Comparison of Phenotypic and Whole-Genome Sequencing-Derived Antimicrobial Resistance Profiles of Legionella pneumophila Isolated in England and Wales from 2020 to 2023
Abstract
1. Introduction
2. Results
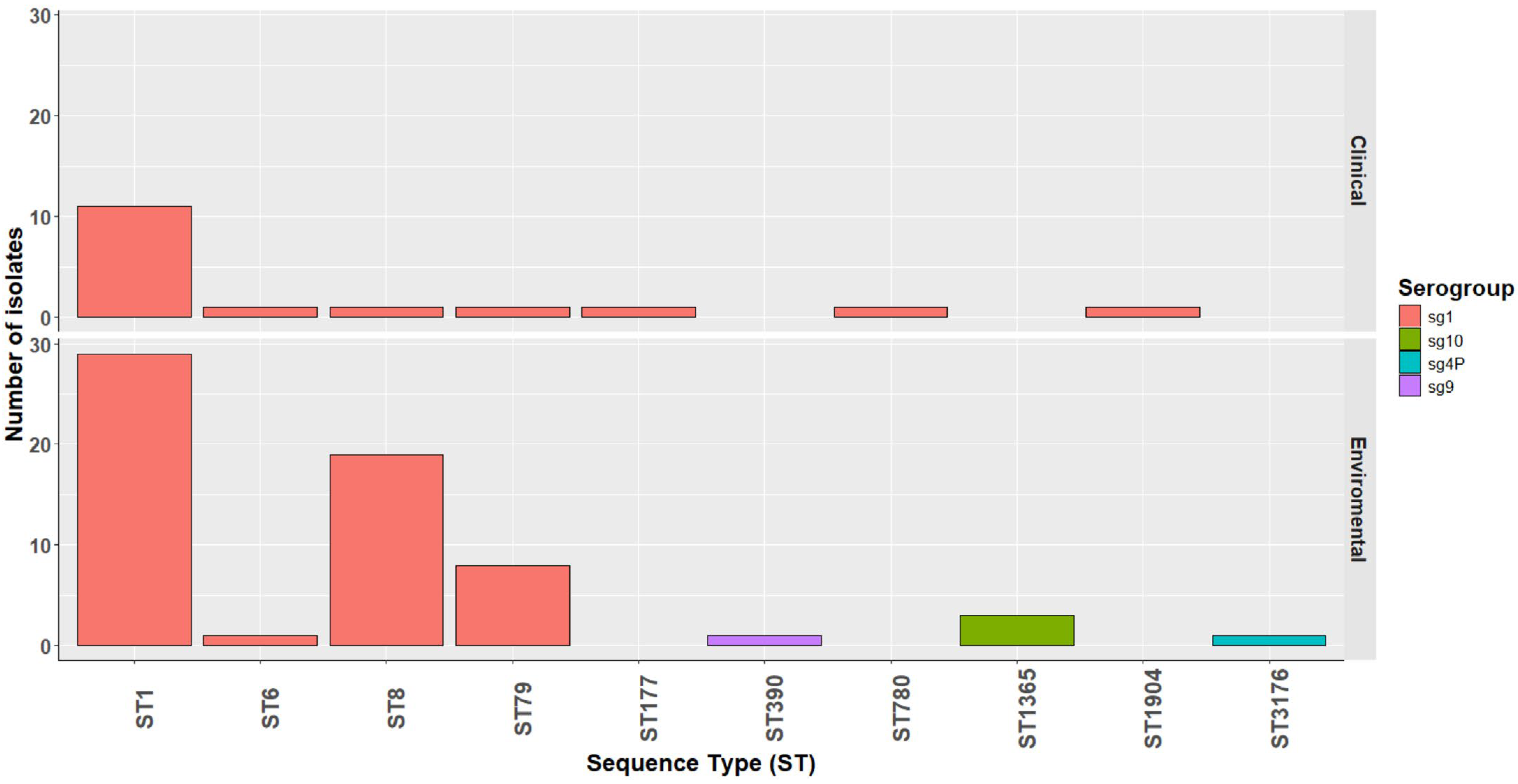
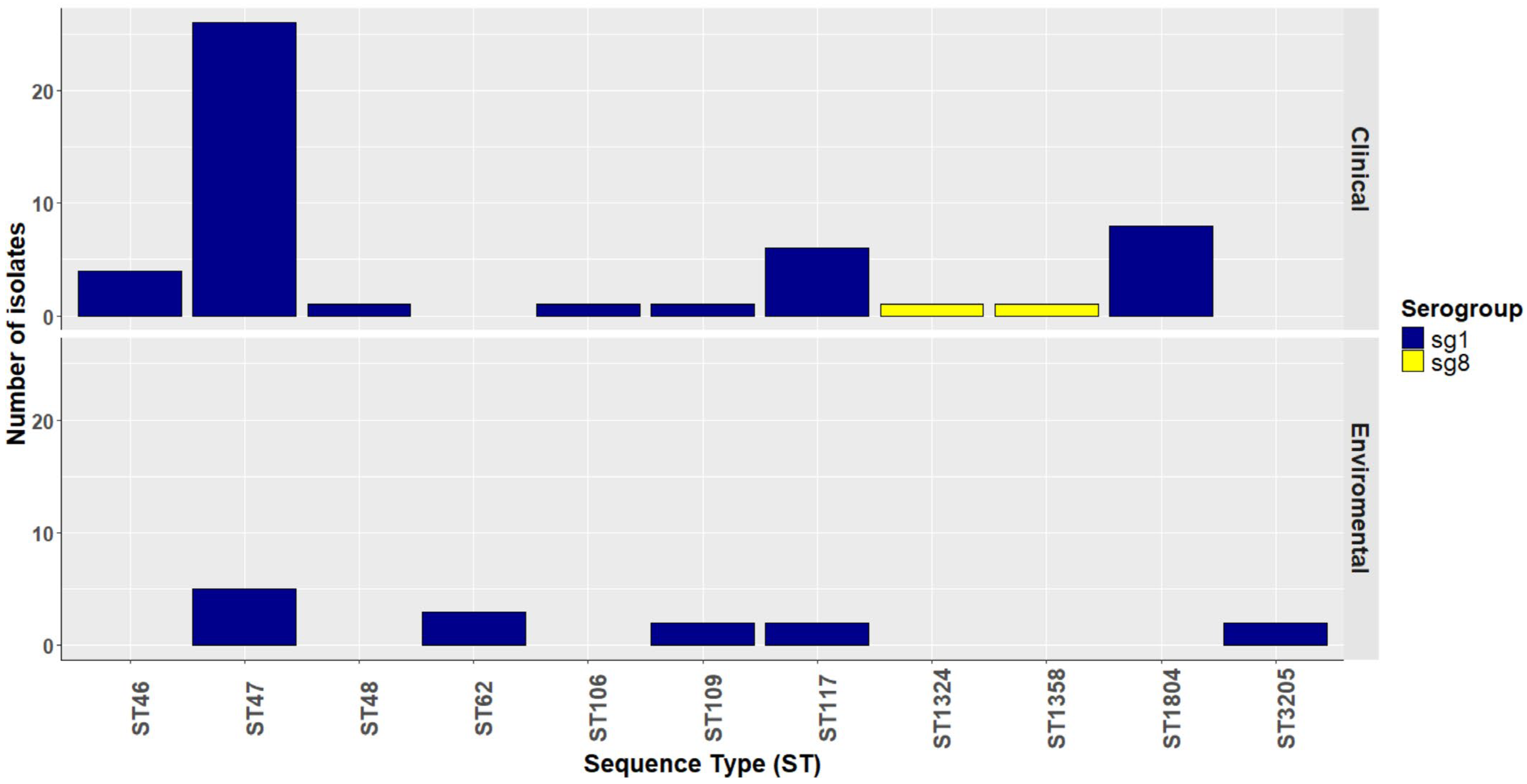
2.1. Source and Epidemiologic Data of Legionella pneumophila Isolates
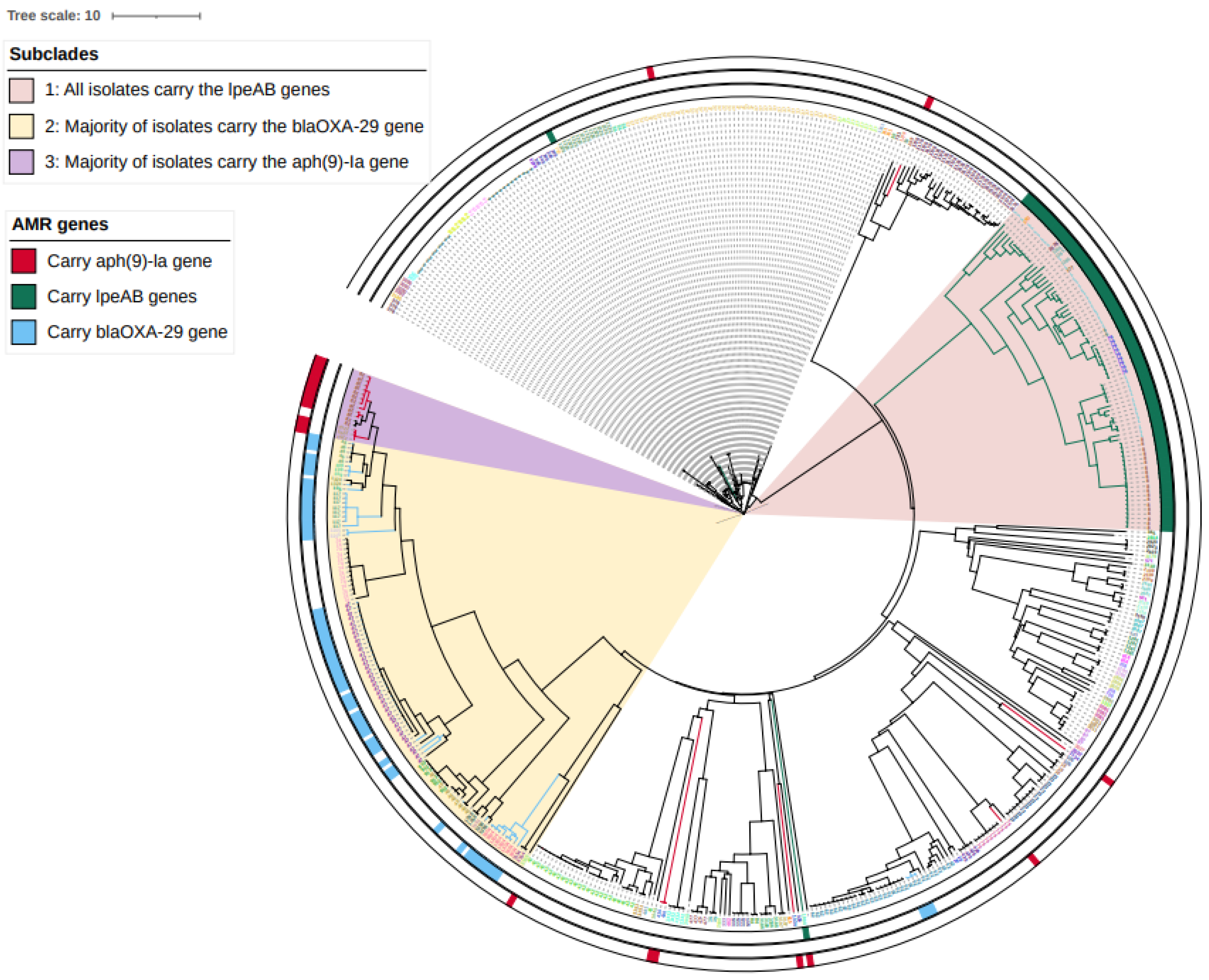
2.2. Prevalence of ARGs and Mutations Arising from WGS Analysis
2.3. Phenotypic Antimicrobial Susceptibility Testing Comparison of Isolates
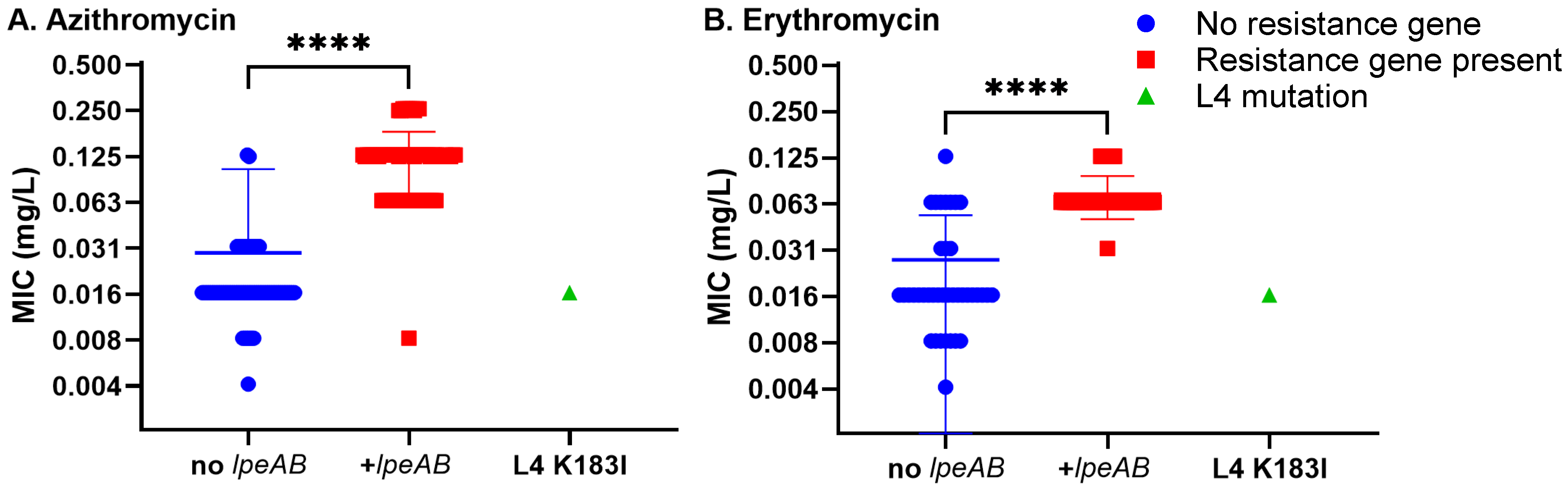
2.3.1. Evaluation of Macrolide Susceptibility
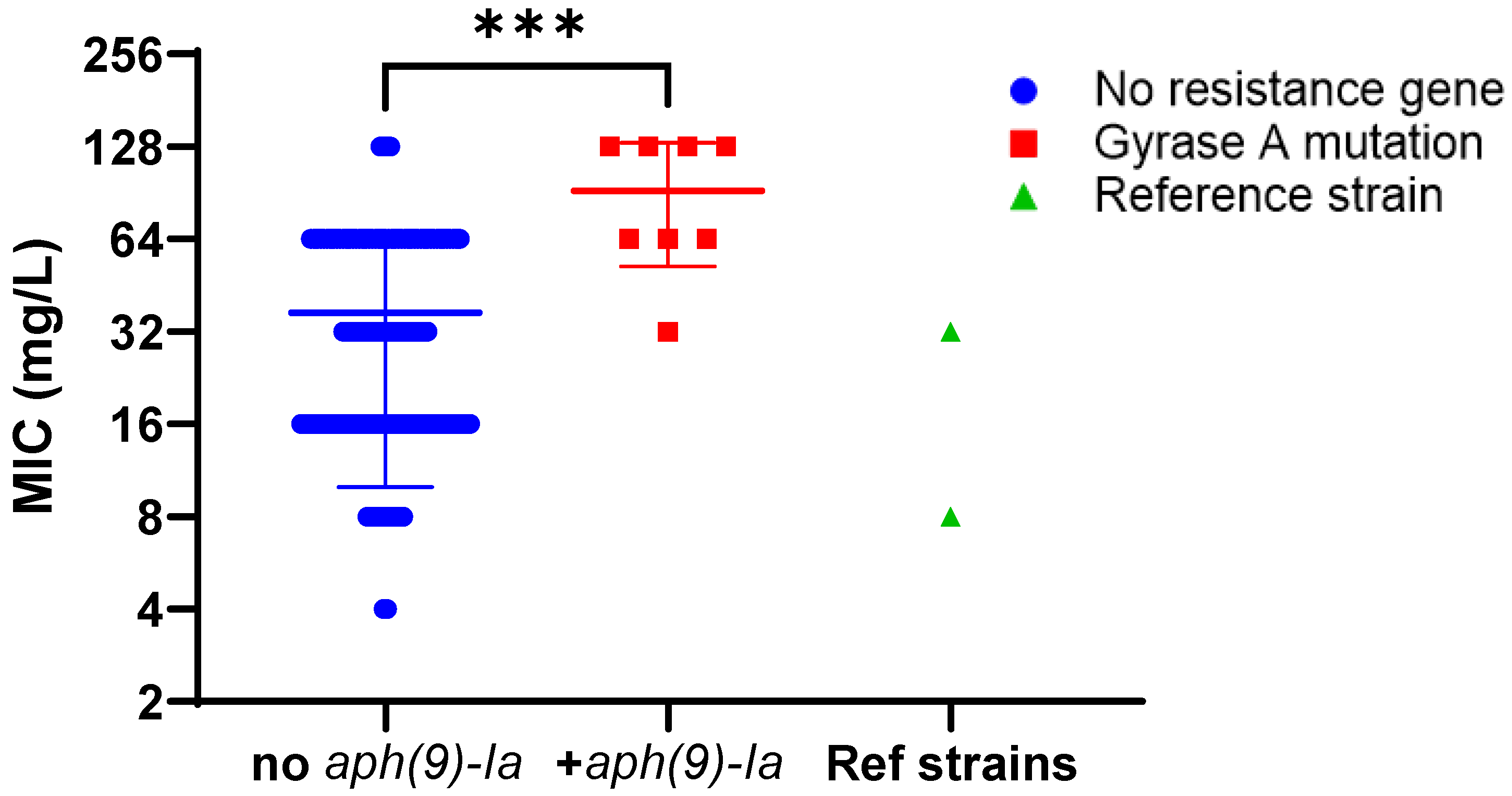
2.3.2. Evaluation of Aminoglycoside Susceptibility
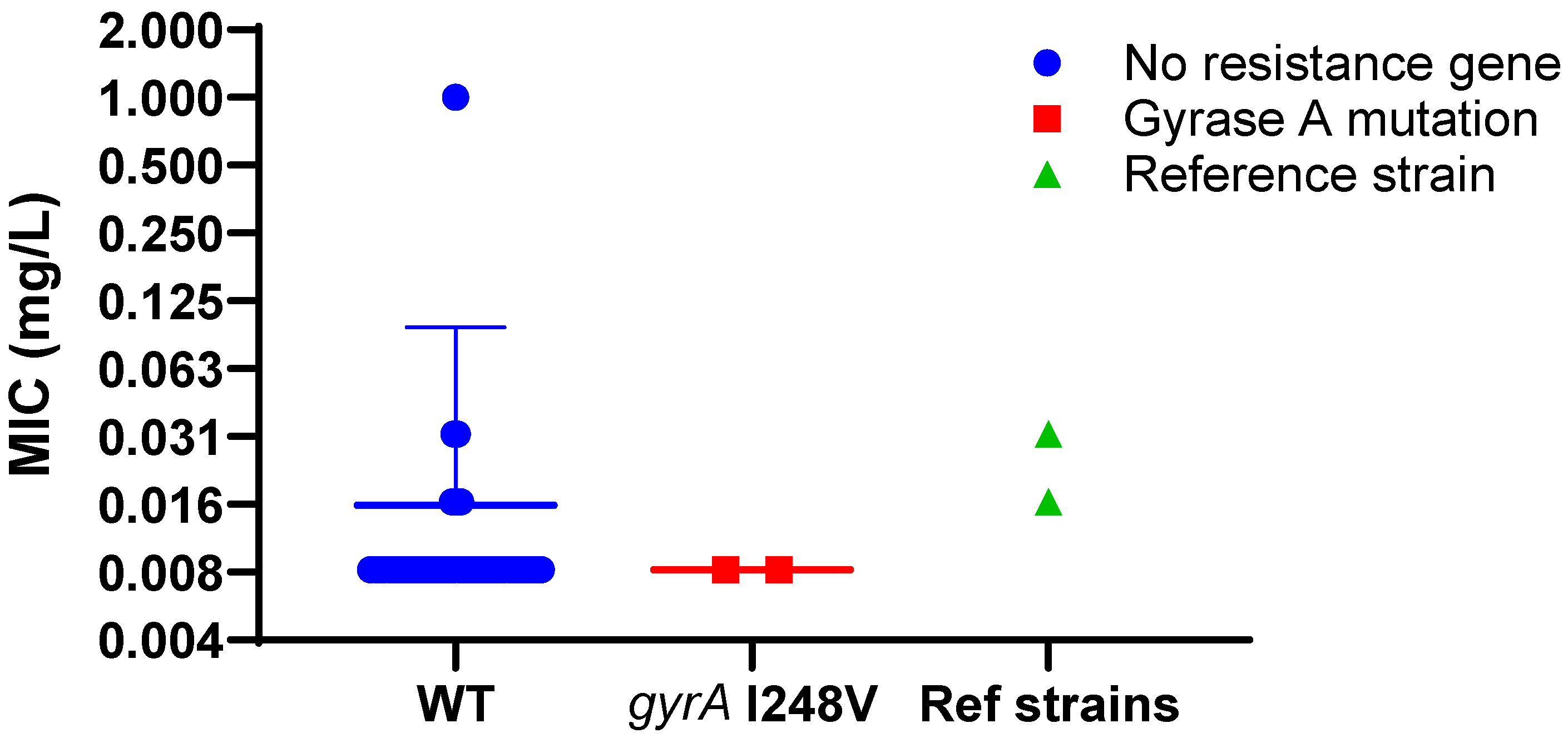
2.3.3. Evaluation of Fluoroquinolone Susceptibility
2.3.4. Evaluation of Tetracycline Susceptibility
2.3.5. Evaluation of β-Lactam Susceptibility
2.4. Genomic Analysis of L. pneumophila Isolates
2.4.1. Detection of 23S rRNA Mutations in High-Azithromycin MIC Isolates
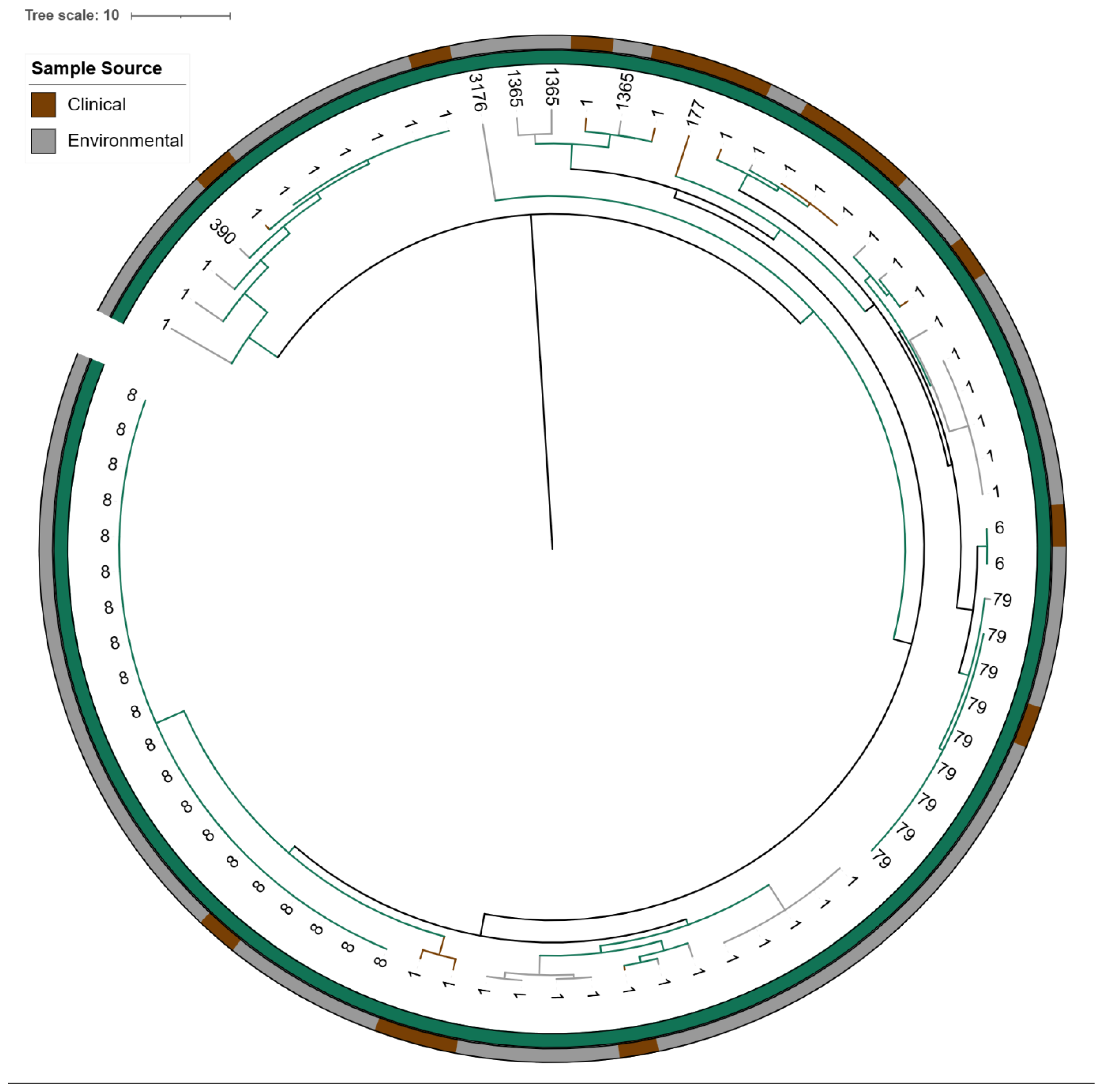
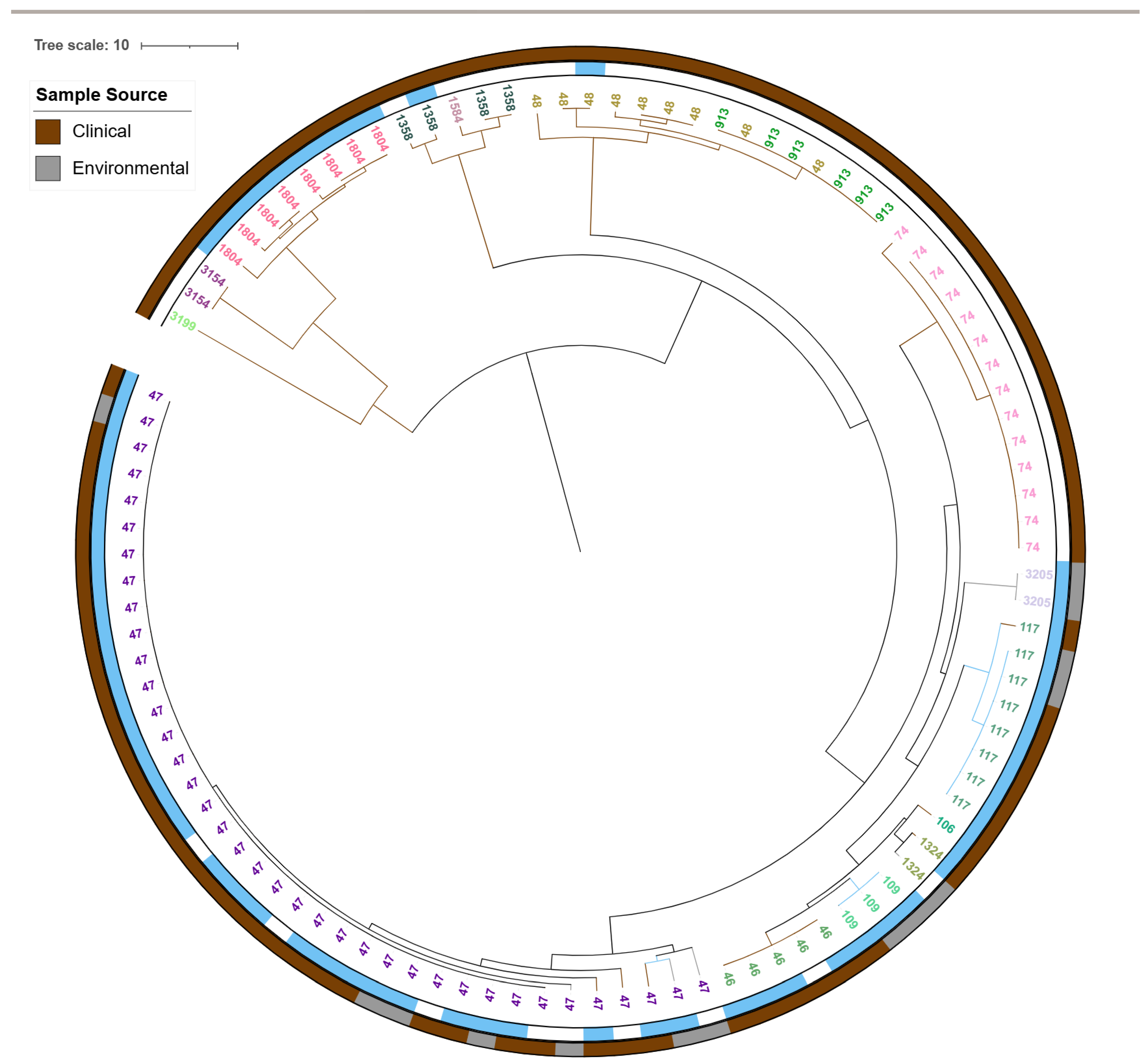
2.4.2. cgMLST-Based Core-Genome Phylogenetic Analysis
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Culture Conditions
4.2. Illumina Sequencing and Read Preparation
4.3. FASTQ Quality Thresholds
4.4. Legionella Species Identification
4.5. AMR Detection in Isolates from England and Wales
- (i)
- macrolide determinants: lpeAB (efflux; lpeA, lpeB), 23S rRNA methylases erm(A), erm(B), erm(C), erm(F), phosphotransferase mph(A) and esterases ere(A), ere(B);
- (ii)
- class D β-lactamase blaOXA-29;
- (iii)
- tetracycline destructase tet(56);
- (iv)
- spectinomycin phosphotransferase aph(9)-Ia.
4.6. AMR Detection in External Macrolide-Resistant Isolates
4.7. Minimum Inhibitory Concentrations (MICs)
4.8. Statistical Analysis
4.9. Genome Assembly
4.10. Core-Genome MLST and Phylogeny
4.11. Data Deposition
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fraser, D.W.; Tsai, T.R.; Orenstein, W.; Parkin, W.E.; Beecham, H.J.; Sharrar, R.G.; Harris, J.; Mallison, G.F.; Martin, S.M.; McDade, J.E.; et al. Legionnaires’ Disease: Description of an Epidemic of Pneumonia. N. Eng. J. Med. 1977, 297, 1189–1197. [Google Scholar] [CrossRef]
- Fields, B.S.; Benson, R.F.; Besser, R.E. Legionella and Legionnaires’ Disease: 25 Years of Investigation. Clin. Microbiol. Rev. 2002, 15, 506–526. [Google Scholar] [CrossRef]
- Zhu, Y.; Liu, D. Legionella. In Molecular Medical Microbiology, 3rd ed.; Elsevier: Amsterdam, The Netherlands, 2023; pp. 1547–1557. ISBN 9780128186190. [Google Scholar]
- Beauté, J. Legionnaires’ Disease in Europe, 2011 to 2015. Eurosurveillance 2017, 22, 30566. [Google Scholar] [CrossRef]
- Newton, H.J.; Ang, D.K.Y.; Van Driel, I.R.; Hartland, E.L. Molecular Pathogenesis of Infections Caused by Legionella Pneumophila. Clin. Microbiol. Rev. 2010, 23, 274–298. [Google Scholar] [CrossRef]
- Vaccaro, L.; Gomes, T.S.; Izquierdo, F.; Magnet, A.; Llorens Berzosa, S.; Ollero, D.; Salso, S.; Alhambra, A.; Gómez, C.; López Cano, M.; et al. Legionella feeleii: Ubiquitous Pathogen in the Environment and Causative Agent of Pneumonia. Front. Microbiol. 2021, 12, 707187. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention Clinical Features of Legionnaires’ Disease and Pontiac Fever. Available online: https://www.cdc.gov/legionella/hcp/clinical-signs/index.html (accessed on 25 August 2025).
- UK Health Security Agency Legionnaires’ Disease: Clinical Case Definitions. Available online: https://www.gov.uk/government/publications/legionnaires-disease-clinical-case-definitions/legionnaires-disease-case-definitions (accessed on 25 August 2025).
- Pappa, O.; Chochlakis, D.; Sandalakis, V.; Dioli, C.; Psaroulaki, A.; Mavridou, A. Antibiotic Resistance of Legionella Pneumophila in Clinical and Water Isolates—A Systematic Review. Int. J. Environ. Res. Public Health 2020, 17, 5809. [Google Scholar] [CrossRef]
- Gilbert, D.N.; Eliopoulos, G.M.; Chambers, H.F.; Saag, M.S.; Pavia, A.T. The Sanford Guide to Antimicrobial Therapy 2020: 50 Years: 1969–2019, 50th ed.; Antimicrobial Therapy: Sperryville, VA, USA, 2020. [Google Scholar]
- Viasus, D.; Gaia, V.; Manzur-Barbur, C.; Carratalà, J. Legionnaires’ Disease: Update on Diagnosis and Treatment. Infect. Dis. Ther. 2022, 11, 973–986. [Google Scholar] [CrossRef] [PubMed]
- Franceschini, N.; Boschi, L.; Pollini, S.; Herman, R.; Perilli, M.; Galleni, M.; Frère, J.M.; Amicosante, G.; Rossolini, G.M. Characterization of OXA-29 from Legionella (Fluoribacter) Gormanii: Molecular Class D β-Lactamase with Unusual Properties. Antimicrob. Agents Chemother. 2001, 45, 3509–3516. [Google Scholar] [CrossRef] [PubMed]
- Avison, M.B.; Simm, A.M. Sequence and Genome Context Analysis of a New Molecular Class D β-Lactamase Gene from Legionella Pneumophila. J. Antimicrob. Chemother. 2002, 50, 331–338. [Google Scholar] [CrossRef]
- Yang, J.L.; Sun, H.; Zhou, X.; Yang, M.; Zhan, X.Y. Antimicrobial Susceptibility Profiles and Tentative Epidemiological Cutoff Values of Legionella Pneumophila from Environmental Water and Soil Sources in China. Front. Microbiol. 2022, 13, 924709. [Google Scholar] [CrossRef]
- Massip, C.; Descours, G.; Ginevra, C.; Doublet, P.; Jarraud, S.; Gilbert, C. Macrolide Resistance in Legionella Pneumophila: The Role of LpeAB Efflux Pump. J. Antimicrob. Chemother. 2017, 72, 1327–1333. [Google Scholar] [CrossRef]
- Sewell, M.; Farley, C.; Portal, E.A.R.; Lindsay, D.; Ricci, M.L.; Jarraud, S.; Scaturro, M.; Descours, G.; Krøvel, A.V.; Barton, R.; et al. Broth Microdilution Protocol for Determining Antimicrobial Susceptibility of Legionella Pneumophila to Clinically Relevant Antimicrobials. J. Microbiol. Methods 2025, 228, 107071. [Google Scholar] [CrossRef]
- Minetti, C.; Barton, R.; Farley, C.; Spiller, O.B.; Rodrigues, R.; Gonçalves, P. Antimicrobial Susceptibility Testing Reveals Reduced Susceptibility to Azithromycin and Other Antibiotics in Legionella Pneumophila Serogroup 1 Isolates from Portugal. Eur. J. Clin. Microbiol. Infect. Dis. 2024, 43, 1297–1308. [Google Scholar] [CrossRef] [PubMed]
- Ginevra, C.; Beraud, L.; Pionnier, I.; Sallabery, K.; Bentayeb, H.; Simon, B.; Allam, C.; Chastang, J.; Ibranosyan, M.; Decroix, V.; et al. Detection of Highly Macrolide-Resistant Legionella Pneumophila Strains from a Hotel Water Network Using Systematic Whole-Genome Sequencing. J. Antimicrob. Chemother. 2022, 77, 2167–2170. [Google Scholar] [CrossRef]
- Michel, C.; Echahidi, F.; De Muylder, G.; Sewell, M.; Boostrom, I.; Denis, O.; Spiller, O.B.; Pierard, D. Occurrence of Macrolides Resistance in Legionella Pneumophila ST188: Results of the Belgian Epidemiology and Resistome Investigation of Clinical Isolates. Int. J. Infect. Dis. 2025, 153, 107786. [Google Scholar] [CrossRef]
- Jia, X.; Ren, H.; Nie, X.; Li, Y.; Li, J.; Qin, T. Antibiotic Resistance and Azithromycin Resistance Mechanism of Legionella Pneumophila Serogroup 1 in China. Antimicrob. Agents Chemother. 2019, 63, e00768-19. [Google Scholar] [CrossRef] [PubMed]
- Almahmoud, I.; Kay, E.; Schneider, D.; Maurin, M. Mutational paths towards increased fluoroquinolone resistance in Legionella pneumophila. J. Antimicrob. Chemother. 2009, 64, 284–293. [Google Scholar] [CrossRef]
- Svetlicic, E.; Jaén-Luchoro, D.; Klobucar, R.S.; Jers, C.; Kazazic, S.; Franjevic, D.; Klobucar, G.; Shelton, B.G.; Mijakovic, I. Genomic Characterization and Assessment of Pathogenic Potential of Legionella Spp. Isolates from Environmental Monitoring. Front. Microbiol. 2023, 13, 1091964. [Google Scholar] [CrossRef]
- Portal, E.; Sands, K.; Portnojs, A.; Chalker, V.J.; Spiller, O.B. Legionella Antimicrobial Sensitivity Testing: Comparison of Microbroth Dilution with BCYE and LASARUS Solid Media. J. Antimicrob. Chemother. 2021, 76, 1197–1204. [Google Scholar] [CrossRef] [PubMed]
- Fong, D.H.; Lemke, C.T.; Hwang, J.; Xiong, B.; Berghuis, A.M. Structure of the Antibiotic Resistance Factor Spectinomycin Phosphotransferase from Legionella Pneumophila. J. Biol. Chem. 2010, 285, 9545–9555. [Google Scholar] [CrossRef][Green Version]
- Descours, G.; Ginevra, C.; Jacotin, N.; Forey, F.; Chastang, J.; Kay, E.; Etienne, J.; Lina, G.; Doublet, P.; Jarraud, S. Ribosomal Mutations Conferring Macrolide Resistance in Legionella Pneumophila. Antimicrob. Agents Chemother. 2017, 61, e02188-16. [Google Scholar] [CrossRef]
- Coluzzi, C.; Rocha, E.P.C. The Spread of Antibiotic Resistance Is Driven by Plasmids Among the Fastest Evolving and of Broadest Host Range. Mol. Biol. Evol. 2025, 42, msaf060. [Google Scholar] [CrossRef]
- Suter, T.M.; Viswanathan, V.K.; Cianciotto, N.P. Isolation of a Gene Encoding a Novel Spectinomycin Phosphotransferase from Legionella Pneumophila. Antimicrob. Agents Chemother. 1997, 41, 1385–1388. [Google Scholar] [CrossRef] [PubMed]
- Kanchgal, P.S.; Selmer, M. Structural Recognition of Spectinomycin by Resistance Enzyme ANT(9) from Enterococcus Faecalis. Antibiotics 2020, 64, e00371-e20. [Google Scholar] [CrossRef] [PubMed]
- Stevanovic, M.; Teuber Carvalho, J.P.; Bittihn, P.; Schultz, D. Dynamical Model of Antibiotic Responses Linking Expression of Resistance Genes to Metabolism Explains Emergence of Heterogeneity during Drug Exposures. Phys. Biol. 2024, 21, 036002. [Google Scholar] [CrossRef] [PubMed]
- Mentasti, M.; Kese, D.; Echahidi, F.; Uldum, S.A.; Afshar, B.; David, S.; Mrazek, J.; De Mendonça, R.; Harrison, T.G.; Chalker, V.J. Design and Validation of a QPCR Assay for Accurate Detection and Initial Serogrouping of Legionella Pneumophila in Clinical Specimens by the ESCMID Study Group for Legionella Infections (ESGLI). Eur. J. Clin. Microbiol. Infect. Dis. 2015, 34, 1387–1393. [Google Scholar] [CrossRef]
- Gaia, V.; Fry, N.K.; Afshar, B.; Lück, P.C.; Meugnier, H.; Etienne, J.; Peduzzi, R.; Harrison, T.G. Consensus Sequence-Based Scheme for Epidemiological Typing of Clinical and Environmental Isolates of Legionella Pneumophila. J. Clin. Microbiol. 2005, 43, 2047–2052. [Google Scholar] [CrossRef]
- Helbig, J.H.; Bernander, S.; Castellani Pastoris, M.; Etienne, J.; Gaia, V.; Lauwers, S.; Lindsay, D.; Luck, P.C.; Marques, T.; Mentula, S.; et al. Pan-European study on culture-proven Legionnaires’ disease: Distribution of Legionella pneumophila serogroups and monoclonal subgroups. Eur. J. Clin. Microbiol. Infect. Dis. 2002, 21, 710–716. [Google Scholar]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality Assessment Tool for Genome Assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef] [PubMed]
- David, S.; Mentasti, M.; Tewolde, R.; Aslett, M.; Harris, S.R.; Afshar, B.; Underwood, A.; Fry, N.K.; Parkhill, J.; Harrison, T.G. Evaluation of an Optimal Epidemiological Typing Scheme for Legionella Pneumophila with Whole-Genome Sequence Data Using Validation Guidelines. J. Clin. Microbiol. 2016, 54, 2135–2148. [Google Scholar] [CrossRef] [PubMed]

| ARG or Mutation | Found in N Isolates | Phenotypically Tested |
|---|---|---|
| lpeAB | 79 | 79 |
| aph(9)-Ia | 22 | 8 |
| blaOXA-29 | 63 | 63 |
| gyrA I248V | 2 | 2 |
| L4 K183I | 1 | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tewolde, R.; Thombre, R.; Farley, C.; Nadarajah, S.; Khan, I.; Sewell, M.; Spiller, O.B.; Afshar, B. Comparison of Phenotypic and Whole-Genome Sequencing-Derived Antimicrobial Resistance Profiles of Legionella pneumophila Isolated in England and Wales from 2020 to 2023. Antibiotics 2025, 14, 1053. https://doi.org/10.3390/antibiotics14101053
Tewolde R, Thombre R, Farley C, Nadarajah S, Khan I, Sewell M, Spiller OB, Afshar B. Comparison of Phenotypic and Whole-Genome Sequencing-Derived Antimicrobial Resistance Profiles of Legionella pneumophila Isolated in England and Wales from 2020 to 2023. Antibiotics. 2025; 14(10):1053. https://doi.org/10.3390/antibiotics14101053
Chicago/Turabian StyleTewolde, Rediat, Rebecca Thombre, Caitlin Farley, Sendurann Nadarajah, Ishrath Khan, Max Sewell, Owen B. Spiller, and Baharak Afshar. 2025. "Comparison of Phenotypic and Whole-Genome Sequencing-Derived Antimicrobial Resistance Profiles of Legionella pneumophila Isolated in England and Wales from 2020 to 2023" Antibiotics 14, no. 10: 1053. https://doi.org/10.3390/antibiotics14101053
APA StyleTewolde, R., Thombre, R., Farley, C., Nadarajah, S., Khan, I., Sewell, M., Spiller, O. B., & Afshar, B. (2025). Comparison of Phenotypic and Whole-Genome Sequencing-Derived Antimicrobial Resistance Profiles of Legionella pneumophila Isolated in England and Wales from 2020 to 2023. Antibiotics, 14(10), 1053. https://doi.org/10.3390/antibiotics14101053






