado-Evans, V.; Heckmann, E.R.; Sanchez, B.C.; Trautner, B.W.; Maresso, A.W.; et al. Phage resistance accompanies reduced fitness of uropathogenic Escherichia coli in the urinary environment. mSphere 2022, 7, e0034522. [Google Scholar] [CrossRef] [PubMed]
- Cieślik, M.; Wójcicki, M.; Migdał, P.; Grygiel, I.; Bajrak, O.; Orwat, F.; Górski, A.; Jończyk-Matysiak, E. Fighting biofilm: Bacteriophages eliminate biofilm formed by multidrug-resistant Enterobacter hormaechei on urological catheters. Med. Microbiol. Immunol. 2025, 214, 33. [Google Scholar] [CrossRef]
- Cadena, M.; Kelman, T.; Marco, M.L.; Pitesky, M. Understanding antimicrobial resistance (AMR) profiles of Salmonella biofilm and planktonic bacteria challenged with disinfectants commonly used during poultry processing. Foods 2019, 8, 275. [Google Scholar] [CrossRef]
- Ali, A.; Zahra, A.; Kamthan, M.; Husain, F.M.; Albalawi, T.; Zubair, M.; Alatawy, R.; Abid, M.; Noorani, M.S. Microbial biofilms: Applications, clinical consequences, and alternative therapies. Microorganisms 2023, 11, 1934. [Google Scholar] [CrossRef]
- Merino, L.; Procura, F.; Trejo, F.M.; Bueno, D.J.; Golowczyc, M.A. Biofilm formation by Salmonella sp. in the poultry industry: Detection, control and eradication strategies. Food Res. Int. 2019, 119, 530–540. [Google Scholar] [CrossRef] [PubMed]
- Nasrollahian, S.; Graham, J.P.; Halaji, M. A review of the mechanisms that confer antibiotic resistance in pathotypes of E. coli. Front. Cell. Infect. Microbiol. 2024, 14, 1387497. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez, D.; Rodríguez-Rubio, L.; Martínez, B.; Rodríguez, A.; García, P. Bacteriophages as weapons against bacterial biofilms in the food industry. Front. Microbiol. 2016, 7, 825. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, P.; Maira-Litran, T.; McBain, A.J.; Rickard, A.H.; Whyte, F.W. The physiology and collective recalcitrance of microbial biofilm communities. Adv. Microb. Physiol. 2002, 46, 203–256. [Google Scholar]
- McMurray, R.L.; Ball, M.E.E.; Tunney, M.M.; Corcionivoschi, N.; Situ, C. Antibacterial activity of four plant extracts extracted from traditional chinese medicinal plants against Listeria monocytogenes, Escherichia coli, and Salmonella enterica subsp. enterica serovar Enteritidis. Microorganisms 2020, 8, 962. [Google Scholar] [CrossRef]
- Krzyżewska, E.; Książczyk, M.; Futoma-Kołoch, B.; Bugla-Płaskocińska, G. Modifications of cell structure of microorganisms induced by biocides. Advanc. Microbiol. 2015, 54, 380–391. [Google Scholar]
- Rather, M.A.; Gupta, K.; Mandal, M. Microbial biofilm: Formation, architecture, antibiotic resistance, and control strategies. Braz. J. Microbiol. 2021, 52, 1701–1718. [Google Scholar] [CrossRef]
- Larsson, D.G.J.; Flach, C.-F. Antibiotic resistance in the environment. Nat. Rev. Microbiol. 2022, 20, 257–269. [Google Scholar] [CrossRef]
- Ding, D.; Wang, B.; Zhang, X.; Zhang, J.; Zhang, H.; Liu, X.; Gao, Z.; Yu, Z. The spread of antibiotic resistance to humans and potential protection strategies. Ecotoxicol. Environ. Saf. 2023, 254, 114734. [Google Scholar] [CrossRef]
- Wang, M.; Zhang, Z.; Sun, Z.; Wang, X.; Zhu, J.; Jiang, M.; Zhao, S.; Chen, L.; Feng, Q.; Du, H. The emergence of highly resistant and hypervirulent Escherichia coli ST405 clone in a tertiary hospital over 8 years. Emerg. Microbes Infect. 2025, 14, 2479048. [Google Scholar] [CrossRef] [PubMed]
- Abbas, S.H.; Khan, S.; Shah, M.; Aslam, J.; Nawaz, H.; Ilyas, N.; Gamaryani, A.; Afridi, S.Q.; Khan, I.; Shah, B.; et al. Public health threats posed by biofilms and innovative strategies for their control. Discoveries 2024, 12, e197. [Google Scholar] [CrossRef] [PubMed]
- Zheng, W.; Cheng, Y.; Shen, H.; Liu, L.; Hu, W.; Qian, H. Research progress on antibacterial applications of bioactive materials in wound infections: Design, challenges, and prospects. Adv. Healthc. Mater. 2025, 14, 2405103. [Google Scholar] [CrossRef]
- Sawa, T.; Moriyama, K.; Kinoshita, M. Current status of bacteriophage therapy for severe bacterial infections. J. Intensiv. Care 2024, 12, 44. [Google Scholar] [CrossRef]
- Gutiérrez, D.; Ruas-Madiedo, P.; Martínez, B.; Rodríguez, A.; García, P. Effective removal of staphylococcal biofilms by the endolysin LysH5. PLoS ONE 2014, 9, e107307. [Google Scholar] [CrossRef]
- Pires, D.P.; Oliveira, H.; Melo, L.D.R.; Sillankorva, S.; Azeredo, J. Bacteriophage-encoded depolymerases: Their diversity and biotechnological applications. Appl. Microbiol. Biotechnol. 2016, 100, 2141–2151. [Google Scholar] [CrossRef] [PubMed]
- Wesołowski, W.; Łukasiak, A.; Bloch, S.; Kuligowska, K.; Neumann, J.; Lewandowska, N.; Węglińska, E.; Węgrzyn, G.; Nejman-Faleńczyk, B. Phage endolysins as promising and effective candidates for use against uropathogenic Escherichia coli. Viruses 2025, 17, 560. [Google Scholar] [CrossRef]
- Mishra, R.; Panda, A.K.; De Mandal, S.; Shakeel, M.; Bisht, S.S.; Khan, J. Natural anti-biofilm agents: Strategies to control biofilm-forming pathogens. Front. Microbiol. 2020, 11, 566325. [Google Scholar] [CrossRef]
- Michno, M.; Sydor, A.; Wałaszek, M.; Sułowicz, W. Microbiology and drug resistance of pathogens in patients hospitalized at the nephrology department in the south of Poland. Pol. J. Microbiol. 2018, 67, 517–524. [Google Scholar] [CrossRef]
- Al-Maddboly, L.A.; El-Salam, M.; Bastos, J.K.; Hasby, E.A.; Kushkevych, I.; El-Morsi, R.M. Anti-biofilm and anti-quorum sensing activities of galloylquinic acid against clinical isolates of multidrug-resistant Pseudomonas aeruginosa in open wound infection: In vitro and in vivo efficacy studies. BMC Microbiol. 2025, 25, 206. [Google Scholar] [CrossRef]
- Roch, E.; Ducrocq, J.; Jacquier, N. Recent advances in the understanding, detection and therapeutic targeting of bacterial recalcitrance. BMC Microbiol. 2025, 25, 488. [Google Scholar] [CrossRef]
- Choi, N.; Choi, E.; Cho, Y.J.; Kim, M.J.; Choi, H.W.; Lee, E.J. A shared mechanism of multidrug resistance in laboratory-evolved uropathogenic Escherichia coli. Virulence 2024, 15, 2367648. [Google Scholar] [CrossRef]
- Boueroy, P.; Chopjitt, P.; Hatrongjit, R.; Morita, M.; Sugawara, Y.; Akeda, Y.; Iida, T.; Hamada, S.; Kerdsin, A. Fluoroquinolone resistance determinants in carbapenem-resistant Escherichia coli isolated from urine clinical samples in Thailand. PeerJ 2023, 11, e16401. [Google Scholar] [CrossRef]
- Futoma-Kołoch, B.; Bugla-Płoskonska, G.; Dudek, B.; Dorotkiewicz-Jach, A.; Drulis-Kawa, Z.; Gamian, A. Outer membrane proteins of Salmonella as potential markers of resistance to serum, antibiotics, and biocides. Curr. Med. Chem. 2019, 26, 1960–1978. [Google Scholar] [CrossRef]
- World Health Organization. The Evolving Threat of Antimicrobial Resistance: Options for Action; World Health Organization: Geneva, Switzerland, 2012. [Google Scholar]
- Robinson, T.P.; Bu, D.P.; Carrique-Mas, J.; Fèvre, E.M.; Gilbert, M.; Grace, D.; Hay, S.I.; Jiwakanon, J.; Kakkar, M.; Kariuki, S.; et al. Antibiotic resistance is the quintessential One Health issue. Trans. R. Soc. Trop. Med. Hyg. 2016, 110, 377–380. [Google Scholar] [CrossRef] [PubMed]
- Sopirala, M.M.; Mangino, J.E.; Gebreyes, W.A.; Biller, B.; Bannerman, T.; Balada-Llasat, J.M.; Pancholi, P. Synergy testing by Etest, microdilution checkerboard, and time-kill methods for pan-drug-resistant Acinetobacter baumannii. Antimicrob. Agents Chemother. 2010, 54, 4678–4683. [Google Scholar] [CrossRef] [PubMed]
- Magiorakos, A.-P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.; Giske, C.; Harbarth, S.; Hindler, J.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [PubMed]
- Pereira, B.M.P.; Wang, X.; Tagkopoulos, I. Biocide-induced emergence of antibiotic resistance in Escherichia coli. Front. Microbiol. 2021, 12, 640923. [Google Scholar] [CrossRef]
- Economou, V.; Gousia, P. Agriculture and food animals as a source of antimicrobial-resistant bacteria. Infect. Drug Resist. 2015, 8, 49–61. [Google Scholar] [CrossRef]
- Reich, F.; Atanassova, V.; Klein, G. Extended-spectrum ß-lactamase- and ampc-producing enterobacteria in healthy broiler chickens, Germany. Emerg. Infect. Dis. 2013, 19, 1253–1259. [Google Scholar] [CrossRef]
- Crofts, T.S.; Gasparrini, A.J.; Dantas, G. Next-generation approaches to understand and combat the antibiotic resistome. Nat. Rev. Microbiol. 2017, 15, 422–434. [Google Scholar] [CrossRef]
- Hussain, H.I.; Aqib, A.I.; Seleem, M.N.; Shabbir, M.A.; Hao, H.; Iqbal, Z.; Kulyar, M.F.-A.; Zaheer, T.; Li, K. Genetic basis of molecular mechanisms in β-lactam resistant gram-negative bacteria. Microb. Pathog. 2021, 158, 105040. [Google Scholar] [CrossRef] [PubMed]
- Blair, J.M.; Webber, M.A.; Baylay, A.J.; Ogbolu, D.O.; Piddock, L.J. Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol. 2015, 13, 42–51. [Google Scholar] [CrossRef]
- Kaplan, J.B. Antibiotic-induced biofilm formation. Int. J. Artif. Organs 2011, 34, 737–751. [Google Scholar] [CrossRef] [PubMed]
- Adamus-Białek, W.; Wawszczaka, M.; Arabskib, M.; Majchrzak, M.; Gulba, M.; Jarych, D.; Parniewski, P.; Głuszeka, S. Ciprofloxacin, amoxicillin, and aminoglycosides stimulate genetic and phenotypic changes in uropathogenic Escherichia coli strains. Virulence 2019, 10, 260–276. [Google Scholar] [CrossRef] [PubMed]
- Rafaque, Z.; Abid, N.; Liaqat, N.; Afridi, P.; Siddique, S.; Masood, S.; Kanwal, S.; Dasti, J.I. In-vitro investigation of antibiotics efficacy against uropathogenic Escherichia coli biofilms and antibiotic induced biofilm formation at sub-minimum inhibitory concentration of ciprofloxacin. Infect. Drug Resist. 2020, 13, 2801–2810. [Google Scholar] [CrossRef]
- Awad, M.M.; Tovmasyan, A.; Craik, J.D.; Batinic-Haberle, I.; Benov, L.T. Important cellular targets for antimicrobial photodynamic therapy. Appl. Microbiol. Biot. 2016, 100, 7679–7688. [Google Scholar] [CrossRef]
- Linhares, I.; Raposo, T.; Rodrigues, A.; Almeida, A. Frequency and antimicrobial resistance patterns of bacteria implicated in community urinary tract infections: A ten-year surveillance study (2000–2009). BMC Infect. Dis. 2013, 13, 19. [Google Scholar] [CrossRef]
- Sabir, N.; Ikram, A.; Zaman, G.; Satti, L.; Gardezi, A.; Ahmed, A.; Ahmed, P. Bacterial biofilm-based catheter-associated urinary tract infections: Causative pathogens and antibiotic resistance. Am. J. Infect. Control. 2017, 45, 1101–1105. [Google Scholar] [CrossRef]
- Rogers, B.A.; Sidjabat, H.E.; Paterson, D.L. Escherichia coli O25b-ST131: A pandemic, multiresistant, community-associated strain. J. Antimicrob. Chemother. 2011, 66, 1–14. [Google Scholar] [CrossRef]
- Croxall, G.; Weston, V.; Joseph, S.; Manning, G.; Cheetham, P.; McNally, A. Increased human pathogenic potential of Escherichia coli from polymicrobial urinary tract infections in comparison to isolates from monomicrobial culture samples. J. Med. Microbiol. 2011, 60, 102–109. [Google Scholar] [CrossRef] [PubMed]
- Córdoba, G.; Holm, A.; Hansen, F.; Hammerum, A.M.; Bjerrum, L. Prevalence of antimicrobial resistant Escherichia coli from patients with suspected urinary tract infection in primary care, Denmark. BMC Infect. Dis. 2017, 17, 670. [Google Scholar] [CrossRef]
- Malekzadegan, Y.; Khashei, R.; Ebrahim-Saraie, H.S.; Jahanabadi, Z. Distribution of virulence genes and their association with antimicrobial resistance among uropathogenic Escherichia coli isolates from Iranian patients. BMC Infect. Dis. 2018, 18, 572. [Google Scholar] [CrossRef]
- Murugalakshmi, T.; Devi, J.; Bindu, N.; Suriakumar, J. Global prevalence and antibiotic resistance patterns of ESBL-producing Escherichia coli in clinical isolates: A meta-analysis of studies from 2015 to 2024. Int. J. Med. Pub. Health 2025, 15, 1241–1247. [Google Scholar]
- Katongole, P.; Nalubega, F.; Florence, N.C.; Asiimwe, B.; Andia, I. Biofilm formation, antimicrobial susceptibility and virulence genes of uropathogenic Escherichia coli isolated from clinical isolates in Uganda. BMC Infect. Dis. 2020, 20, 453. [Google Scholar] [CrossRef] [PubMed]
- Ochońska, D.; Ścibik, Ł.; Brzychczy-Włoch, M. Biofilm formation of clinical Klebsiella pneumoniae strains isolated from tracheostomy tubes and their association with antimicrobial resistance, virulence and genetic diversity. Pathogens 2021, 10, 1345. [Google Scholar] [CrossRef]
- Gatya Al-Mayahie, S.M.; Al-Guranie, D.R.T.; Hussein, A.A.; Bachai, Z.A. Prevalence of common carbapenemase genes and multidrug resistance among uropathogenic Escherichia coli phylogroup B2 isolates from outpatients in Wasit Province/Iraq. PLoS ONE 2022, 17, e0262984. [Google Scholar] [CrossRef]
- Wang, M.-C.; Fan, Y.-H.; Zhang, Y.-Z.; Bregente, C.J.B.; Lin, W.-H.; Chen, C.-A.; Lin, T.-P.; Kao, C.-Y. Characterization of uropathogenic Escherichia coli phylogroups associated with antimicrobial resistance, virulence factor distribution, and virulence-related phenotypes. Infect. Genet. Evol. 2023, 114, 105493. [Google Scholar] [CrossRef] [PubMed]
- Mujahid, F.; Rasool, M.H.; Shafiq, M.; Aslam, B.; Khurshid, M. Emergence of carbapenem-resistant uropathogenic Escherichia coli (ST405 and ST167) strains carrying blaCTX-M-15, blaNDM-5 and diverse virulence factors in hospitalized patients. Pathogens 2024, 13, 964. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Xu, X.; Pu, S.; Huang, S.; Sun, J.; Yang, S.; Zhang, L. Characterization of carbapenemases, extended spectrum β-lactamases, quinolone resistance and aminoglycoside resistance determinants in carbapenem-non-susceptible Escherichia coli from a teaching hospital in Chongqing, Southwest China. Infect. Genet. Evol. 2014, 27, 271–276. [Google Scholar] [CrossRef]
- Dulanto Chiang, A.; Dekker, J.P. Efflux pump-mediated resistance to new beta lactam antibiotics in multidrug-resistant gram-negative Bacteria. Commun. Med. 2024, 4, 170. [Google Scholar] [CrossRef] [PubMed]
- Fydrych, D.; Jeziurska, J.; Wełna, J.; Kwiecińska-Piróg, J. Potential use of selected natural compounds with anti-biofilm activity. Int. J. Mol. Sci. 2025, 26, 607. [Google Scholar] [CrossRef]
- Bunduki, G.K.; Heinz, E.; Phiri, V.S.; Noah, P.; Feasey, N.; Musaya, J. Virulence factors and antimicrobial resistance of uropathogenic Escherichia coli (UPEC) isolated from urinary tract infections: A systematic review and meta-analysis. BMC Infect. Dis. 2021, 21, 753. [Google Scholar] [CrossRef]
- De Gaetano, G.V.; Lentini, G.; Famà, A.; Coppolino, F.; Beninati, C. Antimicrobial resistance: Two-component regulatory systems and multidrug efflux pumps. Antibiotics 2023, 12, 965. [Google Scholar] [CrossRef]
- Jung, K.; Fabiani, F.; Hoyer, E.; Lassak, J. Bacterial transmembrane signalling systems and their engineering for biosensing. Open Biol. 2018, 8, 180023. [Google Scholar] [CrossRef]
- Brüderlin, M.; Böhm, R.; Fadel, F.; Hiller, S.; Schirmer, T.; Dubey, B.N. Structural features discriminating hybrid histidine kinase Rec domains from response regulator homologs. Nat. Commun. 2023, 14, 1002. [Google Scholar] [CrossRef]
- Bhagirath, A.Y.; Li, Y.; Patidar, R.; Yerex, K.; Ma, X.; Kumar, A.; Duan, K. Two component regulatory systems and antibiotic resistance in gram-negative pathogens. Int. J. Mol. Sci. 2019, 20, 1781. [Google Scholar] [CrossRef] [PubMed]
- Spurbeck, R.R.; Stapleton, A.E.; Johnson, J.R.; Walk, S.T.; Hooton, T.M.; Mobley, H.L.T. Fimbrial profiles predict virulence of uropathogenic Escherichia coli strains: Contribution of ygi and yad fimbriae. Infect. Immun. 2011, 79, 4753–4763. [Google Scholar] [CrossRef]
- Li, X.; Zhou, K.; Wang, J.; Guo, J.; Cao, Y.; Ren, J.; Guan, T.; Sheng, W.; Zhang, M.; Yao, Z.; et al. Diagnostic value of the fimbriae distribution pattern in localization of urinary tract infection. Front. Med. 2021, 8, 602691. [Google Scholar] [CrossRef]
- Rahdar, M.; Rashki, A.; Miri, H.R.; Rashki Ghalehnoo, M. Detection of pap, sfa, sfa, foc, and fim adhesin-encoding operons in uropathogenic Escherichia coli isolates collected from patients with urinary tract infection. Jundishapur J. Microbiol. 2015, 8, e22647. [Google Scholar] [CrossRef]
- Mobley, H.L.; Chippendale, G.R.; Tenney, J.H.; Hull, R.A.; Warren, J.W. Expression of type 1 fimbriae may be required for persistence of Escherichia coli in the catheterized urinary tract. J. Clin. Microbiol. 1987, 25, 2253–2257. [Google Scholar] [CrossRef]
- Sarshar, M.; Behzadi, P.; Ambrosi, C.; Zagaglia, C.; Palamara, A.T.; Scribano, D. FimH and anti-adhesive therapeutics: A disarming strategy against uropathogens. Antibiotics 2020, 9, 397. [Google Scholar] [CrossRef]
- Jacobsen, S.M.; Stickler, D.J.; Mobley, H.L.; Shirtliff, M.E. Complicated catheter-associated urinary tract infections due to Escherichia coli and Proteus mirabilis. Clin. Microbiol. Rev. 2008, 21, 26–59. [Google Scholar] [CrossRef]
- Reid, G.; van der Mei, H.C.; Tieszer, C.; Busscher, H.J. Uropathogenic Escherichia coli adhere to urinary catheters without using fimbriae. FEMS Immunol. Med. Microbiol. 1996, 16, 159–162. [Google Scholar] [CrossRef]
- Kanamaru, S.; Kurazono, H.; Ishitoya, S.; Terai, A.; Habuchi, T.; Nakano, M.; Ogawa, O.; Yamamoto, S. Distribution and genetic association of putative uropathogenic virulence factors iron, iha, kpsMT, ompT and usp in Escherichia coli isolated from urinary tract infections in Japan. J. Urol. 2003, 170, 2490–2493. [Google Scholar] [CrossRef] [PubMed]
- Subashchandrabose, S.; Mobley, H.L.T. Virulence and Fitness Determinants of Uropathogenic Escherichia coli. In Urinary Tract Infections: Molecular Pathogenesis and Clinical Management, 2nd ed.; ASM Press: Washington, DC, USA, 2015; pp. 235–261. [Google Scholar]
- Marrs, C.F.; Zhang, L.; Foxman, B. Escherichia coli mediated urinary tract infections: Are there distinct uropathogenic E. coli (UPEC) pathotypes? FEMS Microbiol. Lett. 2005, 252, 183–190. [Google Scholar] [CrossRef] [PubMed]
- Kuan, L.; Schaffer, J.N.; Zouzias, C.D.; Pearson, M.M. Characterization of 17 chaperone-usher fimbriae encoded by Proteus mirabilis reveals strong conservation. J. Med. Microbiol. 2014, 63, 911–922. [Google Scholar] [CrossRef] [PubMed]
- Wurpel, D.J.; Beatson, S.A.; Totsika, M.; Petty, N.K.; Schembri, M.A. Chaperone-usher fimbriae of Escherichia coli. PLoS ONE 2013, 8, e52835. [Google Scholar] [CrossRef]
- Daniels, R.; Vanderleyden, J.; Michiels, J. Quorum sensing and swarming migration in bacteria. FEMS Microbiol. Rev. 2004, 28, 261–289. [Google Scholar] [CrossRef]
- Terlizzi, M.E.; Gribaudo, G.; Maffei, M.E. UroPathogenic Escherichia coli (UPEC) infections: Virulence factors, bladder responses, antibiotic, and non-antibiotic antimicrobial strategies. Front. Microbiol. 2017, 8, 1566. [Google Scholar] [CrossRef]
- Thakur, V.; Uniyal, A.; Tiwari, V. A comprehensive review on pharmacology of efflux pumps and their inhibitors in antibiotic resistance. Eur. J. Pharmacol. 2021, 903, 174151. [Google Scholar] [CrossRef]
- Lorusso, A.B.; Carrara, J.A.; Barroso, C.D.N.; Tuon, F.F.; Faoro, H. Role of efflux pumps on antimicrobial resistance in Pseudomonas aeruginosa. Int. J. Mol. Sci. 2022, 23, 15779. [Google Scholar] [CrossRef] [PubMed]
- Jarmula, A.; Oblak, E.; Wawrzycka, D.; Gutowicz, J. Efflux-mediated antimicrobial multidrug resistance. Postepy Hig. Med. Dosw. 2011, 65, 216–227. [Google Scholar]
- Cernicchi, G.; Felicetti, T.; Sabatini, S. Microbial efflux pump inhibitors: A journey around quinoline and indole derivatives. Molecules 2021, 26, 6996. [Google Scholar] [CrossRef]
- Du, D.; Wang-Kan, X.; Neuberger, A.; Van Veen, H.W.; Pos, K.M.; Piddock, L.J.V.; Luisi, B.F. Author correction: Multidrug efflux pumps: Structure, function and regulation. Nat. Rev. Microbiol. 2018, 16, 577. [Google Scholar] [CrossRef]
- Alenazy, R. Drug efflux pump inhibitors: A promising approach to counter multidrug resistance in gram-negative pathogens by targeting AcrB protein from AcrAB-TolC multidrug efflux pump from Escherichia coli. Biology 2022, 11, 1328. [Google Scholar] [CrossRef] [PubMed]
- Seukep, A.J.; Fokoua-Maxime, C.D.; Mbuntcha, H.G.; Chen, G.; Assob, J.C.N.; Tenniswood, M.; Sarker, S.D.; Kuete, V.; Ming-Quan, G. Bacterial drug efflux pump inhibitors from plants. In Antimicrobial Resistance: Underlying Mechanisms and Therapeutic Approaches; Springer: Berlin/Heidelberg, Germany, 2022; pp. 487–532. [Google Scholar]
- Sharma, A.; Gupta, V.K.; Pathania, R. Efflux pump inhibitors for bacterial pathogens: From bench to bedside. Indian J. Med. Res. 2019, 149, 129–145. [Google Scholar] [CrossRef]
- Sung, K.; Nawaz, M.; Park, M.; Chon, J.; Khan, S.A.; Alotaibi, K.; Khan, A.A. Comprehensive genomic analysis of uropathogenic E. coli: Virulence factors, antimicrobial resistance, and mobile genetic elements. Pathogens 2024, 13, 794. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, N.; Suhani, S.; Purkaystha, A.; Begum, M.K.; Raihan, T.; Alam, M.J.; Islam, K.; Azad, A.K. Identification of AcrAB-TolC efflux pump genes and detection of mutation in efflux repressor AcrR from omeprazole responsive multidrug-resistant Escherichia coli isolates causing urinary tract infections. Microbiol. Insights 2019, 12, 1178636119889629. [Google Scholar] [CrossRef]
- Fattahi, S.; Kafil, H.S.; Nahai, M.R.; Asgharzadeh, M.; Nori, R.; Aghazadeh, M. Relationship of biofilm formation and different virulence genes in uropathogenic Escherichia coli isolates from Northwest Iran. GMS Hyg. Infect. Control 2015, 10, 11. [Google Scholar]
- Høiby, N.; Bjarnsholt, T.; Givskov, M.; Molin, S.; Ciofu, O. Antibiotic resistance of bacterial biofilms. Int. J. Antimicrob. Agents 2010, 35, 322–332. [Google Scholar] [CrossRef]
- Zhurina, M.V.; Gannesen, A.V.; Zdorovenko, E.L.; Plakunov, V.K. Composition and functions of the extracellular polymer matrix of bacterial biofilms. Microbiology 2014, 83, 713–722. [Google Scholar] [CrossRef]
- Poole, K. Bacterial stress responses as determinants of antimicrobial resistance. J. Antimicrob. Chemother. 2012, 67, 2069–2089. [Google Scholar] [CrossRef] [PubMed]
- Soto, S.M. Role of efflux pumps in the antibiotic resistance of bacteria embedded in a biofilm. Virulence 2013, 4, 223–229. [Google Scholar] [CrossRef]
- Martinez, J.L.; Sánchez, M.B.; Martínez-Solano, L.; Hernandez, A.; Garmendia, L.; Fajardo, A.; Alvarez-Ortega, C. Functional role of bacterial multidrug efflux pumps in microbial natural ecosystems. FEMS Microbiol. Rev. 2009, 33, 430–449. [Google Scholar] [CrossRef] [PubMed]
- Lebeaux, D.; Ghigo, J.-M.; Beloin, C. Biofilm-related infections: Bridging the gap between clinical management and fundamental aspects of recalcitrance toward antibiotics. Microbiol. Mol. Biol. Rev. 2014, 78, 510–543. [Google Scholar] [CrossRef]
- Wood, T.K.; Knabel, S.J.; Kwan, B.W. Bacterial persister cell formation and dormancy. Appl. Environ. Microbiol. 2013, 79, 7116–7121. [Google Scholar] [CrossRef]
- Niu, H.; Gu, J.; Zhang, Y. Bacterial persisters: Molecular mechanisms and therapeutic development. Sig. Transduct. Target Ther. 2024, 9, 174. [Google Scholar] [CrossRef]
- Burrowes, B.; Harper, D.R.; Anderson, J.; McConville, M.; Enright, M.C. Bacteriophage therapy: Potential uses in the control of antibiotic-resistant pathogens. Expert Rev. Anti-Infect. Ther. 2011, 9, 775–785. [Google Scholar] [CrossRef]
- Wozniak, A.; Grinholc, M. Combined antimicrobial activity of photodynamic inactivation and antimicrobials-state of the art. Front. Microbiol. 2018, 9, 930. [Google Scholar] [CrossRef] [PubMed]
- Dusane, D.; Hosseinidoust, Z.; Asadishad, B.; Tufenkji, N. Alkaloids modulate motility, biofilm formation and antibiotic susceptibility of uropathogenic Escherichia coli. PLoS ONE 2014, 9, e112093. [Google Scholar] [CrossRef]
- Uma, K.; Huang, X.; Kumar, B.A. Antifungal effect of plant extract and essential oil. Chin. J. Integr. Med. 2017, 23, 233–239. [Google Scholar] [CrossRef] [PubMed]
- Popowski, D.; Pawłowska, K.A.; Deipenbrock, M.; Hensel, A.; Kruk, A.; Melzig, M.F.; Piwowarski, J.P.; Granica, S. Antiadhesive activity of hydroethanolic extract from bean pods of Phaseolus vulgaris (common bean) against uropathogenic E. coli and permeability of its constituents through Caco-2 cells monolayer. J. Ethnopharmacol. 2021, 274, 114053. [Google Scholar] [CrossRef]
- Hendrich, A.B.; Strugała, P.; Dudra, A.; Kucharska, A.Z.; Sokół-Łętowska, A.; Wojnicz, D.; Cisowska, A.; Sroka, Z.; Gabrielska, J. Microbiological, antioxidant and lipoxygenase-1 inhibitory activities of fruit extracts of chosen Rosaceae family species. Adv. Clin. Exp. Med. 2020, 29, 215–224. [Google Scholar] [CrossRef] [PubMed]
- Wojnicz, D.; Kucharska, A.Z.; Sokół-Łętowska, A.; Kicia, M.; Tichaczek-Goska, D. Medicinal plants extracts affect virulence factors expression and biofilm formation by the uropathogenic Escherichia coli. Urol. Res. 2012, 40, 683–697. [Google Scholar] [CrossRef]
- Withington-Wray, D.J.; Roberts, B.L.; Taylor, E.W. The topographical organization of the vagal motor column in the elasmobranch fish, Scyliorhinus canicula L. J. Comp. Neurol. 1986, 248, 95–104. [Google Scholar] [CrossRef]
- Guttenplan, S.B.; Kearns, D.B. Regulation of flagellar motility during biofilm formation. FEMS Microbiol. Rev. 2013, 37, 849–871. [Google Scholar] [CrossRef]
- Zagaglia, C.; Ammendolia, M.G.; Maurizi, L.; Nicoletti, M.; Longhi, C. Urinary tract infections caused by uropathogenic Escherichia coli strains—New strategies for an old pathogen. Microorganisms 2022, 10, 1425. [Google Scholar] [CrossRef]
- Hidalgo, G.; Chan, M.; Tufenkji, N. Inhibition of Escherichia coli cft073 flic expression and motility by cranberry materials. Appl. Environ. Microbiol. 2011, 77, 6852–6857. [Google Scholar] [CrossRef]
- Wojnicz, D.; Tichaczek-Goska, D. Effect of sub-minimum inhibitory concentrations of ciprofloxacin, amikacin and colistin on biofilm formation and virulence factors of Escherichia coli planktonic and biofilm forms isolated from human urine. Braz. J. Microbiol. 2013, 44, 259–265. [Google Scholar] [CrossRef] [PubMed]
- Ranfaing, J.; Dunyach-Remy, C.; Lavigne, J.-P.; Sotto, A. Propolis potentiates the effect of cranberry (Vaccinium macrocarpon) in reducing the motility and the biofilm formation of uropathogenic Escherichia coli. PLoS ONE 2018, 13, e0202609. [Google Scholar] [CrossRef] [PubMed]
- González de Llano, D.; Esteban-Fernández, A.; Sánchez-Patán, F.; Martínlvarez, P.J.; Moreno-Arribas, M.V.; Bartolomé, B. Anti-adhesive activity of cranberry phenolic compounds and their microbial-derived metabolites against uropathogenic Escherichia coli in bladder epithelial cell cultures. Int. J. Mol. Sci. 2015, 16, 12119–12130. [Google Scholar] [CrossRef]
- Scharf, B.; Schmidt, T.J.; Rabbani, S.; Stork, C.; Dobrindt, U.; Sendker, J.; Ernst, B.; Hensel, A. Antiadhesive natural products against uropathogenic E. coli: What can we learn from cranberry extract? J. Ethnopharmacol. 2020, 257, 112889. [Google Scholar] [CrossRef]
- Amalaradjou, M.A.; Narayanan, A.; Baskaran, S.A.; Venkitanarayanan, K. Antibiofilm effect of trans-cinnamaldehyde on uropathogenic Escherichia coli. J. Urol. 2010, 184, 358–363. [Google Scholar] [CrossRef]
- Lou, Z.; Wang, H.; Rao, S.; Sun, J.; Ma, C.; Li, J. p-coumaric acid kills bacteria through dual damage mechanisms. Food Control. 2012, 25, 550–554. [Google Scholar] [CrossRef]
- Ning, Y.; Yan, A.; Yang, K.; Wang, Z.; Li, X.; Jia, Y. Antibacterial activity of phenyllactic acid against Listeria monocytogenes and Escherichia coli by dual mechanisms. Food Chem. 2017, 228, 533–540. [Google Scholar] [CrossRef]
- Borges, A.; Abreu, A.C.; Ferreira, C.; Saavedra, M.J.; Simões, L.C.; Simões, M. Antibacterial activity and mode of action of selected glucosinolate hydrolysis products against bacterial pathogens. J. Food Sci. Technol. 2015, 52, 4737–4748. [Google Scholar] [CrossRef] [PubMed]
- Yao, R.; Mao, X.; Xu, Y.; Qiu, X.; Zhou, L.; Wang, Y.; Pang, B.; Chen, M.; Cao, S.; Bao, L.; et al. Polysaccharides from Vaccaria segetalis seeds reduce urinary tract infections by inhibiting the adhesion and invasion abilities of uropathogenic Escherichia coli. Front. Cell Infect. Microbiol. 2022, 12, 1004751. [Google Scholar] [CrossRef]
- Lewis, A.J.; Richards, A.C.; Mendez, A.A.; Dhakal, B.K.; Jones, T.A.; Sundsbak, J.L.; Eto, D.S.; Rousek, A.A.; Mulvey, M.A. Plant phenolics inhibit focal adhesion kinase and suppress host cell invasion by uropathogenic Escherichia coli. Infect. Immun. 2024, 92, e00080-24. [Google Scholar] [CrossRef]
- Ribeiro-Santos, R.; Andrade, M.; Melo, N.R.D.; Sanches-Silva, A. Use of essential oils in active food packaging: Recent advances and future trends. Trends Food Sci. Technol. 2017, 61, 132–140. [Google Scholar] [CrossRef]
- Fournomiti, M.; Kimbaris, A.; Mantzourani, I.; Plessas, S.; Theodoridou, I.; Papaemmanouil, V.; Kapsiotis, I.; Panopoulou, M.; Stavropoulou, E.; Bezirtzoglou, E.E.; et al. Antimicrobial activity of essential oils of cultivated oregano (Origanum vulgare), sage (Salvia officinalis), and thyme (Thymus vulgaris) against clinical isolates of Escherichia coli, Klebsiella oxytoca, and Klebsiella pneumoniae. Microb. Ecol. Health Dis. 2015, 26, 23289. [Google Scholar] [CrossRef] [PubMed]
- Mangalagiri, N.P.; Panditi, S.K.; Jeevigunta, N.L.L. Antimicrobial activity of essential plant oils and their major components. Heliyon 2021, 7, e06835. [Google Scholar] [CrossRef] [PubMed]
- McMahon, M.A.S.; Blair, I.S.; Moore, J.E.; McDowell, D.A. Habituation to sub-lethal concentrations of tea tree oil (Melaleuca alternifolia) is associated with reduced susceptibility to antibiotics in human pathogens. J. Antimicrob. Chemother. 2007, 59, 125–127. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Futoma-Kołoch, B.; Sarowska, J.; Abd El-Salam, M.; Miñana-Galbis, D.; Drabová, B.; Guz-Regner, K.; Wiśniewska, P.; Kryniewska, V. Current Insights into Antibiotic Resistance in Uropathogenic Escherichia coli and Interventions Using Selected Bioactive Phytochemicals. Antibiotics 2025, 14, 1242. https://doi.org/10.3390/antibiotics14121242
Futoma-Kołoch B, Sarowska J, Abd El-Salam M, Miñana-Galbis D, Drabová B, Guz-Regner K, Wiśniewska P, Kryniewska V. Current Insights into Antibiotic Resistance in Uropathogenic Escherichia coli and Interventions Using Selected Bioactive Phytochemicals. Antibiotics. 2025; 14(12):1242. https://doi.org/10.3390/antibiotics14121242
Chicago/Turabian StyleFutoma-Kołoch, Bożena, Jolanta Sarowska, Mohamed Abd El-Salam, David Miñana-Galbis, Barbora Drabová, Katarzyna Guz-Regner, Paula Wiśniewska, and Vivien Kryniewska. 2025. "Current Insights into Antibiotic Resistance in Uropathogenic Escherichia coli and Interventions Using Selected Bioactive Phytochemicals" Antibiotics 14, no. 12: 1242. https://doi.org/10.3390/antibiotics14121242
APA StyleFutoma-Kołoch, B., Sarowska, J., Abd El-Salam, M., Miñana-Galbis, D., Drabová, B., Guz-Regner, K., Wiśniewska, P., & Kryniewska, V. (2025). Current Insights into Antibiotic Resistance in Uropathogenic Escherichia coli and Interventions Using Selected Bioactive Phytochemicals. Antibiotics, 14(12), 1242. https://doi.org/10.3390/antibiotics14121242






